Abstract
Background
Genetically improved farmed tilapia (GIFT, Oreochromis niloticus) are susceptible to infection by Streptococcus iniae when maintained in modern intensive culture systems. GIFT are commercially important fishes that are cultured widely in southern China. The role of microRNAs (miRNAs) in the regulatory response of GIFT to S. iniae infection has been underestimated and has not yet been well studied. Head kidney has an important immune function in teleost fishes. The main aim of this study was to determine the possible function of miRNAs in head kidney of S. iniae-infected GIFT. MiRNAs are small, non-coding RNAs that regulate gene expression by binding to the 3’-untranslated regions of their target mRNAs. MiRNAs are known to regulate immune-regulated signaling and inflammatory response pathways.
Results
High-throughput deep sequencing of two libraries (control group [CO] and infected group [IN]) of RNA extracted from GIFT head kidney tissues generated 12,089,630 (CO) and 12,624,975 (IN) clean reads. Bioinformatics analysis identified 1736 and 1729 conserved miRNAs and 164 and 165 novel miRNAs in the CO and IN libraries, respectively. Three miRNAs (miR-310-3p, miR-92, and miR-127) were found to be up-regulated and four miRNAs (miR-92d-3p, miR-375-5p, miR-146-3p, and miR-694) were found to be down-regulated in the S. iniae-infected GIFT. The expressions of these miRNAs were verified by quantitative real-time PCR. RNAhybrid and TargetScan were used to identify complementary miRNA and mRNA target sites, and the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes databases were used to annotate and predict potential downstream regulation of biological pathways. Seven target genes, which encode immune-related proteins (complement C3, cytidine deaminase, regulator of G-protein Rgs22, mitogen-activated protein kinase Mapk1, metabotropic glutamate receptorm GluR8, calcium-sensing receptor CaSR, and microtubule-associated protein Map1S) were predicted to play crucial roles in the GIFT response to S. iniae infection.
Conclusions
S. iniae outbreaks have hindered the development of the tilapia industry in China. Understanding the miRNA transcriptome of S. iniae-infected GIFT is important for exploring the immune responses regulated by miRNAs as well as for studying novel regulated networks to prevent and treat S. iniae infections in the future.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-017-3591-z) contains supplementary material, which is available to authorized users.
Keywords: GIFT, microRNA, Deep sequencing, Target prediction, Streptococcus iniae
Background
Genetically improved farmed tilapia (GIFT, Oreochromis niloticus) is a commercially important farmed fish in southern China [1]. Data from the Food and Agriculture Organization show that tilapia output reached 5,446,800 tons in about 100 farming countries during 2014. In China, the tilapia output was estimated to be about 145 million tons in 2014. Because of its intraspecific cross and relatively rapid growth rate, GIFT is highly susceptible to bacterial diseases triggered by Streptococcus iniae and Streptococcus agalactiae when reared in intensive culture systems [2]. Thus, a better understanding of the regulatory response of GIFT to infection will provide crucial information for both immunomodulation and GIFT breeding.
The non-specific immune system in fish plays a major role at all stages of infection [3]. Thus, understanding the immune regulation mechanism may contribute to better disease prevention. MicroRNAs (miRNA) are small non-coding single-stranded RNAs (18–25 nucleotides) that are found in plants, animals and some viruses. MiRNAs function in RNA silencing and post-transcriptional regulation of gene expression by binding to the 3’-untranslated regions of their target mRNAs [4]. MiRNA-mediated regulation has been reported in the innate immune response, inflammatory diseases, cell signal transduction, and targeted therapy [5]; however, the functions of miRNAs have been studied mainly in mammalian and plants [6, 7]. The miRBase database (Release 21.0, June 2014) (http://www.mirbase.org/) contains 28,645 miRNAs in 223 species expressing 35,828 mature miRNAs. Many mature miRNAs are highly conserved across vertebrate taxa [8], but their role in the fish host immune response is known in only a few species, mainly zebrafish (Danio rerio) [9], large yellow croaker (Pseudosciaena crocea) [10], and grass carp (Ctenopharyngodon idella) [11]. Several immune-specific miRNAs that regulate innate and adaptive immune responses in fish have been identified and classified, including miR-122, miR-192, miR-194a, miR-146a/b, and miR-155. For example, miR-122 and miR-194a were markedly decreased in zebrafish infected by Vibrio harveyi, and miR-146a/b and miR-155 down-regulated Toll-like receptor 4 (TLR4) and downstream nuclear factor kB activity in zebrafish with lipopolysaccharide infection [9]. Up-regulated expression of miR-122-3p, miR-214-3p, and miR-181c was detected in infected large yellow croaker treated with polyriboinosinic: polyribocytidylic acid (poly[I:C]) [10]. Further, let-7i and miR-118 were reported to down-regulate TLR4 and nuclear factor interleukin 3-6 (NFIL3-6) in response to an excessive inflammatory response in grass carp infected with Aeromonas hydrophila [11]. Aberrant regulation of some of these miRNAs can disrupt intracellular signaling networks and inflammatory responses, which can result in pathological conditions [12].
In recent years, a growing number of genes related to immunology have been detected and identified in tilapia using various molecular biological techniques [3]. Following the successful completion of the O.niloticus genome project (http://www.ncbi.nlm.nih.gov/genome/?term=Oreochromis%20niloticus), studies on the expression and regulation mechanisms of miRNA-mediated genes in the immune response will grow in importance. However, O. niloticus miRNA transcriptome data have not yet been reported, and changes in miRNA expression profiles in the GIFT immune response to a foreign challenge have not been studied until now. The aim of this study is to identify and analyze the miRNAs and their target genes involved in the immune response of GIFT to S. iniae infection. Two small RNA libraries from GIFT infected with S. iniae for 24 h and uninfected GIFT were sequenced by high-throughput sequencing. MiRNAs were identified and RNAhybrid and TargetScan were used to predict miRNA target genes, and the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were used to explore potential biological processing of downstream targets genes. We identified miRNAs and their target genes involved in the immune response and verified the expression profiles of miRNAs by quantitative real-time PCR (qRT-PCR). Identification of miRNAs involved in immune regulation is an essential prerequisite towards better understanding of their function in intracellular signaling networks and inflammatory responses. The new insights from this study will pave the way for the development of disease control measures and for genetic improvement in GIFT.
Results and discussion
Streptococcal disease caused by S. iniae is unquestionably one of the main bacterial diseases in tilapia. It has been reported to cause huge mortality and economic loss in tilapia culture zones in more than four provinces in China, including Guangdong, Guangxi, Hainan, and Fujian. However, very few studies have described the molecular mechanisms underlying the regulation of the miRNA-mediated immune response in tilapia. The pre-experiment determined that the 96 h semi-lethal S. iniae dose (96 h LD50) in GIFT was 107CFUmL-1 (Additional file 1: Table S1). GIFT infected with a concentration of 107CFUmL-1 could stimulate their own immune systems to deal with bacterial invasion 24 h post- S. iniae infection [2, 13], the survival rate was 90%. In a previous study, we found that C-type lysozyme, heat-shock protein 70 (Hsp70), interleukin-1β (IL-1β), interferon-γ (IFN-γ), and tumor necrosis factor-α (TNF-α) expression levels in the head kidney tissue of S. iniae-infected GIFT for 96 h reached peak values 24 h post-infection [2]. However, prolonging the infection time weakened the fish’s own immune response, survival decreased rapidly at 36 h and 48 h post-infection in the pre-experimental group injected with 107 CFUmL-1. The following symptoms occurred after 24 h in GIFT infected with the 96 h LD50, diseased fish began to rotate intermittently in the water, unbalanced swimming motions, corneal opacity in the white of the eye, enlarged and reddened spleen, liver congestion, hemorrhaging kidneys, enlarged gallbladder, and dark green bile. Hemorrhaging kidneys may have resulted from lymphatic cell reduction and replacement by a large number of red blood cells, and a high rate of macrophage cell infiltration [14]. Macrophages can reduce bacterial invasion by phagocytosis. Head kidney was found to be an important immune-regulated tissue in adult fish during the early stage of bacterial infections [15], and is considered to play a similar role as bone marrow in mammals [16]. Head kidney is composed mainly of granulocytes, macrophages, and T and B lymphocytes in the presence of any acquired specific or non-specific immunoregulation [17]. Therefore, in this study, we aimed to identify miRNAs involved in resistance to pathogens infection and miRNA-mediated regulatory pathways, to better understand the immune-regulated response in tilapia head kidney triggered by a S. iniae challenge.
Two small RNA libraries from the head kidney of GIFT infected with S. iniae for 24 h (IN) and from uninfected GIFT (CO) were built and sequenced. A total of 12,423,338 and 12,845,540 raw reads were obtained from the CO and IN libraries, respectively (Table 1). After removing reads that contained adaptor sequences and low-quality reads, 12,089,630 and 12,624,975 clean reads remained in the CO and IN libraries, respectively. We found that 38.63% (CO) and 39.16% (IN) of the reads had no matches in the searched databases; these unannotated sequences were considered to be unique reads. The total clean reads contained 72.08% (CO) and 64.36% (IN) of the conserved miRNAs, and the unique reads contained 13.94% (CO) and 11.96% (IN) of the conserved miRNAs (Additional files 2 and 3: Tables S2 and S3).
Table 1.
Preliminary analysis of solexa high-throughput sequencing data
| CO library | IN library | |
|---|---|---|
| Raw reads | 12,423,338 | 12,845,540 |
| Clean reads | 12,089,630 | 12,624,975 |
| Total small RNA(18–30 nt) | 541,895 | 640,740 |
| Mapping to genome | 264,661 | 312,600 |
| Known miRNA | 1736 | 1729 |
| Novel miRNA | 164 | 165 |
We found that 264,661 (CO) and 312,600 (IN) of the unique reads and 164 and 165 of the candidate novel miRNAs could be mapped to the O. niloticus (Nile tilapia) genome sequence. The lengths of the read sequences in the two libraries ranged from 10 nt to 44 nt, with the 22-nt, 21-nt, and 23-nt long reads being the most abundant (Fig. 1). Li et al. [18] and Hong et al. [19] also reported that 22-nt long miRNAs were enriched in miRNA transcriptome libraries of both Apostichopus japonicus and Gobiocypris rarus.
Fig. 1.
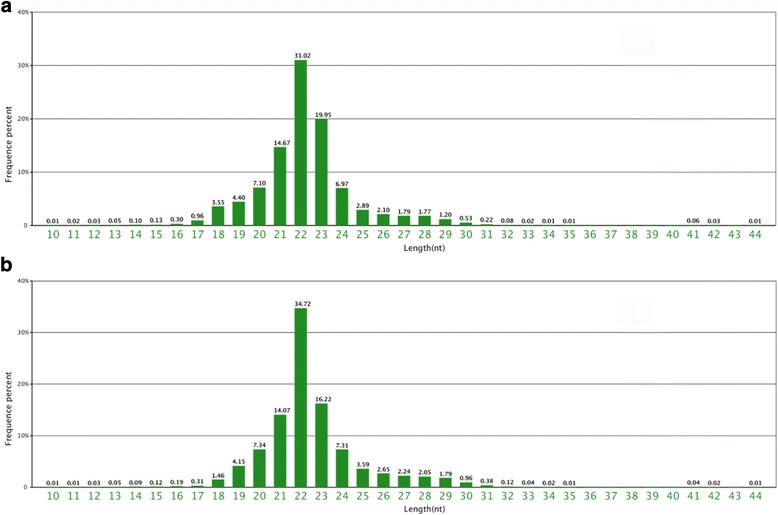
Length size distribution small RNA sequences both the control group (a) and infected group(b)
The numbers of reads in a library have been employed to estimate the abundance of the reads in a sequencing sample. The ten most abundant miRNAs in the CO library were miR-6585-5p, miR-181a, miR-181a-5p, miR-462, miR-92a-3p, miR-26-5p, miR-26a-5p, miR-10b-5p, miR-10b, and miR-26a (Fig. 2a), and they accounted for 35.4% of the reads that mapped to sequences in miRBase. Eight of these miRNAs were among the ten most abundant miRNAs in the IN library (miR-6585-5p, miR-181a, miR-92a-3p, miR-26-5p, miR-26a-5p, miR-10b-5p, miR-10b, and miR-26a) (Fig. 2b), which accounted for 40.2% of the reads that were mapped to sequences in miRBase. Among these miRNAs, miR-181a, miR-92a-3p, miR-26a-5p, miR-10b, and miR-26a have been reported to play fundamental roles in immune-regulated signaling and inflammatory responses in human [20–24].
Fig. 2.
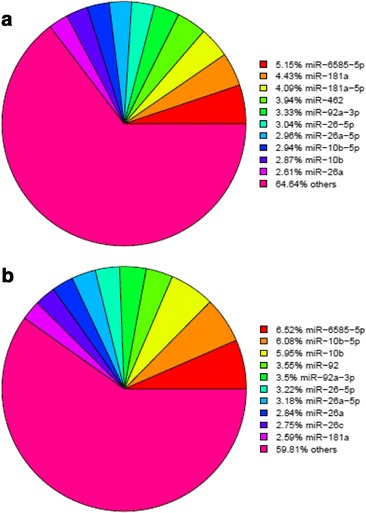
Top 10 lists of most abundant miRNAs both the control group (a) and infected group (b)
We selected and analyzed 19 differentially expressed miRNAs with more than 10 reads between the CO and IN libraries (Additional file 4: Table S4). Among the 19 miRNAs, eight (miR-92, miR-101c, miR-310-3p, miR-127, let-7, miR-599, miR-122-5p, miR-10d-5p) were significantly up-regulated and 11 (miR-92d-3p, miR-146-3p, miR-17-5p, miR-10, miR-694, miR-181b, miR-138, miR-10-5p, miR-33a, miR-375-5p, miR-10b) were significantly down-regulated in the IN library compared with the CO library (Fig. 3). We screened for putative miRNA target genes using RNAhybrid and TargetScan with the O. niloticus genome sequence. The candidate target genes were annotated with GO terms, and 22 biological processes were assigned to the predicted targets (Fig. 4). The heat map indicates that these differentially expressed miRNAs may play important roles in immune-regulation, signaling transmission, and cellular processes (Fig. 4).
Fig. 3.
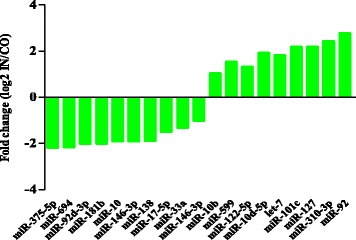
Fold changes of differentially expressed miRNAs between the control group (CO) and infected group (IN). Fold changes were calculated as log2 (IN/CO)
Fig. 4.
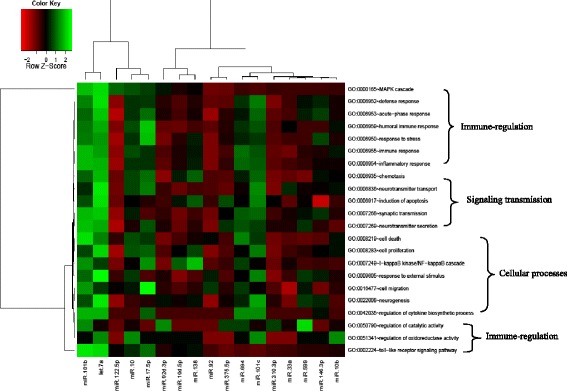
Cluster analysis of differentially expressed miRNAs. The color indicates the log2-fold change from high (red) to low (green), as indicated by the color scale. The names of the miRNAs and the clusters to which they belong are shown in the bottom of the panel. A heat map was constructed based on the 22 biological processes of the miRNA target genes indentified by a gene ontology analysis
To verify the expression levels estimated from the transcriptome sequencing data, we measured the expression levels of eight miRNAs (miR-92d-3p, miR-127, miR-310-3p, miR-146-3p, miR-599, miR-375-5p, miR-92, and miR-694) by qRT-PCR. The eight miRNAs were selected because they had relatively high fold-changes post-infection. The PCR results showed significant up-regulation of miR-310-3p, miR-92, and miR-127 and significant down-regulation of miR-92d-3p, miR-375-5p, miR-146-3p, and miR-694 in the IN group compared with the CO group (Fig. 5), in agreement with the results obtained from the transcriptome data. However, the PCR results did not show up-regulation of miR-599 expression in the IN group (Fig. 5), which contradicts the result obtained from the transcriptome data. In our study, we considered transcripts with more than 10 reads; however, Cristino et al. [25] considered that transcripts with less than 100 reads may be unreliable and only roughly measure relative miRNA abundance. The miR-599 transcript in the CO library was represented by 41 reads, which was the lowest number of reads among the eight selected miRNAs (Additional files 4: Table S4). This might explain the difference between the miR-599 expression levels estimated from the transcriptome sequencing data and by qRT-PCR.
Fig. 5.
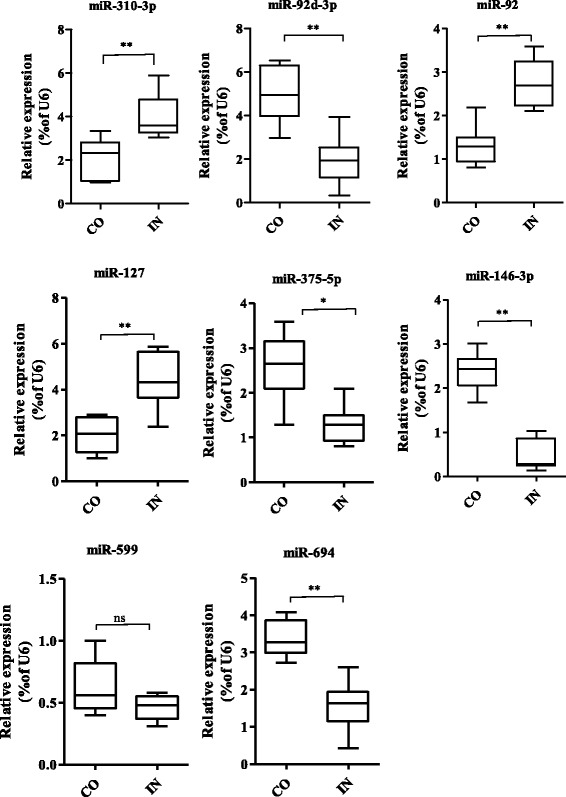
Quantitative real-time PCR validation of eight miRNA expression levels both the control group (CO) and infected group (IN). The levels of the indicated miRNAs expression were showed by Box and whisker plots (n = 10 replicates per group). The values are expressed as the relative ratio with U6 as an internal control. *P < 0.05 and **P < 0.01 by unpaired Student’s T-tests. NS, no significant difference
Previous observations have suggested that miR-146, miR-127, miR-92a, miR-310, miR-375, and miR-694 play vital immune-regulated functions in fish and mammals. For example, a recently study by Zhang et al. [26] found that miR-92a was up-regulated in sea cucumber exposed to Vibrio splendidus and lipopolysaccharides and its target gene was predicted as E3 ubiquitin-protein ligase (SMURF), which might act as a tumor suppressor by modulating the signaling pathway involved in transforming growth factor-beta(TGF-β) and bone morphogenetic protein(BMP) [27]. In breast cancer patients, down-regulated miR-92a levels were found in tumor tissues and serum [28]. Dysregulated miR-127 in transgenic cloned sheep mediated the expression of retrotransposon-like gene 1(Rtl1), a key placental development gene that may cause developmental retardation [29]. MiR-694 was shown to be down-regulated in the liver of Lass II knockout mice and may be involved in the association between liver dysregulation and metabolism dysfunction [30]. MiR-375 over-expression was found to inhibit lung surfactant secretion, possibly by influencing actin dynamicand and injury pancreas in patients [31]. As tumor suppressors or oncogenes, the miR-310/13 cluster might regulate evolutionarily conserved immune signaling networks in cancer [32].
Seven genes involved in immune-related responses were predicted to be the targets of seven miRNAs (miR-92d-3p, miR-127, miR-310-3p, miR-146-3p, miR-375-5p, miR-92, and miR-694). Complement C3 and cytidine deaminase were predicted as potential targets for miR-92d-3p and miR-127, respectively. The levels of complement C3 mRNA were reported to increase during the 24-h acute phase response of large yellow croaker (Larimichthys crocea) [33] and Miiuy croaker (Miichthys miiuy) [34] triggered by experimental infections. Up-regulation of complement C3 has been linked to innate immunity, which plays an important role in mediating tissue injury after stress. In this study, we found markedly up-regulated levels of complement C3 in the IN library (Fig. 6), suggesting its role as a potential modulator of the anti-inflammatory response defense against invading pathogens. Cytidine deaminase is a mutagenic activation-induced DNA/RNA-editing enzyme that has been implicated in human tumorigenesis. Abnormal cytidine deaminase levels were found in gastric epithelial cells infected by Helicobacter pylori, and it was suggested that this might be a mechanism responsible for the accumulation of submicroscopic deletions and somatic mutations [35]. Cytidine deaminase levels in aquatic animals have been little researched. Our results suggest that the down-regulated expression of cytidine deaminase mRNA after S.iniae infection for 24 h, may be associated with infectious diseases and immune-mediated disorders.
Fig. 6.
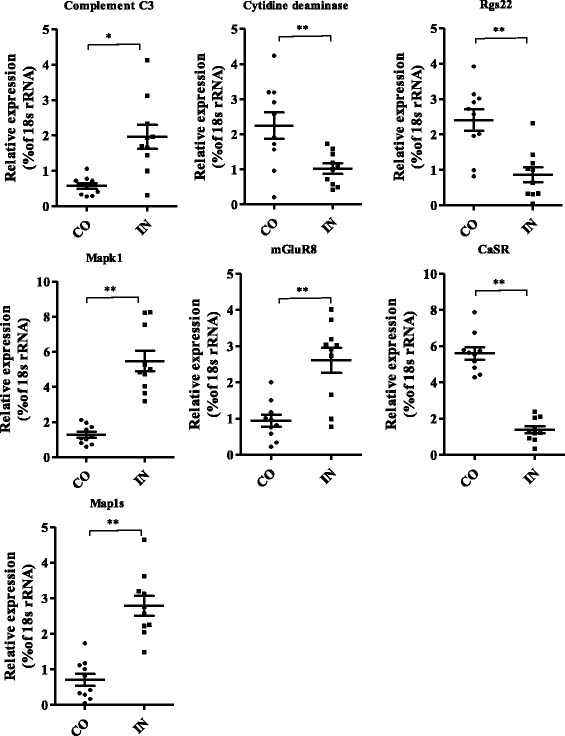
Quantitative real-time PCR validation of seven gene mRNA expression levels both the control group (CO) and infected group (IN). The expression levels of the indicated immune-regulated or signaling transmission genes in the CO and IN groups (n = 10 replicates per group) were normalized to that of 18 s rRNA. Statistical analyses were performed using unpaired Student’s t-tests. *P < 0.05 and **P < 0.01 by unpaired Student’s T-tests
Regulators of G protein signaling (Rgs) are multi-functional GTPase-accelerating proteins that have become a focus of attention in recent years [36]. Down-regulated Rgs2 was reported in androgen-independent prostate tumor cells, which were inhibited by over-expression of Rgs2 in vivo and in vitro [37]. Rgs22 was a predicted target gene of miR-310-3p in our study. MiR-310-3p was down-regulated in the IN library (Fig. 6), suggesting that the reduction of Rgs22 may be associated with an increased inflammation response against infection. Signaling pathways of mitogen-activated protein kinases (Mapks) include extracellular signal-regulated kinase1/2 (ERK1/2), p38, cMYC, and c-Jun N-terminal kinase (JNK), which mediate the activities of several transcription factors [36]. Modulation of the Mapk signaling pathway may regulate the transcription of cell cycle genes, which was shown to be essential for mediating many cellular processes, including inflammatory response, cell stress, cell differentiation, and cell proliferation [38]. TNF-α is a major pro-inflammatory cytokine in cancer cells [39]. Meili et al.[39] and Zhu et al.[40] reported that TNF-α expression affected the downstream Mapk effector pathway. A significant increase in the expression of TNF-α mRNA was found in S. iniae-infected GIFT at 24 h post-infection [2], which might lead to activation of Mapk 1 and the immune response.
Metabotropic glutamate receptors (mGluRs) are widely known for their roles in synaptic signaling. Aberrant glutamate signaling has been shown to be involved in the transformation and maintenance for human cancer, including glioma, melanoma skin cancer, breast cancer, and prostate cancer [41]. In this study, mGluR8 was identified as a potential target of miR-375-5p, and mGluR8 mRNA was found to be up-regulated in the IN library, suggesting that mGluR8 may promote inflammatory cell infiltration via activation of the Mapk signaling pathways [42]. The calcium-sensing receptor (CaSR) is an important mediator of inflammation [43–45], and dysregulation of intestinal barrier integrity was reported to be induced in CaSR knockout mice [45]. Our findings suggest that down-regulation of CaSR mRNA expression may alter the composition of the gut microbiota and disrupt immune homeostasis in S. iniae-infected GIFT at 24 h post-infection.
Microtubule-associated protein 1S (Map1s) was the predicted target gene of miR-694. As a novel member of the microtubule-associatedprotein 1 (Map1) family, Map1s is important in inducing microtubule stabilization, bonding the microtubule organizing center to the centrosome, and regulation of mitotic cell cycle progression [46]. Zou et al. [47] reported that the interaction of suppressor of cytokine signaling 3 (SOCS3) and Map1s, and the integrity of the microtubule cytoskeleton was an important modulator of interleukin-6 (IL-6) signaling. Our results suggest that Map1s may be crucial in the response to pathogenic infection, and the regulation of Map1s by miR-694 may be involved in inflammatory regulating networks.
Conclusions
In this study, we used high-throughput sequencing and analysis to screen for different miRNAs between normal and infected tilapia. We confirmed the expression levels of seven differentially expressed miRNAs and their predicted target genes, which may play essential roles in triggering the immune response of head kidney in S. iniae-infected GIFT at 24 h post-infection. These miRNAs and their putative target genes were associated mainly with autoimmune inflammation and regulation of cytokine signaling. Our findings provide insights into the role of miRNAs in the immune system of GIFT, and will help to form the basis for understanding miRNA-mediated control of gene expression against infection in GIFT. Further in vivo and invitro studies are needed to gain more insight into the association and the regulatory response between miRNAs and their predicted target genes, biological pathways, and molecular pathogenesis in tilapia.
Methods
Sample collection
Healthy adult male GIFT were obtained from the Yixing tilapia farm at the Freshwater Fisheries Research Center of the Chinese Academy of Fishery Sciences (Wuxi, China). The average weight of the fish was 350 ± 19 g. The fish were reared in indoor concrete tanks and acclimated for 2 weeks before treatment. During acclimation, the water temperature was maintained at 28 ± 0.3 °C and the fish were kept under a natural photoperiod and continuous aeration.
S. iniae bacteria (CMS No-005) were provided by the biotechnology laboratory at the Freshwater Fisheries Research Center of the Chinese Academy of Fishery Sciences [48]. The bacteria were inoculated into brain heart infusion (BHI) broth and incubated in a shaker (180 rpm) incubator at 28 °C overnight. The broth was centrifuged at 4000 × g for 5 min, washed twice with 0.65% sterile saline, We used turbidimetric methods to roughly estimate the stock solution concentration. We employed 10-fold serial dilutions to make five concentration gradients [105; 106; 107; 108 and 109 colony forming unit (CFU) per ml] and determined the 96 h LD50. In the pre-experiment, 100 experimental fish were randomly distributed in five 800-L tanks each containing 20 fish. We injected different concentrations of bacteria intraperitoneally. The bacterial concentration was determined by plating 1 ml of the 10-fold serial dilutions onto BHI agar plates. In the formal experiment, the final concentration was 6.35 × 107CFUmL-1. Fifty fish samples were assigned randomly to two groups of 25 samples each. One group was injected intraperitoneally with S. iniae at a concentration of 6.35 × 107CFUmL−1 as the infected group (IN). We injected 300 μL/100 g fish weight of bacterial suspension into the abdominal cavity of the GIFT. The other group was injected with the same volume of 0.65% physiological saline as the control (CO). Fifteen fish from each treatment were anesthetized using 0.01% tricaine methanesulfonate and sacrificed 24 h after injection. The head kidney tissues were collected and immediately stored in liquid nitrogen at −80 °C for transcriptome sequencing and qRT-PCR.
Construction of small RNA libraries by high-throughput sequencing
Total RNA was isolated from head kidney tissues using a miRNeasy kit (Takara, Dalian, China) according to the manufacturer’s protocol. The total RNA quantity and quality were determined using an Agilent 2100 Bioanalyzer (Agilent, Germany). We selected the samples with RNA integrity numbers more than 8.0 for further processing. The samples, which contained equal amounts of RNA extracted from the head kidney tissues of five fish (CO group or IN group), were mixed and pooled to construct the miRNA libraries. The two small RNA libraries were sequenced and built according to standard methods [49] at the Beijing Genomic Institute, Shenzhen, China (BGI, China).
Analysis of basic sequencing data
Low quality reads and reads contaminated with adapter sequences were removed before analysis [11]. The filtered sequences between 18-nt and 30-nt long were aligned to the non-redundant animal miRNA sequences in miRBase (Release 21.0) (http://www.mirbase.org/), allowing no more than two mismatches outside the seed region. Reads that matched the miRNAs from other animals were considered to be conserved miRNAs in O.niloticus. Based on their similarity (identity score) to know miRNAs, we classified the conserved miRNAs into the corresponding miRNA families. The secondary structures of the remaining unidentified miRNAs were predicted using MIREAP (https://sourceforge.net/projects/mireap/) [50] and reads that could form the characteristic hairpin structure were classified as novel miRNAs.
Identification of differentially expressed miRNAs
To determine the differential expression levels of the miRNAs between the CO and IN libraries, the sequencing data were normalized [51] and the fold-change was determined as: fold-change = log 2(IN/CO). The data from the two libraries were statistically analyzed based on the Audic and Claverie method [52].
Prediction of target genes and bioinformatic analysis
We used TargetScan (http://www.targetscan.org/) and RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid) to predict the potential target genes of the differentially expressed miRNAs. The criteria used for target prediction were according to Zhang et al. [53]. We used the O.niloticus transcriptome sequence data (http://www.ncbi.nlm.nih.gov/genome/?term=Oreochromis%20niloticus) to screen and annotate immune-related target genes for further study. The GO (http://www.geneontology.org) and KEGG (http://www.genome.jp/kegg/pathway.html) databases were used to assign terms and pathways to the target genes to determine their potential downstream biological functions.
Quantitative real-time PCR
The head kidney tissues from the 10 remaining fish in each group were used for qRT-PCR. A Mir-X™ miRNA First-Strand Synthesis kit (Takara, Dalian, China) was used to synthesize first-strand cDNA. The 10.0 μL reverse transcription (RT) reaction mixture contained 5.0 μL 2 × mRQ Buffer, 3.75 μL of the RNA sample (0.25-8 μg), and 1.25 μL of mRQ Enzyme. The reaction mixtures were incubated at 37 °C for 60 min, 85 °C for 5 min, followed by hold at 4 °C. The miRNA expression levels were determined using a miRNA SYBR Green qRT-PCR Kit (Takara, Dalian, China) with the provided miRNA universal primer (U6). The 25 μL PCR mixture included 2.0 μL of RT product, 12.5 μL 2 × SYBR Advantage Premix, 9 μL ddH2O, 0.5 μL 50× ROX Dye, 0.5μLmiRNA-specific Primer (10 μM), and 0.5 μL mRQ 3’ Primer. The reactions were performed at 95 °C for 10 s, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 20s, with a final dissociation step at 95 °C for 60 s, 55 °C for 30s, and 95 °C for 30s. The miRNA specific primers (Table 2) were synthesized by Genewiz Inc., Suzhou, China.
Table 2.
Primer design of miRNA
| Name | Primer sequence (5’-3’) |
|---|---|
| miR-92 | AATTGCACTCGTCCCGGC |
| miR-310-3p | TATGGCACTTCTCCCGGCCTG |
| miR-127 | ATCGGATCCGTACTAGTGGTG |
| miR-599 | GAAGTGTTACTGTGGAGGTCT |
| miR-92d-3p | TATGGCACTTATCCCGGCC |
| miR-146-3p | AGCGCTGTGGATCTTCGGTTTTC |
| miR-694 | CTGAAAATGTATGGCGCTGGAG |
| miR-375-5p | ACTTGAGCGCGTGCGATAG |
PrimeScript™ RT Master Mix (Takara, Dalian, China) was used for the RT reaction of the target genes. The 10.0 μL RT reaction mixture included 2.0 μL 5 × PrimeScript RT Master Mix, the RNA sample (≤500 ng), and RNase Free dH2O up to 10 μL. The reactions were incubated at 37 °C for 15 min, 85 °C for 5 s, followed by hold at 4 °C. The qRT-PCR was performed using SYBR® Premix Ex Taq kits(Takara, Dalian, China). The 20 μL PCR reaction mixture contained 2.0 μL of RT product, 10.0 μL 2 × SYBR® Premix Ex Taq II, 0.8 μL each of the forward and reverse primers (10 μM), 0.4 μL 50× ROX Dye, and 6 μL ddH2O. The reaction mixtures were incubated at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, and 60 °C for 30s. The primers are listed in Table 3. The transcript level of 18S rRNA was taken as the reference to calculate expression levels of the target genes. The miRNA and mRNA expression levels were analyzed using the 2−ΔΔCt method [54], and the values related to the lower group represented the n-fold difference. The miRNA and mRNA expression levels were quantified using the ABI 7900HT Fast Real-Time PCR System (ABI, USA) and compared using the relative quantification (RQ) manager software. We used unpaired Student’s T-tests to analyze the qRT-PCR expression results.
Table 3.
Primer design of putative target genes
| Name | Primer sequence (5’-3’) |
|---|---|
| Complement C3 | F: 5’-CAGGCAGGAGGATGTATCGG-3’ R: 3’- TGCCAGCGTCAAGTCTTTTCT-5’ |
| Cytidine deaminase | F: 5’-CAGGCTGCAATGTGGAGAAT-3’ R: 3’-TGGTCCAACAAAGCGGTCTT-5’ |
| Rgs22 | F: 5’-TAGAGCGGCAGCGTTTGTG-3’ R: 3’-TGATCCACTGCATTCCTTGC-5’ |
| Mapk1 | F: 5’-TCAGCCCCTTTGAGCACC-3’ R: 3’-TGCACGGATAATGTCGTTGATT-5’ |
| CaSR | F: 5’-AAAATCTATGATGCTTGTGGCTCC-3’ R: 3’-ATTGCCATGCAAGGGGAG-5’ |
| Map1s | F: 5’-CTCCTCACTGGTCGCTAAAACT-3’ R: 3’-GGGACTGGTAGAAAGGGGTATGT-5’ |
| mGluR8 | F: 5’-TCAGCGAATCCCCACAGAC-3’ R: 3’-TGGGTAGCAGTTCGGGGTC-5’ |
| 18 s rRNA | F: 5’-GGCCGTTCTTAGTTGGTGGA-3’ R: 3’-TTGCTCAATCTCGTGTGGCT-5’ |
Acknowledgements
We thank two anonymous reviewers and the editors for their constructive comments and suggestions to improve the manuscript.
Funding
The study was supported financially by grants from the Special Fund for National Natural Science Foundation of China (Project 31502143), the National Science & Technology Program (2012BAD26B03-1), and the Central Public-interest Scientific Institution Basal Research Fund, Freshwater Fisheries Research Center, CAFS (NO.2017JBFM08).
Availability of data and material
All data supporting the conclusions of this article are included within the article and its Additional files 1, 2, 3 and 4.
Authors’ contributions
XP and QJ conceived and designed the expreriment, QJ and HJ conceived and implemented the database and conducted all bioinformatic analyses together with TYF. BJW and SYL sampled the head kidney at 24 h post-S. iniae. and extracted RNA with HJ. TYF and BJW carried out the functional annotation analysis and verified experiment of miRNA and their target genes. QJ wrote the paper with contributions from TYF, BJW, XP, HJ and SYL. All authors read and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
The study protocols were approved by the Freshwater Fisheries Research Center of the Chinese Academy of Fishery Sciences (Wuxi, China). The fish were maintained in well-aerated water and anesthetized by injecting 0.01% tricaine methanesulfonate (Sigma, USA) before the head kidney tissue was extracted based on the Guide for the Care and Use of Laboratory Animals in China.
Abbreviations
- 96 h LD50
96 h semi-lethal S. iniae dose
- BHI
Brain heart infusion
- BMP
Bone morphogenetic protein
- CaSR
Calcium-sensing receptor
- CFU
Colony forming units
- ERK1/2
Extracellular signal-regulated kinase1/2
- GAP
GTPase-activating protein
- GIFT
Genetically improved farmed tilapia
- GO
Gene ontology
- GPCR
G-protein coupled receptor
- HECT
Homologous to E6AP C-terminus
- HSP70
Heat-shock protein 70
- IFN-γ
Interferon-γ
- IL-1β
Interleukin-1β
- IL-6
Interleukin-6
- JNK
c-Jun N-terminal kinase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LPS
Lipopolysaccharide
- Map1s
Microtubule-associated protein 1S
- Mapk
Mitogen-activated protein kinase
- mGluR
Metabotropic glutamate receptor
- miRNA
microRNA
- NFIL3–6
Nuclear factor interleukin 3–6
- qRT-PCR
Quantitative real-time PCR
- Rgs
Regulators of G protein signaling
- RQ
Relative quantification
- RT
Reverse transcription
- Rtl1
Retrotransposon-like gene 1
- SMURF
E3 ubiquitin-protein ligase
- SOCS3
Cytokine signaling 3
- TGF-β
Transforming growth factor-beta
- TLR4
Toll-like receptor4
- TNF-α
Tumor necrosis factor-α
Additional files
Cumulative mortality of GIFT at the injected dose of 105; 106; 107; 108 and 109 CFU ml-1 respectively for 96 h. (DOCX 13 kb)
Statistics of the small RNA reads in the CO library. The GIFT were injected with the 0.65% physiological saline as the control (CO). The small RNA reads of CO group were analyzed and built by deep-sequencing. (DOCX 13 kb)
Statistics of the small RNA reads in the IN library. The GIFT were was injected intraperitoneally with S. iniae at a concentration of 6.35 × 107CFUmL−1 as the infected group (IN). The small RNA reads of IN group were analyzed and built by deep-sequencing. (DOCX 13 kb)
Some of the miRNAs that were differentially expressed between the CO and IN libraries. Eight (miR-92/101c/310-3p/127/599/122-5p/10d-5p and let-7) were significantly up-regulated and 11 (miR-92d-3p/146-3p/17-5p/10/694/181b/138/10-5p/33a,/375-5p/10b) were significantly down-regulated in the IN library compared with the CO library based on deep-sequencing results. (DOCX 15 kb)
Contributor Information
Jun Qiang, Email: qiangjunn@163.com.
Fanyi Tao, Email: 13921176236@163.com.
Jie He, Email: Hej@ffrc.cn.
Lanyi Sun, Email: Sunyl@ffrc.cn.
Pao Xu, Email: Xup@ffrc.cn.
Wenjin Bao, Email: dangsnow@163.com.
References
- 1.Qiang J, Yang H, Wang H, Kpundeh MD, Xu P. Interactive effects of temperature-dietary protein level on somatotropic gene expression and its interrelationship with growth in juvenile GIFT tilapia. Oreochromis niloticus. Aquaculture. 2012;364–365:263–71. doi: 10.1016/j.aquaculture.2012.08.021. [DOI] [Google Scholar]
- 2.Qiang J, He J, Yang H, Xu P, Habte-Tsion HM, Ma XY, Zhu ZX. The changes in cortisol and expression of immune genes of GIFT tilapia Oreochromis niloticus (L.) at different rearing densities under Streptococcus iniae infection. Aquacult Int. 2016;24:1365–1378. doi: 10.1007/s10499-016-9995-y. [DOI] [Google Scholar]
- 3.Zhu JJ, Li C, Ao QW, Tan Y, Luo YJ, Guo YF, Lan GQ, Jiang HS, Gan X. Trancriptomic profiling revealed the signatures of acute immune response in tilapia (Oreochromis niloticus) following Streptococcus iniae challenge. Fish Shellfish Immunol. 2015;46:346–53. doi: 10.1016/j.fsi.2015.06.027. [DOI] [PubMed] [Google Scholar]
- 4.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–26. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Bergman P, James T, Kular L, Ruhrmann S, Kramarova T, Kvist A, Supic G, Gillett A, Pivarcsi A, Jagodic M. Next-generation sequencing identifies microRNAs that associate with pathogenic autoimmune neuroinflammation in rats. J Immunol. 2013;190:4066–75. doi: 10.4049/jimmunol.1200728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–9. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 7.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–51. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Friedman RC, Farh KKH, Burge CB, Bartel D-P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu TH, Pan CY, Lin MC, Hsieh JC, Hui CF, Chen JY. In vivo screening of zebrafish microRNA responses to bacterial infection and their possible roles in regulating immune response genes after lipopolysaccharide stimulation. Fish Physiol Biochem. 2012;8:1299–310. doi: 10.1007/s10695-012-9617-1. [DOI] [PubMed] [Google Scholar]
- 10.Qi PZ, Guo BY, Zhu AY, Wu CW, Liu CL. Identification and comparative analysis of the Pseudosciaena crocea microRNA transcriptome response to poly(I:C) infection using a deep sequencing approach. Fish Shellfish Immunol. 2014;39:483–91. doi: 10.1016/j.fsi.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Xu X, Shen Y, Fu J, Lu L, Li J. Next-generation sequencing identified microRNAs that associate with motile aeromonad septicemia in grass carp. Fish Shellfish Immunol. 2015;45:94–103. doi: 10.1016/j.fsi.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 12.Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011;30:1977–89. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiang J, Yang H, Wang H, Xu P, Qi ZL, He J. Studies on blood biochemical indices and expression of hepatic HSP70 mRNA of different tilapia strains artificially challenged with Streptococcus iniae. J Fisheries China. 2012;36:958–968. doi: 10.3724/SP.J.1231.2012.27883. [DOI] [Google Scholar]
- 14.Zhu JL, Qi ZL, Li DY, Zou ZY, Xiao W, Yue YR, et al. Pathological changes in tilapia(Oreochromis niloticus) naturally infected by Streptococcus iniae. J Fisheries China. 2014;38:722–730. [Google Scholar]
- 15.Agnewa W, Barnes AC. Streptococcus iniae: An aquatic pathogen of global veterinary significance and a challenging candidate for reliable vaccination. Vet Microbiol. 2007;122:1–15. doi: 10.1016/j.vetmic.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Press CML, Evensen O. The morphology of the immune system in teleost fishes. Fish Shellfish Immunol. 1999;9:309–18. doi: 10.1006/fsim.1998.0181. [DOI] [Google Scholar]
- 17.Chen J, Li C, Huang R, Du FK, Liao LJ, Zhu ZY, Wang YP. Transcriptome analysis of head kidney in grass carp and discovery of immune-related genes. BMC Genomics. 2012;8:108. doi: 10.1186/1746-6148-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li CH, Feng WD, Qiu LH, Xia CG, Su XR, Jin CH, Zhou TT, Zeng Y, Li TW. Characterization of skin ulceration syndrome associated microRNAs in sea cucumber Apostichopus japonicus by deep sequencing. Fish Shellfish Immunol. 2012;33:436–41. doi: 10.1016/j.fsi.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 19.Hong XS, Qin JH, Chen R, Yuan LL, Zha JM, Wang ZJ. Identification and characterization of novel and conserved microRNAs in several tissues of the Chinese rare minnow (Gobiocypris rarus) based on illumina deep sequencing technology. BMC Genomics. 2016;17:283. doi: 10.1186/s12864-016-2606-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123:150–63. doi: 10.1172/JCI64946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Della VSG, Calura E, Di Marino M, Romualdi C, Beltrame L, Malapelle U, Troncone G, De SA, Pepe S, De Placido S, D’Incalci M, Marchini S, Carlomagno C. Analysis of differential miRNA expression in primary tumor and stroma of colorectal cancer patients. Biomed Res Int. 2014;2014:840921. doi: 10.1155/2014/840921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghanbari R, Mosakhani N, Asadi J, Nouraee N, Mowla SJ, Yazdani Y, Mohamadkhani A, Poustchi H, Knuutila S, Malekzadeh R. Downregulation of Plasma MiR-142-3p and MiR-26a-5p in Patients With Colorectal Carcinoma. Iran J Cancer Prev. 2015;8:e2329. doi: 10.17795/ijcp2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M. Role of miR-10b in breast cancer metastasis. Breast Cancer Res. 2010;12:210. doi: 10.1186/bcr2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang C, Yu H, Sun Q, Zhu W, Xu J, Gao N, Zhang R, Liu L, Wu X, Yang X, Meng L, Lu S. Extracellular microRNA-21 and microRNA-26a increase in body fluids from rats with antigen induced pulmonary inflammation and children with recurrent wheezing. BMC Pulm Med. 2016;16:50. doi: 10.1186/s12890-016-0216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cristino AS, Tanaka ED, Rubio M, Piulachs MD, Belles X. Deep sequencing of organ- and stage-specific microRNAs in the evolutionarily basal insect Blattella germanica (L.) (Dictyoptera, Blattellidae) PLoS One. 2011;6:e19350. doi: 10.1371/journal.pone.0019350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang PJ, Li CH, Shao Y, Chen XC, Li Y, Su XR, Li TW. Identification and characterization of miR-92a and its targets modulating Vibrio splendidus challenged Apostichopus japonicus. Fish Shellfish Immunol. 2014;38:383–8. doi: 10.1016/j.fsi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 27.David D, Nair SA, Pillai MR. Smurf E3 ubiquitin ligases at the cross roads of oncogenesis and tumor suppression. Biochim Biophys Acta. 1835;2013:119–28. doi: 10.1016/j.bbcan.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Li M, Guan X, Sun Y, Mi J, Shu X, Liu F, Li C. miR-92a family and their target genes in tumorigenesis and metastasis. Exp Cell Res. 2014;323:1–6. doi: 10.1016/j.yexcr.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 29.Ni W, Shuang Y, Cao Y, Li CY, Wei JC, Wang DW, Qiao J, Zhao XX, Hu SW, Quan RZ. Aberrant expression of miR-127, miR-21 and miR-16 in placentas of deceased cloned sheep. Res Vet Sci. 2016;105:200–4. doi: 10.1016/j.rvsc.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 30.Chen YY. The effect of hepatocyte-specific Lass 2 knockout mice on microRNA-694 and lipid metabolism. In: Ph.D. Thesis. Zhenjiang: Jiangsu University. 2015;1–44 (In Chinese).
- 31.Yu L, Todd NW, Xing LX, Xie Y, Zhang H, Liu ZQ, Fang HB, Zhang J, Katz RL, Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870–8. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pancratov R, Peng F, Smibert P, Yang S, Jr, Olson ER, Guha-Gilford C, Kapoor AJ, Liang FX, Lai EC, Flaherty MS, DasgGupta R. The miR-310/13 cluster antagonizes β-catenin function in the regulation of germ and somatic cell differentiation in the Drosophila testis. Development. 2013;140:2904–16. doi: 10.1242/dev.092817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang HL, Qi PZ, Guo BY, Li JJ, He JY, Wu CW, Gu Y. Molecular characterization and expression analysis of a complement component C3 in large yellow croaker (Larimichthys crocea) Fish Shellfish Immunol. 2015;42:272–9. doi: 10.1016/j.fsi.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 34.Sun YY, Wang RX, Xu TJ. Conserved structural complement component C3 in miiuy croaker Miichthys miiuy and their involvement in pathogenic bacteria induced immunity. Fish Shellfish Immunol. 2013;35:184–7. doi: 10.1016/j.fsi.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto Y, Marusaw H, Kinoshit K, Niw Y, Sakai Y, Chib T. Up-regulation of activation-induced cytidine deaminase causes genetic aberrations at the CDKN2b-CDKN2a in gastric cancer. Gastroenterology. 2010;139:1984–94. doi: 10.1053/j.gastro.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 36.Dohlman HG, Thorner J. RGS proteins and signaling by heterotrimeric G proteins. J Biol Chem. 1997;272:3871–74. doi: 10.1074/jbc.272.7.3871. [DOI] [PubMed] [Google Scholar]
- 37.Wolff DW, Xie Y, Deng C, Gatalica Z, Yang M, Wang B, Wang J, Lin MF, Abel PW, Tu Y. Epigenetic repression of regulator of G-protein signaling 2 promotes androgen-independent prostate cancer cell growth. Int J Cancer. 2012;130:1521–31. doi: 10.1002/ijc.26138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Liu HT. LIUMAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 39.Meili N, Christen V, Fent K. Nodularin induces tumor necrosis factor-alpha and mitogen-activated protein kinases (MAPK) and leads to induction of endoplasmic reticulum stress. Toxicol Appl Pharm. 2016;300:25–33. doi: 10.1016/j.taap.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Zhu WW, Kong GQ, Ma MM, Li Y, Huang X, Wang LP, Peng ZY, Zhang XH, Liu XY, Wang XZ. Camel milk ameliorates inflammatory responses and oxidative stress and downregulates mitogen-activated protein kinase signaling pathways in lipopolysaccharide-induced acute respiratory distress syndrome in rats. J Dairy Sci. 2016;99:53–6. doi: 10.3168/jds.2015-10005. [DOI] [PubMed] [Google Scholar]
- 41.Yu LJ, Wall B, Wangari-Talbot J, Chen S. Metabotropic glutamate receptors in cancer. Neuropharmacology. 2016. http://dx.doi.org/10.1016/j.neuropharm.2016.02.011. [DOI] [PMC free article] [PubMed]
- 42.Zhang ZC, Hu F, Liu YF, Ma B, Chen XL, Zhu K, Shi Y, Wei T, Xing Y, Gao YN, Lu HX, Liu Y, Kang QY. Activation of type 5 metabotropic glutamate receptor promotes the proliferation of rat retinal progenitor cell via activation of the PI-3-K and MAPK signaling pathways. Neuroscience. 2016;322:138–51. doi: 10.1016/j.neuroscience.2016.02.030. [DOI] [PubMed] [Google Scholar]
- 43.Hendy GN, Canaff L. Calcium-sensing receptor, proinflammatory cytokines and calcium homeostasis. Semin Cell Devl Biol. 2016;49:37–43. doi: 10.1016/j.semcdb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Klein GL, Castro SM, Garofalo RP. The calcium sensing receptor as a mediator of inflammation. Semin Cell Devl Biol. 2016;49:52–6. doi: 10.1016/j.semcdb.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Owen JL, Cheng SX, Ge Y, Sahay B, Mohamadzadeh M. The role of the calcium-sensing receptor in gastrointestinal inflammation. Semin Cell Devl Biol. 2016;49:44–51. doi: 10.1016/j.semcdb.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orban-Nemeth Z, Simader H, Badurek S, Trancikova A, Propst F. Microtubule-associated protein 1S, a short and ubiquitously expressed member of the microtubule-associated protein 1 family. J Biol Chem. 2005;280:2257–65. doi: 10.1074/jbc.M408984200. [DOI] [PubMed] [Google Scholar]
- 47.Zou TT, Ouyang L, Chen L, Dong W, Qiao H, Liu YL, Qi YP. The role of microtubule-associated protein 1S in SOCS3 regulation of IL-6 signaling. FEBS Lett. 2008;582:4015–22. doi: 10.1016/j.febslet.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 48.Qiang J, Yang H, Wang H, Kpundeh MD, Xu P. Interacting effects of water temperature and dietary protein level on hematological parameters in Nile tilapia juveniles, Oreochromis niloticus (L.) and mortality under Streptococcus iniae infection. Fish Shellfish Immunol. 2013;34:8–16. doi: 10.1016/j.fsi.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Tan TT, Chen M, Harikrishna JA, Khairuddin N, Shamsudin MIM, Zhang GJ, Bhassu S. Deep parallel sequencing reveals conserved and novel miRNAs in gill and hepatopancreas of giant freshwater prawn. Fish Shellfish Immunol. 2013;35:1061–9. doi: 10.1016/j.fsi.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 50.Yuan C, Wang X, Geng R, He X, Qu L, Chen YL. Discovery of cashmere goat (Capra hircus) microRNAs in skin and hair follicles by Solexa sequencing. BMC Genomics. 2013;14:511. doi: 10.1186/1471-2164-14-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu E, Zhao F, Xu G, Hou H, Zhou L, Li X, Sun ZS, Wu JY. mirTools: microRNA profiling and discovery based on high-throughput sequencing. Nucleic Acids Res. 2010;38(Web Server issue):W392–7. doi: 10.1093/nar/gkq393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Audic S, Claverie JM. The significance of the digital expression profiles. Genome Res. 1997;7:986–95. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 53.Zhang DD, Lu KL, Dong ZJ, Jiang GZ, Xu WN, Liu WB. The effect of exposure to a high-fat diet on microRNA expression in the liver of blunt snout bream (Megalobrama amblycephala) PLoS One. 2014;9(5):e96132. doi: 10.1371/journal.pone.0096132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2-△△CT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]


