Abstract
Adenovirus serotype 5 (Ad5) vaccine vectors elicit robust CD8+ T cell responses, but these responses typically exhibit a partially exhausted phenotype. However, the immunologic mechanism by which Ad5 vectors induce dysfunctional CD8+ T cells has not previously been elucidated. Here we demonstrate that, following immunization of B6 mice, Ad5 vectors elicit antigen-specific IL-10+CD4+ T cells with a distinct transcriptional profile in a dose-dependent fashion. In rhesus monkeys, we similarly observed upregulated expression of IL-10 and PD-1 by CD4+ T cells following Ad5 vaccination. These cells markedly suppressed vaccine-elicited CD8+ T cell responses in vivo and IL-10 blockade increased the frequency and functionality of antigen-specific CD8+ T cells as well as improved protective efficacy against challenge with recombinant Listeria monocytogenes. Moreover, induction of these inhibitory IL-10+CD4+ T cells correlated with IL-27 expression and IL-27 blockade substantially improved CD4+ T cell functionality. These data highlight a role for IL-27 in the induction of inhibitory IL-10+CD4+ T cells, which suppress CD8+ T cell magnitude and function following Ad5 vector immunization. A deeper understanding of the cytokine networks and transcriptional profiles induced by vaccine vectors should lead to strategies to improve the immunogenicity and protective efficacy of viral vector-based vaccines.
Introduction
Recombinant Ad5 vector-based vaccines are being developed for multiple pathogens including Ebola (1), influenza (2), hepatitis C (3), and Trypanosoma cruzi (4), however an Ad5 vector-based vaccine for HIV-1 showed no efficacy in humans (5, 6). Ad5 vectors, particularly at high doses, have been reported to induce partially exhausted CD8+ T cell responses with decreased functionality (7, 8). Ad5-elicited CD8+ T cells exhibit markedly elevated expression of PD-1 with reduced production of IL-2 and TNF-α, and also show impaired anamnestic expansion upon second antigen exposure (7–9). Understanding the mechanisms that negatively regulate the functionality of vaccine-elicited T cell responses is critical to the rational design of improved vaccines.
CD4+ T cells play a major role in orchestrating the generation of antigen-specific CD8+ T cell responses following Ad vector immunization (10–13) and dictate viral clearance following LCMV or SIV infection (14, 15). Moreover, Ad5 vectors have been shown to elicit CD4+ T cells that co-express interferon γ (IFN-γ) and interleukin 10 (IL-10) (16). However, the functional significance of these IL-10-producing CD4+ T cells remains unclear. In particular, the extent to which these IL-10+CD4+ T cells may modulate Ad5 vaccine-elicited CD8+ T cell magnitude or function has not previously been explored. Furthermore, the pathways that regulate these IL-10-producing cells remain unknown.
To define the mechanism responsible for the distinct immune phenotype elicited by Ad5 vectors, we evaluated CD4+ T and CD8+ T cell responses following Ad5 immunization of mice and rhesus monkeys. We found that Ad5 vectors induced antigen-specific inhibitory IL-10+CD4+ T cells that markedly suppressed the frequency, functionality, and protective efficacy of antigen-specific CD8+ T cell responses in mice. Moreover, induction of these inhibitory IL-10+CD4+ T cells was dependent on IL-27. Taken together, these data demonstrate that understanding the negative immunologic regulatory pathways of vaccine vectors may pave the way for designing improved vaccines.
Results
Ad5 immunization induces CD8+ T cells with a dysfunctional phenotype in a dose-dependent manner
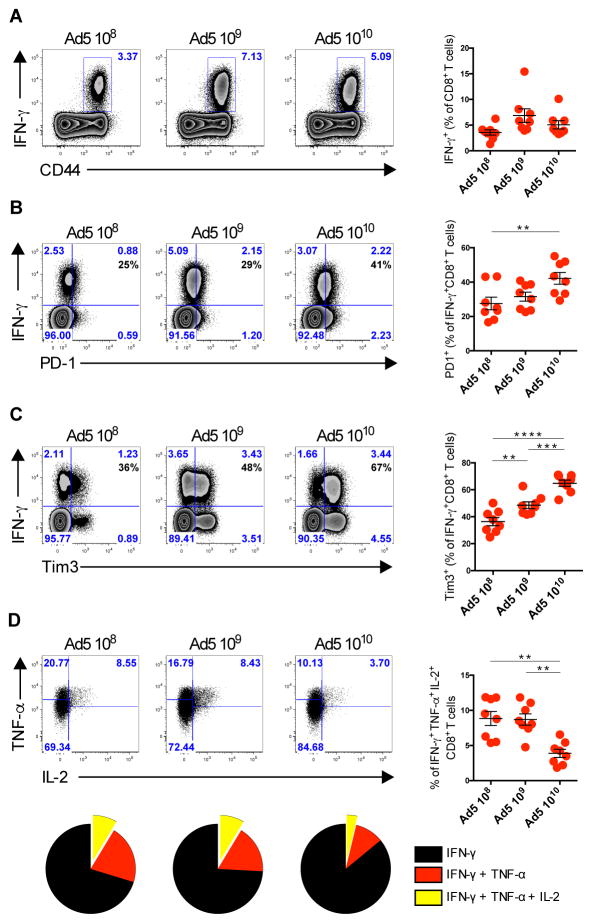
We initiated studies to evaluate the phenotype of antigen-specific CD8+ T cells elicited by Ad5 vectors. We compared the phenotype of CD8+ T cells on day 10, which is the peak of CD4+ T cell responses (fig. S1A), following immunization with escalating doses (108, 109, or 1010) of viral particles (vp) of Ad5 expressing SIVmac239Gag (Ad5-Gag) (Fig. 1, A–D). We did not observe any significant difference in the frequency of Gag-specific IFN-γ+CD8+ T cells at these doses (Fig. 1A). However, the frequency of Gag-specific IFN-γ+CD8+ T cells expressing the inhibitory markers PD-1 and TIM-3 was increased in Ad5-Gag immunized mice in a dose-dependent manner (Fig. 1, B and C). We next evaluated the cytokine polyfunctionality (measured by the co-expression of IFN-γ, TNF-α, and IL-2) by these Gag-specific CD8+ T cells. The frequency of polyfunctional CD8+ T cells was reduced in mice vaccinated with the highest dose of Ad5-Gag (1010 vp) (Fig. 1D), consistent with previous reports from our laboratory and others (7, 9, 17).
Fig. 1. Characterization of Gag-Specific CD8+ T Cells Following increasing doses of Ad5 vaccination.
(A–D) B6 mice were immunized intramuscularly with escalating doses from 108 vp to 1010 vp of Ad5-Gag. Splenic Gag-specific CD8+ T cells were evaluated on day 10 post-immunization. (A) Representative flow plots of IFN-γ production, (B) PD-1 expression, (C) TIM-3 expression, and (D) co-expression of INF-γ, TNF-α, and IL-2 by CD8+ T cells gated on INF-γ+CD8+ T cells. Data are representative of three independent experiments with 5 to 8 animals per group. Each dot represents an individual mouse. p values were calculated using Mann-Whitney U. Mean ± SEM is shown.
We next evaluated whether the phenotype observed in antigen-specific CD8+ T cells persisted over time. We observed different kinetics of CD8+ T cell response (fig. S1B) as previously described (7, 8). Interestingly, Ad5-Gag-elicited CD8+ T cells showed a continuous elevated expression of PD-1 when compared with CD8+ T cells from Ad26-Gag immunized mice, up to day 60 (fig. S1C). Ad5 immunization generated CD8+ T cells that exhibited a short-lived effector phenotype, whereas Ad26 induced CD8+ T cells appearing with long-lived memory CD8+ T cells (fig. S1D and E). Moreover, on day 60 both PD-1 and TIM-3 were elevated on Ad-5-elicited CD8+ T cells (fig. S1F), and these cells also show decreased polyfunctionality, as measured by IL-2 and TNF-α co-expression when compared to Ad26 CD8+ T cells (fig. S1G). These data show that Ad5-elicited CD8+ T cells are dysfunctional as compared with Ad26-elicited CD8+ T cells, and such dysfunction persists for at least 60 days.
Ad5 vaccination induces CD8+ T cells with a dysfunctional transcriptional profile
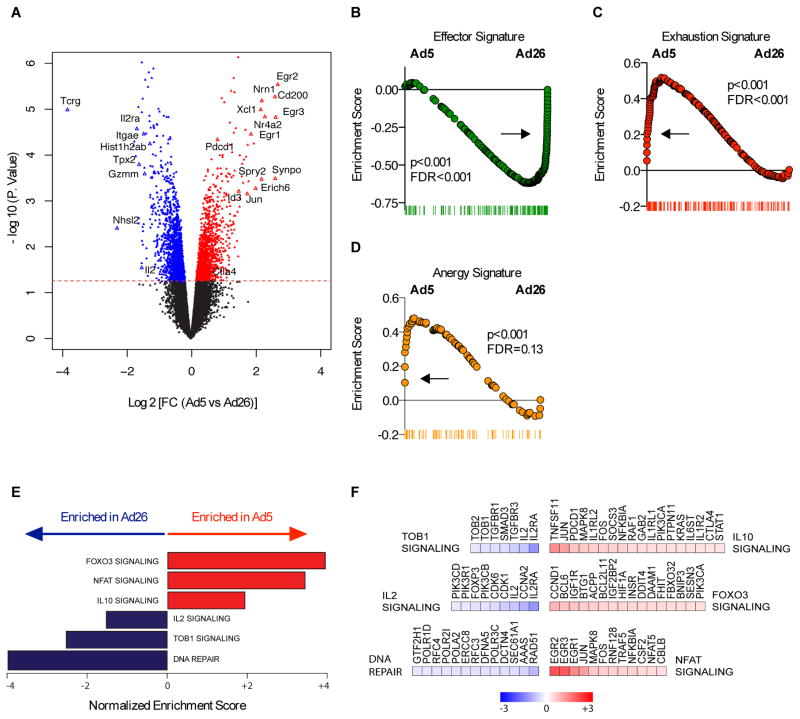
To elucidate the mechanism responsible for the dysfunctional CD8+ T cell phenotype elicited by Ad5 vector immunization, a global transcriptional profile was performed in T cell receptor (TCR) transgenic ovalbumin-specific (OT-I) CD8+ T cells 10 days post-immunization with Ad5 vectors expressing OVA (fig. S1A). Ad26-OVA was utilized as a control, as Ad26 vectors do not induce this exhausted phenotype (7). The transcriptional profile revealed that both Ad5-OVA and Ad26-OVA vaccination induced distinct gene expression profiles in CD8+ and CD4+ T cells when compared to naïve CD8+ and CD4+ T cells from OT-I and OT-II mice respectively (fig. S2, and Table S1–S6).
Ad5 immunization induced OVA-specific CD8+ T cells that expressed higher levels of inhibitory molecules including Pdcd1 (encodes PD-1) and Ctla4 (encodes CTLA-4) as compared to Ad26 (Fig. 2A). In addition, following Ad5-OVA immunization, the expression was increased for EGR family genes (Egr1, Egr2 and Egr3) (Fig. 2A), which regulate the immune system (18). Of note, Egr2 is a downstream gene of Foxo3A and TGF-β, two key regulators of the immune response (19). In contrast, genes that decreased upon Ad5 immunization included Il2 and Il2rα (Fig. 2A), which are important in the development of CD8 T cell responses (20).
Fig. 2. Transcriptional profiling of CD8+ T cells elicited by Ad5 vectors.
(A) Volcano plots showing the global transcriptional changes in CD8+ T cell responses elicited by Ad5 compared to Ad26 vectors. Each dot represents one gene. The log2 fold change in Ad5 versus Ad26 is represented on the x-axis. The y-axis shows the -log10 of the p value. A p-value cut-off of 0.05 is indicated by the horizontal red line. Genes significantly increased (red) or decreased (blue) in Ad5 compared to Ad26. (B to D) GSEA enrichment score of genes within the (B) Effector signature (C) Exhaustion signature, and (D) Anergy signature. (E) Bar plots showing the GSEA normalized enrichment scores of selected enriched pathways that significantly increased (red bars) or decreased (blue bars) in Ad5. (F) Heatmaps of log2 fold change of the significant (T test; p-value <0.05) leading edge genes within each pathway represented in (E). Color gradient ranging genes from dark blue (decreased in Ad5) to dark red (increased in Ad5).
We next performed gene set enrichment analysis (GSEA) (21) to identify the biological pathways that are modulated following Ad5 immunization as compared to control Ad26 immunization in OVA-specific CD8+ T cells. First, we tested for the enrichment of published gene expression signatures of CD8+ T cells in various differentiation states (Fig. 2, B–E). We found that following Ad5 immunization, CD8+ T cells exhibited a decreased effector signature (Fig. 2B), with a phenotype highlighted by increased exhaustion (FDR=0.001; p<0.001) (Fig. 2C) and anergy (FDR=0.13; p< 0.001) (Fig. 2D) signatures. Next we tested for the enrichment of canonical gene sets from MsigDB and ingenuity (IPA) databases. GSEA analysis revealed that Ad5-elicited CD8+ T cells induced pathways such as IL-10, NFAT, and FOXO3A signaling, suggesting the induction of regulatory and exhaustion pathways upon Ad5 immunization (Fig. 2, E and F).
In particular, we observed an enrichment of IL-10 target genes in Ad5-induced CD8+ T cells as compared to control Ad26-induced CD8+ T cells (Fig. 2F). IL-10 is an inhibitory cytokine that regulates several transcriptional and chromatin repressors and plays an important role in impairing antiviral immunity during chronic viral infections such as HIV-1 (22) and suppresses viral clearance in animal models (23, 24). Of note, IL-10 acts directly through inhibition of proliferation, differentiation, cytokine production (IL-2 / IFN-γ) and tyrosine phosphorylation downstream of CD28 signaling in T cells (25), and indirectly through suppression of critical APC functions by inhibiting cytokine production and preventing MHC / co-stimulatory molecules upregulation upon maturation (25). We observed decreased IL-2 signaling and Tob1 signaling-mediated T cell activation (Fig. 2, E and F). These data suggest that Ad5 vectors induce distinct transcriptomic pathways that result in T cell anergy, exhaustion, and suboptimal activation and differentiation.
IL-10 blockade enhances antigen-specific CD8+ T cell functionality and protective efficacy
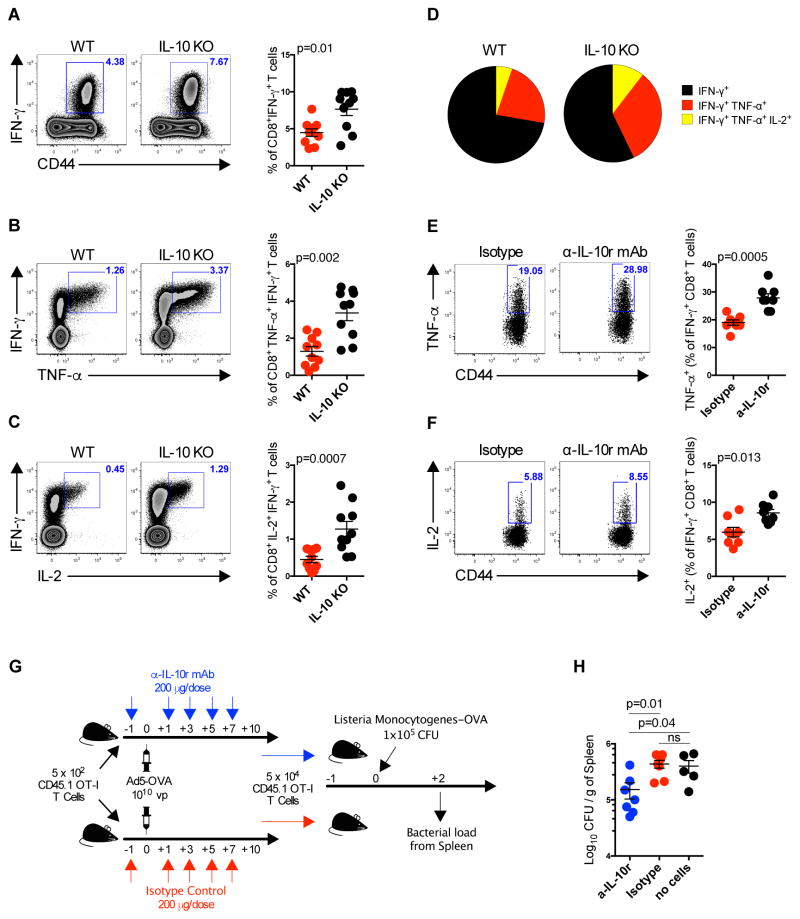
We next evaluated if blocking IL-10 would improve antigen-specific CD8+ T cell responses elicited by Ad5 vectors. We immunized transgenic IL-10KO mice with Ad5-Gag and measured Gag-specific CD8+ T cell responses. We observed a 1.7-fold increase in the frequency of IFN-γ expression by Gag-specific CD8+ T cells in IL-10KO as compared to wild type mice (p=0.01) (Fig. 3A). We also observed a 2.6-fold increase in the frequency of TNF-α+IFN-γ+ (p=0.002) (Fig. 3B), and a 2.8-fold increase in IL-2+IFN-γ+ (p=0.0007) (Fig. 3C) Gag-specific CD8+ T cells in Ad5-Gag immunized IL-10KO as compared to wild type control mice. Gag-specific CD8+ T cell cytokine polyfunctionality (p=0.001) (Fig. 3D) was also enhanced in IL-10KO mice. Consistent with these findings, anti-IL-10 receptor blockade by monoclonal antibody (mAb) administration similarly led to an increased frequency of TNF-α+ (p=0.0005) (Fig. 3E) and IL-2+ (p=0.013) (Fig. 3F) CD8+ T cells as compared with isotype control antibody administration following Ad5-Gag immunization.
Fig. 3. IL-10 producing CD4+ T Cells Impair Antigen-specific CD8+ T Cells Responses Following Ad5 Immunization.
(A to D) IL-10 knockout (IL-10KO) or wild-type B6 mice were immunized intramuscularly with 1010 vp of Ad5-Gag, and splenic Gag-specific CD8+ T cells were evaluated on 10 days post-immunization. (A) Representative flow plots and group summary of IFN-γ+ CD8+ T cells. (B) Co-expression of INF-γ and TNF-α or (C) IL-2 by CD8+ T cells. (D) Pie chart showing the CD8+ T cells cytokine polyfunctionality distribution. (E and F) B6 mice were immunized intramuscularly with 1010 vp of Ad5-Gag, and treated IP every other day from day -1 until day 7 with 200 μg of anti-IL-10r mAb or isotype control. Gag-specific CD8+ T cells were evaluated 10 days post immunization for (E) expression of TNF-α or (F) IL-2, gated on IFN-γ+ CD8+ T cells. (G) Experimental outline of the recombinant Listeria monocytogenes-OVA challenge. (H) Listeria monocytogenes titers in spleens at day 2 following challenge. Data are representative of two independent experiments with at least 5 animals per group. Each dot represents an individual mouse. p values were calculated using Mann-Whitney U. Mean ± SEM is shown.
To assess the functional relevance of IL-10 blockade, we assessed the protective efficacy of antigen-specific CD8+ T cells with or without IL-10 blockade. 5x102 naïve OT-I CD8+ T cells from a CD45.1 congenic mouse were adoptively transferred into CD45.2 naïve mice and immunized with 1010 vp of Ad5-OVA. Mice were divided into two experimental groups: 1) treated with 200 μg/dose of anti-IL-10 receptor (IL-10r) mAb every other day from day -1 through day 7; 2) treated with isotype control mAb. Ten days post-immunization, 5x104 OT-I CD8+ T cells were FACS sorted and adoptively transferred to congenic naïve mice to equalize number of antigen-specific CD8+ T cells prior to challenge. One-day post-transfer, mice were challenged intravenously with 1x105 CFU of recombinant Listeria monocytogenes expressing OVA (Fig. 3G). OT-I CD8+ T cells from mice treated with anti-IL-10r mAb afforded a significant reduction of L. monocytogenes bacterial loads (p=0.01) (Fig. 3H), whereas the same number of OT-I CD8+ T cells from animals treated with isotype control mAb were unable to reduce bacterial loads as compared to control mice. These data demonstrate that IL-10 blockade augments vaccine-elicited functional CD8+ T cells and improves protective efficacy in this model.
IL-10+CD4+ T cells are immunomodulatory and impair CD8+ T cell responses
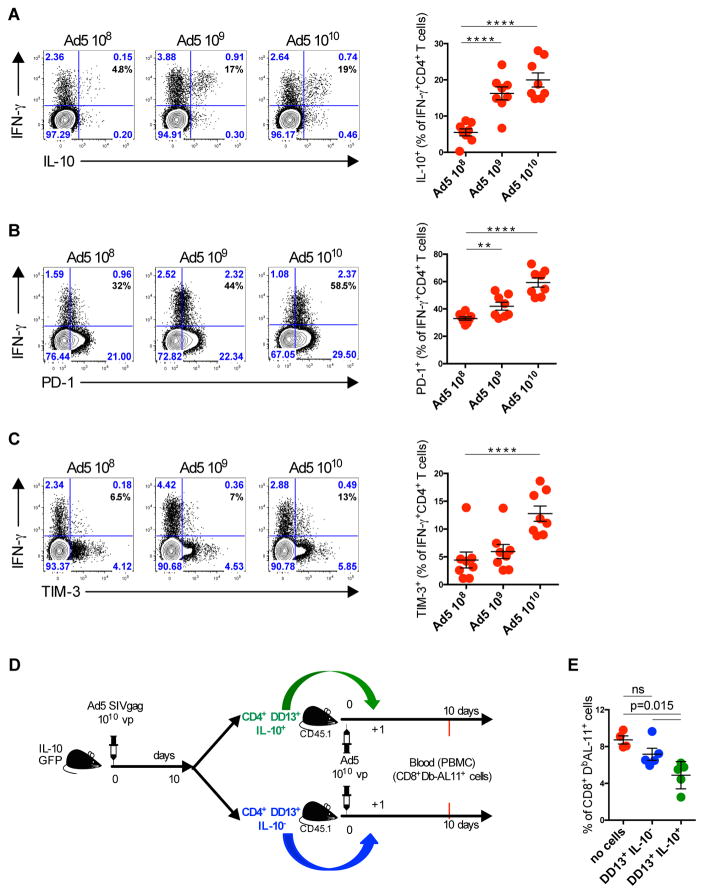
We next explored whether CD4+ T cells could be the source of the IL-10 that suppresses CD8+ T cell responses. We first compared the phenotype of CD4+ T cells on day 10, reflecting the peak of the CD4+ T cell response following immunization with 108, 109, or 1010 vp of Ad5-Gag. The frequency of Gag-specific IFN-γ+CD4+ T cells expressing IL-10 and the inhibitory markers PD-1 and TIM-3 were increased in Ad5-Gag immunized mice in a dose dependent manner (Fig. 4, A–C). When compared with Gag-specific IFN-γ+CD4+ T cells from mice immunized with 108 vp of Ad5-Gag, IFN-γ+CD4+ T cells from mice immunized with 1010 vp of Ad5-Gag showed a 4, 2, and 2-fold higher expression of IL-10, PD-1 and TIM-3, respectively (p=0.0001) (Fig. 4, A–C).
Fig. 4. Characterization of Gag-Specific CD4+ T Cells Following Ad5 vaccination.
(A–C) B6 mice were immunized intramuscularly with escalating doses from 108 vp to 1010 vp of Ad5-Gag. Splenic Gag-specific CD4+ T cells were evaluated on day 10 post-immunization. (A) Representative flow plots of IL-10 production, (B) PD-1 expression, and (C) TIM-3 expression. (D and E) Adoptive transfer of Gag-specific (I-AbDD13) CD45.2+ CD4+DD13+IL-10eGFP+ or CD4+DD13+IL-10eGFP− T cells to CD45.1 mice one day following immunization with 1010 vp of Ad5-Gag. Data are representative of three independent experiments with 5–8 animals per group. Each dot represents an individual mouse (n=5 to 8). p values were calculated using Mann-Whitney U. Mean ± SEM is shown.
To characterize further the IL-10+ population induced by Ad5 vaccination, we compared IFN-γ+IL-10+ (green) and IFN-γ+IL-10− (purple) (fig. S3) Gag-specific CD4+ T cells following Ad5-Gag immunization. IFN-γ+IL-10+ CD4+ T cells did not express the conventional regulatory CD4+ T cell (Treg) transcription factor Foxp3 but did express the Th1-driving transcription factor T-bet (fig. S3). Gag-specific IFN-γ+IL-10+ CD4+ T cells expressed more Tbet (p=0.007), IRF4 (p=0.007), and CTLA-4 (p=0.0002) than Gag-specific IFN-γ+IL-10− CD4+ T cells. BCL-6 and CD226 expression were significantly downregulated on these cells (p=0.007 and p=0.001, respectively).
We next evaluated whether Gag-specific IL-10+IFN-γ+CD4+ T cells generated following Ad5-Gag immunization suppress Gag-specific CD8+ T cell responses. To address this, we immunized CD45.2 transgenic IL-10GFP knock-in mice with Ad5-Gag. Ten days post immunization, Gag-specific IL-10+ or IL-10− CD4+ T cells were FACS sorted using an I-Ab/DD13 tetramer (26) and 5x104 Gag-specific cells were adoptively transferred intravenously into CD45.1 congenic mice that were immunized with Ad5-Gag one day prior (Fig. 4D). Mice that received DD13+IL-10+ CD4+ T cells, but not those that received DD13+IL-10− CD4+ T cells, showed a significant decrease in Gag-specific Db/AL11+CD8+ T cells (p=0.015) (Fig. 4E). These data demonstrate that IL-10-producing CD4+ T cells suppress CD8+ T cell responses following Ad5 immunization.
CD4+ T cells phenotype observed following Ad5 immunization is not correlated with antigen load or persistence
To confirm that the differences observed between Ad5 and Ad26 immunization were not an artifact arising from the fact that mice do not express human receptor CD46, which is the primary attachment receptor of Ad26 (27), we repeated these experiments in human CD46 transgenic (hCD46tg) mice. A comparable phenotype of Gag-specific CD4+ T cells in terms of IFN-γ, IL-10, PD-1, and TIM-3 expression was observed in hCD46tg and wild-type mice following Ad26-Gag immunization (fig. S4A–4D). To address whether differences in antigen expression led to the observed phenotypic differences, we used an in vivo imaging system (IVIS) to detect luciferase transgene expression in mice immunized with Ad5 or Ad26 expressing luciferase (fig. S5A). We observed comparable luciferase expression at the injection site three hours post-immunization. Twenty-four hours post-immunization, luciferase expression was 7.5-fold lower in Ad5-Luc as compared with Ad26-Luc immunized mice (p=0.02). In addition, a 5.1-fold higher luciferase expression in Ad26 as compared to Ad5 immunized mice persisted out until 21 days (p=0.02) (fig. S5A). Thus, the dysfunctional phenotype observed following Ad5 immunization was not due to elevated or prolonged transgene expression in Ad5 immunized mice.
We also determined whether Ad5 and Ad26 differ with respect to the phenotype of Gag-specific CD4+ T cell, due to liver tropism of Ad5 (28). To address this, we used a chimeric Ad5 vector that has its hypervariable regions (HVRs) replaced with those from the non-hepatotropic Ad48 vector, referred to as Ad5HVR48 (29). No differences in IL-10, PD-1, or TIM-3 expression by Gag-specific IFN-γ+CD4+ T cells between Ad5HVR48-Gag and Ad5-Gag immunized mice were observed (fig. S5B–5D). Taken together, these data suggest that the distinct CD4+ T cell phenotype observed following Ad5 immunization is not associated with differences in receptor usage, liver tropism, or transgene persistence.
Altered CD4+ T cell phenotype in rhesus monkeys following Ad5 immunization
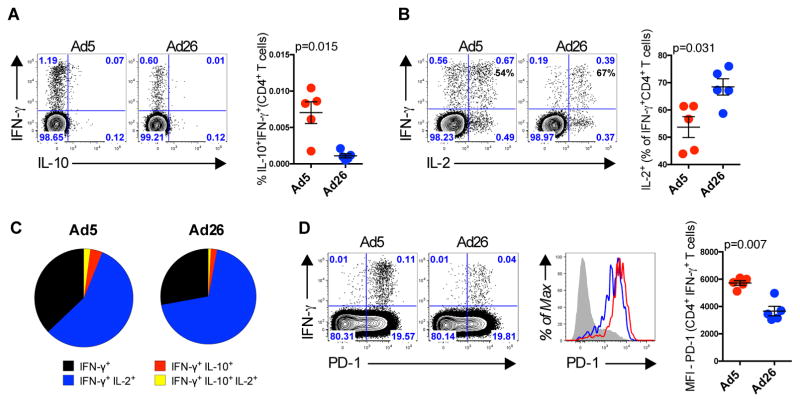
Finally, we investigated whether these findings in mice were translatable to non-human primates. PBMCs were collected from 10 rhesus macaques 2 weeks following immunization with 1010 vp of Ad5-Gag or Ad26-Gag (n=5/group). We observed an 8-fold increase in the frequency of Gag-specific IL-10+IFN-γ+ CD4+ T cells (p=0.015) (Fig. 5A) in Ad5-Gag as compared to control Ad26-Gag immunized macaques. We also observed a lower frequency of Gag-specific CD4+ T cells co-producing IL-2 and IFN-γ (p=0.031) (Fig. 5B), and reduced Gag-specific CD4+ T cell cytokine polyfunctionality (Fig. 5C) in Ad5-Gag immunized as compared to Ad26-Gag immunized animals. Additionally, Gag-specific IFN-γ+CD4+ T cells from Ad5-Gag immunized monkeys had elevated PD-1 expression (p=0.007) as compared to those from Ad26-Gag immunized animals (Fig. 5D). Taken together, these data demonstrate that the frequency of dysfunctional CD4+ T cells is increased following Ad5 immunization in both mice and rhesus monkeys.
Fig. 5. CD4 T Cells From Rhesus Monkeys Upregulate the Expression of IL-10 and PD-1 Following Ad5 Immunization.
(A to D) Rhesus Monkeys were immunized intramuscularly with 1010 vp of Ad5-Gag or Ad26-Gag, and Gag-specific CD4+ T cells from PBMCs were evaluated 2 weeks post immunization. Representative flow plots are shown gated on CD4+CD69+ T cells. (A) Co-expression of IFN-γ and IL-10 and (B) co-expression of IFN-γ and IL-2. (C) Pie chart showing the CD8+ T cells cytokine polyfunctionality distribution. (D) Co-expression of IFN-γ and PD-1 gated on CD4+CD69+ T cells. Histogram plots showing the expression of PD-1 by IFN-γ+ CD4+ T cells. Group summary graph showing the IFN-γ mean fluorescence intensity (MFI). Each dot represents an individual monkey (n=5 per group). p values were calculated using Mann-Whitney U. Mean ± SEM is shown.
CD4+ T cells are transcriptionally dysfunctional following Ad5 immunization
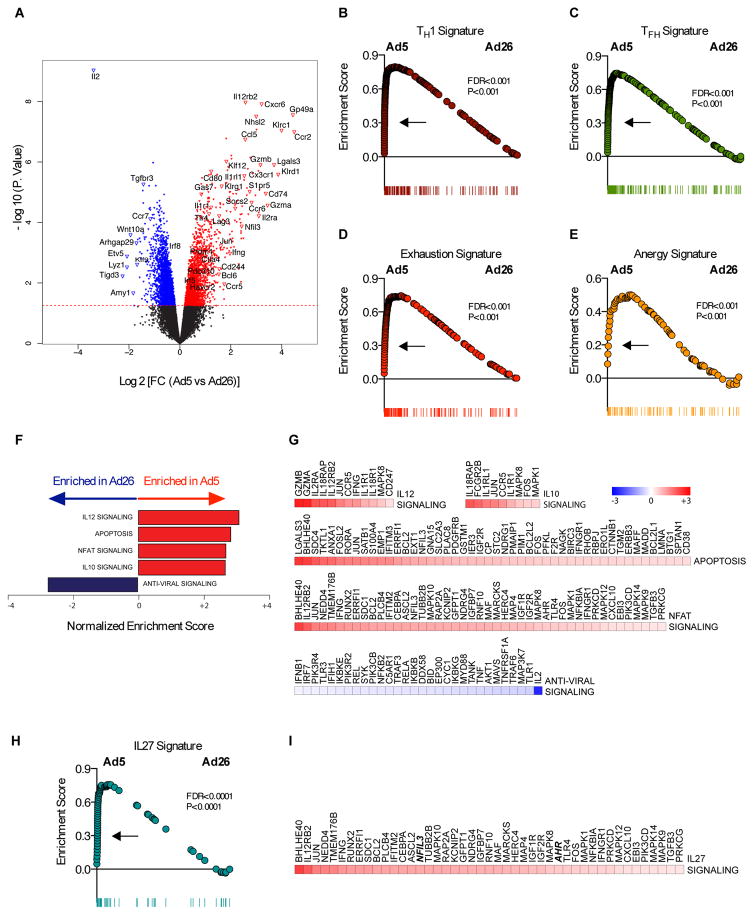
We next evaluated the transcriptional profile of Ad5-elicited CD4+ T cells. Ad5-OVA immunization induced OVA-specific CD4+ T cells that expressed higher levels of genes associated with effector function (Ccr2, Gzma, Gzmb, Il2ra, Il12rb2, Klrg1, Ifng, and Ccr5), certain transcription factors (Socs2, Jun, Nfil3, Bcl6, and Prdm1), and inhibitory markers (Lag3, Cd80, Ctla4, Cd244, and Havcr2), whereas CD4+ T cells from Ad26-OVA immunized mice expressed higher levels of effector genes (Ccr7, and Il2), and the transcription factor Irf8 (Fig. 6A).
Fig. 6. Transcriptional profiling of CD4+ T cell elicited by Ad5 vectors.
(A) Volcano plots showing the global transcriptional changes in CD4+ T cells elicited by Ad5 compared to Ad26 vectors. Each dot represents one gene. The log2 fold change in Ad5 versus Ad26 is represented on the x-axis. The y-axis shows the -log10 of the p value. A p-value cut-off of 0.05 is indicated by the horizontal red line. Genes significantly increased (red) or decreased (blue) in Ad5 compared to Ad26. (B to E, and H) represent the distribution of the GSEA enrichment score of genes within (B) TH1, (C) TFH, (D) exhaustion, (E) anergy, and (H) IL-27 signatures. (F) Barplots showing the GSEA normalized enrichment scores of selected enriched pathways that significantly increased (red bars) or decreased (blue bars) in Ad5 versus Ad26. (G) heatmaps of log2 fold change of the significant (T test; p-value <0.05) leading edge genes within each pathway represented in (F), and (I) leading genes represented in the IL-27 pathway (I). Color gradient genes ranging from dark blue (decreased in Ad5) to dark red (increased in Ad5).
GSEA pathway analysis revealed that following Ad5-OVA immunization, OVA-specific CD4+ T cells exhibit a significant enrichment in gene signatures for TH1 differentiation (FDR<0.001; p=0.001) (Fig. 6B), TFH differentiation (FDR<0.001; p<0.001) (Fig. 6C), exhaustion (FDR<0.001; p<0.001) (Fig. 6D), and anergy (FDR<0.001; p<0.001) (Fig. 6E) (18, 30–32). Moreover, Ad5-elicited CD4+ T cells had increased expression of NFAT target genes, analogous to what was observed in Ad5-elicited CD8+ T cells (Fig. 6, F and G). Apoptosis signaling was also increased, highlighting potential survival defects of these cells upon Ad5 immunization. Strikingly, we observed an enrichment of IL-10 signaling upon Ad5 immunization (Fig. 6F), analogous to what was seen with CD8+ T cells. These data suggest that Ad5 vectors induce highly dysregulated CD4+ T cell responses that do not fully match classical definitions.
We next investigated which pathways regulated the production of IL-10 by Ad5-induced CD4+ T cells. Both IL-12 and IL-27 pathways have been reported to be involved in IL-10 production (33, 34). We observed that both IL-12 and IL-27 signaling (FDR < 0.0001) (Fig. 6, F–I) were induced following Ad5 immunization as compared to Ad26 immunization. Taken together, these data demonstrate that Ad5 vaccination elicited CD4+ T cells with a dysregulated phenotype, including expression of several inhibitory molecules as well as IL-12 and IL-27 pathways.
IL-10 expression by CD4+ T cells following Ad5 immunization is not IL-12-dependent
IL-12 has been reported to play a pivotal role during the expansion of TH1 responses (34). IL-12 has also been reported to be associated with the generation of IL-10-producing CD4+ T cells in vitro (35). To test whether IL-12 was involved in the generation of IL-10+CD4+ T cells following Ad5 immunization, mice lacking the receptor β for IL-12 (IL-12rβ2 KO) were immunized with Ad5 expressing SIV-Gag. Absence of IL-12 signaling had no detectable effect on induction of Gag-specific IFN-γ+CD4+ T cells post-immunization (fig. S6A). Moreover, in IL-12rβ2 KO mice, we observed a slight increase in the expression of IL-10, PD-1, and TIM-3 (p=0.015) (fig. S6, B–D) by Gag-specific IFN-γ+CD4+ T cells compared to wild type control mice following Ad5-Gag immunization. These data demonstrate that IL-12 signaling is largely dispensable for the generation of IL-10+CD4+ T cells following Ad vaccination.
IL-10 expression by CD4+ T cells following Ad5 immunization is IL-27-dependent
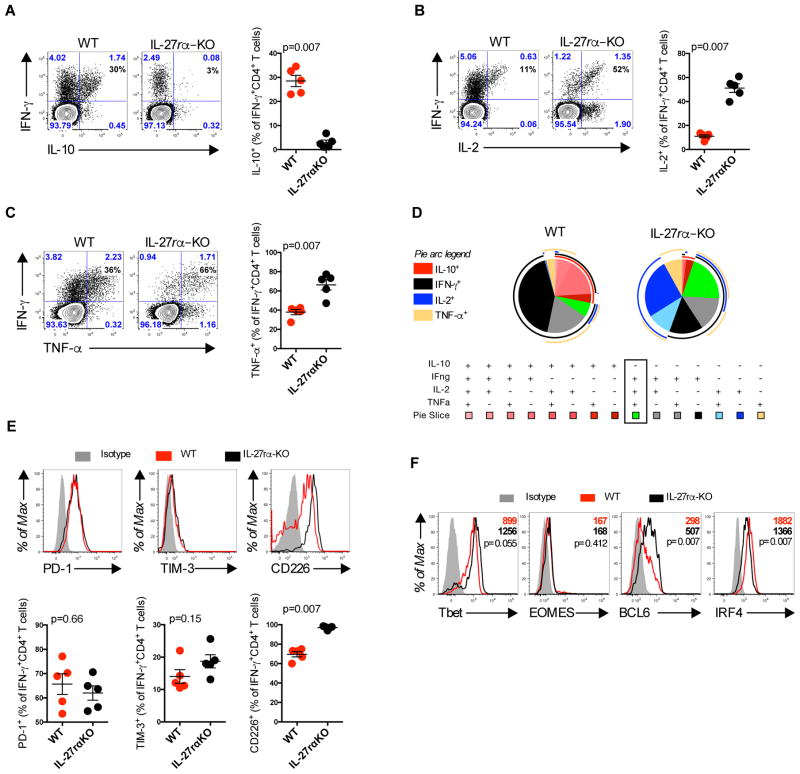
IL-27 has been shown to be associated with induction of IL-10 production by T cells in several models of autoimmune disease and infection (36). Thus, we next examined whether elevated IL-27 expression correlated with the dysfunctional phenotype observed in CD4+ T cells following Ad5 immunization. Mice lacking the alpha-chain of the IL-27 receptor (IL-27rαKO) were immunized and 10 days post-immunization CD4+ T cells were evaluated by ICS. Strikingly, production of IL-10 (p=0.007) by Gag-specific IFN-γ+CD4+ T cells was dramatically impaired in Ad5-Gag immunized IL-27rαKO as compared to wild type mice (Fig. 7A). In contrast, expression of IL-2 and TNF-α by Gag-specific IFN-γ+CD4+ T cells was 2 and 1.3-fold elevated, respectively, in Ad5-Gag immunized IL-27rαKO as compared to wild-type mice (p=0.007) (Fig. 7, B and C). Moreover, the lack of IL-27 signaling improved CD4+ T cell cytokine polyfunctionality (Fig. 7D).
Fig. 7. IL-27-dependent expression of IL-10 by CD4+ T cells.
(A–F) IL-27rα knockout or B6 mice were immunized intramuscularly with 1010 vp of Ad5-Gag. Splenic Gag-specific CD4+ T cells were evaluated 10 days post immunization. (A) IL-10 production, (B) IL-2 production, and (C) TNF-α production by Gag-specific CD4+CD44+T cells. (D) Pie charts representing the proportion of IL-10, IFN-γ, IL-2, and TNF-α cytokine production by CD4+CD44+ T cells measured by flow cytometry. External pie arcs represent the distribution of each individual cytokine. (E) Expression of PD-1, TIM-3, and CD226 by IFN-γ+ CD4+ T cells. (F) Expression of Tbet, EOMES, BCL6, and IRF4 in IFN-γ+ CD44+CD4+ T cells quantified by mean fluorescence intensity (MFI). Data are representative of three independent experiments with 5 animals per group. Each dot represents an individual mouse. p values were calculated using Mann-Whitney U. Mean ± SEM is shown.
We next tested the impact of IL-27 signaling on PD-1 TIM-3, and CD226 expression by Gag-specific IFN-γ+CD4+ T cells. No changes in the expression of PD-1 or TIM-3 were observed following Ad5-Gag immunization in IL-27RrαKO as compared to wild type mice. Interestingly, Gag-specific IFN-γ+CD4+ T cells up-regulated the expression of CD226 in Ad5-Gag immunized IL-27rαKO as compared to wild type mice (Fig. 7E). Additionally, Gag-specific IFN-γ+CD4+ T cells up-regulated Tbet (p=0.055) and BCL6 (p=0.007) and down-regulated IRF4 (p=0.007) in Ad5-Gag immunized IL-27rαKO as compared to wild type mice (Fig. 6F). These data suggest that IL-27 blockade enhances CD4+ T cell effector phenotype.
Consistent with these findings, the absence of IL-27 using IL-27/35 double knockout mice (EBI3 subunit protein KO) or blockade with anti-IL-27 (p28 subunit protein) monoclonal antibody (aIL-27mAb) yielded similar results as compared to IL-27rαKO mice following Ad5-Gag immunization (fig. S7). These data demonstrate that Ad5 vectors trigger IL-27 expression that is correlated with upregulation of IL-10 by Gag-specific IFN-γ+CD4+ T cells.
Discussion
In this study, we demonstrate that Ad5 vectors induce high frequencies of suppressive IL-10+CD4+ T cells in both mice and rhesus monkeys. These IL-10+CD4+ T cells are dependent on IL-27, suppress vaccine-elicited CD8+ T cell magnitude and function, and inhibit protective efficacy in vivo. These data suggest that the cytokine networks and transcriptomic profiles induced by Ad5 vectors underlie the unique dysfunctional phenotype of CD8+ T cells induced by Ad5. These data raise the possibility that IL-10 or IL-27 inhibition could be explored as novel therapeutic strategies to improve the immunogenicity and protective efficacy of viral vector-based vaccines with phenotypes similar to those seen in this study.
IL-27 was initially classified as a pro-inflammatory cytokine due its ability to induce the production of IFN-γ by T cells (33). Indeed, following subunit vaccination, IL-27 is required for triggering CD4+ and CD8+ T cell responses (37). A role for IL-27 as a negative regulator of T cell responses has also been reported in studies showing the antagonistic effect of IL-27 on TH1, TH2, and TH17 CD4+ T cell responses in vivo (33). CD4+ T cells from IL-27 receptor deficient mice chronically infected with Mycobacterium tuberculosis (TB) maintain elevated PD-1 but display increased IL-2 expression compared to cells from WT animals and this resulted in bacterial clearance (38). In the present study, we show that IL-27 blockade improves Ad5-elicited CD4+ T cell phenotype by inhibiting IL-10 expression and increasing expression of IL-2, TNF-α, and CD226. Our data also corroborate a previous report showing that CD226 expression is regulated by IL-27 (39). IL-27 has already been correlated with limiting the size and duration of T cell responses in several models of pathogen infections and autoimmunity (40). These data reflect a novel role for IL-27 in triggering dysfunctional T cells following Ad vector vaccination. These data further suggest that IL-27 blockade could represent a strategy to prevent the generation of suppressive IL-10+CD4+ T cells following vaccination.
The potential therapeutic benefit of blocking IL-10 was described as a strategy to recover T cell function and improve viral clearance in a model of persistent lymphocitic choriomeningitis virus (LCMV) infection (23). In our experiments, we observed a suppressive effect of antigen-specific IL-10+CD4+ T cells on antigen-specific CD8+ T cells following vaccination, and blockade of IL-10 also improved CD8+ T cell cytokine polyfunctionality with increased co-expression of IFN-γ, IL-2, and TNF-α following Ad5 vaccination. We observed these effects despite the fact that IL-10 levels were substantially reduced in our model as compared with mice chronically infected with LCMV clone-13.
We have previously reported that antigen-specific CD8+ T cells from Ad5 immunized mice fail to upregulate the IL-7 receptor (CD127) and CD62L, and that these cells have sustained elevated PD-1 expression in comparison with other Ad serotypes (7, 8). In this study, we show that Ad5-elicited CD4+ T cells also upregulate the expression of PD-1 and TIM-3. In addition, the transcriptional profile of Ad5-elicited CD8+ T cells and CD4+ T cells reveals a dysfunctional phenotype, with upregulation of several inhibitory markers and an enrichment of transcriptional signatures for anergy and exhaustion for both cell populations. Thus, it is clear that Ad5 vectors, particularly at high does, induce both CD4+ T cell and CD8+ T cell responses that are phenotypically, functionally, and transcriptionally similar to exhausted cells observed during chronic viral infection. These data also raise the possibility that the observed increase in HIV-1 acquisition during the STEP trial (HVTN502), could be associated with the Ad5 induction of CD4+ T cells with phenotype similar to those used by HIV-1 as reservoirs (41). However, it has been also reported that besides the partially exhausted phenotype induced by Ad5, CD8+ T cells are still able to confer protection in different viral challenge models (42, 43).
Despite identifying that IL-27 plays a major role inducing the phenotype observed in CD4+ T cells, the direct effect of IL-27 on CD8+ T cells remains to be elucidated. Our study show that IL-10 derived from CD4+ T cells is enough to suppress CD8+ T cells in mice, but future studies need to be performed to clarify whether the same mechanism will translate for both monkeys and humans.
Together, our data in mice suggest that Ad5 vectors elicit dysfunctional CD4+ and CD8+ T cell responses by a mechanism that involves IL-27 induction of suppressive IL-10+CD4+ T cells. Although this model may be specific for Ad5 vectors, it is likely that other cytokine networks and transcriptomic pathways dictate the phenotype of T cell responses elicited by other Ad serotypes and other viral vectors as well. Future therapeutic strategies can be developed to exploit these pathways to enhance the immunogenicity and protective efficacy of viral vector-based vaccines.
Materials and Methods
Study design
The present study aimed to investigate the role of IL-10 producing CD4+ T cells impacting the efficacy of adenovirus 5 vector-vaccine following immunization. Our approach was conducted using intracellular flow cytometry, micro-array analysis, cellular adoptive transfer and bacterial challenge experiments as described in the figure legends. Age and sex-matched mice and monkeys were randomly divided in experimental groups containing at least 5 animals per group, and repeated at least two times with the exception of the monkey study (Fig. 5).
Animals, Immunizations, and Antibody Treatment
C57BL/6, B6.SJL-ptprca (CD45.1+), B6.129S1-Il12rb2tm1Jm/J (IL-12rβ KO), B6N.129P2-Il27ratm1Mak/J (IL-27rαKO), B6.(Cg)-Il10tm1.1Karp/J (IL-10 eGFP), B6.129P2-Il10tm1Cgn/J (IL-10 KO), B6.129X1-Ebi3tm1Rsb/J (IL-27/IL-35 KO), C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I), B6.Cg-Tg(TcraTcrb)425Cbn/J (OT-II), and B6.FVB-Tg(CD46)2Gsv/J (hCD46Tg) animals were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were immunized intramuscularly in the quadriceps with 108 – 1010 viral particles of each vector in a volume of 100 μl divided equally between the two legs with the previously described E1/E3 deleted Ad5, Ad26 or Ad5HVR48 vectors expressing SIVmac239Gag, OVA or Luciferase (Luc). All animal experimental procedures were performed in accordance with the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee guidelines. The monoclonal anti-IL-10r antibody clone 1B1.3A (BioXcell) or control IgG were administered intraperitoneally (i.p.) (200 μg per mouse) every other day from day −1 until day 7 (44). Anti-IL-27 monoclonal antibody clone MM27-7B1 (Biolegend) or control IgG were administered i.p. (500 μg per mouse) on days −2, 0 and +2.
Flow cytometry, Intracellular Cytokine Staining and Cell Sorting
Lymphocytes were isolated from spleen were than stained, and analyzed by flow cytometry as described (10). Antigen-specific CD4+ and CD8+ T cells were examined with MHC class I or class II tetramers. MHC class I and MHC class II tetramers were provided by the NIH Tetramer Facility (Emory University, GA). For ICS staining cells were re-stimulated in vitro for 1 hour at 37 °C with 1 μg/ml of an overlapping SIVmac239gag peptide pool. After this incubation, Brefeldin-A and Monensin (BioLegend) were added and samples incubated for an additional 6 hours at 37 °C. Cells were subsequently washed, stained, and permeabilized with Cytofix/Cytoperm (BD Biosciences) or Foxp3 Fixation/Permeabilization kit (eBioscience). Cells were acquired on an LSR II flow cytometer (BD Biosciences) and data was analyzed using FlowJo v.9.8.3 (Treestar). Cell sorting was performed using FACSAria3 Special Order (BD Biosciences) with purity > 98%. Surface antibodies: anti-CD4 (RM4–5), CD8α (53–6.7), CD11b (M1/70), CD11c (N418), CD44 (IM7), CD45.1 (A20), CD45.2 (104), CD226 (10E5), PD-1 (RMP1–30), and TIM-3 (RMT3–23). Intracellular antibodies: anti-IL-2 (JES6-5H4), IL-10 (JES5-16E3), IL-12 (C15.6), IL-27 (MM27-7B1), BCL-2 (BCL/10C4), BCL-6 (K112-91), CTLA-4 (UC10-4F10-11), Eomes (Dan11mag), Foxp3 (FJK-16S), IFN-γ (XMG1.2), IRF4 (3E4), T-bet (4B10), and TNF-α (MP6-XT22). All antibodies were purchased from BD Biosciences, eBioscience, or BioLegend. Rhesus monkey staining antibodies: CD3 (SP34-2), CD4 (L200), CD8 (RPA-T8), CD69 (FN50), IL-2 (MQ1-17H12), IFN-γ (B27) (BD Bioscience), and IL-10 (JES3-9D7) (eBioscience). Vital dye exclusion: (LIVE/DEAD®) was purchased from Life Technologies.
Adoptive Cell Transfer
For microarray analysis, one day prior to immunization with Ad5-OVA or Ad26-OVA, 5x103 CD4+ (OT-II) and 1x103 CD8+ (OT-I) T cells were purified “untouched” using the negative selection CD4 or CD8 T cell isolation kit (STEMCELL), following manufacturer’s instructions, and transferred into a CD45.1+ congenic mice (fig. S2A). For the in vivo suppression assay, 5x104 of IL-10+ tetramer+ or IL-10− tetramer+ CD4+ T cells were transferred one-day post immunization with 1010 vp of Ad5-Gag into CD45.1+ congenic mice (fig. 4D). For recombinant Listeria monocytogenes challenge study, 5x102 OT-I cells were isolated using T cell isolation kit (STEMCELL) and transferred to naïve mice. Ten days post immunization OT-I CD45.1 T cells were FACS sorted and 5x104 cell were transferred a day prior Listeria monocytogenes challenge.
Microarray Data Acquisition and Analysis
Gene expression profiling was performed as previously described (45, 46) and data were uploaded (GSE73118). Briefly, OVA-specific CD4+ T cells were sorted to ≥98% purity on a FACS Aria (BD Biosciences). Sorted cells were stored at −80 °C in 1 ml of TRIzol (Life Sciences). RNA extraction was performed using the RNAdvance Tissue Isolation kit (Agencourt) as per manufacturer’s instructions. The cDNA synthesis was performed using the Ovation Pico WTA v2 kit (NuGEN) following manufacturer’s instructions. Proper amplification of cDNA was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies) and performed by the Harvard Biopolymers Facility. cDNA was subsequently fragmented and biotinylated using an Encore Biotin Module 4200 (NuGEN). cDNA was hybridized to Mouse Genome 430 v2.0 chip (Affymetrix) by the Microarray Core of Dana Farber Cancer Institute. Analysis of gene array output data was conducted using the R statistical language (http://www.r-project.org/) and the Linear Models for Microarray Data (LIMMA) statistical package from Bioconductor (https://www.bioconductor.org/). Gene intensities were log2 transformed before being normalized using the quantile normalization method. Differentially expressed gene modulated by Ad26 or Ad5 was performed in CD8 and CD4. The moderated t-test implemented in the R Limma package was used to assess the statistical significance (P < 0.05) of differential expression of genes between Ad5 and Ad26. We used Gene Set Enrichment Analysis (GSEA) (21) to identify enriched biological pathways that are modulated between Ad5 and Ad26 in both CD4 and CD8 cells. GSEA is a statistical approach that analyzes the distribution of a defined set of genes (the “gene set”) against a separate rank-ordered gene list. We used the pre-ranked list option of GSEA and tested for the enrichment of C2, C7 MsigDB gene sets (http://software.broadinstitute.org/gsea/msigdb). Genes were ranked by their fold change (FC) between Ad5 and Ad26 from the highest FC to the lowest FC. The ranked list was submitted to GSEA and for each geneset enrichment score (ES) was calculated reflecting the degree of over-representation at the top (or bottom) of the ranked list. GSEA allows for the identification of a core list of genes (leading edge genes) that contribute to the ES. A gene-based permutation test (1,000 permutations) was used to normalize the ES (NES) to correct for the size of the gene set, assess the statistical significance (nominal p value) of the ES and calculates the probability of the gene set to be false discovery (FDR). We discarded gene sets with FDR > 5%. Gene sets normalized enrichment scores in all figures were plotted using R (21).
In vivo Bioluminescence
Mice were initially shaved using Nair™ hair removal (Church & Dwight Co). Animals were immunized intramuscularly (quadriceps) with 1010 vp of Ad5 or Ad26 expressing luciferase. In the selected time-points 3, 6, 12, 24 hours and 21 days animals were injected i.p. with 150 μl (150 mg/Kg) of XenoLight RediJect D-Luciferin Ultra (PerkinElmer). After 10 minutes, imaging was performed using the IVIS Series 100 (Xenogen) with an exposure time of 2 minutes. Images and luminescence measurements were performed using Living Image software (version 2.50.1; Xenogen) (47).
Statistical Analysis
Statistical analysis was performed using a two-tailed nonparametric Mann-Whitney U test or ordinary one-way ANOVA with multiple comparison test using Prism version 6.0c (GraphPad Software). Data are presented as mean ± SEM.
Supplementary Material
Acknowledgments
We thank Lily R. Parenteau and Stephen Blackmore for technical support and the Center for Virology and Vaccine Research Flow Cytometry core facility for cell sorting. We also thank the NIH Tetramer Core Facility (Emory University) for the kind provision of the MHC class I and II monomers used in these studies.
Funding: We acknowledge support from NIH grants AI060354, AI078526, AI096040 (D.H.B.) and AI007245, AI07387 (P.P.), and the Bill & Melinda Gates Foundation grant OPP1033091 (D.H.B.).
Footnotes
Author Contributions: R.A.L. and D.H.B. conceived the study and designed. R.A.L. and N.M.P. performed and analyzed the experiments. M.A. analyzed the micro-array experiments. M.J.I., E.N.B., A.B.Z., P.A., D.N., C.A.B., E.B., P.P.M, K.E.S. helped with the experiments and manuscript preparation. R.A.L. prepared the figures. D.H.B. supervised the study. R.A.L. and D.H.B wrote the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Micro-array data have been deposited at the National Center for Biotechnology Information under Gene Expression Omnibus accession number GSE73118 and are available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73118.
References and Notes
- 1.Sullivan NJ, et al. CD8+ cellular immunity mediates rAd5 vaccine protection against Ebola virus infection of nonhuman primates. Nat Med. 2011;17:1128–1131. doi: 10.1038/nm.2447. [DOI] [PubMed] [Google Scholar]
- 2.Alexander J, et al. Pre-clinical evaluation of a replication-competent recombinant adenovirus serotype 4 vaccine expressing influenza H5 hemagglutinin. PLoS ONE. 2012;7:e31177. doi: 10.1371/journal.pone.0031177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes E, et al. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci Transl Med. 2012;4:115ra1. doi: 10.1126/scitranslmed.3003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira IR, et al. A human type 5 adenovirus-based Trypanosoma cruzi therapeutic vaccine re-programs immune response and reverses chronic cardiomyopathy. PLoS Pathog. 2015;11:e1004594. doi: 10.1371/journal.ppat.1004594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchbinder SP, et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammer SM, et al. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N Engl J Med. 2013;369:2083–2092. doi: 10.1056/NEJMoa1310566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penaloza-MacMaster P, et al. Alternative serotype adenovirus vaccine vectors elicit memory T cells with enhanced anamnestic capacity compared to Ad5 vectors. J Virol. 2013;87:1373–1384. doi: 10.1128/JVI.02058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan WG, et al. Comparative analysis of simian immunodeficiency virus gag-specific effector and memory CD8+ T cells induced by different adenovirus vectors. J Virol. 2013;87:1359–1372. doi: 10.1128/JVI.02055-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, et al. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature. 2009;457:87–91. doi: 10.1038/nature07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Provine NM, et al. Longitudinal requirement for CD4+ T cell help for adenovirus vector-elicited CD8+ T cell responses. J Immunol. 2014;192:5214–5225. doi: 10.4049/jimmunol.1302806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holst PJ, Bartholdy C, Stryhn A, Thomsen AR, Christensen JP. Rapid and sustained CD4(+) T-cell-independent immunity from adenovirus-encoded vaccine antigens. J Gen Virol. 2007;88:1708–1716. doi: 10.1099/vir.0.82727-0. [DOI] [PubMed] [Google Scholar]
- 12.Yang TC, et al. On the role of CD4+ T cells in the CD8+ T-cell response elicited by recombinant adenovirus vaccines. Mol Ther. 2007;15:997–1006. doi: 10.1038/sj.mt.6300130. [DOI] [PubMed] [Google Scholar]
- 13.Provine NM, et al. Immediate Dysfunction of Vaccine-Elicited CD8 + T Cells Primed in the Absence of CD4 + T Cells. 2016;197:1–17. doi: 10.4049/jimmunol.1600591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol. 1994;68:8056–8063. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortiz AM, et al. Depletion of CD4+ T cells abrogates post-peak decline of viremia in SIV-infected rhesus macaques. J Clin Invest. 2011;121:4433–4445. doi: 10.1172/JCI46023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darrah PA, et al. IL-10 production differentially influences the magnitude, quality, and protective capacity of Th1 responses depending on the vaccine platform. Journal of Experimental Medicine. 2010;207:1421–1433. doi: 10.1084/jem.20092532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinn KM, et al. Antigen expression determines adenoviral vaccine potency independent of IFN and STING signaling. J Clin Invest. 2015;125:1129–1146. doi: 10.1172/JCI78280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Safford M, et al. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat Immunol. 2005;6:472–480. doi: 10.1038/ni1193. [DOI] [PubMed] [Google Scholar]
- 19.Gao B, Kong Q, Kemp K, Zhao YS, Fang D. Analysis of sirtuin 1 expression reveals a molecular explanation of IL-2-mediated reversal of T-cell tolerance. Proc Natl Acad Sci USA. 2012;109:899–904. doi: 10.1073/pnas.1118462109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012;12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 21.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brockman MA, et al. IL-10 is up-regulated in multiple cell types during viremic HIV infection and reversibly inhibits virus-specific T cells. Blood. 2009;114:346–356. doi: 10.1182/blood-2008-12-191296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks DG, et al. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12:1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gassa A, et al. IL-10 Induces T Cell Exhaustion During Transplantation of Virus Infected Hearts. Cell Physiol Biochem. 2016;38:1171–1181. doi: 10.1159/000443067. [DOI] [PubMed] [Google Scholar]
- 25.Couper KN, Blount DG, Riley EM. IL-10: the master regulator of immunity to infection. J Immunol. 2008;180:5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, et al. Modulation of DNA vaccine-elicited CD8+ T-lymphocyte epitope immunodominance hierarchies. J Virol. 2006;80:11991–11997. doi: 10.1128/JVI.01348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, et al. Adenovirus serotype 26 utilizes CD46 as a primary cellular receptor and only transiently activates T lymphocytes following vaccination of rhesus monkeys. J Virol. 2012;86:10862–10865. doi: 10.1128/JVI.00928-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waddington SN, et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Roberts DM, et al. Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature. 2006;441:239–243. doi: 10.1038/nature04721. [DOI] [PubMed] [Google Scholar]
- 30.Crawford A, et al. Molecular and transcriptional basis of CD4+ T cell dysfunction during chronic infection. Immunity. 2014;40:289–302. doi: 10.1016/j.immuni.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yusuf I, et al. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2010;185:190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida H, Hunter CA. The immunobiology of interleukin-27. Annu Rev Immunol. 2015;33:417–443. doi: 10.1146/annurev-immunol-032414-112134. [DOI] [PubMed] [Google Scholar]
- 34.Hsieh CS, et al. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 35.Saraiva M, et al. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity. 2009;31:209–219. doi: 10.1016/j.immuni.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunter CA, Kastelein R. Interleukin-27: balancing protective and pathological immunity. Immunity. 2012;37:960–969. doi: 10.1016/j.immuni.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pennock ND, Gapin L, Kedl RM. IL-27 is required for shaping the magnitude, affinity distribution, and memory of T cells responding to subunit immunization. Proc Natl Acad Sci USA. 2014;111:16472–16477. doi: 10.1073/pnas.1407393111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torrado E, et al. Interleukin 27R regulates CD4+ T cell phenotype and impacts protective immunity during Mycobacterium tuberculosis infection. Journal of Experimental Medicine. 2015;172:4672. doi: 10.1084/jem.20141520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villegas-Mendez A, et al. IL-27 receptor signalling restricts the formation of pathogenic, terminally differentiated Th1 cells during malaria infection by repressing IL-12 dependent signals. PLoS Pathog. 2013;9:e1003293. doi: 10.1371/journal.ppat.1003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kastelein RA, Hunter CA, Cua DJ. Discovery and Biology of IL-23 and IL-27: Related but Functionally Distinct Regulators of Inflammation. 2007;25:221–242. doi: 10.1146/annurev.immunol.22.012703.104758. http://dx.doi.org.ezp-prod1.hul.harvard.edu/10.1146/annurev.immunol.22.012703.104758. [DOI] [PubMed] [Google Scholar]
- 41.Fromentin R, et al. CD4+ T Cells Expressing PD-1, TIGIT and LAG-3 Contribute to HIV Persistence during ART. PLoS Pathog. 2016;12:e1005761. doi: 10.1371/journal.ppat.1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang TC, et al. The CD8+ T cell population elicited by recombinant adenovirus displays a novel partially exhausted phenotype associated with prolonged antigen presentation that nonetheless provides long-term immunity. J Immunol. 2006;176:200–210. doi: 10.4049/jimmunol.176.1.200. [DOI] [PubMed] [Google Scholar]
- 43.Steffensen MA, et al. Qualitative and quantitative analysis of adenovirus type 5 vector-induced memory CD8 T cells: not as bad as their reputation. J Virol. 2013;87:6283–6295. doi: 10.1128/JVI.00465-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujisaki J, et al. In vivo imaging of Treg cells providing immune privilege to the haematopoietic stem-cell niche. Nature. 2011;474:216–219. doi: 10.1038/nature10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quigley M, et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med. 2010;16:1147–1151. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnitz RA, Imam S, Yates K, Haining WN. Isolation of RNA and the synthesis and amplification of cDNA from antigen-specific T cells for genome-wide expression analysis. Methods Mol Biol. 2013;979:161–173. doi: 10.1007/978-1-62703-290-2_13. [DOI] [PubMed] [Google Scholar]
- 47.Geiben-Lynn R, Greenland JR, Frimpong-Boateng K, Letvin NL. Kinetics of recombinant adenovirus type 5, vaccinia virus, modified vaccinia ankara virus, and DNA antigen expression in vivo and the induction of memory T-lymphocyte responses. Clin Vaccine Immunol. 2008;15:691–696. doi: 10.1128/CVI.00418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.