Abstract
Objectives
Atypical Antipsychotics (AAPs) carry a significant risk of cardiometabolic side effects including insulin resistance. It is thought that the insulin resistance resulting from the use of AAP may be associated with changes in DNA methylation. We aimed to identify and validate a candidate gene associated with AAP-induced insulin resistance by using a multi-step approach that included an epigenome-wide association study (EWAS) and validation with site-specific methylation and metabolomics data.
Methods
Bipolar subjects treated with AAPs or lithium monotherapy were recruited for a cross-sectional visit to analyze peripheral blood DNA methylation and insulin resistance. Epigenome-wide DNA methylation was analyzed in a discovery sample (n=48) using the Illumina 450K BeadChip. Validation analyses of the epigenome-wide findings occurred in a separate sample (n=72) using site-specific methylation with pyrosequencing and untargeted metabolomics data. Regression analyses were conducted controlling for known confounders in all analyses and a mediation analysis was performed to investigate if AAP-induced insulin resistance occurs through changes in DNA methylation.
Results
A differentially methylated probe associated with insulin resistance was discovered and validated in the Fatty Acyl CoA Reductase 2 (FAR2) gene of Chromosome 12. Functional associations of this DNA methylation site on untargeted phospholipid-related metabolites were also detected. Our results identified a mediating effect of this FAR2 methylation site on AAP-induced insulin resistance.
Conclusions
Going forward, prospective, longitudinal studies assessing comprehensive changes in FAR2 DNA methylation, expression, and lipid metabolism before and after AAP treatment are required to assess its potential role in the development of insulin resistance.
Keywords: antipsychotic, insulin, bipolar, methylation, mediation
INTRODUCTION
It is estimated that approximately 1–2% of the United States’ population has bipolar disorder (1). Psychopharmacologic treatment of bipolar disorder utilizes several different medication classes including mood stabilizers, antidepressants and benzodiazepines. The atypical antipsychotics (AAPs) have become an important maintenance pharmacotherapeutic option in the past 10 years (2). Although the efficacy of AAPs in bipolar disorder has been shown in randomized, placebo-controlled trials, they also carry a significant risk of weight gain, diabetes, metabolic syndrome, and cardiovascular disease (3). This may be particularly concerning in severe mental illnesses, like bipolar disorder, which have elevated rates of metabolic co-morbidity, and in particular, insulin resistance prior to AAP treatment (4, 5). Physical health may be linked to psychiatric health. Within bipolar disorder, patients with endocrine abnormalities, in particular insulin resistance, have worse psychiatric outcomes (6–9). The impact of AAP cardiometabolic side effects on long-term psychiatric outcomes is not known, however, given the connection between cardiometabolic and psychiatric health, as well as the high rates of cardiovascular mortality in bipolar disorder, it is important to understand how these side effects occur (10). Thus, there is a need to identify the mechanisms by which AAPs increase the risk of insulin resistance so that the beneficial effects of these medications can be used to treat psychiatric symptoms without sacrificing a patient’s physical health or compounding the problem of metabolic illness within this vulnerable population.
Pharmacoepigenetics may aid in understanding the complex mechanisms of AAP-induced side effects by examining the interface of medication use and gene expression. Previous work has identified associations between one-carbon metabolism and AAP-induced insulin resistance and metabolic syndrome in bipolar disorder as well as psychopharmacology-specific effects on the epigenome (11–13). It may be plausible that an epigenetic mechanism underlies these side effects since the endpoint of one-carbon metabolism is the production of methyl donors that are used for various methylation reactions in the body including that of DNA methylation. This hypothesis was supported through identified associations between a global measure of DNA methylation and AAP-induced insulin resistance within bipolar disorder (14). In the only candidate gene study to date, Moons and colleagues found no associations between AAP-induced metabolic side effects and genetic or epigenetic variability in the Insulin-like growth factor 2 (IGF2) gene (15).
The metabolome, which represents all of the small molecules in the cell that arise as a consequence of metabolism, is directed by upstream processes including genetic and epigenetic variability. Although it can be relatively stable, AAPs may have significant effects on the metabolome (16–19). Metabolomics can be a useful tool in studying AAP-induced side effects due to its ability to represent the echo of complex lifestyles of individuals interacting with genetic and environmental factors like medications (20). The changes in the metabolome secondary to AAP-induced insulin resistance may be the functional consequence of epigenetic changes, therefore combining these two molecular approaches may aid in understanding the pathways involved in AAP-induced insulin resistance (21, 22).
Despite previous work implicating DNA methylation changes in AAP-induced insulin resistance at the global level, it is not known which regions or genes may be differentially methylated in AAP-induced insulin resistance and whether it is a potential mediator of the drug-induced side effect. Within this study, we aimed to identify differentially methylated genes associated with AAP-induced insulin resistance in a multi-step, systematic approach (Supplementary Figure 1). In the first step we employed an Epigenome-Wide Association Study (EWAS) to discover a candidate gene (or genes). In the next steps, we validated the discovery finding using site-specific and metabolomics analyses in a separate sample. Finally, within the validation analyses we tested the hypothesis that AAP-induced insulin resistance is mediated by our candidate gene’s DNA methylation. Although completion of the multi-step design wasn’t guaranteed, we detail below our successful identification of a novel candidate gene whose differential methylation may mediate AAP-induced insulin resistance.
METHODS
Subject Recruitment
Samples and data for both the discovery and validation samples came from an ongoing study on the rates of metabolic syndrome in bipolar disorder and schizophrenia (11). Subjects were included if they had a Diagnostic and Statistical Manual of Mental Disorders, 4th edition, text revision (DSM-IV-TR) of Bipolar I disorder via the structured clinical interview for DSM diagnoses (SCID) and were currently treated with an AAP or lithium monotherapy for at least 3 months (23). Subjects were excluded if they had a documented metabolic disturbance (e.g., obesity, diabetes or cardiovascular disease) prior to starting their AAP or lithium therapy, were currently treated with medications for diabetes, or had an active substance abuse diagnosis (smoking was allowed). Subjects underwent a medical and medication history, anthropometric, vital sign, psychiatric symptom, and insulin resistance (Homeostatic Model Assessment of Insulin Resistance (HOMA-IR)) measurements (24). A fasting (8–10 hours) blood draw within 3 hours of the subject’s normal time of awakening was obtained for epigenetic and metabolomics assessment. Current smoking was defined as smoking 5 or more cigarettes per day. For the purposes of description, AAP use was stratified based on a general metabolic side effect risk profile to include the following groups: Group 1 included olanzapine and clozapine which have the highest propensity to cause metabolic side effects, Group 2 included quetiapine and risperidone which carry a moderate risk of metabolic side effects and Group 3 included aripiprazole and ziprasidone which are considered to have the lowest risk of all AAPs of causing metabolic side effects. Such categorization (e.g., low, intermediate and high risk) has been used in previous studies and is supported by meta-analyses and consensus expert opinion (25–29). All subjects underwent informed consent before participating and the study was approved by the University of Michigan Medical Institutional Review Board (UM-MED IRB).
Discovery using an Epigenome-Wide Analysis Study (EWAS)
From the above cohort, 48 subjects currently treated with AAPs were randomly chosen for the discovery step. Genomic DNA was obtained from whole blood, bisulfite converted, and submitted to the University of Michigan DNA sequencing core for analysis using the Illumina Human Methylation 450K BeadChip (30). The core returned raw image (IDAT) files for analysis by the investigators.
Validation – Site-Specific Epigenetic Analysis
Validation of the top gene-related finding from the EWAS (detailed in results) occurred in the validation sample using site-specific methylation with pyrosequencing. The validation sample (n=72 on AAP and lithium monotherapy) was chosen from the same pool of samples described in the recruiting process above, however, there was no overlap. Primers were created to measure the methylation of the Chr12:29302232 site (GR37/HG19) identified from the EWAS using the Qiagen Assay Design 2.0 Software (Supplementary Table I). Duplicate reactions were ran on a Pyrosequencer MD96 (31) and duplicates resulting in a coefficient of variation greater than 2% methylation difference were discarded. Two samples were removed from validation analysis due to unreliable methylation results. Percent methylation at Chr12:29302232 was standardized using a 6-point standard curve (32).
Validation – Metabolomics Analysis
An untargeted metabolomics analysis in the validation sample was conducted using the Michigan Regional Comprehensive Metabolomics Resource Core (MRC)2 at the University of Michigan as described elsewhere (33). The core processed all samples and resulting features were stored and further processed using in-house developed software and finally returned as a single, normalized (via internal standards), and pre-processed (auto-scaled, log transformed) data set. The core assisted in narrowing the returned data set to metabolites related to our discovery gene findings using their in-house pathway software as well as commercially available platforms. Metabolites were chosen if they were found in phospholipid pathways (this pathway was chosen based on the discovery EWAS finding) by entering metabolite identifiers into Kegg Reaction, Metaboanalyst and Metscape Databases.
Statistical Analysis
Demographic and clinical characteristics are described using means with standard deviation or percentages. The discovery and validation samples were compared using student t-tests for continuous variables and chi-square or Fisher’s Exact tests for categorical data. Raw IDAT data files from the Illumina 450K BeadChip were downloaded from the DNA sequencing core and processed using available bioinformatics pipelines in R Statistical software 3.1 (34, 35). These pipelines included the CHAMP, Minfi and COMBAT R packages (36–39). No samples were removed due to missing data points. Cellular composition was inferred by the method of Houseman and colleagues using the FlowSorted. Blood.450k R package in order to be included as a covariate in the analysis as it has been shown that cellular heterogeneity can be an important confounder in EWAS and particularly in studying medication effects in bipolar patients taking medications (12, 40, 41). To assess the relationship between HOMA-IR and 450K methylation, differentially methylated probes were identified using a linear model (“lm” in Limma package) with the covariates of age, race, gender, AAP group, smoking, folate, BMI, and cellular composition (42). Covariates of age, race, gender, BMI, folate, and smoking were chosen based on our previous work and from work showing that they may be considered biologically relevant predictors of methylation status in addition to HOMA-IR (14, 43). Furthermore, individual antipsychotics have different propensities to cause insulin resistance therefore the 3 AAP groups listed above were also included in the model. The use of linear modeling with Limma and its benefits in analyzing microarray data has been described elsewhere (44) and our modeling improved the overall fit (see QQ plot in Supplementary Figure 2). Correction for multiple testing was done using the Bonferroni method with an epigenome-wide significance level of p<1.1×10−7 (0.05/450,000 sites).
For the validation analyses the correlation between CpG methylation (at the site identified from the EWAS) and HOMA-IR were analyzed in the validation sample using linear regression (32). Covariates for linear regressions were chosen based on the literature, previous results (as described above), and the 10% rule to keep covariates that change the association between independent and dependent variables of interest by more than 10% when removed. Cellular composition data was not available for the validation sample and thus could not be included as a covariate. A p-value of less than 0.05 was considered significant for the site-specific validation analyses. All genomic coordinates in this study are described using the Human Genome Version 19 (HG19).
Metabolite data was analyzed by linear regression to assess their association with Chr12:29302232 methylation. Dependent variables were the relative metabolite levels. Covariates were chosen in the same manner described above for the site-specific analyses. A total of 13 metabolites were analyzed, and a False Discovery Rate (using the online tool from http://users.ox.ac.uk/npike/fdr/) corrected p-value of less than 0.05 was considered statistically significant.
Finally, to test the hypothesis that AAP-induced insulin resistance may be explained by medication-induced changes in the candidate gene’s methylation we conducted a mediation analysis. We first analyzed the association of AAP use with our candidate gene’s methylation by evaluating the effect of medication (AAP or lithium monotherapy, Supplementary Figure 1, Step 4). If this association was significant, we then conducted a mediation analysis according to the method of Ditlevsen and colleagues (45). An overview of the mediation analysis is provided in Supplementary Figure 3.
RESULTS
Population Characteristics
Forty-eight Bipolar I subjects currently treated with an AAP were included in the discovery cohort. A total of 72 Bipolar I subjects treated with AAPs (n=54, 75%) and lithium monotherapy (n=18, 25%) were included in the validation analyses (Table I). The sets were comparable in terms of demographic and clinical variables and did not significantly differ based on age, race, HOMA-IR or medication use (all p>0.07). The validation sample did have a higher percentage of female participants (p=0.08), African-American participants (p=0.22), and a higher body mass index (BMI) (p=0.09).
Table I.
Demographic and Clinical Characteristics of Discovery and Validation Samples
| Discovery Sample (n=48) | Validation Sample (n=72) | |
|---|---|---|
| Age (years ± S.D.) | 45.1 ± 11.2 | 42.3 ± 12.1 |
| % Female | 54 | 69.4 |
| Race/Ethnicity (% Caucasian; % African-American) | 94; 4 | 80;11 |
| BMI (kg/m2± S.D.) | 29.7 ± 6.29 | 32.3 ± 9.19 |
| HOMA-IR | 6.53 ± 4.92 | 5.24 ± 2.96 |
| % smoker | 27 | 32 |
| Folate (ng/ml) | 18.9 ± 5.98 | 18.7 ± 6.00 |
| % Group 1 Antipsychotic | 16 | 11 |
| % Group 2 Antipsychotic | 65 | 40 |
| % Group 3 Antipsychotic | 19 | 24 |
| % Lithium Monotherapy | 25 |
means ± standard deviation (S.D.) or % of variables for each population. No significant differences were identified between the Discovery and Validation cohorts. Race/ethnicity is categorized as “Caucasian”, “African-American”, and “Other”. Second generation antipsychotic use was stratified by metabolic risk profile. Group 1 antipsychotic included olanzapine and clozapine, which have the highest risk to cause metabolic side effects. Group 2 antipsychotic included quetiapine and risperidone which carry a moderate risk and Group 3 antipsychotic included aripiprazole and ziprasidone which have the lowest risk to cause metabolic side effects.
Discovery Sample EWAS
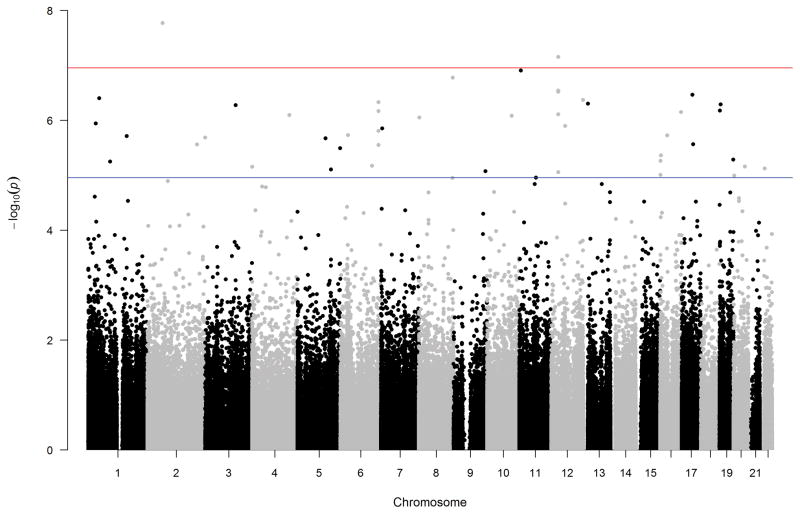
After correction for multiple testing, only the top 2 hits, cg25329211 and cg10171063, remained significant from the EWAS analysis based on HOMA-IR. Probe cg25329211 is located in an intergenic shelf region on chromosome 2 and probe cg10171063 is located within a gene called Fatty Acyl CoA Reductase 2 (FAR2) on chromosome 12 at position 29302232 (herein referred to as Chr12:29302232 methylation). The FAR2 enzyme catalyzes the rate-limiting reduction of fatty acyl CoAs to fatty alcohols (46). The top 10 differentially methylated probes from EWAS analysis are found in Table II. A Manhattan plot for the EWAS analysis can be found in Figure 1. Although not significant after Bonferroni correction, three additional probes within the same CpG island (chr12:29302035-29302954) of FAR2 were in the top 10 most significant sites associated with HOMA-IR.
Table II.
Top 10 Differentially Methylated Probes from EWAS Analysis based on HOMA-IR
| CpG Probe | Chromosome | Gene | Base pair | Feature | Log2 Fold Change | Original P- value | Bonferroni Corrected P- value |
|---|---|---|---|---|---|---|---|
| cg25329211 | 2 | NA | 64375257 | IGR – Shelf | −0.004 | 4.56E-08 | 0.01 |
| cg10171063 | 12 | FAR2 | 29302232 | Exon 1 - Island | −0.005 | 6.98E-08 | 0.02 |
| cg12503835 | 8 | TSNARE1 | 143324660 | Body – Open Sea | −0.02 | 1.69E-07 | 0.06 |
| cg04488853 | 11 | CCKBR | 6293288 | 3′UTR - shore | −0.02 | 2.49E-07 | 0.08 |
| cg10902825 | 12 | FAR2 | 29302179 | Exon1 - Island | −0.005 | 2.57E-07 | 0.09 |
| cg11390073 | 12 | FAR2 | 29302515 | Intron 1 - Island | −0.009 | 3.00E-07 | 0.10 |
| cg24677744 | 12 | FAR2 | 29302016 | Exon1 - Island | −0.008 | 3.01E-07 | 0.10 |
| cg20697767 | 17 | RPRML | 45056325 | Exon 1 - Island | 0.02 | 3.41E-07 | 0.12 |
| cg21725878 | 12 | NA | 132956918 | IGR – Island | −0.01 | 3.86E-07 | 0.13 |
| cg15076218 | 1 | PIK3R3 | 46599002 | TSS1500 - island | 0.01 | 4.02E-07 | 0.14 |
Lists the top 10 differentially methylated probes (CpG probes) from the EWAS analysis based on HOMA-IR while adjusting for age, race, gender, AAP group, smoking, folate, BMI and cellular composition. Location, gene (if applicable), feature of probe and Log2 Fold Change are listed along with original and Bonferroni adjusted p-values. The top two hits remained significant after multiple testing correction.
Figure 1. Manhattan Plot of EWAS based on HOMA-IR.
A Manhattan plot of the p-values based on HOMA-IR while adjusting for age, race, gender, antipsychotic type, smoking, folate, Body Mass Index (BMI) and cellular composition. The x-axis is broken down by chromosome and the y-axis is the –log10 p-value. The two probes that met the EWAS significance level (upper horizontal line) were in Chromosome 2 (probe cg25329211) and chromosome 12 (probe cg10171063). The lower horizontal line is the suggestive EWAS significance threshold cutoff. Note the two hits above the significance threshold of 1.1×10−7 in chromosomes 2 and 12 corresponding to the top two hits in Table II.
Validation Sample – Replication of FAR2 Methylation Association with Insulin Resistance
Due to the enrichment of the FAR2 gene in the top EWAS results, and being the only statistically significant hit located in a gene coding region, we aimed to replicate the association of decreased methylation or hypomethylation at the Chr12:29302232 (cg10171063) with increased HOMA-IR (indicating an insulin resistant state) in an additional sample of bipolar subjects assessed for insulin resistance and currently treated with AAPs or lithium monotherapy. The average detected methylation in the validation sample at this site was 58.3 ± 16.1 and the association of decreased Chr12:29302232 methylation and increased HOMA-IR was replicated (crude beta = −0.08; crude p=0.0016; adjusted beta = −0.06; adjusted p=0.007) while adjusting for age, race, gender, BMI, and smoking status (Table III). Using the same model, while stratifying the validation sample based on medication use (AAP or lithium monotherapy), only showed this negative association between Chr12:29302232 methylation and HOMA-IR in AAP treated subjects (crude beta = −0.09; crude p=0.0047; adjusted beta = −0.07; adjusted p=0.018), while no association was observed in subjects treated with lithium (crude beta = −0.005; crude p=0.9; adjusted beat = −0.008; adjusted p=0.8). Although not done in our primary validation analyses due to a limited sample size, stratifying the sample by AAP metabolic risk (i.e., low, medium and high) showed that within all 3 groups the same negative correlation between Chr12:29302232 methylation and HOMA-IR was observed. The relationship between Chr12:29302232 hypomethylation and higher HOMA-IR in the olanzapine and clozapine group (high metabolic risk) did not reach statistical significance (beta=−0.11; p=0.06) which may be due to the small sample size in this group. The medium risk group and low risk group still had significant negative correlations between Chr12:29302232 methylation and HOMA-IR (beta=−0.09; p=0.047 and beta=−0.07; p=0.014, respectively).
Table III.
Validation Analyses
| DEPENDENT VARIABLE | CHR12:29302232 CRUDE BETA COEFFICIENTS |
CRUDE P-VALUE | CHR12:29302232 ADJUSTED BETA COEFFICIENTS |
ADJUSTED P- VALUE |
|---|---|---|---|---|
| HOMA-IR | −0.08 | 0.0016* | −0.06 | 0.0070* |
| ARACHIDONOYLGLYCEROPHOSPHOETHANOLAMINE | 15762.91 | 0.36 | 11314.09 | 0.57 |
| HEXADECANEDIOATE | 0.0069 | 0.47 | 0.0017 | 0.86 |
| LINOLEOYLGLYCEROPHOSPHOETHANOLAMINE | −248848.80 | 0.045* | −393919.30 | 0.033* |
| OCTADECANEDIOATE | 0.016 | 0.0395* | 0.010 | 0.21 |
| OLEOYLGLYCEROPHOSPHOETHANOLAMINE | 19886.00 | 0.76 | −2079.11 | 0.98 |
| PALMITOLEIC ACID | 0.0031 | 0.42 | 0.004 | 0.41 |
| PALMITOLEOYLGLYCEROPHOSPHOCHOLINE | −0.018 | 0.047* | −0.020 | 0.049* |
| PALMITOYL SPHINGOMYELIN | −0.015 | 0.035* | −0.018 | 0.028* |
| PALMITOYLCARNITINE | 0.0018 | 0.84 | 0.0033 | 0.74 |
| PALMITOYLPLASMENYLETHANOLAMINE | 0.004 | 0.41 | 0.003 | 0.62 |
| PALMITOYLGLYCEROPHOSPHOINOSITOL | −0.0013 | 0.88 | −0.0066 | 0.50 |
| STEAROYLGLYCEROPHOSPHOCHOLINE | −0.0036 | 0.57 | −0.0021 | 0.82 |
| STEAROYLGLYCEROPHOSPHOETHANOLAMINE | −0.0040 | 0.62 | −0.0089 | 0.34 |
Validation analyses investigating associations between CHR12:29302232 methylation and insulin resistance as measured by HOMA-IR and adjusted for age, race, gender, body mass index (BMI), and smoking status. Additional functional analyses between Chr12:29302232 and an untargeted metabolomics set related to phospholipid metabolites adjusting for age, race, gender, BMI and folate levels. The units of each metabolite are relative arbitrary units established by the metabolomics core processing methods. Presented p-values for the functional metabolite analyses were corrected for multiple testing using the false discovery rate method (a total of 13 metabolites/tests).
P-value < 0.05
Validation Sample – Metabolomics Analyses for Functional Effects of FAR2 Methylation
Our top gene-related finding from the EWAS analysis identified differential methylation of a lipid enzyme gene associated with HOMA-IR (FAR2). We restricted our metabolomics analyses in the validation sample to identified metabolites related to pathways that could be associated with FAR2 in order to reduce Type I error. A total of 13 metabolites related to these pathways were identified from the known subset of metabolites. These metabolites, as well as crude and adjusted beta values and the p-values, for the regressions with Chr12:29302232 methylation are listed in Table III. The levels of linoleoylglycerophosphoethanolamine, palmitoleoylglycerophosphocholine and palmitoylsphingomyelin were negatively correlated to Chr12:29302232 methylation meaning that hypermethylation at this site was associated with lower metabolite levels.
Validation Sample – Testing of Atypical Antipsychotic Effects on FAR2 Methylation and HOMA-IR
In the next step of our approach (Supplementary Figure 1) we investigated the effect of AAP use on FAR2 Methylation and HOMA-IR. A linear regression adjusting for age, race, and gender confirmed the association between AAP use and FAR2 methylation (crude beta: −5.27; p-value: 0.037; adjusted beta: −5.28; adjusted p-value: 0.040). Our data also confirmed the well-studied and known effect of AAP use on insulin resistance. A regression adjusting for age, gender, race, bmi and smoking identified a significant association between AAP use and HOMA-IR (crude beta: 0.922; p-value:0.022; adjusted beta:0.751; adjusted p-value: 0.028).
Validation Sample – Mediation Analyses
A mediation analysis aids in understanding to what degree, if any, FAR2 methylation may be the mediating factor between AAP use and HOMA-IR. The coefficients from sequential modeling described above found that there were significant associations between 1) AAP use and HOMA-IR, 2) AAP use and FAR2 DNA methylation, and 3) FAR2 methylation and HOMA-IR. The association between AAP use and HOMA-IR was not significant when accounting for FAR2 DNA methylation suggesting a complete mediation effect by FAR2 methylation. Calculations using the crude beta coefficients from these consecutive regressions (see supplementary Figure 3) estimated a percent mediation of 40% indicating that 2/5ths of the risk of AAP-induced insulin resistance may be mediated by hypomethylation at the FAR2 gene. Thus, the direct mediation effect is 60%.
DISCUSSION
To our knowledge, this is the first study to use an epigenome-wide platform to investigate epigenetic associations with an AAP-induced metabolic side effect. Two differentially methylated probes were identified in the discovery set, one of which was found within a gene with possible biological relevance (i.e., lipid metabolism) to insulin resistance. This methylation site, within the Fatty Acyl CoA Reductase 2 (FAR2) gene on chromosome 12 at position 29302232, was successfully replicated in a separate, validation sample using site-specific methylation analyses and found to be a mediator of the association between AAP use and insulin resistance.
Findings from EWAS and Methylation Validation
The top hit, probe cg25329211 (Chr2: 64375257), is located within an intergenic shelf region. The significance of such methylation sites are not fully known. However, these locations may be important for regulating alternative promoters, preventing unneeded expression from intergenic regions, and for splicing events (47, 48). Further work is needed to investigate the relevance of this finding and its relationship to AAP-induced insulin resistance, if any. It may be possible that this site could serve as a biomarker regarding the risk for AAP-induced insulin resistance despite its current unknown functional significance.
The second CpG site from the EWAS that remained significant after correction for multiple testing was located at Chr12:29302232 (cg10171063). Given the association of this site with a gene, we chose to focus our validation work here. We were able to confirm the association of hyopmethylation at this CpG site with insulin resistance as measured by HOMA-IR in our validation sample. The methylation site of Chr12:29302232 is found within a CpG Island of Exon 1 of the FAR2 gene (also known as the male sterility domain-containing protein 1 (MLSTD1) gene) within 150bp of a transcription start site. This gene encodes a peroxisomal protein that utilizes fatty acyl CoA substrates for conversion to fatty alcohols (46). The production of fatty alcohols is the rate-limiting step to the downstream formation of wax monoesters and ether phospholipids. Ether phospholipids include both an ether bond to an alkyl moiety and an ester bond to a fatty acid, and are thought to be important constituent of cell membranes and possibly cell signaling (49). One of the most common classes of ether lipids is called plasmalogens which contain a phospholipid with a glycerol chain attachment (50, 51). Plasmalogens are found within many human tissues with particularly high concentrations in the central nervous system and cardiovascular system accounting for 15–20% of total phospholipid content in cell membranes (50, 52). Plasmalogens may have an important role in the cardiometabolic health of certain populations (53, 54).
Phospholipid Changes and AAP Use
Changes in phospholipids including the plasmalogens have been identified following AAP treatment in several metabolomics studies in schizophrenia subjects (16–19, 33). In two lipidomic studies, by Kaddurah-Doak and McEvoy and colleagues, significant elevations in phospholipid levels were found following AAP treatment in several different populations of schizophrenia patients including first episode, treatment naïve, and recurrent episode patients compared to controls. Within both studies, increases in the phospholipid, phosphatidylethanolamine, was identified after treatment with AAPs. Xuan and colleagues found that the myo-inositol, which is required for the synthesis of membrane inositol phospholipids, was a top influencing metabolite that defined AAP treatment response in their untargeted metabolomics study of risperidone effects in schizophrenia subjects. Finally, Oresic and colleagues conducted a lipidomic investigation in twin pairs discordant for schizophrenia (74% of schizophrenia twins were on an AAP) and found varying levels of lipids depending on the subspecies being analyzed (55). With their study they specifically identified increases in sphingomyelins in schizophrenia subjects which may add support between our identified association between palmitoylsphingomyelin and Chr12:29302232 methylation. Finally, although not statistically significant in their study, Oresic identified relatively higher levels of ether phospholipids, which are directly controlled by the activity of our FAR2 candidate gene, in schizophrenia subjects compared to their discordant twins and healthy controls.
Within our study, insulin resistance was associated with hypomethylation of FAR2. Our study identified possible functional effects of Chr12:29302232 methylation using an untargeted metabolomics analysis of phospholipid-related metabolites. We identified an inverse relationship between Chr12:29302232 methylation and three metabolites, linoleoylglycerophosphoethanolamine, palmitoleoylglycerophosphocholine and palmitoylsphingomyelin, where, hyopmethylation was associated with higher metabolite levels. This relationship may follow the general notion that decreased methylation of a gene results in increased expression or downstream product associated with that gene or vice versa. It may further suggest that methylation in this particular CpG island of FAR2 may be important for FAR2 gene regulation. It also may be plausible that differences in methylation of a rate-limiting enzymes like that of FAR2 could have substantial effects on the lipid balances occurring in the body.
The plasmalogen phospholipids have been characterized as polyunsaturated fatty acid (PUFA) “sinks” due to their higher likelihood to have linkage to both the n-3 and n-6 PUFAs compared to the diacyl alternative (56). Our metabolites included two phospholipids with PUFA attachments (arachidonoylglycerophosphoethanolamine and linoleoylglycerophosphoethanolamine). Higher relative linoleoylglycerophosphoethanolamine levels was found to be significantly associated with lower Chr12:29302232 methylation. The intersection of PUFA and phospholipid metabolism continues to be a theme in the literature regarding both positive and negative AAP outcomes in bipolar disorder and schizophrenia (16, 18, 19, 55, 57) and further exploration may be warranted.
DNA methylation as a mediator of AAP use and insulin resistance
Finally, our work showed a mediating effect by DNA methylation at FAR2 on AAP-induced insulin resistance. This may give preliminary and partial support to our overall hypothesis that AAPs cause changes in DNA methylation at particular locations in the genome which subsequently alter RNA, protein and metabolite regulation to cause insulin resistance. We speculate that the lowering of FAR2 methylation by AAPs leads to increased production of certain lipid species, leading to a lipid imbalance and increased insulin resistance. Lipid imbalances have been implicated in insulin resistance pathophysiology (58). Ultimately, the molecular cause of AAP-induced insulin resistance is likely multifactorial and may include several different pathways. It could also be highly dependent on the particular population being studied. Indeed, genetic variation of lipid metabolizing enzymes from other pathways have been linked to AAP-induced insulin resistance (59). Our work and others have suggested that AAPs may have significant effects on the epigenome which could have implications on both therapeutic and adverse effects.
Strengths and Limitations
The findings from this study should be interpreted cautiously in light of two main limitations which include: 1) a limited sample size and 2) lack of longitudinal sampling. Psychiatric polypharmacy is well known in bipolar disorder and although we were able to overcome this in our sample, it did limit the sample size which reduces power when correcting for multiple comparisons. Previous work has identified an optimal sample size of at least 112 cases and controls to achieve 80% power in detecting methylation differences in epigenome-wide studies (60). The current study did not reach this suggested sample size and it did not include controls and thus could have insufficient power to detect true findings. Additionally, the lack of longitudinal samples does not allow us to infer causal changes but only single time-point methylation and metabolite associations in a population already on AAPs or lithium for at least 3 months. Such a study design carries treatment length and dosage variability that should be taken into account when interpreting the results. Thus, replication in longitudinal formats and other AAP-treated populations should be pursued to investigate the validity of our identified gene as well as other top hits from our study and/or other candidate pathways involved in AAP-induced insulin resistance.
A few other limitations should be considered. We did not have the ability to assess changes in FAR2 gene expression (with RNA) in relation to AAP-induced insulin resistance as RNA was not available. We also only validated a single DNA methylation site and studies in the future should expand the methylation analyses to include other areas of the FAR2. Our metabolite data only measured relative levels, and so the lipidomic pathways that may be influenced by this gene’s methylation require further, targeted exploration. Finally, our work also included both obese and non-obese subjects. BMI is a well-known risk factor of insulin resistance and this was seen in our study samples (data not shown). Nevertheless, when adjusting analyses for BMI our findings were still significant. Additionally, we describe the effects of obesity on Chr12:29302232 methylation and metabolite levels in Supplementary Table II for descriptive purposes however detected effects were minimal. Obesity remains a significant independent predictor of insulin resistance and so future studies will need to separate the effects of AAP-induced obesity and insulin resistance on gene methylation and metabolite changes with either prospective studies or studies in non-obese populations
This study had several strengths including an epigenome-wide analysis coupled with validation analyses that utilized site-specific methylation and metabolomics data in two samples of bipolar subjects with well characterized medication regimens. Within the top ten differentially methylated genes associated with HOMA-IR from our EWAS analysis, four were found within the same CpG island of the FAR2 gene and all were found to have lower methylation with higher HOMA-IR. This should be interpreted cautiously as only the top two genes from this list remained significant after multiple testing corrections, but our unbiased discovery approach followed by validation does strengthen this candidate gene finding. Although the sample size was limited and cross-sectional, the ability to replicate our finding and the possible biological relevance of the FAR2 gene does strengthen the findings in their present form. We cannot rule out a reverse causation effect on our identified gene methylation meaning that insulin resistance causes changes in gene methylation. However, our multi-step approach in the validation sample of identifying mediation effect reduces the chance of reverse causation. Finally, this work was done in the peripheral blood, which is heterogeneous in cell type. Our EWAS analysis used available strategies to control for the heterogeneity but we were unable to control for this in our validation sample, where the DNA had already been collected. Due to this heterogeneity, future work will need to translate the peripheral blood findings to a target tissue. There are several target tissues that may be good candidates for investigating the epigenetic and molecular mechanisms of AAP-induced insulin resistance which our group is currently undertaking.
CONCLUSIONS
This study identified and validated a differentially methylated CpG site in a novel gene associated with AAP-induced insulin resistance. We were able to identify a mediating effect of this CpG site on AAP-induced insulin resistance as well as possible functional effects of lipid metabolites. To date, we still do not fully understand the precise mechanism of AAP-induced insulin resistance. Identification of the mechanism of AAP-induced insulin resistance could lead to future personalized intervention strategies such as biomarkers that will allow us to screen patients for the risk of becoming insulin resistant or potentially allow to design drug therapy to prevent AAP-induced insulin resistance itself. The interventions based on such knowledge will allow clinicians to utilize AAPs for their beneficial, psychiatric effects, while minimizing or removing the detrimental effects to cardiometabolic and physical health which will ultimately improve a psychiatric patient’s morbidity and mortality. Going forward, targeted, prospective epigenetic analyses in candidate insulin-sensitive tissues are needed to fully understand the interaction of genes and medications in the severely mentally ill.
Supplementary Material
Acknowledgments
We would like to acknowledge Dr. Melvin McInnis, M.D., Wally Prechter and the Prechter Longitudinal Study of Bipolar Disorder at the University of Michigan for their assistance with access to patient populations.
List of Abbreviations
- BMI
body mass index
- EWAS
epigenome-wide association study
- FAR2
Fatty Acyl CoA Reductase 2
- HOMA-IR
homeostatic model assessment of insulin resistance
- AAP
second generation antipsychotic
Footnotes
Authors’ Contributions
Author KB conceived and designed the study, prepared samples for analysis, conducted site-specific assays, performed statistical analyses and wrote the article. Author JG assisted in the epignome-wide analyses, site-specific methylation analyses, statistical analyses and manuscript writing. Author DD assisted in statistical analyses, interpretation and manuscript writing. Author VE provided samples, assisted in study design, provided equipment for site-specific methylation analyses, interpretation of results and manuscript writing.
Disclosures
The authors have no conflicts of interest to disclose. This work and the authors were supported by: The Rachael Upjohn Clinical Scholars Grant, Michigan Institute of Clinical and Health Research (MICHR) Pilot Grant (UL1RR024986), NIMH (R01 MH082784), NIH-NCCR, the Chemistry Core of the Michigan Diabetes Research and Training Center (P30DK020572, P30DK092926), the Washtenaw Community Health Organization (WCHO, Ann Arbor, Michigan), The Brain and Behavior Research Foundation (formerly NARSAD, Great Neck, New York), University of Michigan National Institutes of Environmental Health Sciences (NIEHS) Core Center P30 ES017885 and the Prechter Longitudinal Study of Bipolar Disorder (Ann Arbor, Michigan).
References
- 1.Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Archives of general psychiatry. 2005;62(6):593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 2.Pillarella J, Higashi A, Alexander GC, Conti R. Trends in use of second-generation antipsychotics for treatment of bipolar disorder in the United States, 1998–2009. Psychiatric services (Washington, DC) 2012;63(1):83–6. doi: 10.1176/appi.ps.201100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rummel-Kluge C, Komossa K, Schwarz S, Hunger H, Schmid F, Lobos CA, et al. Head-to-head comparisons of metabolic side effects of second generation antipsychotics in the treatment of schizophrenia: a systematic review and meta-analysis. Schizophrenia research. 2010;123(2–3):225–33. doi: 10.1016/j.schres.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McIntyre RS, Konarski JZ, Misener VL, Kennedy SH. Bipolar disorder and diabetes mellitus: epidemiology, etiology, and treatment implications. Annals of clinical psychiatry : official journal of the American Academy of Clinical Psychiatrists. 2005;17(2):83–93. doi: 10.1080/10401230590932380. [DOI] [PubMed] [Google Scholar]
- 5.Guo JJ, Keck PE, Jr, Corey-Lisle PK, Li H, Jiang D, Jang R, et al. Risk of diabetes mellitus associated with atypical antipsychotic use among Medicaid patients with bipolar disorder: a nested case-control study. Pharmacotherapy. 2007;27(1):27–35. doi: 10.1592/phco.27.1.27. [DOI] [PubMed] [Google Scholar]
- 6.Kemp DE, Gao K, Chan PK, Ganocy SJ, Findling RL, Calabrese JR. Medical comorbidity in bipolar disorder: relationship between illnesses of the endocrine/metabolic system and treatment outcome. Bipolar disorders. 2010;12(4):404–13. doi: 10.1111/j.1399-5618.2010.00823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calkin CV, Ruzickova M, Uher R, Hajek T, Slaney CM, Garnham JS, et al. Insulin resistance and outcome in bipolar disorder. The British journal of psychiatry : the journal of mental science. 2014 doi: 10.1192/bjp.bp.114.152850. [DOI] [PubMed] [Google Scholar]
- 8.Fiedorowicz JG, Solomon DA, Endicott J, Leon AC, Li C, Rice JP, et al. Manic/hypomanic symptom burden and cardiovascular mortality in bipolar disorder. Psychosomatic medicine. 2009;71(6):598–606. doi: 10.1097/PSY.0b013e3181acee26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sylvia LG, Shelton RC, Kemp DE, Bernstein EE, Friedman ES, Brody BD, et al. Medical burden in bipolar disorder: findings from the Clinical and Health Outcomes Initiative in Comparative Effectiveness for Bipolar Disorder study (Bipolar CHOICE) Bipolar disorders. 2015;17(2):212–23. doi: 10.1111/bdi.12243. [DOI] [PubMed] [Google Scholar]
- 10.Laursen TM, Wahlbeck K, Hallgren J, Westman J, Osby U, Alinaghizadeh H, et al. Life expectancy and death by diseases of the circulatory system in patients with bipolar disorder or schizophrenia in the Nordic countries. PloS one. 2013;8(6):e67133. doi: 10.1371/journal.pone.0067133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellingrod VL, Taylor SF, Dalack G, Grove TB, Bly MJ, Brook RD, et al. Risk factors associated with metabolic syndrome in bipolar and schizophrenia subjects treated with antipsychotics: the role of folate pharmacogenetics. Journal of clinical psychopharmacology. 2012;32(2):261–5. doi: 10.1097/JCP.0b013e3182485888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houtepen LC, van Bergen AH, Vinkers CH, Boks MP. DNA methylation signatures of mood stabilizers and antipsychotics in bipolar disorder. Epigenomics. 2016;8(2):197–208. doi: 10.2217/epi.15.98. [DOI] [PubMed] [Google Scholar]
- 13.Melka MG, Laufer BI, McDonald P, Castellani CA, Rajakumar N, O’Reilly R, et al. The effects of olanzapine on genome-wide DNA methylation in the hippocampus and cerebellum. Clinical epigenetics. 2014;6(1):1. doi: 10.1186/1868-7083-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burghardt KJ, Goodrich JM, Dolinoy DC, Ellingrod VL. DNA methylation, insulin resistance and second-generation antipsychotics in bipolar disorder. Epigenomics. 2015;7(3):343–52. doi: 10.2217/epi.15.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moons T, De Hert M, Kenis G, Viechtbauer W, van Os J, Gohlke H, et al. No association between genetic or epigenetic variation in insulin growth factors and antipsychotic-induced metabolic disturbances in a cross-sectional sample. Pharmacogenomics. 2014;15(7):951–62. doi: 10.2217/pgs.14.46. [DOI] [PubMed] [Google Scholar]
- 16.Kaddurah-Daouk R, McEvoy J, Baillie R, Lee D, Yao J, Doraiswamy P, et al. Metabolomic mapping of atypical antipsychotic effects in schizophrenia. Molecular psychiatry. 2007;12(10):934–45. doi: 10.1038/sj.mp.4002000. [DOI] [PubMed] [Google Scholar]
- 17.Kaddurah-Daouk R, McEvoy J, Baillie R, Zhu H, Yao JK, Nimgaonkar VL, et al. Impaired plasmalogens in patients with schizophrenia. Psychiatry research. 2012;198(3):347–52. doi: 10.1016/j.psychres.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 18.McEvoy J, Baillie RA, Zhu H, Buckley P, Keshavan MS, Nasrallah HA, et al. Lipidomics reveals early metabolic changes in subjects with schizophrenia: effects of atypical antipsychotics. PloS one. 2013;8(7):e68717. doi: 10.1371/journal.pone.0068717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xuan J, Pan G, Qiu Y, Yang L, Su M, Liu Y, et al. Metabolomic profiling to identify potential serum biomarkers for schizophrenia and risperidone action. Journal of proteome research. 2011;10(12):5433–43. doi: 10.1021/pr2006796. [DOI] [PubMed] [Google Scholar]
- 20.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annual review of pharmacology and toxicology. 2008;48:653–83. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 21.Xue SS, He JL, Zhang X, Liu YJ, Xue FX, Wang CJ, et al. Metabolomic analysis revealed the role of DNA methylation in the balance of arachidonic acid metabolism and endothelial activation. Biochimica et biophysica acta. 2015;1851(10):1317–26. doi: 10.1016/j.bbalip.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D. Methods of integrating data to uncover genotype-phenotype interactions. Nature reviews Genetics. 2015;16(2):85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- 23.First MBSR, Gibbon M, Williams JBW. Research version, Non-Patient Edition. New York: New York State Psychiatric Institute, Biometrics Research; 2002. Structured Clinical Interview for DSMIV Axis I Disoders. [Google Scholar]
- 24.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–9. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 25.Lett TA, Wallace TJ, Chowdhury NI, Tiwari AK, Kennedy JL, Muller DJ. Pharmacogenetics of antipsychotic-induced weight gain: review and clinical implications. Molecular psychiatry. 2012;17(3):242–66. doi: 10.1038/mp.2011.109. [DOI] [PubMed] [Google Scholar]
- 26.Consensus development conference on antipsychotic drugs and obesity and diabetes. Diabetes care. 2004;27(2):596–601. doi: 10.2337/diacare.27.2.596. [DOI] [PubMed] [Google Scholar]
- 27.De Hert M, Detraux J, van Winkel R, Yu W, Correll CU. Metabolic and cardiovascular adverse effects associated with antipsychotic drugs. Nature reviews Endocrinology. 2012;8(2):114–26. doi: 10.1038/nrendo.2011.156. [DOI] [PubMed] [Google Scholar]
- 28.MDEH, Correll CU, Bobes J, Cetkovich-Bakmas M, Cohen D, Asai I, et al. Physical illness in patients with severe mental disorders. I. Prevalence, impact of medications and disparities in health care. World psychiatry : official journal of the World Psychiatric Association (WPA) 2011;10(1):52–77. doi: 10.1002/j.2051-5545.2011.tb00014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newcomer JW. Second-generation (atypical) antipsychotics and metabolic effects: a comprehensive literature review. CNS drugs. 2005;19(Suppl 1):1–93. doi: 10.2165/00023210-200519001-00001. [DOI] [PubMed] [Google Scholar]
- 30.Lahiri DK, Nurnberger JI., Jr A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic acids research. 1991;19(19):5444. doi: 10.1093/nar/19.19.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. BioTechniques. 2003;35(1):152–6. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- 32.Goodrich JM, Sanchez BN, Dolinoy DC, Zhang Z, Hernandez-Avila M, Hu H, et al. Quality control and statistical modeling for environmental epigenetics: A study on in utero lead exposure and DNA methylation at birth. Epigenetics : official journal of the DNA Methylation Society. 2015;10(1):19–30. doi: 10.4161/15592294.2014.989077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burghardt KJ, Evans SJ, Wiese KM, Ellingrod VL. An Untargeted Metabolomics Analysis of Antipsychotic Use in Bipolar Disorder. Clinical and translational science. 2015 doi: 10.1111/cts.12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris TJ, Beck S. Analysis pipelines and packages for Infinium HumanMethylation450 BeadChip (450k) data. Methods (San Diego, Calif) 2015;72:3–8. doi: 10.1016/j.ymeth.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marabita F, Almgren M, Lindholm ME, Ruhrmann S, Fagerstrom-Billai F, Jagodic M, et al. An evaluation of analysis pipelines for DNA methylation profiling using the Illumina HumanMethylation450 BeadChip platform. Epigenetics : official journal of the DNA Methylation Society. 2013;8(3):333–46. doi: 10.4161/epi.24008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 2007;8(1):118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 37.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics (Oxford, England) 2013;29(2):189–96. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, et al. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics (Oxford, England) 2014;30(3):428–30. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics (Oxford, England) 2014;30(10):1363–9. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome biology. 2014;15(2):R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern SE, Williams K, Ferrannini E, DeFronzo RA, Bogardus C, Stern MP. Identification of individuals with insulin resistance using routine clinical measurements. Diabetes. 2005;54(2):333–9. doi: 10.2337/diabetes.54.2.333. [DOI] [PubMed] [Google Scholar]
- 44.Berkeley C. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. E-book. 2004 doi: 10.2202/1544-6115.1027. available at http://www bepress com/sagmb/vol3/iss1/art3. [DOI] [PubMed]
- 45.Ditlevsen S, Christensen U, Lynch J, Damsgaard MT, Keiding N. The mediation proportion: a structural equation approach for estimating the proportion of exposure effect on outcome explained by an intermediate variable. Epidemiology (Cambridge, Mass) 2005;16(1):114–20. doi: 10.1097/01.ede.0000147107.76079.07. [DOI] [PubMed] [Google Scholar]
- 46.Cheng JB, Russell DW. Mammalian wax biosynthesis I. Identification of two fatty acyl-coenzyme A reductases with different substrate specificities and tissue distributions. Journal of Biological Chemistry. 2004;279(36):37789–97. doi: 10.1074/jbc.M406225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–7. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell research. 2013;23(11):1256–69. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paltauf F. Ether lipids in biomembranes. Chemistry and physics of lipids. 1994;74(2):101–39. doi: 10.1016/0009-3084(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 50.Braverman NE, Moser AB. Functions of plasmalogen lipids in health and disease. Biochimica et biophysica acta. 2012;1822(9):1442–52. doi: 10.1016/j.bbadis.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 51.Wanders RJ, Brites P. Biosynthesis of ether-phospholipids including plasmalogens, peroxisomes and human disease: new insights into an old problem. Clinical Lipidology. 2010;5(3):379–86. [Google Scholar]
- 52.Nagan N, Zoeller RA. Plasmalogens: biosynthesis and functions. Progress in lipid research. 2001;40(3):199–229. doi: 10.1016/s0163-7827(01)00003-0. [DOI] [PubMed] [Google Scholar]
- 53.Pietiläinen KH, Sysi-Aho M, Rissanen A, Seppänen-Laakso T, Yki-Järvinen H, Kaprio J, et al. Acquired obesity is associated with changes in the serum lipidomic profile independent of genetic effects–a monozygotic twin study. 2007 doi: 10.1371/journal.pone.0000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laurila P-P, Surakka I, Sarin A-P, Yetukuri L, Hyötyläinen T, Söderlund S, et al. Genomic, transcriptomic, and lipidomic profiling highlights the role of inflammation in individuals with low high-density lipoprotein cholesterol. Arteriosclerosis, thrombosis, and vascular biology. 2013;33(4):847–57. doi: 10.1161/ATVBAHA.112.300733. [DOI] [PubMed] [Google Scholar]
- 55.Oresic M, Seppanen-Laakso T, Sun D, Tang J, Therman S, Viehman R, et al. Phospholipids and insulin resistance in psychosis: a lipidomics study of twin pairs discordant for schizophrenia. Genome medicine. 2012;4(1):1. doi: 10.1186/gm300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas S, Byers D, Palmer FSC, Spence M, Cook H. Incorporation of polyunsaturated fatty acids into plasmalogens, compared to other phospholipids of cultured glioma cells, is more dependent on chain length than on selectivity between (n– 3) and (n– 6) families. Biochimica et Biophysica Acta (BBA)-Lipids and Lipid Metabolism. 1990;1044(3):349–56. doi: 10.1016/0005-2760(90)90079-d. [DOI] [PubMed] [Google Scholar]
- 57.Evans SJ, Ringrose RN, Harrington GJ, Mancuso P, Burant CF, McInnis MG. Dietary intake and plasma metabolomic analysis of polyunsaturated fatty acids in bipolar subjects reveal dysregulation of linoleic acid metabolism. Journal of psychiatric research. 2014;57:58–64. doi: 10.1016/j.jpsychires.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: unravelling the mechanism. Lancet. 2010;375(9733):2267–77. doi: 10.1016/S0140-6736(10)60408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burghardt KJ, Gardner KN, Johnson JW, Ellingrod VL. Fatty Acid desaturase gene polymorphisms and metabolic measures in schizophrenia and bipolar patients taking antipsychotics. Cardiovascular psychiatry and neurology. 2013;2013:596945. doi: 10.1155/2013/596945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsai PC, Bell JT. Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. International journal of epidemiology. 2015 doi: 10.1093/ije/dyv041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.