Abstract
Background
The recent Ebola outbreak in West Africa led to the use of a variety of different platform technologies for assaying antibodies because of the difficulties of handling the live virus. The same types of method could be applied rapidly to other infections when they emerge. There is a need to compare quantitative results of different assays, which means that the assays must measure similar parameters and give comparable results.
Methods
A collaborative study was carried out to establish an International Reference Reagent through WHO. Nine samples were sent to 16 laboratories and the results from 22 different assays compared to those obtained by neutralisation assays using the wild type virus.
Findings
Quantitative correlation with the wild type neutralisation assays was very variable but generally poor, with only five of the twenty-two assays giving a correlation coefficient of 0.7 or greater; the five best assays included methods based on wild type and VSV pseudotype neutralisation and ELISA. They could be applicable to other rapidly emerging diseases. The remaining assays including neutralisation of lentiviral pseudotypes need further development.
Interpretation
The assay platform should be chosen with care to ensure that it is fit for purpose. Many of the assays were not suitable for quantitation of antibody levels, a finding that is not surprising given the urgency with which they had to be implemented but some may be of generic value. Antibody titres in samples from a vaccine trial were comparable to those from convalescent patients or lower.
Funding
Funding was from the UK DoH and the Wellcome Tust.
Keywords: Ebola, Antibodies, Assay platforms, Pseudotype neutralisation, Quantitation
1. Background
The need for rapid responses to emerging diseases has assumed greater significance in the wake of the outbreak of Ebola virus disease in West Africa and the current concern over Zika virus. One approach is to develop quantitative platform technologies that can be generally applied for diagnostic and other purposes, including methods for assaying antibody levels to assess the potency of immunological therapeutics such as convalescent plasma and immunoglobulin. Such methods would also be applied to evaluate clinical trials and in clinical diagnosis or serological surveys. The validation and comparison of such assays is outside the normal commercial and regulatory process because by definition they are a response to an emergency where time is of the essence, so there is little information on how the methods compare in a quantitative manner. This paper describes a comparison of a range of assays for antibody to Ebola virus (EBOV) emerging from a project to establish reference reagents under the auspices of WHO.
WHO recorded twenty-four outbreaks of Ebola disease in Africa from 1976 to 2013 with a global total of 1716 cases. The latest Ebola epidemic in Western Africa started in 2014 and up until December 20th 2015 it had resulted in 15,249 cases and 11,315 deaths making it the largest and most significant on record [1]. There were no fully validated commercial assays available because previous outbreaks were sporadic and small scale. Assays for antibody that did not involve working with live Ebola virus had been developed; they included the use of pseudotype neutralisation and ELISA based on expressed recombinant glycoprotein. The relationship between the results of different assays is not clear and given that for example the antibody content of therapeutic materials such as whole plasma or immunoglobulins may be central to their efficacy this is a matter of concern. This paper describes a comparison of assays and analytes used in the collaborative study leading to the establishment of the first reference reagent for antibodies to Ebola virus by WHO in October 2015. The full data are to be found in the report to ECBS available on the WHO website [2].
While the objective of the WHO collaborative study was to identify the most suitable sample to serve as the reference, the study also provided the opportunity to compare the performance of the different platforms using the same samples.
2. Materials and methods
2.1. Samples
The nine samples included in the study are listed in Table 1 and were similar to materials that might be used in therapy.
Table 1.
Samples distributed in the collaborative study.
| EBOV Ab sample code | Sample name | Preparation |
|---|---|---|
| 9 | Tc Bovine IgG (negative) | 1 mg/mL in sterile buffer# |
| 36 | NHSBT EBOV Ab Negative Plasma | SD-extracted |
| 28 | NHSBT EBOV Convalescent Ab | SD-extracted |
| 43 | Norwegian EBOV Convalescent Ab | SD-extracted |
| 79 | American Red Cross EBOV Convalescent Ab | SD-extracted |
| 31 | Tc Bovine IgG (immunized with recombinant rGPZaire2014) | 1 mg/mL in sterile buffer# |
| 88 | Tc Bovine IgG (immunized with Zaire95 + Sudan GP DNA) | 1 mg/mL in sterile buffer# |
| 58 | Vaccinees Plasma Pool (high) | Plasma pool |
| 64 | Vaccinees Plasma Pool (low) | Plasma pool |
Abbreviations: NHSBT = National Health Service Blood and Transplant; SD = Solvent-detergent.
# PBS-Ca2 + -Mg2+; 5% human serum albumin.
Full details are given in the WHO report2. Three were plasma preparations from repatriated convalescent patients, one from Norway (sample 43), one from the USA (sample 79) and one from the UK (NHSBT) (sample 28). Plasma from a normal blood donor in the UK was used as one negative control (sample36). No plasma from West Africa was available at the time of the study. Unlike most patients in West Africa all of the repatriated patients had received sophisticated nursing care and chemotherapeutic or immunological treatments such as Zmapp, a cocktail of human monoclonal antibodies to the Ebola virus glycoprotein, and it is conceivable that the antibodies measured could have been influenced by this; for instance some of the therapeutic antibodies might have persisted despite the time between onset of disease and donation of the plasma. In immunoblots all possessed antibodies to viral proteins in addition to the glycoprotein indicating that antibodies resulting from infection were present. The reference reagent established by WHO is sample 79. The patient who donated sample 79 plasma did not receive monoclonal antibody but did receive other treatment.
Other samples included high and low titre pools of serum obtained from participants in a vaccine trial involving chimp adeno3 vectored EBOV Mayinga glycoprotein followed by vaccinia (MVA) vectored glycoprotein from EBOV Sudan, and Tai forest and Marburg virus. Unfortunately insufficient material was available for these samples to be assayed by all methods. Finally material was obtained from transchromosomal bovines expressing the genes required to produce human immunoglobulin. The animals were immunized with DNA encoding the glycoprotein gene from Zaire 95 and Sudan strains (sample 88) or with a virus like particle formed from the Zaire 2014 Ebola glycoprotein (sample 31), or unimmunised (sample 9). The bovine derived material was highly purified human IgG and immunoglobulin treatment could also be an option for therapy or short term prophylaxis.
2.2. Assay methods
The range of assays used is given in detail elsewhere [2] and is summarised in outline in Table 2. Four participants performed neutralisation assays using infectious Ebola virus under high containment. Six laboratories performed neutralisation assays using pseudoviruses in which the glycoprotein was expressed on another particle; three involved lentiviral pseudotypes, two vesicular stomatitis virus pseudotypes and one an Ebola virus like particle. The readout of the assays varied. Eight laboratories used different versions of ELISA; one (laboratory 5) used two separate formats and one (laboratory 16) four separate formats. All were directed against the glycoprotein, the likely target of biologically active, neutralising, antibodies. An indirect immunofluorescence assay using wild type virus (IFA) (laboratory 14) and a western blot assay (laboratory 4) were also used in the study. The data from these two laboratories were not strictly quantitative and are not considered further here although the western blot data were used to confirm that the presence of antibodies to specific antigens.
Table 2.
Median titres of antibody preparations.
| Laboratory | sample numbers |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 9 | 36 | 28 | 43 | 79 | 31 | 88 | 58 | 64 | |
| Neutralisation with wild type | |||||||||
| 2 (Zaire)a | < | < | < | < | 80 | < | < | < | < |
| 9 (Makona)a | < | < | 30 | < | < | < | < | 160 | < |
| 11a (Zaire)a | < | < | 45 | 27 | 181 | 91 | 64 | – | – |
| 12b (Makona)a | < | < | 30 | 20 | 160 | 20 | 20 | 20 | < |
| Pseudoneutralisation | |||||||||
| 6 (lentivirus) | < | Not reliable | |||||||
| 8 (EbolaVLP) | < | 24 | 66 | 106 | 129 | 22 | 22 | < | < |
| 15 (lentivirus) | < | 70 | 36,450 | 14,580 | 43,740 | 1620 | 540 | – | – |
| 17 (lentivirus) | < | 34 | 197 | 123 | 257 | 59 | 56 | < | < |
| 7 (non-replicating VSV) | < | < | 623 | 669 | 3395 | 1365 | 499 | 469 | 157 |
| 16b (replicating VSV) | < | < | 88 | 65 | 320 | 119 | 50 | – | – |
| ELISA (relative to 79) | |||||||||
| 1 | – | – | 0.49 | 0.63 | 1 | 0.85 | 0.8 | 0.69 | 0.3 |
| 3 | – | – | 0.36 | 0.46 | 1 | 0.77 | 0.45 | 0.45 | 0.14 |
| 4 | – | – | 0.45 | 0.78 | 1 | 0.68 | 0.52 | 0.65 | 0.14 |
| 5a | – | – | 0.29 | 0.5 | 1 | 1.33 | 0.65 | 1.06 | 0.26 |
| 5c | – | – | 0.81 | 0.07 | 1 | 0.21 | 0.27 | 0.41 | 0.12 |
| 10 | – | – | – | – | 1 | – | 0.16 | 0.24 | 0.08 |
| 12a | – | – | 0.41 | 0.59 | 1 | 1.01 | 0.69 | – | – |
| 13 | – | – | 0.88 | 0.61 | 1 | 0.04a | 0.34 | 0.81 | 0.7 |
| 16a | – | – | 0.61 | 0.68 | 1 | 0.63 | 1.43 | 3.1 | 0.51 |
| 16c | – | 1.19 | 0.27 | 0.22 | 1 | 0.03 | 0.51 | 0.26 | 0.08 |
| 16d | – | 8.81 | 0.43 | 0.5 | 1 | 0 | 0 | 0.37 | 0.51 |
| 16e | – | 12.81 | 0.66 | 0.74 | 1 | 0 | 0 | 0.6 | 0.69 |
Sample not assayed: –; antibody not detected:<.
Sample 9, 31, 88: human immunoglobulins from transchromosomal bovine.
Sample 36, 28, 43, 79: human samples from healthy donors (36) or convalescent patients.
Sample 58, 64: human samples from clinical trial.
Sample 79 was established as the first International reference reagent for Ebola antibodies by WHO in October 2015.
Strain used.
2.3. Expression of results
Most laboratories performed three separate assays and generally each assay included replicates. However insufficient data were available from this study to perform meaningful statistical analysis on intra-laboratory variation. It was also clear that some of the assays were modified between runs to improve sensitivity so that agreement between separate runs was sometimes poor. Given the rapidity with which some of the assays were implemented, full scale validation, as required for a commercial kit, was not to be expected. Results for the neutralisation and pseudo-neutralisation assays were expressed as the median of the 50% end point titres submitted. The results from the ELISA were obtained from dilution series of each samples and were in very different formats. The potencies were therefore determined by parallel line analysis and expressed relative to the results obtained with the highest titre human convalescent plasma sample (sample 79, which was later established by WHO as the first reference reagent). The samples taken from sources unimmunised or unexposed to Ebola did not give dose response curves and were scored as negative except for the human sample 36 when assayed by assayed 16c, d and e. Scatter plots for the results of different assays were generated. The small number of data points meant that the calculated correlation coefficients did not achieve statistical significance. In many cases the points were not evenly distributed over the range of readouts with positive signals being clustered at the high end. The correlation coefficients were therefore calculated with and without the negative sample results to clarify the extent to which the assays were quantitative when testing positive samples. The results including all samples showed the qualitative value of the assays while those including just the known positive samples indicated whether they could be used quantitatively.
3. Results
The results of all assays are summarised in Table 2.
3.1. Neutralisation with live Ebola virus
Of the four laboratories performing neutralisation assays with the wild type virus, two (11a and 12b) submitted comparable results. Laboratory 2 found all samples negative except for sample 79 which had the highest titre in other assays. This is consistent with the view that the assay is reliable but less sensitive for these samples. The fourth laboratory, laboratory 9, gave results that were inconsistent with the other three laboratories or with results from other assay methods; also, in one run there was difficulty in maintaining the cell sheet, suggesting that there may have been issues with assaying the samples by this method.
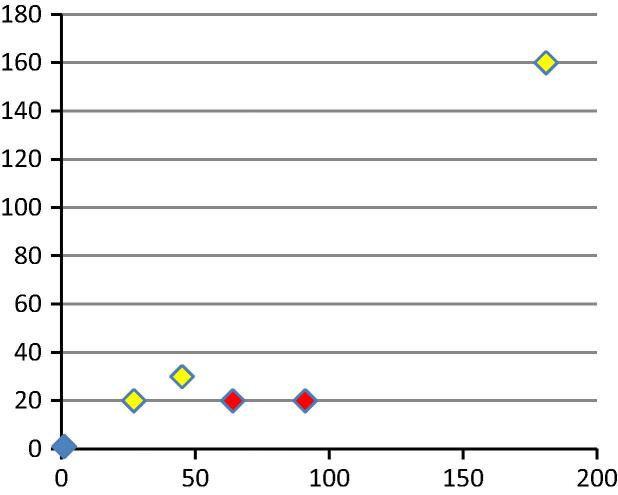
Fig. 1 shows findings for laboratories 11a and 12b, who used neutralisation assays with wild type virus; the correlation coefficient was 0.97 when all points were considered and 0.78 when the negative samples were omitted. The purified human immunoglobulins derived from the transchromosomal bovine were poorly detected by laboratory 12b although they were positive. This is believed to be because of the inaccuracies inherent in measuring low antibody levels with an assay with high variation from run to run. The wild type neutralisation assays were taken as the gold standard as they measure a biological response with the natural antigen. Results from laboratory 11a were more consistent from run to run and between replicates than those from laboratory 12b and were therefore used as the reference data set. Unfortunately laboratory 11a had not been supplied with the vaccine trial plasma samples which were limited in amounts. However, laboratory 12b detected antibody at low titre in sample 58 but not 64, which was consistent with the pre-evaluation testing of the pools as high and low titre respectively at NIBSC.
Fig. 1.
Scatter plot of titres from wild type neutralisation assays from laboratories 11a (x axis) and 12b (y axis). Human samples are shown in yellow, bovine immunoglobulins in red. The correlation coefficient (r) was 0.78 for all positive samples and 0.97 if known negative samples were included.
3.2. Pseudo-neutralisation assays
Of the six laboratories performing pseudo-neutralisation assays, results from laboratory 6 were not consistent between assay runs or within replicates in an assay run, and are not considered further. Laboratory 15 submitted results with titres that were greater by ten to twenty fold than any other laboratory. It is strongly suspected that this was a result of technical dilution error and the results are not considered further here although they ranked the samples in a similar order to other assays.
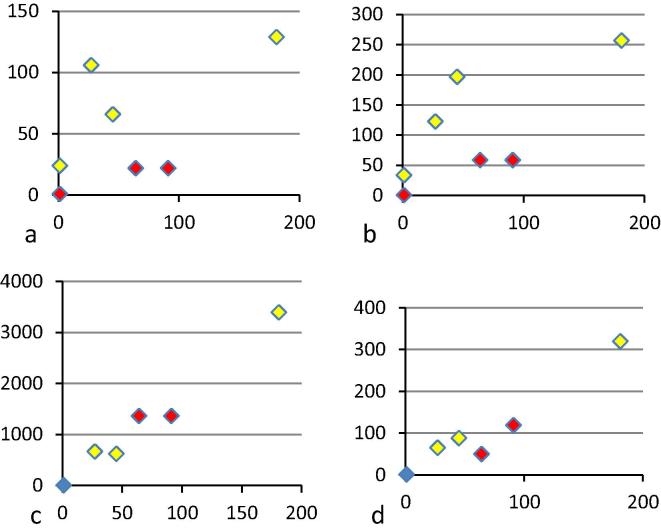
Fig. 2 shows the correlation of results of the wild type neutralisation assay from laboratory 11a with the four laboratories whose pseudo-neutralisation data were analysed. There was a good quantitative relationship between the neutralisation data from laboratory 11a and the results from laboratories 7 (r = 0.99 for all points or 0.84 if only positive samples are considered) and 16b (r = 0.99 for all samples and 0.96 if the negative samples are excluded) who used different versions of a vesicular stomatitis virus (VSV) pseudotype-based neutralisation assay. Laboratory 7 used a non-replicating virus possessing a marker gene to detect infection while laboratory 16b used a replication competent virus that possessed a different marker gene. Of the other two laboratories using pseudo-neutralisation assays laboratory 8 used an Ebola virus like particle and laboratory 17a pseudotyped lentiviral vector. Both showed a poor quantitative correlation with the results from laboratory 11a (r = 0.68 and 0.77 for all points and −0.03 and 0.18 for all positive samples respectively). Both assays also detected neutralising activity in the healthy donor control plasma sourced in the United Kingdom (Tables 1 and 2). This sample is most unlikely to contain antibodies to Ebola virus. Moreover, as the plasma came from a UK blood donor and had been tested, it is most unlikely that it contained specific antibodies to any component of the HIV vector used by laboratory 17. The non-specific neutralisation remains unexplained. These two laboratories also gave low relative titres for the immunoglobulins sourced from transchromosomal cows. This contrasts with the results from the VSV systems. It is possible that the Ebola virus like particle and lentiviral systems are sensitive to plasma factors; for instance the immunoglobulin preparations will be devoid of complement which could play a specific role in neutralisation in the presence of antibody or a non-specific role in its absence.
Fig. 2.
Scatter plot of titres from four laboratories using pseudotype neutralisation (y axis) against results from wild type neutralisation assays from laboratory 11a (x axis). (a) Laboratory 8, (r = 0.68 for all samples; r = −0.03 if known negative samples are excluded), (b) laboratory 17 (r = 0.77; 0.18), (c) laboratory 7 (r = 0.99; 0.84), (d) laboratory 16b (r = 0.99; 0.96). Human samples are shown in yellow, bovine immunoglobulins in red.
While the vaccine trial sera were not assayed by laboratory 11a, laboratory 7 detected a higher titre in sample 58 than in sample 64 as should have been the case based on the pooling performed at NIBSC. Neither laboratory 8 nor 17 detected antibody in either sample and laboratory 16b did not test them.
3.3. ELISA methods
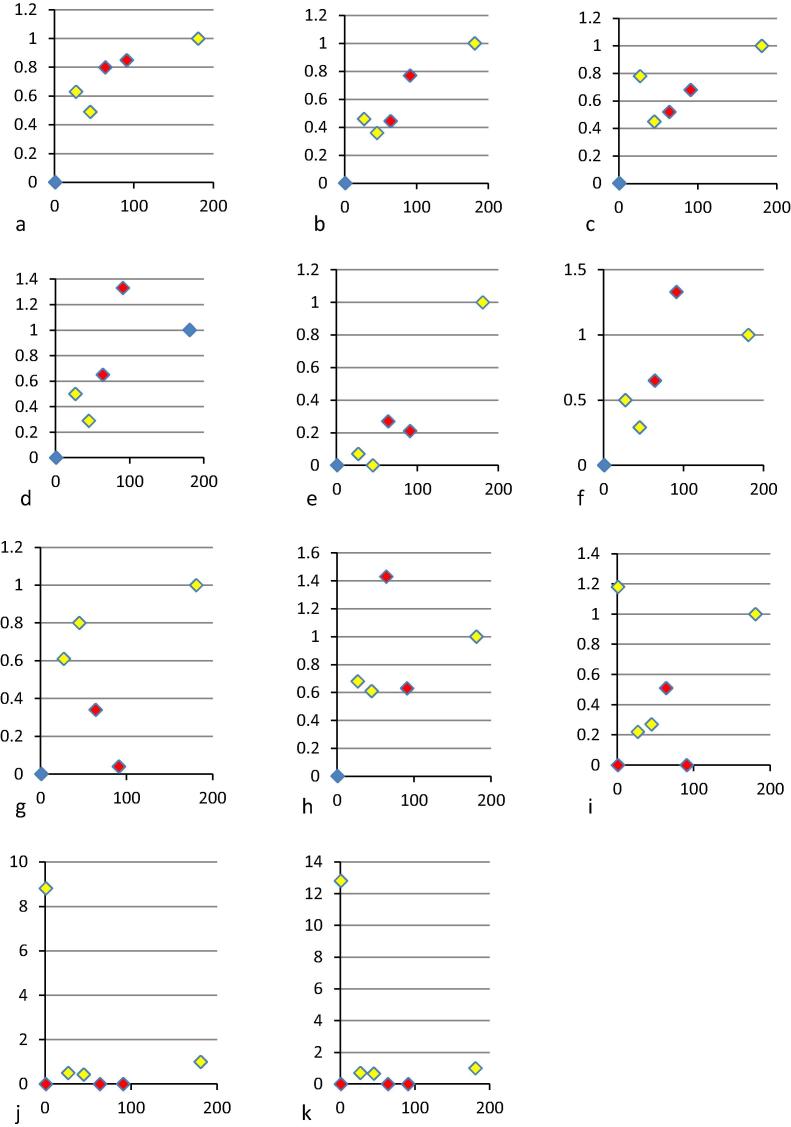
The ELISAs involved assaying dilutions of the samples with results plotted to generate a dose response curve. As the read-out varied with the assay the data were not directly comparable. The relative potencies were therefore scored by parallel line analysis relative to the curves generated for sample 79 in the same assay. While in many cases the negative samples gave a signal by ELISA, apart from laboratory 16 none gave a dose response curve covering sufficient dilutions to allow this type of analysis. Most assays therefore correctly scored the negative samples as negative. The high figures from laboratory 16 c, d and e remain unexplained. It may be of interest that, while assay 16a is intended for the detection of anti-EBOV, assays 16c, 16d and 16e were designed for the detection of antibodies against SUDV, BDBV and MARV, respectively. Fig. 3 shows the correlation between the data from the ELISA and the neutralisation assay from laboratory 11a. In general for all assays where there is neutralising antibody there is also ELISA reactivity. It is notable that the vaccine trial plasma do show the appropriate quantitative relationship in most of the ELISA methods, sample 64 being lower in titre than sample 58. The two pools were created based on the ELISA at NIBSC. Finally all the ELISA data were expressed as a potency relative to sample 79, the human plasma sample established as a reference reagent by WHO, which may influence the findings. For instance certain assays may detect antibodies in whole plasma such as sample 79 differently to the purified immunoglobulins in the bovine samples. All ELISA methods in the study were directed against the glycoprotein.
Fig. 3.
Scatter plot of titres from eleven laboratories using ELISA against results from wild type neutralisation assays from laboratory 11a (x axis). ELISA titres (y axis) were determined by parallel line analysis and expressed relative to sample 79 because of differences in the readout of the assays. (a) Laboratory 1 (r = 0.98 for all samples, r = 0.81 if known negative samples are excluded), (b) laboratory 3 (r = 0.99; 0.85), (c) laboratory 4 (r = 0.97; 0.45), (d) laboratory 5a (r = 0.98; 0.71), (e) laboratory 5c (r = 0.93; 0.65), (f) laboratory 12a (r = 0.98; 0.77), (g) laboratory 13 (r = 0.8; −0.13), (h) laboratory 16a (r = 0.97; 0.36), (i) laboratory 16 c (r = 0.25; 0.21), (j) laboratory 16d (r not calculated), (k) laboratory 16e (r not calculated). Human samples are shown in yellow, bovine immunoglobulins in red.
The correlation coefficients with the data from laboratory 11a ranged from good (for labs 1 and 3 respectively: 0.98 and 0.99 for all samples and 0.81, 0.85 for all positive samples) to poor (for laboratories 13 and 16c: 0.8 and 0.25 for all samples and −0.13, 0.21 for all positive samples). It is not clear why this is the case.
The materials prepared from transchromosomal cows did not give anomalous titres in the scatter plots compared to the other samples.
4. Discussion
The samples included in this study were similar to those that might be used in a therapeutic intervention. Transfusions of convalescent plasma have been used in trials of potential therapeutic interventions in Ebola patients in the recent outbreak [3] and showed little beneficial effect. The amount of antibody present is being measured and the data presented here suggest that care should be taken when selecting the assay. If assays perform poorly with these sample types it is arguable that they are not fit for measuring the potency of plasma for transfusion. This is also an important issue for generic platform technologies applicable to other emerging diseases.
The conclusions from this analysis are that neutralisation assays of wild type virus gave credible results in three of the four laboratories concerned, but results from the fourth laboratory were unreliable for unknown reasons. In addition the methods differ in sensitivity with laboratory 2 having a less sensitive assay than the other two. The inclusion of the WHO International reference reagent would help in defining assay sensitivity and in confirming the reproducibility of the assay used. Where there are questions concerning the titre of plasma transfused in clinical studies of efficacy [3] the choice of assay and the possibility of comparing results from one study to the next are important factors. Data from laboratory 11a were chosen as the reference set as they involved a biological assay on the native virus and gave highly reproducible results consistent with other assays. It was notable that the end point titre observed with the most potent sample (the human convalescent plasma 79, later established as the first reference reagent by WHO) was less than 200. In contrast neutralising antibody levels in plasma after measles or polio infections or immunisations are usually in the thousands. Similarly, where the samples were assayed, the post immunisation titres of vaccinees were barely above the limit of detection.
Correlation between the wild type and pseudotype assays was best for the VSV system (0.84 and 0.96 for the two assays using variants of this platform where only known positives were considered and 0.99 and 0.99 when negative samples were included). The lentiviral and other platforms were problematic. The correlation between ELISA and biological assays was variable, although all those investigated here were against the glycoprotein that is assumed to be the target of a biologically significant neutralising response. Only laboratories 1, 3, 5a and 12a gave r values over the non-stringent figure of 0.7 when compared to laboratory 11a when only positive samples were considered while other assays were far poorer. When all samples were included all assays other than 13 and 16c had r values of 0.97 or greater. Most assays are therefore probably suitable to demonstrate whether the individual has been infected with Ebola. However they are not equally suitable for measuring the quantitative level achieved which would be required to establish a protective level or to assess the potency of immunoglobulin preparations.
Improving the rapidity of the response to an emerging infection has been identified as a key issue for Ebola and other emerging diseases such as MERS, SARS or Zika. ELISA and pseudotype neutralisation assays are platforms that could be rapidly adapted to viruses other than Ebola if needed by cloning the relevant genes into expression vectors or pseudotype vectors. This study raises questions of reliability and relevance that need to be addressed. Likewise the transchromosomal bovine-produced human immunoglobulin could be produced rapidly under controlled conditions to generate a reference or therapeutic material. It performed satisfactorily in most assays in the current study.
This study raises questions of the reliability and relevance of the recombinant DNA based antibody assays that need to be addressed. In particular the neutralisation of the lentiviral pseudotypes here is questionable both from the point of view of specificity, as a number of negative samples were scored positive, and the poor quantitative correlation with the wild type neutralisation assays. In contrast the assays based on VSV gave a good correlation.
The inclusion of the International reference reagent established by WHO enables the comparison of results from different assays and monitors the performance of the test on a day to day basis. We believe that its inclusion in assays would greatly improve comparability and the reliability of results. The use of these platform technologies depends on their fitness for purpose and the work described here identifies further issues to explore.
Conflicts of interest
None.
Funding source
The work was supported by the Wellcome Trust (grant 110002) and from NIBSC funds from the UK Department of Health. NIBSC is part of the MHRA.
References
- 1.WHO Ebola virus disease outbreak. <www.who.int/csr/disease/ebola/en/>; 2016. [accessed 22nd January 2016].
- 2.WHO/ECBS. <http://apps.who.int/iris/bitstream/10665/197777/1/WHO_BS_2015.2280_eng.pdf>; 2015. [accessed 10th March 2016].
- 3.van Griensven J., Edwards Tansy, de Lamballerie Xavier. Evaluation of convalescent plasma for Ebola virus disease in guinea. N Engl J Med. 2016;374:33–42. doi: 10.1056/NEJMoa1511812. [DOI] [PMC free article] [PubMed] [Google Scholar]