Abstract
Generating monoclonal antibodies from single B cells is a valuable tool for characterizing the specificity and functional properties of humoral responses. We and others developed protocols that have facilitated major advances in our understanding of B cell development, tolerance, and effector responses to HIV and influenza. Here, we demonstrate various refinements and dramatically reduce the time required to produce recombinant antibodies. Further, we present new methods for cloning and isolating antibodies from cells with lower immunoglobulin mRNA levels that may be resistant to traditional techniques. Together, these refinements significantly increase single-cell antibody expression efficiency and are easily integrated into established and novel pipelines.
Keywords: monoclonal antibodies, single-cell PCR, human antibodies, gibson assembly, antibody expression
Technical Note
Antibodies are the primary mediator of humoral immunity, so they are a recurrent focus of scientific inquiry and medical discovery. In recent years, characterization of humoral responses by production of monoclonal antibodies from single cells has enabled rapid advances in B cell biology due to the high throughput and efficiency of these methods1–3. These include the isolation of large numbers of broadly neutralizing antibodies against diverse pathogens like HIV and influenza, which are revolutionizing vaccine and therapeutic design. Single cells from a population of interest are isolated using flow cytometry then immunoglobulin genes from each cell are cloned into a vector for protein expression. The resulting antibodies are used for downstream assays to study their specificity, function, and repertoire characteristics.
A popular use for this involves screening panels of monoclonal antibodies, allowing a clonal assessment of the specificities present in a population of interest. Antibodies derived from plasmablasts, the temporary effector cells that peak 7 days after an immune response, present an opportunity to study acute infection and vaccination4,5. Memory B cells give access to affinity-matured antibodies, especially those formed during chronic or repeated infections6,7. An advantage of this method over traditional serum or hybridoma methods is that rare single cells with desirable antibodies can be isolated. Characterization of individual monoclonal antibodies yields data on specificity, binding conformation, reactivity breadth and potency, and protective capacity. Studies using these methods have shed light on potential targets for universal vaccine development and provide benchmarks for evaluating future therapeutics.
Here, we show several refinements for single-cell cloning and antibody expression that eliminate bottlenecks by replacing various single-sample manipulations with high-throughput alternatives. For large-scale projects, these changes are easily implemented and noticeably increase the pace at which antibodies can be generated. Further, our methods enable high-throughput isolation and characterization of antibodies from cells with relatively low immunoglobulin expression levels such as naïve or memory B cells. Limitations that are inherent to generating antibodies from single cells remain, including cell fragility (plasmablasts cannot be frozen and thawed) and the time-consuming nature of sorting cells and transfecting individual antibodies. Additionally, the lack of PCR error correcting mechanisms necessitate redundancy measures and the occasional redo. Instructions and primers for creating both fully human and chimeric (murine variable region and human constant region) antibodies are included.
Methods
This section describes the steps that are substantially different from our previously published protocol3. A complete protocol, which includes other modifications and quality-of-life changes, is included as supplementary material. Table 1 compares the bench time required for each step for our old protocol and this new protocol.
Table 1.
Comparison of the bench time required to perform the steps in the protocol.
| Smith, Nature Methods, 2009 |
Step | New protocol |
|---|---|---|
| Unchanged | Blood draw and flow cytometry | Unchanged |
| Unchanged | cDNA prep | Unchanged (RNA bead purification: add 45 minutes/plate) |
| Unchanged | PCR | Unchanged |
| 1 day, 100 sequences | Prepare expression vector | 15 minutes, 96 sequences |
| 1 hour, 24 sequences | Transformation | 1 hour, 96 sequences |
| Unchanged | Plasmid DNA preparation | Unchanged |
| Unchanged | Cell culture and transfection | Unchanged |
| 1 day, 24 antibodies | Antibody purification | 45 minutes, 24 antibodies |
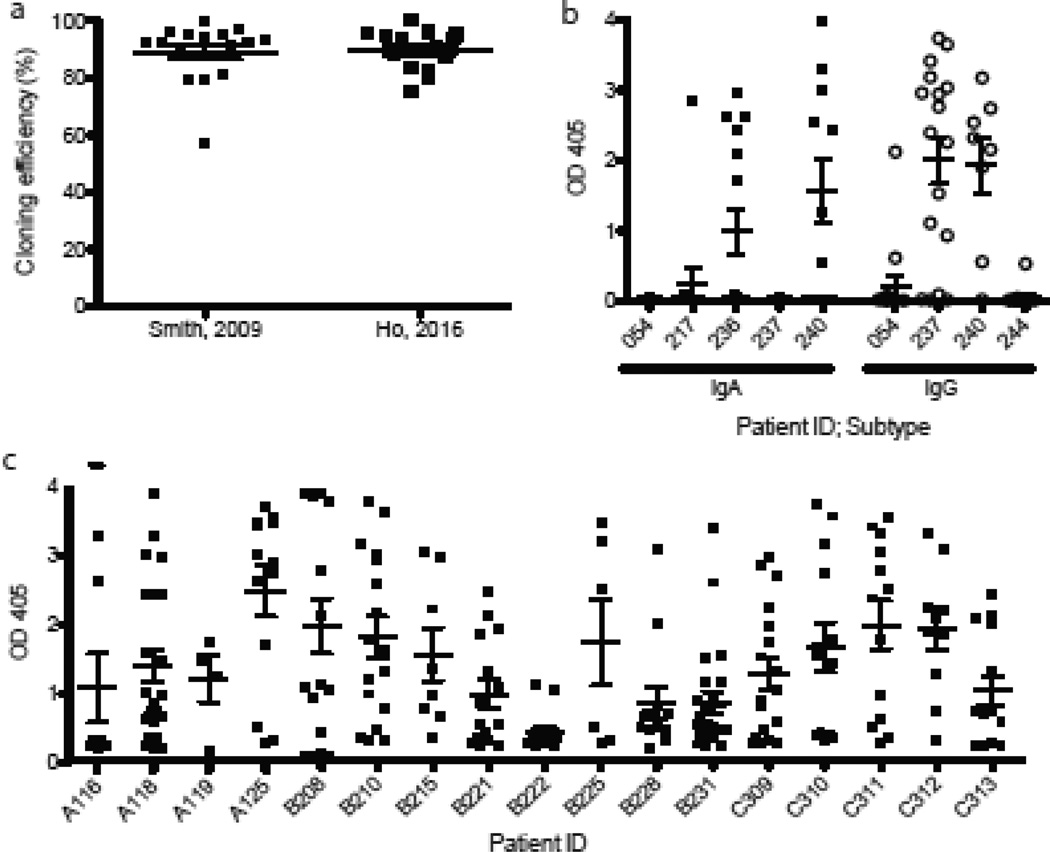
We applied this procedure to human plasmablasts isolated seven days after seasonal influenza vaccination and pneumococcal vaccination. We validated the cloning efficiency of the new protocol (Figure 1a) and confirmed the production of vaccine-specific antibodies by ELISA (Figures 1b, 1c).
Figure 1.
a, Comparison of cloning efficiency (the proportion of sequences with a consensus miniprep sequence after one attempt) for old and new protocols; each data point represents a separate plate. b, ELISA vaccine binding data for 115 influenza vaccine-induced IgG and IgA monoclonal antibodies, grouped by subtype and subject ID. c, ELISA vaccine binding data for 231 pneumococcal vaccine-induced IgG monoclonal antibodies, grouped by subject ID.
Peripheral blood samples were collected in accordance with the University of Chicago Institutional Review Board (#09-043-A) and informed consent was obtained from all subjects.
cDNA preparation
To prepare template cDNA from lysed cells, a non-specific cDNA synthesis kit is used rather than immunoglobulin-specific primers. With specific primers, for each sample plate only one of the light chain genes (lambda or kappa) can be reverse transcribed, and antibodies that use the other gene are lost. With non-specific primers, both lambda and kappa chain DNA can be recovered from the same plate.
Important: use RNase-free precautions.
Direct cDNA synthesis from sorted and lysed cells
This is efficient for most cell types, especially plasmablasts. Non-plasmablast human cells may experience slightly diminished PCR efficiency.
Note: Sort cells into 10 µl/well of catch buffer (for ten half-plates: 5 ml RNase-free water, 50 µl 1M Tris pH 8.0, 125 µl RNasin; make fresh each time)
-
1.
Create the master mix.
cDNA reaction 1 well 5x Buffer Mix 3 µl Maxima Enzyme Mix 1.5 µl 5% IGEPAL 1.5 µl -
2.
Aliquot 6 µl master mix into each well with catch buffer, for a total reaction volume of 16 µl. Run the following program. Store plates at −20°C.
-
25°C for 10 min
50°C for 30 min
85°C for 5 min
4°C forever
-
cDNA synthesis after RNA purification
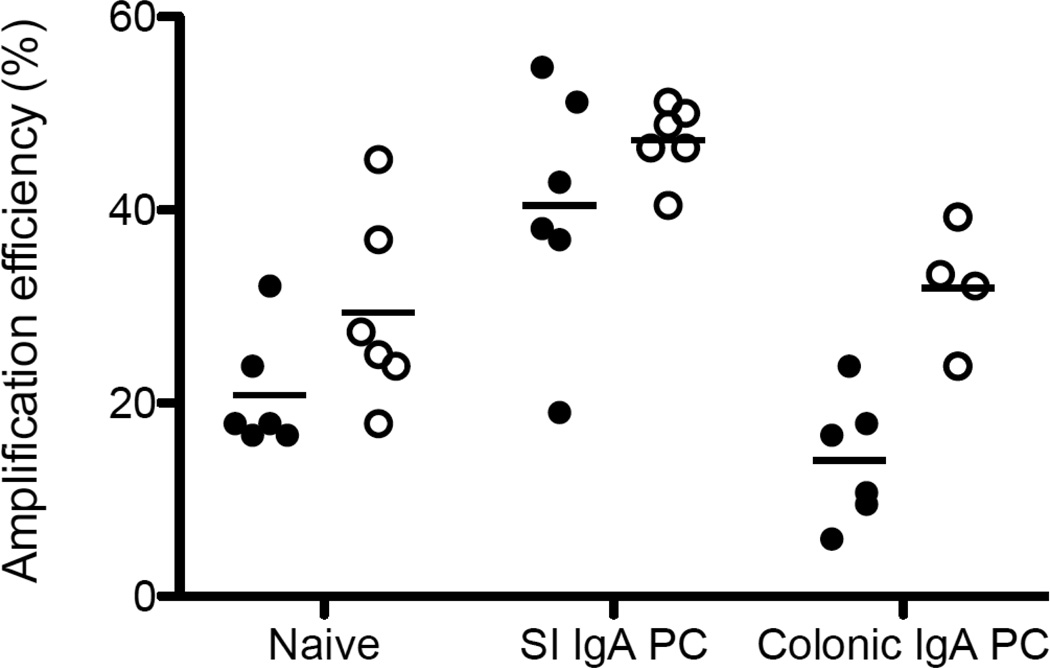
Purification with SPRI beads removes debris and other contaminants, increasing the efficiency of cDNA synthesis and subsequent PCRs8. A variety of murine non-plasmablast cells amplify very inefficiently; SPRI purification increases the yield of naïve cells, small intestinal IgA plasma cells, and colonic IgA plasma cells by 41%, 17%, and 127% respectively (Figure 2). However, this technique may be useful for all plates that have low PCR efficiency.
Note: Do not sort cells into catch buffer. Instead, use 5 µl/well of TCL buffer and 1% beta-mercaptoethanol (vol/vol). Plates can be flash-frozen on dry ice and stored at −80°C.
-
1.
Warm RNase-free SPRI beads to room temperature.
-
2.
Add 10 µl nuclease-free water to each sample, then add 33 µl SPRI beads to each sample. Cover plate to prevent contamination and incubate at room temperature for 10 min.
-
3.
Place the plate on top of the magnetic plate and incubate, covered, at room temperature for 5 minutes.
-
4.
Wash the plate 3x with 75 µl 80% EtOH, incubating 30 sec each cycle. Upon final aspiration, air dry for 8 min and remove from the magnetic plate.
-
5.
Begin cDNA synthesis by resuspending beads in 10 µl/well reverse transcription mix #1, and thermocycle.
-
cDNA reaction #1 1 well 10mM dNTPs 1.25 µl Oligo d(T)18V 1 µl Template RNA pellet - Nuclease-free water to 10 µl -
65°C for 5 min
4°C forever
-
-
6.
Incubate on ice for 1 min, then add reverse transcription mix #2; aliquot 10 µl/well and cycle. Store plates at −20°C.
-
cDNA reaction #2 1 well 5x SuperScript IV RT Buffer 4 µl 100 mM DTT 1 µl RNaseOUT 0.5 µl SuperScript IV Reverse Transcriptase 0.25 µl Nuclease-free water to 10 µl -
50°C for 1 hr
80°C for 10 min
4°C forever
-
Figure 2.
PCR amplification efficiency for various murine cell types, expressed as the percentage of wells that have a successfully amplified heavy and light chain; each data point represents a plate.
Cloning
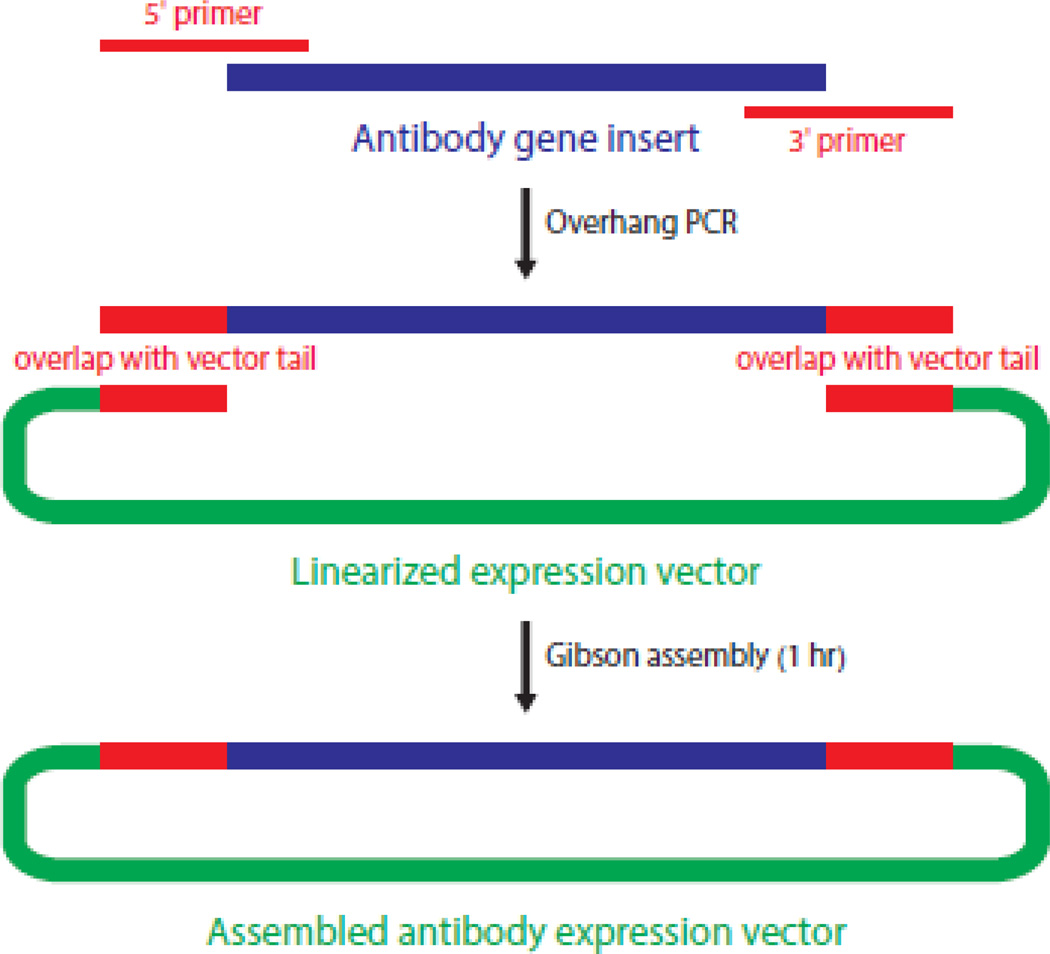
After cDNA preparation, two rounds of PCR are performed to amplify immunoglobulin genes for sequencing. The products of the first round are used as templates in a cloning PCR reaction to generate DNA for insertion into expression vectors. We designed sequence-specific primers that each contain an overhanging region that overlaps with the proper expression vector. To join the insert with the vector, we use Gibson assembly, a rapid procedure for assembling DNA fragments with overlapping ends9 (Figure 3). Overall, this process is much quicker than the traditional digest-ligation method and slightly more reliable, reducing the number of sequences that must be redone (data not shown).
Figure 3.
Schematic for cloning antibody genes into expression vectors. PCR with gene-specific overhang primers frames the antibody gene on both sides with approximately 20–25 bp of overlap with the expression vector. Gibson assembly is then used to join the two fragments.
Materials:
-
QIAquick gel extraction kit (Qiagen, 28706)
FastDigest BshTI (AgeI) (Thermo, FD1464)
FastDigest SalI (Thermo, FD0644)
FastDigest Pfl23II (BsiWI) (Thermo, FD0854)
FastDigest Xho1 (Thermo, FD0694)
FastAP (Thermo, EF0654)
GeneJET Gel Extraction Kit (Thermo, K0692)
Linearize plasmids to prepare expression vectors for assembly
(Vector sequences can be found on NCBI GenBank: accession numbers FJ475055, FJ475056 and FJ517647 for heavy, kappa, and lambda, respectively; the vectors are available upon request)
-
1.
Reaction setup
Note: 25 ng gel-purified vector is needed per assembly reaction
Vector digest Plasmid with heavy, kappa, or lambda vector 5 µg 10x FastDigest buffer 5 µl Chain-specific restriction enzyme 2.5 µl BshTI (AgeI) 2.5 µl H2O to 50 µl -
Chain-specific restriction enzymes:
-
Heavy: SalI
Kappa: Pfl23II (BsiWI)
Lambda: Xho1
-
-
2.
Incubate at 37°C for 15 minutes, then add 2.5 µl FastAP. Repeat.
-
3.
Incubate at 37°C for 10 minutes.
-
4.
Heat-inactivate at 80°C for 10 minutes.
-
5.
Run on a 1.2% agarose gel (100V for 5 min, then 75V for 75 min) and cut bands with the digested vectors from the gel for extraction.
-
6.
Purify by following the protocol outlined in the GeneJET Gel Extraction Kit with one exception: elute the final product in 40 µl EB.
Perform overlap PCR with Gibson assembly primers
Important: to avoid contamination, avoid contact with the interior of Eppendorf tubes and limit airflow over open racks. Wipe pipettes and surfaces with DNA off before use.
-
1.
Create a master mix for each combination of 5' and 3' primers (for details on choosing primers, refer to the Supplementary Methods). The template is the unpurified product of the 1st PCR step. Include a template-less negative control for each master mix of primers to ensure there is no contamination.
Human cloning PCR 1 well 5' primer (10 µM) 1 µl 3' primer (10 µM) 1 µl 2X Green DreamTaq MasterMix 12.5 µl Nuclease-free H2O 9 µl 1st PCR product 1.5 µl -
Thermocycle:
-
94°C for 4 min
Repeat 40x:
-
94°C for 30 sec
58°C for 30 sec
72°C for 45 sec
-
-
72°C for 8 min
4°C forever
-
The above PCR details are optimized for human primers; see the supplementary methods for mouse cloning PCR instructions.
-
2.
Run 2 µl PCR product directly on a 1% agarose gel to confirm a successful reaction and to check for contamination in the negative control wells.
Perform Gibson assembly
-
1.
Reaction setup (it is not necessary to purify the cloning PCR products: Gibson assembly and bacterial transformation are equally effective when using unpurified PCR products)
Gibson assembly 1 well NEBuilder 2X MasterMix 5 µl Linearized heavy, kappa, or lambda vector 25 ng Cloning PCR product 1 µl H2O to 10 µl -
2.
Incubate the reaction in a thermocycler at 50°C for 1 hr. The assembled product can be stored at −20°C before transformation.
Supplementary Material
Acknowledgments
The authors thank Carole Henry for technical discussions and assistance. This project was funded in parts with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under CEIRS contract HHSN272201400006C (P.C.W.) and NIH grants U19AI109946, P01AI097092, U19AI057266, U19AI05766-11, U19AI082724, and U19AI090023. J.J.B. was supported by NIH grant F30AI124476, NIH Medical Scientist Training Program grant T32GM007281, and the University of Chicago Digestive Diseases Research Core Center grant P30DK42086.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wardemann H. Predominant Autoantibody Production by Early Human B Cell Precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 2.Tiller T, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith K, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 2009;4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corti D, et al. A Neutralizing Antibody Selected from Plasma Cells That Binds to Group 1 and Group 2 Influenza A Hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 5.Henry Dunand CJ, et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe. 2016;19:800–813. doi: 10.1016/j.chom.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muellenbeck MF, et al. Atypical and classical memory B cells produce Plasmodium falciparum neutralizing antibodies. J. Exp. Med. 2013;210:389–399. doi: 10.1084/jem.20121970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 8.Trombetta JJ, et al. In: Current Protocols in Molecular Biology. Ausubel FM, et al., editors. John Wiley & Sons, Inc.; 2014. pp. 4.22.1–4.22.17. [Google Scholar]
- 9.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.