Abstract
The mitochondrial genomes of chlamydomonadalean green algae are renowned for their highly reduced and conserved gene repertoires, which are almost fixed at 12 genes across the entire lineage. The sizes of these genomes, however, are much more variable, with some species having small, compact mitochondrial DNAs (mtDNAs) and others having expanded ones. Earlier work demonstrated that the halophilic genus Dunaliella contains extremely inflated organelle genomes, but to date the mtDNA of only one isolate has been explored. Here, by surveying mtDNA architecture across the Chlamydomonadales, we show that various Dunaliella species have undergone massive levels of mitochondrial genomic expansion, harboring the most inflated, intron-dense mtDNAs available from chlorophyte green algae. The same also appears to be true for their plastid genomes, which are potentially among the largest of all plastid-containing eukaryotes. Genetic divergence data are used to investigate the underlying causes of such extreme organelle genomic architectures, and ultimately reveal order-of-magnitude differences in mitochondrial versus plastid mutation rates within Dunaliella.
Keywords: Chlamydomonas, intron, mitochondrial DNA, plastid DNA, Polytoma
Introduction
The mitochondrial genomes of chlamydomonadalean green algae (Chlorophyta, Chlorophyceae) are somewhat of a contradiction (Leliaert et al. 2012). On the one hand, they have the smallest gene contents of any known organelle genomes from the Archaeplastida (Plantae sensu lato), encoding 7–8 proteins, 2 rRNAs, and 1–3 tRNAs (Smith, Hamaji, et al. 2013; Smith, Hua, et al. 2013), and they can also be very small (<13.5 kb) and compact (>80% coding) (Smith, Hua, et al. 2010). On the other hand, certain chlamydomonadalean mitochondrial DNAs (mtDNAs) are distended with repeats and introns, and composed almost entirely of noncoding nucleotides (Smith and Lee 2010).
One species with a particularly bloated mitochondrial genome is Dunaliella salina—a unicellular biflagellate, which lives in hypersaline environments, can accumulate large amounts of β-carotene, and is a prime candidate for biofuel production (Oren 2005). Complete mtDNA sequencing of D. salina CCAP 19/18, isolated from the Hutt Lagoon in Western Australia, revealed an unprecedentedly high intron density for a green alga (∼1.5 intron per gene) as well as vast, repeat-rich intergenic regions (Smith, Lee, et al. 2010). An equally expanded genome was also uncovered in the plastid, implying that similar forces are shaping both organelle DNAs (Smith, Lee, et al. 2010).
Various studies have used chlamydomonadalean algae, including the model organism Chlamydomonas reinhardtii and its close multicellular relative Volvox carteri, to explore the evolution of genome size (e.g., Smith and Lee 2010). These investigations suggest that mutation rate, DNA maintenance machineries, and random genetic drift have a major role in fashioning organelle chromosomes (Smith and Lee 2010; Hua et al. 2012; Smith, Hamaji, et al. 2013). Such studies, however, have yet to be applied to Dunaliella species, and it is still unknown if other members of the genus have inflated organelle genomes.
Here, we survey mitochondrial genome size and content within and outside the Dunaliella lineage. We show that although the mtDNA gene repertoire is nearly fixed across the Chlamydomonadales, there is an approximately 4-fold variation in genome size and an 18-fold variation in intron content, with Dunaliella species having among the most expanded mtDNAs of all explored green algae. The same is likely true for their plastid genomes as well. The levels of organelle DNA divergence between distinct D. salina strains are used to investigate the potential forces underpinning such massive levels of genomic expansion.
Sequencing New Chlamydomonadalean Mitochondrial Genomes
As part of an ongoing, collaborative initiative, we have been sequencing and characterizing organelle genomes from diverse chlamydomonadalean species (Hamaji et al. 2013; Smith, Hamaji, et al. 2013). Among these species are two distinct Dunaliella isolates, which were collected from the La Rinconada hypersaline pond in the Atacama Desert in northern Chile: D. salina CONC-001 and Dunaliella viridis CONC-002 (Gómez-Silva et al. 1990; González et al. 1999; Gómez and González 2005). Closely related to Dunaliella are two other algae that we have also been investigating: the freshwater flagellate Chlamydomonas leiostraca SAG 11-49 and the free-living, nonphotosynthetic unicell Polytoma uvella UTEX 964, which are a model duo for studying the loss of photosynthesis (Figueroa-Martinez et al. 2015).
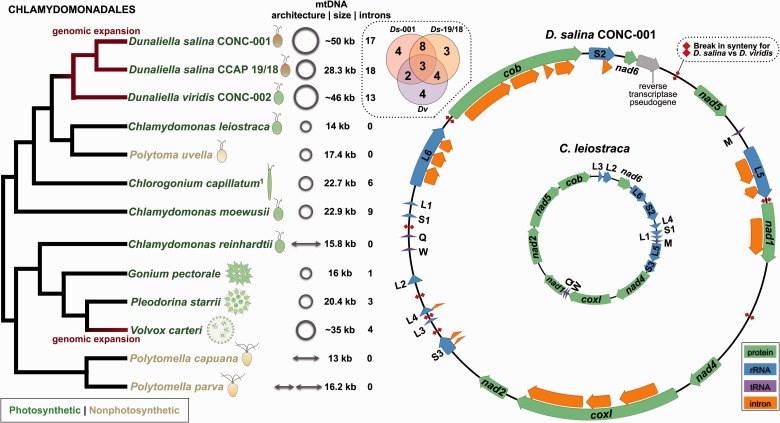
Next-generation sequencing of total cellular DNA from these algae followed by mitochondrial genome assembly yielded both expected and unexpected results. At first glance, all four mtDNAs appear similar to one another and to those of various other chlamydomonadaleans: they map as single circular chromosomes (Bendich 1993), have identical gene compliments (representing 7 proteins, 3 tRNAs, and 2 rRNAs), and contain sections of overlapping gene order (fig. 1). Moreover, in each of the algae the mitochondrial large and small subunit rRNA genes are fragmented and scrambled into six and three coding modules, respectively (fig. 1), which is a common theme throughout the order, with species from the Reinhardtinia clade (Nakada et al. 2008) displaying even greater levels of rRNA gene fragmentation (Smith, Hamaji, et al. 2013). Three of the four genomes also have notably high guanine and cytosine (GC) contents: 39% (C. leiostraca), 46.7% (D. salina CONC-001), 47.1% (D. viridis CONC-002), and 55% (P. uvella). These elevated GC values are not entirely unexpected: The Chlamydomonadales is known to harbor species with exceptionally high mtDNA GC compositions, including the colorless alga Polytomella capuana (57.2%) as well as some members of the Lobochlamys genus (∼50–65%; Smith 2012). That said, D. salina CCAP 19/18, unlike its Chilean counterparts, has a low mitochondrial GC content (34.4%), underscoring that organelle nucleotide content can differ drastically even among closely related species and strains.
Fig. 1.—
Mitochondrial genomic architecture and expansion within the Chlamydomonadales. Tree of chlamydomonadalean algae, showing mitochondrial genome conformation, size, intron content, and expansion (red); branching order based on the phylogenetic analyses of Nakada et al. (2008), González et al. (2009), Smith, Hua, et al. (2013), and Figueroa-Martinez et al. (2015), as well as that in supplementary figure S1, Supplementary Material online. Venn diagram highlighting shared and unique introns (based on insertion sites) among the three available Dunaliella mtDNAs. Mitochondrial genome maps for Dunaliella salina CONC-001 (outer) and Chlamydomonas leiostraca (inner). Dunaliella salina CONC-001 and D. salina CCAP 19/18 have identical mtDNA gene orders and contents (not including introns, intronic ORFs, or pseudogenes), and so do C. leiostraca and Polytoma uvella. Breakpoints in mitochondrial gene synteny between D. salina and D. viridis are marked with a double-diamond symbol (red). Note: the mitochondrial genome size and intron number for C. reinhardtii and V. carteri can vary due to optional introns in some strains (Smith, Hamaji, et al. 2013). Superscript 1 indicates Chlorogonium capillatum SAG 12-2e was formerly called Chlorogonium elongatum SAG 12-2e.
Further inspection revealed even more differences among the mitochondrial genomes. The D. viridis CONC-002 and D. salina CONC-001 mtDNAs, with respective lengths of approximately 46 and 50 kb, are around 3–4-times larger than those of C. leiostraca (14 kb) and P. uvella (17.4 kb), and on average greater than 2-times larger than other available chlamydomonadalean mtDNAs, despite mitochondrial gene content being almost identical across the entire lineage. What’s more, the C. leiostraca and P. uvella mtDNAs contain no introns, are densely packed (≤30% noncoding), and have matching gene orders, whereas D. viridis CONC-002 has 13 introns and D. salina CONC-001 has 17, and both species are distended with noncoding mtDNA (>70%) and have differing gene orders (fig. 1). In fact, the mtDNA of the Chilean D. salina described here is almost twice as large as the previously reported mtDNA of the Australian D. salina CCAP 19/18 (∼50 kb vs. 28.3 kb), which, like the Chilean strain, also has an abundance of introns (18) (fig. 1; Smith, Lee, et al. 2010).
In all, 48 putative introns are distributed among the three sequenced Dunaliella mitochondrial genomes, representing approximately 65% of all identified mitochondrial introns from the Chlamydomonadales. When looking at the location of the Dunaliella introns, 28 have unique insertion sites (situated within four different protein-coding genes and five different rRNA-coding modules), 17 are found in at least two of the isolates, 11 are located in only a single isolate, and 8 contain an intronic open reading frame (ORF; fig. 1). All but one of the introns appear to be of group I affiliation, and the decaying remnants of intronic ORFs were uncovered in the intergenic DNA of both D. salina CONC-001 and D. viridis CONC-002 (fig. 1), suggesting a complex history of intron loss and gain throughout the evolution of Dunaliella mitochondria.
Genomic Upheaval within Dunaliella Mitochondria
The mitochondrial genomes of Dunaliella algae seem to be in a state of upheaval (fig. 2). They are the most bloated and intron-rich mtDNAs observed from the Chlorophyta (fig. 2A), have undergone substantial genomic rearrangements, and are riddled with short, simple repeats, which have spread throughout the intergenic, intronic, and, in some cases, coding regions (figs. 2B and C). These repeats differ in sequence among the three Dunaliella mtDNAs, but can be folded into similar hairpin structures (Smith, Lee, et al. 2010), and in some respects resemble the palindromic organelle repeats from V. carteri (Smith and Lee 2009). Repeat-like insertions were also uncovered within coding regions of the Dunaliella mtDNAs, resulting in elongated exonic sequences relative to those from other closely related mtDNAs (fig. 2B). All of this is in stark contrast to the compact and “ordered” mtDNAs of C. leiostraca and P. uvella, which are devoid of introns and have few repeats (figs. 1, 2A, and 2C).
Fig. 2.—
Mitochondrial genomic upheaval in Dunaliella. (A) Noncoding content (x axis) versus intron abundance (y axis) for chlamydomonadalean mtDNAs. Noncoding statistics were calculated following the methods of Smith, Lee, et al. (2010). Chlamydomonas leiostraca, Polytoma uvella, and the three Dunaliella isolates are marked on plot. (B) Insertions within Dunaliella salina CONC-001 (red) and D. viridis (blue) mtDNA protein-coding genes relative to those of D. salina CCAP 19/18. These insertions are absent from the C. leiostraca mtDNA. (C) Dot plot similarity matrices of chlamydomonadalean mitochondrial genomes. Each matrix contains an mtDNA sequence plotted against itself (size of the genome is marked in the bottom right corner). Dots within the matrix highlight regions of nucleotide sequence similarity. The main diagonal represents the mtDNA on the x axis matching against its partner on the y axis. Dots adjacent to the main diagonal correspond to repetitive DNA. Plots were generated with JDotter (Brodie et al. 2004), using a plot size of 1,000 bases/pixel and a sliding window size of 50.
Given what we know about the Chlamydomonadales, the most recent common ancestor of the group likely had a compact, circular-mapping, intron-poor mtDNA (fig. 1). However, at some point after the divergence of the Dunaliella and Polytoma/C. leiostraca lineages, the former experienced severe mtDNA inflation (fig. 1), characterized by the proliferation of intronic and repetitive DNA (figs. 2A and C). Mitochondrial genomic expansion is also observed in other chlorophyte lineages, including the volvocine line (Smith, Hamaji, et al. 2013) and certain members of the Sphaeropleales (Fučíková et al. 2014), but it is not as pronounced as that within Dunaliella, and is mostly a product of increases in intergenic sequence rather than a combination of repeats, introns, and other kinds of genomic embellishments.
Although all three of the available Dunaliella mitochondrial genomes are expanded, there is, nonetheless, an impressive amount of variation in size, intron, and noncoding content among them (28–50 kb; 13–18 introns per genome; ∼60–75% noncoding DNA; figs. 1 and 2A). As is apparent from self-similarity dot plots (fig. 2C), repeat content scales positively with mitochondrial genome size across the Dunaliella genus, and the order as a whole (Smith, Hamaji, et al. 2013). Other types of genomic embellishments also go up in abundance relative to mtDNA size. For example, the Chilean D. salina has more nonstandard ORFs and pseudogenes than its Australian counterpart (fig. 1). The same, however, cannot be said for overall intron number, which again is highest in the Australian isolate, reinforcing that intron abundance alone does not account for the inflated architectures of the Dunaliella mitochondrial genomes.
Coexpansion of the Chloroplast Genomes
Using the same datasets employed for the mitochondrial genome assemblies, we explored the plastid genomic architectures of D. salina CONC-001 and D. viridis CONC-002 (those of C. leiostraca and P. uvella, which are relatively compact, will be described elsewhere). The plastid DNAs (ptDNAs) of CONC-001 and CONC-002 appear to be equally or even more expanded than the neighboring mitochondrial genomes. De novo assemblies of paired-end Illumina reads from each of the algae gave dozens of short (∼0.5–17 kb) ptDNA contigs (table 1). For both Dunaliella isolates, the plastid contigs were AT-rich, had one to a few genes apiece, harbored many introns, and contained extensive repeats, which were found in almost all of the identified intronic and intergenic regions (table 1). These repeats prevented the assembly of larger contigs and the bridging of smaller ones, which is a recurring problem in green algal plastid genomics—and one that is hampering the assembly of the D. salina CCAP 19/18 nuclear genome (United States Department of Energy Joint Genome Institute, DOE JGI). For example, palindromic repeats hindered the assembly of the approximately 525 kb plastid chromosome of V. carteri (Smith and Lee 2009), and the recently sequenced ptDNA of the ulvophyte Acetabularia acetabulum, which is approximately 1 Mb and repeat-rich, resulted in a highly fragmented assembly (>60 contigs; de Vries et al. 2013).
Table 1.
Dunaliella salina CONC-001 and D. viridis CONC-002 Plastid Genome Assembly Statistics
| Plastid DNA Contig Statistics | D. salina CONC-001 | D. viridis CONC-002 |
|---|---|---|
| Total number | 80 | 72 |
| Size range (kb) | 0.6–8.9 | 0.5–16.8 |
| Average size (kb) | 2.2 | 2.4 |
| Average read coverage/base | 43.5 | 31.2 |
| Average number genes/contiga | 0.8 | 1.2 |
| Proportion of genes identified (%)b | 65 | 88 |
| Overall intron count | 32 | 13 |
| Accumulative size (kb) | 178.3 | 171.8 |
| Predicted plastid genome size (kb)c | >370 | >280 |
aNumber is <1 because in some cases a single gene is distributed over multiple contigs.
bPercentage of plastid-encoded genes identified in the CONC-001 and CONC-002 assemblies relative to the genes found in the completely sequenced D. salina CCAP 19/18 ptDNA. Does not include nonstandard genes, such as intronic ORFs; duplicate genes, such as the rRNAs, were counted only once.
cAssuming missing genes are found on single approximately 2 kb contigs and an average gap between contigs of approximately 1 kb.
Based on the number, proportion, and density of genes identified on the ptDNA contigs as well as the accumulative size of these contigs (table 1), we estimate that the Chilean D. salina and D. viridis plastid genomes are at least 370 kb and 280 kb, respectively, and are possibly much larger, which makes them giants among all available ptDNAs. This is consistent with the previously published D. salina CCAP 19/18 ptDNA (assembled with Sanger sequencing reads; Smith, Lee, et al. 2010), which at approximately 265 kb and greater than 65% noncoding, is one of the largest ptDNAs ever observed. Like the corresponding mitochondrial genomes, the CONC-001 and CONC-002 ptDNAs are intron-rich (CONC-001 likely has >30 introns), and repeats have infiltrated both the intronic and the intergenic regions. But unlike the mtDNAs, the plastid introns are mostly of group II affiliation and the plastid repeats do show sequence similarities across the three Dunaliella ptDNAs.
Together, these data are an excellent illustration of how mitochondrial and plastid genomes can arrive at similar extremes in a single organism or cell—in this case, the coexpansion of mtDNA and ptDNA within members of the Dunaliella genus. Similar observations have come from other eukaryotes, including V. carteri for which both the mtDNA and the ptDNA have uncharacteristically long intergenic regions and large amounts of repetitive DNA (Smith and Lee 2009, 2010). The trend can also go in the opposite direction. For instance, the mitochondrial and plastid genomes of many prasinophyte algae, such as Ostreococcus tauri, are paragons of compactness (Robbens et al. 2007), as are those of the red alga Cyanidioschyzon merolae (Ohta et al. 2003). Convergent evolution between mtDNA and ptDNA can be seen throughout the eukaryotic tree, and in many cases both organelle genomes have independently evolved the same features and taken on similar genomic embellishments. However, when this is observed, the intensity of genomic embellishment is typically more pronounced in mitochondria than in plastids (Barbrook et al. 2010; Smith and Keeling 2015). But this does not appear to be true for Dunaliella: the ptDNA has been pushed to an equivalent or greater extreme than the mtDNA, at least in terms of noncoding content.
Genetic Divergence: High in the Mitochondrion, Low in the Plastid
To better understand the evolution of expanded Dunaliella organelle DNAs, we studied the levels of genetic divergence between them. Using a maximum-likelihood approach, we measured the rates of organelle nucleotide substitution between D. salina CONC-001 and CCAP 19/18, which revealed huge, unparalleled differences in mtDNA versus ptDNA divergence for green algae (table 2 and supplementary table S1, Supplementary Material online).
Table 2.
mtDNA and ptDNA Substitution Rates for Two Geographically Distinct Isolates of Dunaliella salina: CONC-001 (Chile) and CCAP 19/18 (Australia)
| Substitutions per Site |
Substitution Rate Ratios (pt:mt) | ||
|---|---|---|---|
| ptDNA | mtDNA | ||
| Synonymous sites | |||
| Average (SD) | 0.09 (0.32) | 1.164 (0.52) | 1:12.9 |
| Concatenation | 0.074 | 0.989 | 1:13.4 |
| Nonsynonymous sites | |||
| Average (SD) | 0.005 (0.02) | 0.043 (0.02) | 1:8.6 |
| Concatenation | 0.004 | 0.041 | 1:10.3 |
| dN/dS (SD) | 0.084 (0.18) | 0.042 (0.02) | –– |
| rRNAsa | 0.056 | 0.107 | 1:1.9 |
| Introns | >>0.1 | Unalignable | –– |
| Intergenic regions | >>0.1 | Unalignable | –– |
Note.—SD, standard deviation; dN/dS, ratio of nonsynonymous to synonymous substitutions per site, based on averages of individual loci not concatenated datasets. The substitution rate statistics for the individual loci within mitochondrial and plastid genomes are shown in supplementary table S1, Supplementary Material online.
aFor mtDNA and ptDNA includes the concatenation of all rRNA-coding regions.
Substitution rates between CONC-01 and CCAP 19/18 were 2–13 times greater in the mtDNA than in the ptDNA (table 2). The average number of substitutions per synonymous site (dS) for mtDNA protein-coding genes (∼1.2) was about 13 times that of ptDNA proteins (0.09). Concatenated gene data sets gave an identical trend (table 2 and supplementary table S1, Supplementary Material online). The rates of substitution at nonsynonymous sites (dN) followed a similar pattern: dN of the mtDNA (0.04) was approximately an order of magnitude greater than that of the ptDNA (0.005); however, both organelle genomes had low dN/dS ratios (<0.1; table 2), indicating strong purifying selection at nonsynonymous sites. Substitution rates for rRNA genes were also greater for the mtDNA than for the ptDNA (0.1 vs. 0.06; table 2).
We tried measuring substitutions within noncoding DNA as well. The mitochondrial introns and intergenic regions from CONC-001 and CCAP 19/18 were unalignable, implying very high levels of substitution (>>1 per site), much higher than those observed for the mtDNA synonymous sites. Although many of the plastid noncoding regions were also unalignable, we were able to align ten complete ptDNA intergenic spacers, which when concatenated (4.6 kb) harbored approximately 0.15 substitutions per site (table 2), which is greater than that observed for plastid synonymous sites. Like with the coding data, these findings are consistent with a higher rate of nucleotide substitution in the mitochondrion as compared with the plastid. Similar overall conclusions come from substitution rate analyses of D. salina versus D. viridis, but the levels of substitution are saturated making it difficult to gauge the relative rates of substitution between the mtDNA and ptDNA, and between synonymous and noncoding sites.
Organelle substitution rate statistics are available for a number of plastid-bearing eukaryotes, including various green algae (supplementary table S2, Supplementary Material online; Hua et al. 2012). Compared with other species, the levels of synonymous-site divergence between CONC-001 and CCAP 19/18 are high for the mtDNA and low for the ptDNA. For example, the average synonymous-site divergence between Chlamydomonas globosa SAG 7.73 (formerly called C. incerta) and C. reinhardtii is approximately 0.30 for both mitochondrial- and plastid-located genes (Hua et al. 2012), which contrast sharply with the values from D. salina: approximately 1.2 for the mtDNA and 0.09 for the ptDNA. Moreover, the relative levels of dS between the mitochondrial and plastid compartments of D. salina (13:1) are among the highest yet observed from green, red, or glaucophyte algae (supplementary table S2, Supplementary Material online).
What do these extreme differences in dS mean? At the very least, the high levels of substitution within the mtDNA suggest that D. salina CONC-001 and CCAP 19/18 represent distinct populations or “species” (González et al. 2009). They also point toward major differences in the organelle mutation rates. If synonymous nucleotide positions are assumed to be neutrally evolving, then the synonymous-site divergence between species or distinct populations can provide an entrée into mutation rate (Kimura 1983). For D. salina, there is a 13-fold difference in dS between mitochondrial-located versus plastid-located genes (table 2), indicating that there is much higher mutation rate in the mitochondrion than the plastid. These findings could also be a sign of high and low absolute mutation rates within mitochondrion and plastid, respectively, but this is speculative as we do not know how long ago CONC-001 and CCAP 19/18 shared a common ancestor. If these two compartments do have drastically different mutational patterns, then it would, on the face of it, conflict with their similarly expanded genomic architectures.
Unraveling the Roots of Organelle Genomic Expansion
In many respects, the Dunaliella organelle genomes have parallel architectures to land plant mtDNAs, which are renowned for their expansive intergenic regions, large densities of introns and repeats, and overall high levels of sequence upheaval (Sloan et al. 2012). Similar to the Dunaliella organelle genomes, land plant mtDNAs boast impressive variations in synonymous substitution rates, both within and among genomes (Sloan et al. 2012; Richardson et al. 2013; Zhu et al. 2014). For instance, the enormous, multichromosomal mtDNAs of Silene conica and S. noctiflora have extraordinarily high synonymous substitution rates (Sloan et al. 2012), whereas the tulip tree has one of the most mutationally quiescent mitochondrial genomes of any eukaryote (Richardson et al. 2013). Land plant mtDNAs, as with Dunaliella, also show vastly different rates of substitution in coding versus noncoding regions, a feature that that has provided major insights into the process of organelle genome expansion (Christensen 2013).
After finding that the modes of molecular evolution differ between coding and noncoding regions in Arabidopsis mtDNA, Christensen (2013) proposed that land plants employ two types of mtDNA repair, each of which has shaped mitochondrial genomic architecture: “Within genes, a bias toward gene conversion would keep measured mutation rates low, whereas in noncoding regions, break-induced replication (BIR) explains the expansion[s] and rearrangements. Both processes are types of double-strand break repair, but enhanced second-strand capture in transcribed regions versus BIR in nontranscribed regions can explain the two seemingly contradictory features of plant mitochondrial genome evolution—the low mutation rates in genes and the striking expansions of noncoding sequences.”
The same argument can be made for the Dunaliella mitochondrial and plastid chromosomes. Both organelle DNAs have much lower substitution rates within synonymous sites as compared with the noncoding regions. This is especially apparent for the ptDNA, which has an average dS of only 0.09 but for which most of the intergenic regions are unalignable (table 2). If the intergenic regions in the Dunaliella organelle genomes are repaired via BIR it would help explain the large amounts of genomic expansion and rearrangements observed between them (fig. 1) as well as the widespread intergenic turmoil (fig. 2B and C). Indeed, BIR within organelle systems is known to be inaccurate and cause rearrangements, chimeric genes, and expansions (Davila et al. 2011). Conversely, accurate repair of coding organelle DNA in Dunaliella, by homologous recombination or gene conversion, for example, would account for the comparatively low synonymous substation rates, particularly in the plastid.
Nevertheless, there is still an order of magnitude variation in dS for the mitochondrial versus plastid protein-coding genes of D. salina, indicating that the efficiency of DNA repair, be it by BIR or gene conversion or homologous recombination, differs greatly between these compartments. In plants and algae, virtually all of the organelle DNA maintenance machineries are nuclear encoded, and their proficiency are known to vary between species and compartments (Sloan and Taylor 2012). A Dunaliella nuclear genome sequence is not yet available, but work is presently underway by the DOE JGI to generate one. Investigations of nuclear-encoded, organelle-targeted DNA repair proteins will likely give further insights into the evolution of the Dunaliella organelle genomes. There is also the potential that the extremely salty habitats in which many Dunaliella species reside is in some way impacting the molecular evolution of their organelle DNAs. Whatever the root cause of their inflated architectures, the Dunaliella mitochondrial and plastid genomes are veritable heavyweights among green algal organelle genomes.
Methods and Materials
Dunaliella salina CONC-001 and D. viridis CONC-002 (Microalgae Culture Collection, Universidad de Concepción, Chile; updated acronyms CCM-UDEC 001 and CCM-UDEC 002, respectively) were grown in J/1 medium, supplemented with 15% NaCl w/v, at 18 °C under a 14-h light/10-h dark cycle, and harvested as previously described (González et al. 1999; Gómez and González 2005). Total DNA from each isolate was extracted using the Qiagen DNeasy Plant Mini Kit (Qiagen, Venlo, Limburg, NL) with liquid nitrogen disruption. Illumina sequencing libraries were prepared with the Nextera DNA Sample Preparation Kit (Illumina, San Diego, CA) and sequenced on an Illumina MiSeq platform (v2 chemistry; 250 × 250 paired-end sequencing reads).
Chlamydomonas leiostraca SAG 11-49 (Culture Collection of Algae at the University of Göttingen, Germany) was grown in Volvox medium (Provasoli and Pintner 1960) at 18 °C under a 14-h light/10-h dark cycle and with constant shaking (200 revolutions per minute [rpm]). Polytoma uvella UTEX 964 (Culture Collection of Algae at the University of Texas at Austin) was cultured in Polytoma medium under the same conditions, but without shaking. Cells were harvested in logarithmic growth by centrifugation at 4,500 rpm for 10 min, and whole-genomic DNA from each strain was extracted by standard phenol–chloroform methods and ethanol precipitation. Library preparation and Illumina sequencing (HiSeq 2000) were performed at the Roy J. Carver Center for Genomics at the University of Iowa (100 × 100 paired-end sequencing reads).
The organelle genomes of all four algae were assembled de novo with Ray v2.2.0 (Boisvert et al. 2010), using k-mers of 21, 27, 31, and 37, and separately with CLC Genomics Workbench v6.0.4 (Qiagen, Prismet, DK), using a word size of 20, bubble size of 50, and paired-end scaffolding. The resulting Ray and CLC contigs were scanned for organelle sequences using BLAST-based methods and the mitochondrial and plastid genomes of chlamydomonadalean algae (fig. 1) as queries. Hits to organelle DNA were assembled into larger contigs using read-mapping approaches with Geneious v7.1.4 (Biomatters Ltd., Auckland, NZ). Organelle introns were identified with RNAweasel (http://megasun.bch.umontreal.ca/RNAweasel/) and through alignments with other chlamydomonadalean organelle DNAs.
Organelle genes were aligned with MUSCLE (Edgar 2004), implemented through Geneious, using default settings. Synonymous and nonsynonymous substitutions were measured with the CODEML program of PAML v4.3 (Yang 2007), employing the maximum likelihood method and the F3x4 codon model. Substitutions in nonprotein-coding regions were estimated with BASEML of PAML, using the HKY85 model. The mitochondrial genome data described here are deposited in GenBank under accession numbers KP691601 (D. salina CONC-001), KP691602 (D. viridis CONC-002), KP696389 (C. leiostraca), and KP696388 (P. uvella). The D. salina CONC-001 protein-coding ptDNA genes used to measure substitution rates are in supplementary table S2, Supplementary Material online.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by Discovery Grants to D.R.S. and A.R.-P. from the Natural Sciences and Engineering Research Council (NSERC) of Canada. A.R.-P. is a Fellow of the Canadian Institute for Advanced Research, Integrated Microbial Biodiversity Program. P.M.D. is supported by Medical Research Council (SA) and National Health Laboratory Service (SA) grants. Funding for Dunaliella sequencing was supported by a NASA (#NNX13AH41G) grant to Prof. R.E. Michod (University of Arizona) and P.M.D.
Literature Cited
- Barbrook AC, Howe CJ, Kurniawan DP, Tarr SJ. Organization and expression of organellar genomes. Philos Trans R Soc Lond B Biol Sci. 2010;365:785–797. doi: 10.1098/rstb.2009.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendich AJ. Reaching for the ring: the study of mitochondrial genome structure. Curr Genet. 1993;24:279–290. doi: 10.1007/BF00336777. [DOI] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 2010;17:1519–1533. doi: 10.1089/cmb.2009.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie R, Roper RL, Upton C. JDotter: a Java interface to multiple dotplots generated by dotter. Bioinformatics. 2004;20:279–281. doi: 10.1093/bioinformatics/btg406. [DOI] [PubMed] [Google Scholar]
- Christensen AC. Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol Evol. 2013;5:1079–1086. doi: 10.1093/gbe/evt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila JI, et al. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 2011;9:64. doi: 10.1186/1741-7007-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, et al. Is ftsH the key to plastid longevity in sacoglossan slugs? Genome Biol Evol. 2013;5:2540–2548. doi: 10.1093/gbe/evt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Martinez F, Nedelcu AM, Smith DR, Reyes-Prieto A. When the lights go out: the evolutionary fate of free-living colorless green algae. New Phytol. Forthcoming 2015 doi: 10.1111/nph.13279. DOI: 10.1111/nph.13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fučíková K, Lewis PO, González-Halphen D, Lewis LA. Gene arrangement convergence, diverse intron content, and genetic code modifications in the mitochondrial genomes of Sphaeropleales (Chlorophyta) Genome Biol Evol. 2014;6:2170–2180. doi: 10.1093/gbe/evu172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez PI, González MA. The effect of temperature and irradiance on the growth and carotenogenic capacity of seven strains of Dunaliella salina (Chlorophyta) cultivated under laboratory conditions. Biol Res. 2005;38:151–162. doi: 10.4067/s0716-97602005000200005. [DOI] [PubMed] [Google Scholar]
- Gómez-Silva B, Olivares H, Rodriguez L. Microalgae from northern Chile I. La Rinconada, a hypersaline aquatic habitat in the Atacama Desert. Est Oceanol. 1990;9:73–76. [Google Scholar]
- González MA, Gómez P, Montoya R. Comparison of PCR-RFLP analysis of the ITS region with morphological criteria of various strains of Dunaliella. J Appl Phycol. 1999;10:573–80. [Google Scholar]
- González MA, Gómez PI, Polle JEW. Taxonomy and phylogeny of the genus Dunaliella. In: Ben-Amotz A, Polle JEW, Subba Rao DV, editors. The alga Dunaliella: biodiversity, physiology, genomics and biotechnology. Enfield (NH): Science Publishers; 2009. pp. 15–44. [Google Scholar]
- Hamaji T, et al. Mitochondrial and plastid genomes of the colonial green alga Gonium pectorale give insights into the origins of organelle DNA architecture within the Volvocales. PLoS One. 2013;8:e57177. doi: 10.1371/journal.pone.0057177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Smith DR, Borza T, Lee RW. Similar relative mutation rates in the three genetic compartments of Mesostigma and Chlamydomonas. Protist. 2012;163:105–115. doi: 10.1016/j.protis.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge, UK: Cambridge University Press; 1983. [Google Scholar]
- Leliaert F, et al. Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci. 2012;31:1–46. [Google Scholar]
- Nakada T, Misawa K, Nozaki H. Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol Phylogenet Evol. 2008;48:281–291. doi: 10.1016/j.ympev.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Ohta N, et al. Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2003;10:67–77. doi: 10.1093/dnares/10.2.67. [DOI] [PubMed] [Google Scholar]
- Oren A, 2005 A hundred years of Dunaliella research: 1905–2005. Saline Syst. 1:1–14. doi: 10.1186/1746-1448-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provasoli L, Pintner IJ. Artificial media for fresh-water algae: problems and suggestions. In: Tyron CA, Hartman RT, editors. The ecology of algae. Pittsburgh (PA): University of Pittsburgh Press; 1960. pp. 84–96. [Google Scholar]
- Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 2013;11:29. doi: 10.1186/1741-7007-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbens S, et al. The complete chloroplast and mitochondrial DNA sequence of Ostreococcus tauri: organelle genomes of the smallest eukaryote are examples of compaction. Mol Biol Evol. 2007;24:956–968. doi: 10.1093/molbev/msm012. [DOI] [PubMed] [Google Scholar]
- Sloan DB, et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10:e1001241. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Taylor DR. Evolutionary rate variation in organelle genomes: the role of mutational processes. In: Bullerwell C, editor. Organelle genetics. Berlin: Springer; 2012. pp. 123–146. [Google Scholar]
- Smith DR. Updating our view of organelle genome nucleotide landscape. Front Genet. 2012;3:175. doi: 10.3389/fgene.2012.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Hamaji, et al. Organelle genome complexity scales positively with organism size in volvocine green algae. Mol Biol Evol. 2013;30:793–797. doi: 10.1093/molbev/mst002. [DOI] [PubMed] [Google Scholar]
- Smith DR, Hua J, Archibald JM, Lee RW. Palindromic genes in the linear mitochondrial genome of the nonphotosynthetic green alga Polytomella magna. Genome Biol Evol. 2013;5:1661–1667. doi: 10.1093/gbe/evt122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Hua J, Lee RW. Evolution of linear mitochondrial DNA in three known lineages of Polytomella. Curr Genet. 2010;56:427–438. doi: 10.1007/s00294-010-0311-5. [DOI] [PubMed] [Google Scholar]
- Smith DR, Keeling PJ. Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci U S A. Forthcoming 2015 doi: 10.1073/pnas.1422049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Lee RW. The mitochondrial and plastid genomes of Volvox carteri: bloated molecules rich in repetitive DNA. BMC Genomics. 2009;10:132. doi: 10.1186/1471-2164-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Lee RW. Low nucleotide diversity for the expanded organelle and nuclear genomes of Volvox carteri supports the mutational-hazard hypothesis. Mol Biol Evol. 2010;27:2244–2256. doi: 10.1093/molbev/msq110. [DOI] [PubMed] [Google Scholar]
- Smith DR, Lee RW, et al. The Dunaliella salina organelle genomes: large sequences, inflated with intronic and intergenic DNA. BMC Plant Biol. 2010;10:83. doi: 10.1186/1471-2229-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhu A, Guo W, Jain K, Mower JP. Unprecedented heterogeneity in the synonymous substitution rate within a plant genome. Mol Biol Evol. 2014;31:1228–1236. doi: 10.1093/molbev/msu079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.