Abstract
A number of studies have revealed that nuclear paraspeckle assembly transcript 1 (NEAT1), a long noncoding RNA (lncRNA), was aberrantly regulated in various cancers. High NEAT1 expression was associated with a poor prognosis and an increased risk of lymph node metastasis (LNM) in cancer patients. This meta-analysis was conducted to identify the potential value of NEAT1 as a biomarker for cancer prognosis. We searched the electronic databases PubMed, Web of Science, and China National Knowledge Infrastructure (up to November 13, 2016) to collect all relevant studies to explore the association between the expression of lncRNA NEAT1 and overall survival (OS) and LNM. The results showed that cancer patients with high NEAT1 expression had a poorer OS than those with low NEAT1 expression (hazard ratio: 1.88, 95% confidence interval [CI]: 1.41–2.50, P=0.000). Subgroup analyses by cancer type and sample size indicated that digestive system cancer patients with high NEAT1 expression experienced an increased risk of developing LNM (odds ratio: 1.96, 95% CI: 1.25–3.06, P=0.003). In conclusion, the present meta-analysis showed that high expression of NEAT1 might potentially serve as a reliable biomarker for poor clinical outcome in various cancers. However, owing to the limited size of samples, further clinical studies are required to verify our findings.
Keywords: NEAT1, lncRNA, prognosis, lymph node metastasis, meta-analysis
Introduction
Long noncoding RNAs (lncRNAs) are a class of RNAs larger than 200 nucleotides and lack protein coding capability.1 Recently, large-scale studies have shown that these kinds of nonprotein-coding transcripts play critical roles in cellular regulatory processes, such as transcriptional regulation, epigenetic modification, and human disease.2 Moreover, with the advances in next-generation sequencing, accumulating evidence has shown the dysregulated expression of numerous lncRNAs in human cancers.3 Meanwhile, emerging reports have demonstrated that lncRNAs may act as oncogenes and tumor suppressor genes through affecting cell proliferation, apoptosis, migration, and genomic stability.4,5 The aberrant expression profiles of lncRNAs in different cancer types indicated that this class of molecules could be applied to tumor diagnosis, prognosis, and therapies.6–8
Nuclear paraspeckle assembly transcript 1 (NEAT1) gene is located on chromosome 11q13.1 and has six transcripts.9 The transcript NEAT1-001, also named lncRNA NEAT1, owns two isoforms: 3.7-kb-long NEAT1_1 and 23-kb-long NEAT1_2.10 Previous studies revealed that lncRNA NEAT1 functions as an essential structural component of a nuclear domain called paraspeckles.11,12 Cumulatively, further studies identified that NEAT1 was aberrantly expressed in a number of solid tumors, including colorectal cancer, gastric cancer, and other human malignancies.13–22 Moreover, regulating the expression of NEAT1 in vitro significantly affected the proliferation, migration, and invasion of tumor cells, suggesting that this lncRNA could exert functions in cancer progression. To explore the correlation of NEAT1 with cancer prognosis and lymph node metastasis (LNM), we collected all relevant studies and conducted this quantitative meta-analysis.
Materials and methods
Search strategy
We carefully searched the online PubMed, Web of Science, and China National Knowledge Infrastructure (CNKI) databases (updated to November 13, 2016). The search terms were as the following: “NEAT1 OR Nuclear paraspeckle assembly transcript 1” AND “cancer OR tumor OR neoplasm”. A manual review of the references of relevant articles was also performed to obtain other potential studies. Our meta-analysis collected all relevant studies to explore the association of lncRNA NEAT1 with overall survival (OS) and LNM.
Selection criteria
Inclusion criteria were as follows: 1) the expression levels of NEAT1 in primary cancerous tissues were measured, 2) patients were divided into high and low expression groups based on the expression levels of NEAT1, 3) articles investigating the correlation between NEAT1 expression and OS in any cancer types, 4) hazard ratios (HRs) with 95% confidence intervals (CIs) were reported or sufficient data were available to evaluate the HRs and 95% CIs, and 5) the number of patients with LNM in each group was recorded to calculate the odds ratios (ORs) with 95% CIs. Exclusion criteria were the following: 1) editorials, letters, expert opinions, reviews, and case reports; 2) studies without usable data; and 3) duplicate publications.
Data extraction
Two investigators (TC and HW) extracted data independently from the eligible studies, and disagreements were resolved by the third investigator (PY). The following information was listed as follows: the first author, year of publication, country, cancer type, sample size of patients, number of high NEAT1 expression group and low expression group, number of patients with LNM in each group, detection method of NEAT1, cutoff values, survival analysis method, and the sources of HRs (95% CI). In two studies, we extracted the relevant numerical value to calculate HRs with their 95% CIs from the Kaplan–Meier survival curve using Engauge Digitizer version 4.1.23
Statistical analysis
The effect of NEAT1 on survival outcome was evaluated by the HRs (95% CIs), and the relationship between NEAT1 and LNM was presented as the ORs (95% CIs). The I2 statistic was used to measure the statistical heterogeneity among studies. The random-effects model was adopted if significant heterogeneity was observed (I2>50% or P<0.05). Otherwise, the fixed-effects model was chosen.24,25 We also performed sensitivity analysis to estimate the stability of our results. Publication bias was evaluated by using funnel plots, Begg’s, and Egger’s test.26 P<0.05 was considered to be statistically significant, and all the P-values were two-tailed. Statistical analyses of HRs for OS and the ORs for LNM were conducted by STATA 12.0 software (StataCorp LP, College Station, TX, USA).
Results
Eligible studies and characteristics
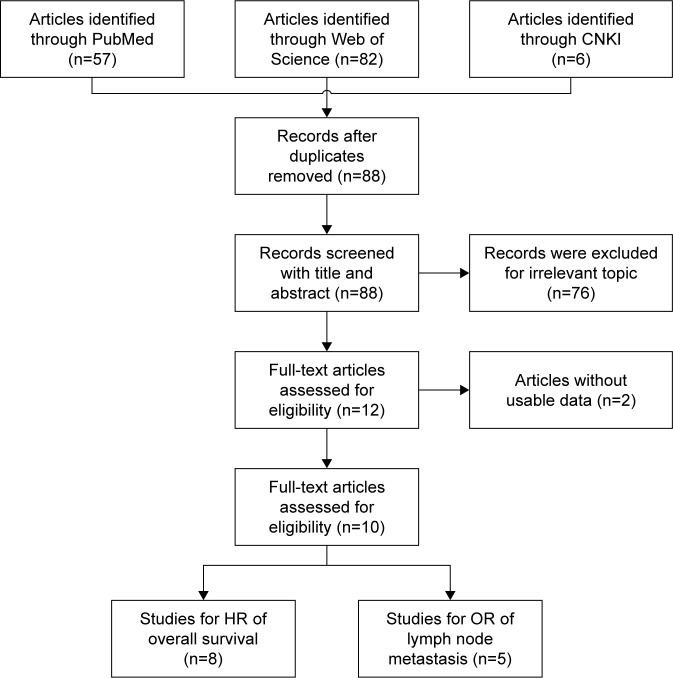
Initially, 145 articles were obtained by searching from PubMed, Web of Science, and CNKI databases. Among these studies, 57 duplicates were removed and 76 records were excluded due to irrelevant contents. By further reviewing remaining 12 articles, another 2 studies lacking available HRs or ORs (95% CI) were excluded. Finally, 10 articles containing 3,085 subjects were included in the meta-analysis. The flow diagram of the selection process is shown in Figure 1.
Figure 1.
Flow diagram of studies selection process.
Abbreviations: CNKI, China National Knowledge Infrastructure cancer; HR, hazard ratio; OR, odds ratio.
The main characteristics are summarized in Table 1. Nine included studies came from the People’s Republic of China, and one study was conducted in UK and Canada. Ten different types of cancers were evaluated, including esophageal squamous cell carcinoma, colorectal cancer, breast cancer, ovarian cancer, gastric cancer, glioma, endometrial endometrioid adenocarcinoma, nasopharyngeal carcinoma, non-small cell lung cancer, and bladder cancer. Quantitative real-time polymerase chain reaction was used in eight studies,13–18,20,21 and in situ hybridization was performed in one research to assess NEAT1 expression.19 In addition, using RNA microarrays, Choudhry et al categorized patients with breast cancer according to NEAT1 expression.22 HRs and 95% CIs were directly extracted from six studies, whereas these necessary statistical variables were calculated by survival curves in two studies.
Table 1.
Characteristics of NEAT1 studies included in the meta-analysis
| First author | Year | Country | Cancer type | Sample size | NEAT1 expression
|
NEAT1 detection method | Cutoff | Outcome | Survival analysis | HR estimate | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High with LNM | High without LNM | Low with LNM | Low without LNM | ||||||||||
| Chen et al13 | 2015 | China | ESCC | 96 | 32 | 20 | 18 | 26 | qRT-PCR | Youden index | OS | Multivariate, univariate | Reported |
| Li et al14 | 2015 | China | CRC | 239 | 76 | 34 | 71 | 58 | qRT-PCR | FC=2 | OS | Multivariate, univariate | Reported |
| Choudhry et al22 | 2015 | UK and Canada | BC1 | 2,000 | – | – | – | – | RNA microarrays | Median | OS | Univariate | Reported |
| Chen et al15 | 2016 | China | OC | 149 | – | – | – | – | qRT-PCR | Median | OS | Multivariate, univariate | Reported |
| Fu et al16 | 2016 | China | GC | 140 | – | – | – | – | qRT-PCR | Median | OS | Multivariate, univariate | Reported |
| He et al17 | 2016 | China | Glioma | 94 | – | – | – | – | qRT-PCR | Median | OS | Multivariate, univariate | Reported |
| Li et al18 | 2016 | China | EEC | 75 | 24 | 10 | 4 | 37 | qRT-PCR | Median | – | – | – |
| Lu et al19 | 2016 | China | NPC | 131 | – | – | – | – | ISH | SI=6 | OS | Univariate | Survival curve |
| Sun et al20 | 2016 | China | NSCLC | 96 | 35 | 32 | 22 | 7 | qRT-PCR | FC=2 | OS | Univariate | Survival curve |
| Chen et al15 | 2016 | China | BC2 | 65 | 3 | 45 | 0 | 17 | qRT-PCR | Youden index | – | – | – |
Abbreviations: BC1, breast cancer; BC2, bladder cancer; CRC, colorectal cancer; EEC, endometrial endometrioid adenocarcinoma; ESCC, esophageal squamous cell carcinoma; FC, fold change; GC, gastric cancer; HR, hazard ratio; ISH, in situ hybridization; LNM, lymph node metastasis; NEAT1, nuclear paraspeckle assembly transcript 1; NPC, nasopharyngeal carcinoma; NSCLC, non-small cell lung cancer; OC, ovarian cancer; OS, overall survival; qRT-PCR, quantitative reverse transcription polymerase chain reaction.
OS is associated with NEAT1 expression
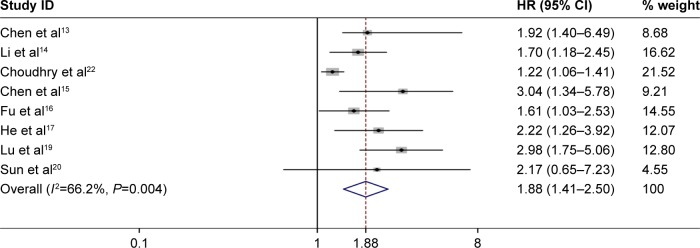
Eight studies consisting of 2,945 patients reported the OS according to NEAT1 expression levels, and the median sample size was 135.5 (range 94–2,000) in this meta-analysis. Because there was significant heterogeneity across our included studies (I2=66.2%, P=0.004; Figure 2), the random-effects model was applied to estimate the pooled HRs and the respective 95% CIs. Our result revealed that high level of NEAT1 expression could predict poor OS in various cancers (pooled HR: 1.88, 95% CI: 1.41–2.50, P=0.000; Figure 2).
Figure 2.
Forest plot for the association between NEAT1 expression levels with OS.
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; HR, hazard ratio; NEAT1, nuclear paraspeckle assembly transcript 1; OS, overall survival.
Subsequently, we performed subgroup analyses according to the following categories: cancer type (digestive system and nondigestive system), survival analysis (multivariate analysis and univariate analysis), and sample size (<135.5 and ≥135.5) to further evaluate the association between NEAT1 expression and OS. As the results shown in Table 2, compared with the total pooled HR, increased NEAT1 exhibited a strong correlation with poor OS in a majority of the subgroups (all P<0.01), except in the subgroup of univariate analysis (HR: 1.88, 95% CI: 0.93–3.84, P=0.08). In addition, significant heterogeneity was still observed in the subgroups of nondigestive system, univariate analysis, and sample size ≥135.5 (all I2>50%, P<0.05).
Table 2.
Subgroup analysis of the pooled HRs of OS
| Subgroup factor | Divided standard | Study number | Pooled HR (95% CI) | I2 value (%) | P-value for heterogeneity |
|---|---|---|---|---|---|
| Cancer type | Digestive system | 3 | 1.69 (1.30–2.21) | 0.0 | 0.929 |
| Nondigestive system | 5 | 2.11 (1.27–3.50) | 78.6 | 0.001 | |
| Survival analysis | Multivariate analysis | 5 | 1.87 (1.49–2.36) | 0.0 | 0.603 |
| Univariate analysis | 3 | 1.88 (0.93–3.84) | 81.5 | 0.004 | |
| Sample size | Number <135.5 | 4 | 2.42 (1.74–3.38) | 0.0 | 0.787 |
| Number ≥135.5 | 4 | 1.59 (1.16–2.19) | 65.7 | 0.033 |
Abbreviations: CI, confidence interval; HR, hazard ratio; OS, overall survival.
Relationship between NEAT1 and LNM
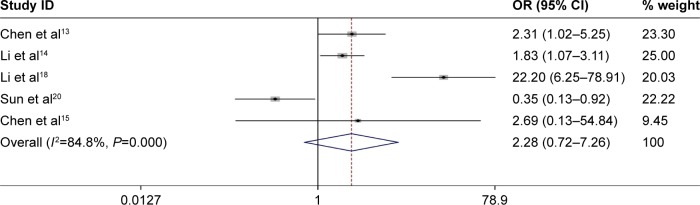
Five studies reported the number of patients with LNM based on different NEAT1 expression levels in a total of 571 individuals. The median sample size was 96 (range 65–239). As shown in Figure 3, the random-effects model was adopted for the significant heterogeneity (I2=84.8%, P=0.000). Our results failed to identify any significant association between NEAT1 expression and LNM (pooled OR: 2.28, 95% CI: 0.72–7.26, P=0.161; Figure 3). To obtain further insight into the NEAT1 prognostic value in LNM, subgroup analysis was performed with cancer type and sample size (Table 3). In summary, our data revealed that NEAT1 over-expression was significantly related to high incidence of LNM in subgroups of digestive system (OR: 1.96, 95% CI: 1.25–3.06, P=0.003) and sample size <96 (OR: 12.16, 95% CI: 1.85–80.00, P=0.009) without significant heterogeneity (all I2≤50%, P≥0.05). No other subgroups exhibited any significant relationships between NEAT1 expression and incidence of LNM.
Figure 3.
Forest plot for the association between NEAT1 expression levels with LNM.
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; LNM, lymph node metastasis; NEAT1, nuclear paraspeckle assembly transcript 1; OR, odds ratio.
Table 3.
Subgroup analysis of the pooled ORs of LNM
| Subgroup factor | Divided standard | Study number | Pooled OR (95% CI) | I2 value (%) | P-value for heterogeneity |
|---|---|---|---|---|---|
| Cancer type | Digestive system | 2 | 1.96 (1.25–3.06) | 0.0 | 0.637 |
| Nondigestive system | 3 | 2.71 (0.12–60.88) | 92.3 | 0.000 | |
| Sample size | Number <96 | 2 | 12.16 (1.85–80.00) | 38.6 | 0.202 |
| Number ≥96 | 3 | 1.20 (0.44–3.25) | 80.4 | 0.006 |
Abbreviations: CI, confidence interval; LNM, lymph node metastasis; OR, odds ratio.
Sensitivity analysis and publication bias
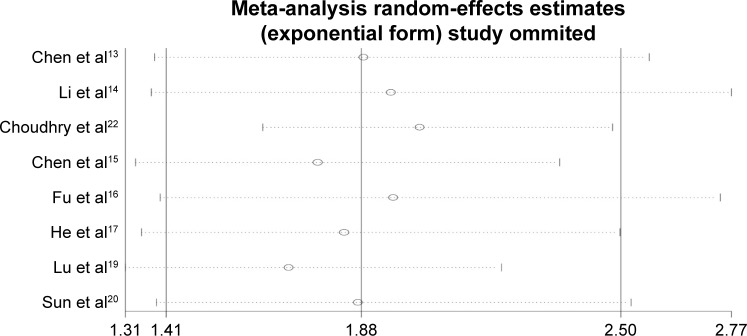
Sensitivity analysis suggested that the association between NEAT1 expression and OS was not significantly influenced by deleting any individual study (Figure 4).
Figure 4.
Sensitivity analysis of effect of individual studies on the pooled HRs for NEAT1 and OS of patients.
Abbreviations: HR, hazard ratio; NEAT1, nuclear paraspeckle assembly transcript 1; OS, overall survival.
In the pooled analyses of OS, Begg’s funnel plot with pseudo 95% CIs indicated an asymmetry (Figure 5). Consistently, the results of Egger’s test also revealed that obvious publication bias was present in our analysis (P=0.005).
Figure 5.
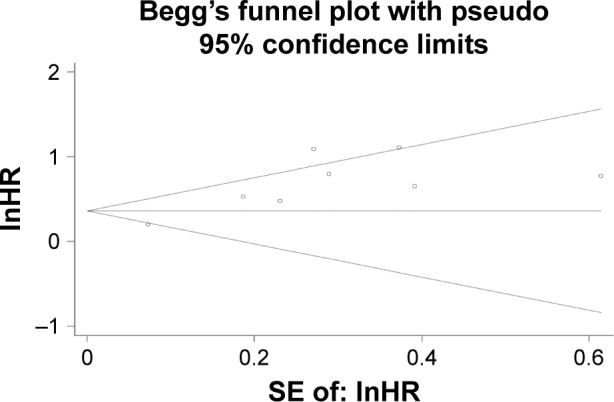
Funnel plot analysis of publication bias for OS.
Abbreviations: lnHR, regular hazard ratio; OS, overall survival; SE, standard error.
Discussion
Recently, aberrant expressions of lncRNAs have been identified in different types of tumors. It was demonstrated by emerging studies that lncRNAs were associated with cancer development and progression.4 In addition, several lncRNAs, such as HOTAIR and MALAT1, have been commonly detected in various tumors to explore their roles in carcinogenesis. HOTAIR, a lncRNA with regulatory functions of transcription, could regulate the chromatin methylation state through binding with PRC2 and acts as an oncogene in different cancer cells, such as gastric, breast, colorectal, and cervical cancer cells.27 LncRNA MALAT1 was first discovered as a prognostic marker for lung cancer metastasis and also has been linked to other human tumor entities and metastasis.28 Thus, the studies of lncRNA have provided a new insight into cancer genomics, but the related mechanisms remain to be explored.
Paraspeckles are formed through the recruitment of Drosophila behavior/human splicing family proteins including P54nrb, PTB-associated splicing factor (PSF), and PSPC1 to the central scaffolding RNA, NEAT1. As an architectural lncRNA in nuclear paraspeckles, NEAT1 RNA is essential for paraspeckle integrity and functions.12 Adriaens et al found that DNA damage agent could induce NEAT1 expression and paraspeckle formation in a p53-dependent manner, contributing to the chemo-resistance in cancer cell.29 Likewise, Choudhry et al showed that hypoxia-induced NEAT1 stimulated the formation of nuclear paraspeckles, accompanied by increased tumorigenesis in breast cancer.22 Especially, these nuclear bodies may suppress the protein synthesis of adenosine-to-inosine edited transcripts through the nuclear retention of target mRNAs. Moreover, NEAT1 has been proposed to sequester paraspeckle proteins into the nuclear bodies, limiting the transcriptional activation or inactivation activity of these proteins in the nucleoplasm.30 For example, Imamura et al found that NEAT1 induction could relocate PSF from the IL8 promoter to the paraspeckles, leading to transcriptional activation of IL8.31 Recent studies also demonstrated that NEAT1 were associated with the initiation and progression of various cancers. Fu et al showed that the overexpression of NEAT1 could promote the cell migration and invasion in vitro and act as an unfavorable prognosis factor in gastric cancer.16 Lu et al revealed that NEAT1 regulated EMT transition and radioresistance through miR-204/ZEB1 axis in nasopharyngeal carcinoma.19 Sun et al found that NEAT1 significantly accelerated cell metastasis through regulation of miR-377-3p-E2F3 pathway in non-small cell lung cancer.20 Moreover, elevated NEAT1 levels were extraordinarily associated with LNM in colorectal cancer and esophageal squamous cell carcinoma.13,14 Therefore, NEAT1 is considered to be an oncogene and a promising indicator of prognosis in human cancers.
Our present study attempted to investigate the association between lncRNA NEAT1 and the clinical prognosis in human cancers. Compared to the study of Yang et al,32 our analysis included all the studies in which the detection methods of NEAT1 expression were not limited to quantitative reverse transcription polymerase chain reaction. Moreover, we excluded the study in which NEAT1 expression was detected not only in primary tumor tissues but also in whole blood.33 We found that high NEAT1 expression was associated with poor OS in different types of cancers (pooled HR: 1.88, 95% CI: 1.41–2.50, P=0.000) with significant heterogeneity (I2=66.2%, P=0.004). However, based on the entire data set, we could not confirm any association between NEAT1 expression and LNM (pooled OR: 2.28, 95% CI: 0.72–7.26, P=0.161). Then we performed subgroup analysis to precisely assess the prognostic effect of NEAT1 on OS and LNM, respectively. The results indicated that high NEAT1 expression exhibited a significant correlation with poor OS in a majority of the subgroups (all P<0.01) except in the subgroup of univariate analysis (HR: 1.88, 95% CI: 0.93–3.84, P=0.08), suggesting that elevated NEAT1 might serve as a reliable molecular marker for poor prognosis in various cancers. Meanwhile, we found the study of Choudhry et al was the only research existing in all the following subgroups (nondigestive system, univariate analysis, and sample size ≥135.5),22 in which significant heterogeneity was still observed. After omitting their study, our analysis showed no significant heterogeneity and no obvious publication bias when exploring the relationship between NEAT1 expression and OS (data not shown), which might partly explain the sources of heterogeneity and publication bias. Although Choudhry et al performed their study with high quality, similar studies with larger sample sizes and long-term follow-up may be required to further confirm our current findings. Notably, our data revealed that the patients with high NEAT1 expression might suffer higher incidence of LNM in subgroup of digestive system cancers (OR: 1.96, 95% CI: 1.25–3.06, P=0.003) without significant heterogeneity (I2=0.0%, P=0.637). Although NEAT1 overexpression was also significantly related to high incidence of LNM in subgroup of sample size <96 (OR: 12.16, 95% CI: 1.85–80.00, P=0.009), considering that meta-analyses including a limited number of patients are prone to overestimate the effects,34 it is necessary to conduct larger size and better designed studies to confirm the role of NEAT1 expression level in cancer LNM.
Limitations
Nevertheless, it should be noticed that there were several limitations in our meta-analysis. First, the cutoff values dividing the NEAT1 expression were not unified in different studies. Second, the number of patients and studies included was small. Third, many included studies reported positive results instead of publishing negative results. Fourth, we only recruited papers written in English. Finally, studies included in our analysis were mostly from the People’s Republic of China, which might limit the results, we found, to the cases of Chinese cancer patients.
Conclusion
In summary, we have demonstrated that high NEAT1 expression was significantly correlated with poor OS in multiple cancers. In addition, our results showed that elevated NEAT1 expression was significantly associated with higher incidence of LNM in digestive system neoplasms. Further studies exploring the relationship between clinicopathological features and NEAT1 expression levels are required to verify its clinical prognostic value in human cancers.
Acknowledgments
This study was supported by the Science and Technology projects of Jiangsu Province, Bureau of Traditional Chinese Medicine (YB2015171).
Footnotes
Author contributions
TC and Z-YH designed the research; PY and HW performed the research and analyzed the data; TC and HW wrote the paper. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 2.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Maruyama R, Suzuki H. Long noncoding RNA involvement in cancer. BMB Rep. 2012;45(11):604–611. doi: 10.5483/BMBRep.2012.45.11.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 6.Meseure D, Drak Alsibai K, Nicolas A, Bieche I, Morillon A. Long noncoding RNAs as new architects in cancer epigenetics, prognostic biomarkers, and potential therapeutic targets. Biomed Res Int. 2015;2015:320214. doi: 10.1155/2015/320214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva A, Bullock M, Calin G. The clinical relevance of long non-coding RNAs in cancer. Cancers (Basel) 2015;7(4):2169–2182. doi: 10.3390/cancers7040884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71(1):3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aken BL, Ayling S, Barrell D, et al. The Ensembl gene annotation system. Database (Oxford) 2016;2016:1–19. doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakravarty D, Sboner A, Nair SS, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemson CM, Hutchinson JN, Sara SA, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long non-coding RNA. J Cell Biol. 2009;186(5):637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Kong J, Ma Z, Gao S, Feng X. Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am J Cancer Res. 2015;5(9):2808–2815. [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Li Y, Chen W, et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6(29):27641–27650. doi: 10.18632/oncotarget.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen ZJ, Zhang Z, Xie BB, Zhang HY. Clinical significance of upregulated lncRNA NEAT1 in prognosis of ovarian cancer. Eur Rev Med Pharmacol Sci. 2016;20(16):3373–3377. [PubMed] [Google Scholar]
- 16.Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142(7):1571–1579. doi: 10.1007/s00432-016-2152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He C, Jiang B, Ma J, Li Q. Aberrant NEAT1 expression is associated with clinical outcome in high grade glioma patients. APMIS. 2016;124(3):169–174. doi: 10.1111/apm.12480. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Wei D, Yang C, Sun H, Lu T, Kuang D. Overexpression of long noncoding RNA, NEAT1 promotes cell proliferation, invasion and migration in endometrial endometrioid adenocarcinoma. Biomed Pharmacother. 2016;84:244–251. doi: 10.1016/j.biopha.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Lu Y, Li T, Wei G, et al. The long non-coding RNA NEAT1 regulates epithelial to mesenchymal transition and radioresistance in through miR-204/ZEB1 axis in nasopharyngeal carcinoma. Tumour Biol. 2016;37(9):11733–11741. doi: 10.1007/s13277-015-4773-4. [DOI] [PubMed] [Google Scholar]
- 20.Sun C, Li S, Zhang F, et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7(32):51784–51814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.XianGuo C, ZongYao H, Jun Z, et al. Promoting progression and clinicopathological significance of NEAT1 over-expression in bladder cancer. Oncotarget. 2016 Jun 15; Epub. [Google Scholar]
- 22.Choudhry H, Albukhari A, Morotti M, et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34(34):4482–4490. doi: 10.1038/onc.2014.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 25.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 26.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai B, Song XQ, Cai JP, Zhang S. HOTAIR: a cancer-related long non-coding RNA. Neoplasma. 2014;61(4):379–391. doi: 10.4149/neo_2014_075. [DOI] [PubMed] [Google Scholar]
- 28.Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91(7):791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 29.Adriaens C, Standaert L, Barra J, et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med. 2016;22(8):861–868. doi: 10.1038/nm.4135. [DOI] [PubMed] [Google Scholar]
- 30.West JA, Mito M, Kurosaka S, et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J Cell Biol. 2016;214(7):817–830. doi: 10.1083/jcb.201601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imamura K, Imamachi N, Akizuki G, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53(3):393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Yang C, Li Z, Li Y, et al. Long non-coding RNA NEAT1 overexpression is associated with poor prognosis in cancer patients: a systematic review and meta-analysis. Oncotarget. 2016 Dec 1; doi: 10.18632/oncotarget.13737. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y, Yang L, Zhao J, et al. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Cancer. 2015;14:191. doi: 10.1186/s12943-015-0455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorlund K, Imberger G, Walsh M, et al. The number of patients and events required to limit the risk of overestimation of intervention effects in meta-analysis – a simulation study. PLoS One. 2011;6(10):e25491. doi: 10.1371/journal.pone.0025491. [DOI] [PMC free article] [PubMed] [Google Scholar]