Abstract
Background
ADAMTS14 is a member of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs), which are proteolytic enzymes with a variety of further ancillary domain in the C-terminal region for substrate specificity and enzyme localization via extracellular matrix association. However, whether ADAMTS14 genetic variants play a role in hepatocellular carcinoma (HCC) susceptibility remains unknown.
Methodology/Principal findings
Four non-synonymous single-nucleotide polymorphisms (nsSNPs) of the ADAMTS14 gene were examined from 680 controls and 340 patients with HCC. Among 141 HCC patients with smoking behaviour, we found significant associations of the rs12774070 (CC+AA vs CC) and rs61573157 (CT+TT vs CC) variants with a clinical stage of HCC (OR: 2.500 and 2.767; 95% CI: 1.148–5.446 and 1.096–6.483; P = 0.019 and 0.026, respectively) and tumour size (OR: 2.387 and 2.659; 95% CI: 1.098–5.188 and 1.055–6.704; P = 0.026 and 0.034, respectively), but not with lymph node metastasis or other clinical statuses. Moreover, an additional integrated in silico analysis proposed that rs12774070 and rs61573157 affected essential post-translation O-glycosylation site within the 3rd thrombospondin type 1 repeat and a novel proline-rich region embedded within the C-terminal extension, respectively.
Conclusions
Taken together, our results suggest an involvement of ADAMTS14 SNP rs12774070 and rs61573157 in the liver tumorigenesis and implicate the ADAMTS14 gene polymorphism as a predict factor during the progression of HCC.
Introduction
Hepatocellular carcinoma (HCC) is one of the most frequent malignancies of the liver and represents the second and sixth leading cause of cancer related death among males and females in 2012, respectively [1]. HCC carcinogenesis is a multistep and complex etiologic process [2], well-documented environmental and potentially preventable risk factors include major etiologic agents such as chronic hepatitis B virus (HBV) and hepatitis C virus (HCV) infection [3]. HBV accounts for at least 60% of HCC among Asian patients. Currently, the incidence of HCC shows that genetic mutations and environmental factors may increase the severity of the hepatic inflammation that lead to cell differentiation, proliferation and the deposition of connective tissue, which are necessary for the pathophysiology of hepatocarcinogenesis [4–6].
ADAMTS14 [7–9], which is located on chromosome 10q22.1, belongs to the ADAMTS (also known as a disintegrin and metalloproteinase domain with thrombospondin motifs) zinc ion-dependent proteinase family [10, 11] that contains an N-terminal catalytic domain and a C-terminal ancillary domain which defines substrate specificity including thrombospondin type 1 repeats (TSR) domain [12] and an unique proline-rich region (PRR) [13, 14] that was found to be essential for recognition of its procollagen substrates. The complete human ADAMTS is a name given to a family of 19 secreted, multi-domain proteolytic enzymes, and many of them appear to binds directly to extracellular matrix (ECMs) components, suggesting that the interaction with glycoconjugates through their TSR domain or their other region is responsible for the physiological processes of tumour-induced angiogenesis, invasion and organgenesis [15].
The TSR domain consists of approximately 60 amino acids, has a divalent cation-dependent modular structure, and has been reported to be a critical mediator of cell-matrix interactions. The majority of TSR domains contain six conserved cysteine residues as a long right-handed spiralling antiparallel β sheet and a groove-like structure that might be the “recognition” face, which mediate interactions with ECM components; the domains have been postulated to have roles involved in anti-angiogenesis activity [16]. Furthermore, a novel PRR domain embedded within the C-terminal extension stabilizes the molecule due to the rigidity conferred by its pyrrolidine ring, which leads to steric hindrance in peptidic backbones [13]. The PRR forms a left-handed extended helix of 3.0 residues per turn. PRRs play a major role through intramolecular interactions with other elements in the molecule, and they serve this structure as recognition sites with PRR-recognizing domains, such as Src-homology 3 (SH3) [17] and WW [18] (named after two highly conserved tryptophan residues in the domain) domains.
Although the development of HCC may take 20 to 50 years, early detection of this cancer is seldom accomplished due to the lack of reliable markers. Excessive angiogenesis has been demonstrated to result in a poor diagnosis for HCC, as well as being essential for development of a solid tumour in size. ADAMTS14 markedly up-regulated expression in human mammary tumours and non-neoplastic breast tissue [19]. In addition, the genetic association of non-synonymous single nucleotide polymorphisms (nsSNPs) in ADAMTS14 proteases to influence osteoarthritis phenotypes and tumour microenvironment has been reported [20, 21]. To understand the roles in tumour pathobiology [22], here, we conducted a hypothesis-driven case-control study to identify four nsSNPs (rs10823607, rs12774070, rs4747096, and rs61537157; Table 1) in ADAMTS14 that are damaging for protein function, and these were studied with the environmental carcinogens to evaluate HCC susceptibility.
Table 1. Variants, position, function, amino acid and changes of observed ADAMTS14 sequence.
| Chromosome # | Exon (contiguous position) | |||
|---|---|---|---|---|
| 10:70,741,007 | 10:70,753,879 | 10:70,758,253 | 10:70,760,503 | |
| cDNA position & nucleotide change ‡ | c.1769T>C | c.2809C>A | c.3146A>G | c.3322C>T |
| Protein position † | 590 | 936 | 1,049 | 1,108 |
| dbSNP (rs number) | rs10823607 | rs12774070 | rs4747096 | rs61573157 |
| Function | Non-synonymous | Non-synonymous | Non-synonymous | Non-synonymous |
| dbSNP allele | CTG>CGG | CTG>ATG | GAA>GGA | CCA>TCA |
| Protein residue | Leu>Pro | Leu>Met | Glu>Gly | Pro>Ser |
Materials and methods
Description of the enrolled participants
This hospital-based case control study recruited 340 (249 men and 91 women; mean age = 63.20 ± 11.68 years) HCC patients between 2007 and 2014 at the Chung Shan Medical University Hospital, Taiwan. During the same study period, 680 ethnic group-matched individuals (498 men and 182 women; mean age = 62.43±4.14 years) were enrolled as the controls; these subjects received a physical examination at the same hospital. The TNM classification of the American Joint Committee on Cancer (AJCC) was used for staging of hepatocellular carcinoma [23, 24]. Vascular invasion was defined by the presence of adjacent thrombus to the tumor in portal vein. The Child-Pugh grade is used to assess the prognosis of chronic liver disease. Before commencing the study, approval was obtained from the Institutional Review Board of Chung Shan Medical University Hospital, and informed written consent was obtained from each individual (CSMUH No:CS15099).
nsSNPs selection and genotyping
Genomic DNA was isolated from peripheral blood using the QIAamp DNA blood mini kit (Qiagen, Valencia, CA, USA) as described in detail previously [25]. DNA was dissolved in TE buffer and used as the template in polymerase chain reactions. Genotyping of four nsSNPs (rs10823607, rs12774070, rs4747096, and rs61537157; Table 1) in ADAMTS14 with minor allele frequencies >5% in the HapMap Chinese Han Beijing (CHB) population was performed by the TaqMan SNP genotyping assay (Applied Biosystems, Foster City, CA, USA) [26]. Moreover, four selected nsSNPs show alleles of that region with recombination rate plotted and haplotype display in Figure A in S1 File. rs10823607 (529th), rs12774070 (621th), rs474096 (669th), and rs61573157 (689th) indicate the independent examined loci that are located on the LD map of haplotype blocks 22, 26, 31, and 32, respectively (Figure A in S1 File).
Bioinformatics analysis
Several semi-automated bioinformatics tools to assess whether SNPs or their linked genetic variants were associated with a putative function that might affect patient outcomes were utilized [27, 28]. We briefly the in silico functional predictions the structural models obtained by de novo prediction methods including the architecture of secondary structure arrangements, disorder content, transmembrane helices, coiled-coils and signal peptides using the Ginzu protocol to have demonstrated the utility for determined biological insight, either through protein functional sites recognition or functional annotation by fold identification [29, 30]. In additional, the development of potential active sites and post-translation modification of N-glycosylation sites were received from UniProtKB database [31] and determined using SignalP-NN [32]. Moreover, the heterogeneity at amino acid level potential function that the model the primary contribution to structural specificity was determined using software BioEdit Entropy algorithm [33]. The potential interaction of membranes with proteins determined by amphiphilic character obtained the structural information using the 3D hydrophobic moments of helical wheel projections [34] and the Wenxiang diagram coordinate system that represented the disposition of hydrophobic and hydrophilic residues that provided an α-helix can be viewed as a 2D diagram generated by its conical projection (http://www.jci-bioinfo.cn/wenxiang2) [35].”
Statistical analysis
The Mann-Whitney U test and Fisher’s exact test were used to compare differences in the distribution of age and demographic characteristics between the controls and HCC patients. ORs with 95% confidence intervals (CIs) were estimated using logistic regression models. AORs with 95% CIs were used to assess the association between genotype frequencies with HCC risk and clinical factors. P values less than 0.05 were considered significant. The data were analysed with the SPSS 12.0 statistical software (SPSS Inc., Chicago, IL, USA).
Results
Study population
A total of 1,020 participants, including 340 HCC cases and 680 controls were successfully genotyped for further analysis. The demographic characteristics including mean age, gender, alcohol consumption, tobacco consumption and disease stage are shown in Table 2. No significant differences between groups existed for age (P = 0.124), gender (P = 1.00) and tobacco consumption (P = 0.442) in the healthy controls and patients with HCC (Table 2).
Table 2. The distributions of demographical characteristics and clinical parameters in 680 controls and 340 patients with HCC.
| Variable | Controls (N = 680) | Patients (N = 340) | P value † |
|---|---|---|---|
| Age (yrs) | Mean ± S.D. | Mean ± S.D. | P = 0.124 |
| 62.43 ± 4.14 | 63.20 ± 11.68 | ||
| Gender | n (%) | n (%) | |
| Male | 498 (73.2%) | 249 (73.2%) | P = 1.000 |
| Female | 182 (26.8%) | 91 (26.8%) | |
| Alcohol consumption | |||
| No | 578 (85.0%) | 211 (62.1%) | P<0.001 |
| Yes | 102 (15.0%) | 129 (37.9%) | |
| Tobacco consumption | |||
| No | 415 (61.0%) | 199 (58.5%) | P = 0.442 |
| Yes | 265 (39.0%) | 141 (41.5%) | |
| Stage | |||
| I+II | 226 (66.5%) | ||
| III+IV | 114 (33.5%) | ||
| Tumor T status | |||
| ≤ T2 | 229 (67.4%) | ||
| > T2 | 111 (32.6%) | ||
| Lymph node status | |||
| N0 | 330 (97.1%) | ||
| N1+N2 | 10 (2.9%) | ||
| Metastasis | |||
| M0 | 322 (94.7%) | ||
| M1 | 18 (5.3%) | ||
| Vascular invasion | |||
| No | 276 (81.2%) | ||
| Yes | 64 (18.8%) |
† Mann-Whitney U test or Fisher’s exact test was used between controls and patients with HCC.
Frequency distribution of ADAMTS14 alleles and their associations with HCC
Table 3 summarized the basic characteristics of ADAMTS14 nsSNPs in the study population. In these controls, the genotypic frequency of ADAMTS14 SNP rs10823607 met the Hardy-Weinberg equilibrium (P = 0.571, χ2 value: 0.32.). The frequencies of ADAMTS14 SNPs rs12774070, rs4747096, and rs61573157 were also in the Hardy-Weinberg equilibrium (P = 0.157, χ2 value: 2.00; P = 0.528, χ2 value: 0.40; and P = 0.872, χ2 value: 0.03, respectively). According to the adjusted odds ratios (AORs) with their 95% confidence interval (CI) with multiple logistic regression model for HCC of ADAMTS14 gene polymorphism, we failed to individually detect any significant association of these ADAMTS14 variants with the occurrence of HCC between the two study groups compared with their corresponding wild-type homozygotes after adjusting for confounding factors (Table 3).
Table 3. Distribution frequency of ADAMTS14 genotypes in 680 controls and 340 patients with HCC.
| Variable | Controls (N = 680) n (%) |
Patients (N = 340) n (%) |
OR (95% CI) | AOR (95% CI) |
|---|---|---|---|---|
| rs10823607 | ||||
| CC | 578 (85.0%) | 290 (85.3%) | 1.00 | 1.00 |
| CT | 99 (14.6%) | 47 (13.8%) | 0.946 (0.650–1.376) | 0.933 (0.633–1.367) |
| TT | 3 (0.4%) | 3 (0.9%) | 1.993 (0.400–9.936) | 2.249 (0.434–11.661) |
| CT+TT | 102 (15.0%) | 50 (14.7%) | 0.977 (0.677–1.410) | 0.969 (0.663–1.417) |
| rs12774070 | ||||
| CC | 501 (73.7%) | 251 (73.8%) | 1.00 | 1.00 |
| CA | 160 (23.5%) | 82 (24.1%) | 1.023 (0.753–1.390) | 0.984 (0.716–1.353) |
| AA | 19 (2.8%) | 7 (2.1%) | 0.735 (0.305–1.772) | 0.894 (0.365–2.187) |
| CA+AA | 179 (26.3%) | 89 (26.2%) | 0.992 (0.738–1.334) | 0.976 (0.718–1.326) |
| rs4747096 | ||||
| AA | 270 (39.7%) | 139 (40.9%) | 1.00 | 1.00 |
| AG | 323 (47.5%) | 166 (48.8%) | 0.998 (0.757–1.317) | 1.010 (0.758–1.345) |
| GG | 87 (12.8%) | 35 (10.3%) | 0.781 (0.502–1.216) | 0.833 (0.528–1.315) |
| AG+GG | 410 (60.3%) | 201 (59.1%) | 0.952 (0.730–1.242) | 0.973 (0.739–1.281) |
| rs61573157 | ||||
| CC | 562 (82.6%) | 282 (82.9%) | 1.00 | 1.00 |
| CT | 112 (16.5%) | 57 (16.8%) | 1.014 (0.715–1.439) | 0.991 (0.690–1.423) |
| TT | 6 (0.9%) | 1 (0.3%) | 0.332 (0.040–2.772) | 0.453 (0.054–3.790) |
| CT+TT | 118 (17.4%) | 58 (17.1%) | 0.980 (0.694–1.384) | 0.967 (0.677–1.383) |
The odds ratios (ORs) and with their 95% confidence intervals (CIs) were estimated by logistic regression models. The adjusted odds ratios (AORs) with their 95% confidence intervals (CIs) were estimated by multiple logistic regression models after controlling for alcohol consumption. * P value < 0.05 as statistically significant.
Effects of interactions between ADAMTS14 variants and smoking behavior in HCC
Tables 4 and 5 show the results of additive and multiple interaction analysis between the two nsSNPs, rs12774070 and rs61573157, with smoking environmental risk factors for HCC. Among 141 HCC patients with smoking behavior in our study, significant associations of rs12774070 (CA+AA vs CC) and rs61573157 (CT+TT vs CC) variants with clinical stage of HCC (OR: 2.500 and 2.767; 95% CI: 1.148–5.446 and 1.096–6.483; P = 0.019 and 0.026, respectively) and tumor size (OR: 2.387 and 2.659; 95% CI: 1.098–5.188 and 1.055–6.704; P = 0.026 and 0.034, respectively), but not lymph node metastasis and other clinical status, were observed (Tables 4 and 5). Therefore, these polymorphic markers may further improve through the analysis of environmental/behavioral agents and background liver function. However, no significant difference in the serum levels of these biomarkers was detected between patients who possessed at least on polymorphic allele and those who did not have any of the ADAMTS14 nsSNPs examined. (Table A in S1 File)
Table 4. Odds Ratio (OR) and 95% Confidence Interval (CI) of clinical status and ADAMTS14 rs12774070 genotypic frequencies in 141 HCC patients with tobacco consumption.
| Variable | Genotypic frequencies | OR (95% CI) | P value | |
|---|---|---|---|---|
| CC (N = 105) | CA+AA (N = 36) | |||
| Clinical Stage | ||||
| Stage I/II | 75 (71.4%) | 18 (50.0%) | 1.00 | P = 0.019 * |
| Stage III/IV | 30 (28.6%) | 18 (50.0%) | 2.500 (1.148–5.446) | |
| Tumor size † | ||||
| ≦ T2 | 74 (70.5%) | 18 (50.0%) | 1.00 | P = 0.026 * |
| > T2 | 31 (29.5%) | 18 (50.0%) | 2.387 (1.098–5.188) | |
| Lymph node metastasis | ||||
| No | 103 (98.1%) | 33 (91.7%) | 1.00 | P = 0.072 |
| Yes | 2 (1.9%) | 3 (8.3%) | 4.682 (0.750–29.234) | |
| Distant metastasis | ||||
| No | 101 (96.2%) | 33 (91.7%) | 1.00 | P = 0.281 |
| Yes | 4 (3.8%) | 3 (8.3%) | 2.295 (0.488–10.790) | |
| Vascular invasion | ||||
| No | 88 (83.8%) | 29 (80.6%) | 1.00 | P = 0.654 |
| Yes | 17 (16.2%) | 7 (19.4%) | 1.249 (0.471–3.313) | |
| Child-Pugh grade | ||||
| A | 83 (79.0%) | 25 (69.4%) | 1.00 | P = 0.240 |
| B or C | 22 (21.0%) | 11 (30.6%) | 1.660 (0.709–3.887) | |
| HBsAg | ||||
| Negative | 63 (60.0%) | 20 (55.6%) | 1.00 | P = 0.640 |
| Positive | 42 (40.0%) | 16 (44.4%) | 1.200 (0.559–2.578) | |
| Anti-HCV | ||||
| Negative | 50 (47.6%) | 22 (61.1%) | 1.00 | P = 0.162 |
| Positive | 55 (52.4%) | 14 (38.9%) | 0.579 (0.267–1.252) | |
| Liver cirrhosis | ||||
| Negative | 19 (18.1%) | 7 (19.4%) | 1.00 | P = 0.857 |
| Positive | 86 (81.9%) | 29 (80.6%) | 0.915 (0.349–2.399) | |
The ORs with analyzed by their 95% CIs were estimated by logistic regression models. Child-Pugh grades indicate the severity of cirrhosis: A = 5–6 points, B = 7–9 points and C = 10–15 points.
† >T2 indicated the multiple tumor more than 5 cm or tumor involving a major branch of the portal or hepatic vein(s).
* P value < 0.05 as statistically significant.
Table 5. Odds Ratio (OR) and 95% Confidence Interval (CI) of clinical status and ADAMTS14 rs61573157 genotypic frequencies in 141 HCC patients with tobacco consumption.
| Variable | Genotypic frequencies | OR (95% CI) | P value | |
|---|---|---|---|---|
| CC (N = 119) | CT+TT (N = 22) | |||
| Clinical Stage | ||||
| Stage I/II | 83 (69.7%) | 10 (45.5%) | 1.00 | P = 0.027 * |
| Stage III/IV | 36 (30.3%) | 12 (54.5%) | 2.767 (1.096–6.483) | |
| Tumor size † | ||||
| ≦ T2 | 82 (68.9%) | 10 (45.5%) | 1.00 | P = 0.034 * |
| > T2 | 37 (31.1%) | 12 (54.5%) | 2.659 (1.055–6.704) | |
| Lymph node metastasis | ||||
| No | 116 (97.5%) | 20 (90.9%) | 1.00 | P = 0.126 |
| Yes | 3 (2.5%) | 2 (9.1%) | 3.867 (0.607–24.617) | |
| Distant metastasis | ||||
| No | 114 (95.8%) | 20 (90.9%) | 1.00 | P = 0.332 |
| Yes | 5 (4.2%) | 2 (9.1%) | 2.280 (0.413–12.572) | |
| Vascular invasion | ||||
| No | 101 (84.9%) | 16 (72.7%) | 1.00 | P = 0.164 |
| Yes | 18 (15.1%) | 6 (27.3%) | 2.104 (0.726–6.097) | |
| Child-Pugh grade | ||||
| A | 93 (78.2%) | 15 (68.2%) | 1.00 | P = 0.310 |
| B or C | 26 (21.8%) | 7 (31.8%) | 1.669 (0.616–4.524) | |
| HBsAg | ||||
| Negative | 72 (60.5%) | 11 (50.0%) | 1.00 | P = 0.358 |
| Positive | 47 (39.5%) | 11 (50.0%) | 1.532 (0.615–3.817) | |
| Anti-HCV | ||||
| Negative | 58 (48.7%) | 14 (63.6%) | 1.00 | P = 0.199 |
| Positive | 61 (51.3%) | 8 (36.4%) | 0.543 (0.212–1.391) | |
| Liver cirrhosis | ||||
| Negative | 22 (18.5%) | 4 (18.2%) | 1.00 | P = 0.973 |
| Positive | 97 (81.5%) | 18 (81.8%) | 1.021 (0.314–3.315) | |
The ORs with analyzed by their 95% CIs were estimated by logistic regression models. Child-Pugh grades indicate the severity of cirrhosis: A = 5–6 points, B = 7–9 points and C = 10–15 points.
† >T2 indicated the multiple tumor more than 5 cm or tumor involving a major branch of the portal or hepatic vein(s).
* P value < 0.05 as statistically significant.
In silico characteristic nsSNPs on structural features of human ADAMTS14
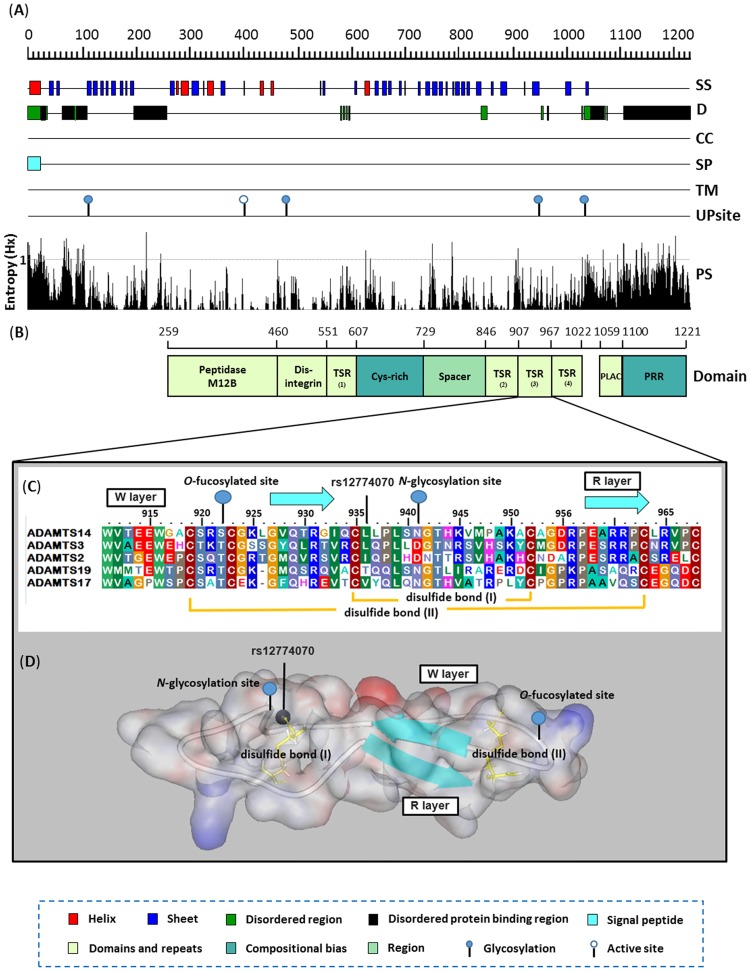
Based on current computational protein annotation methods that predict general features a 22-amino acid signal peptide at the hydrophobic N-terminus of ADAMTS14 is activated in the trans-Golgi network and leads to secretion, while no other obvious transmembrane and coil-coils regions of high hydropathy were present according to TMHMM [36] and COILS [37] predictions (Fig 1A). Otherwise, the basic ADAMTS14 structure comprises a characteristic peptidase M12B catalytic domain and a carboxyl-terminal ancillary domain containing a disintegrin-like module, Cys-rich domain, Spacer, four TSRs and a unique sequence including PLAC (protease and lacunin) [38] that is embedded in a PRR domain (Fig 1B).
Fig 1. Proteome annotation diagram of ADAMTS14 nsSNP rs12774070 by in silico approaches.
(A) The ADAMTS14 protein is first evaluated by sequence-based annotated methods with secondary structure (PSIPRED [SS]), disordered region (DISOPRED [D]), coiled region (COILS [CC]), signal peptides (SignalP [SP]) and transmembrane helices (TMHMM [TM]). In additional, active sites and glycoprotein sites are retrieved from UniProtKB database [UP sites]. (B) Indicating the level of conservation of residues by PSI-BLAST for each position using BioEdit Entropy algorithm [PS]. (C) Schematic representation of the full-length human ADMDTS14 protein, domain symbols are drawn approximately to scale. Amino acids are colored according to residue type: blue, positive; red, negative; light blue, small; green, hydrophobic; light green, aromatic; brown, cysteine and gray, polar. The rectangles represent the key domain structures, Peptidase M12B (IPR013273), Disintegrin (IPR018358), Cys-rich (IPR006586), Spacer (IPR010294), PLAC (IPR010909), Pro-rich (IPR026086) and four TSR (IPR000884) processed by InterPro database. (D) The residues are conserved throughout five procollagen aminopropeptidase subfamily of ADAMTS proteases, including ADAMTS2 (NP_055059.2), ADAMTS3 (NP_055058.2), ADAMTS14 (NP_631894.2), ADAMTS17 (NP_620688.2) and ADAMTS19 (NP_598377.4), as shown by alignment of the protein sequences with Clustal Omega software. Numbering is for human ADAMTS14. The two tryptophans (Trp911 and Trp916) and two arginines (Arg960 and Arg961) that form W layers and R layers, respectively. Protein surface diagram depicts the homology model of 3rd TSR domain of ADAMTS14. The ribbon indicates the Cα carbon of the 3rd TSR domain characterized in this study. The blue ribbon, green sphere, red spheres and yellow sticks indicate the β-strands structure, rs12774070, potential N-linked, O-linked glycosylation sites and disulfide bonds, respectively.
Fig 1C shows the amino acid position 941 of ADAMTS14 (the position of the amino acid substitution corresponding to rs12774070). The 3rd TSR domain is characterized by potential post-translational O-linked and N-linked glycosylation sites [39] with the sequences Cxx(S/T)CG and Nx(S/T) (residues 919–924 and 941–943; where C is cysteine, S is serine, T is threonine, N is asparagine and x is any amino acid) in a 20-residue stretch, respectively (Fig 1C). The ancillary domain is probably independent of the biological function and is more involved with the ECM association and the localization of the protease and its interaction partners for substrate specificity. Considering the essential role of ADAMTS14 in various models of disease, we performed in silico profiling of evolution-based and homology three-dimensional molecular model of the 3rd TSR domain (amino acid residues 911–968) of human ADAMTS14 (Fig 1D). Based on the structural data provided, site-specific information such as for variant rs12774070 near the essential N-glycosylation site within its 3rd TSR domain that can be regulated by its substrate recognition and cellular localization and which seems to involve a reduced affinity of ADAMTS14 associated with ECM components can be appreciated (Fig 1D).
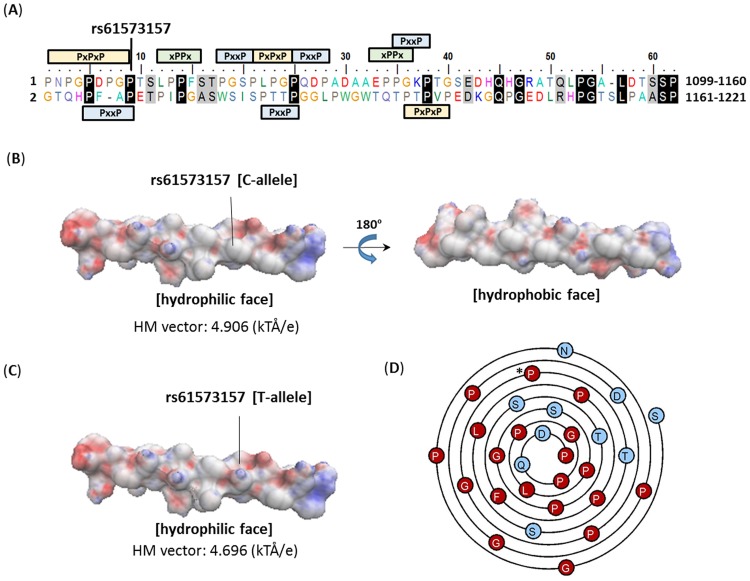
We further analyzed another affected variant (rs61573157), which has a unique PRR relative to the other ADAMTS family members, formed by a 122 amino acid stretch after the PLAC domain that is 25.4% proline and which contains a duplication of a 60-amino acid PRR with an amphipathic helix structure (Fig 2A). Searches of the human genome sequences reveal that at least four different mammalian proteins contain a homologous, similar PRR domain including TPRX1, ARHGAP26, DMRTC2 and COLEC12 (Figure B in S1 File).
Fig 2. Amphipathic helix structure of a duplication of a 60-amino acid proline-rich domain at C-terminal ADAMTS14.
(A) Alignment of a 60-amino acid duplication proline-rich repeats of the amphipathic helix (residues 1099–1160 and 1161–1121) present in with Clustal Omega software. The lines about the repeat 1 and below the repeat 2 show the three types of consensus linear motifs (PxPxP, PxxP and xPPx, where P is proline and x is any amino acid), respectively. The shading show the conserved residues and numbering is for the two repeats. Amino acids are colored according to residue type: blue, positive; red, negative; light blue, small; green, hydrophobic; light green, aromatic; brown, cysteine; and gray, polar. (B) C-allele and (C) T-allele of polymorphic ADAMTS14 nsSNP rs61573157 regular α-helical conformation of Proline-rich domain. Electrostatic potential in the solvent-accessible surface representations colored code where red and blue represent the net negative and positive charges, respectively. The two structure through the axis of rotation 180 degree to across the opposite of helices. White color represents the total neutral positions. 3D hydrophobic moment vectors were calculated in silico in http://www.ibg.kit.edu/HM/. (D) Wenxiang diagram represents that characterizes the disposition of hydrophobic (red-filled circles) and hydrophilic (blue-filled circles) residues in α-helices. The black star symbol indicated the nsSNP rs61573157.
To understand how the rs61573157 variant could affect the PRR structure, we constructed a modelling structure that was obtained from high resolution NMR data using 3D hydrophobic moment (HM) vector tool to characterize the surface polarity of the PRR domain. The helix-formed rods have amphipathic properties leading to the formation of a hydrophobic and a hydrophilic face with electrostatic surface (Fig 2B). Variant rs61573157 (P1108S) maps on to the hydrophilic face of the amphipathic helix of the PRR domain and is thought to constitute a potential binding site for ECM components. In addition, the liver-cancer associated risk [T]-allele creates the average HM vector of the ADAMTS14 PRR domain compared to the [C]-allele, which results in a decreased length of HM vector [HM vector change: 4.5%, from 4.696 (kTÅ/e) to 4.906 (kTÅ/e)] (Fig 2B and 2C). Additionally, the proline residue is a very unusual amino acid, because the side-chain is cyclized back on to the backbone amide position that is extended with a helix conformation as the adaptor role. Wenxiang diagrams show the amphipathic characteristics and the residue distribution with most hydrophobic residues in one-half of the molecule and most hydrophilic residues in the other half (Fig 2D). These data suggested that the risk-associated variant rs61573157 located in the ancillary domain probably alters the specificity of their substrate-binding preferences and enzyme localization via ECM association and may affect the clinical characteristics of tumor stage and size in HCC patients with smoking habits (Tables 4 and 5).
Discussion
Several studies suggested that chromosome 10q21 involved multiple genetic events have been implicated in the progression of multiple cancers [40–42]. Recent studies have reported that ADAMTS can exhibit both oncogenic and tumor-protective effects on tumor angiogenesis, an indispensable process vital for cancer progression and metastasis [43, 44]. Possible anti-angiogenic mechanism mediated by the TSR domains involved the cleavage of large thrombospondin 1 (TSP1) and TSP2 by ADAMTS1 after being released from the ECM and are an established anti-angiogenic activity, suggestion that the TSR domain in proteinase is important for their anti-angiogenic function [45]. Therefore, we hypothesize that genetic variants of ADAMTS14 may influence clinical outcomes in localized HCC patients with smoking behaviors. Our data reveal progressive clinical stage and increased tumor size among HCC smoking patients with at least one polymorphic A-allele of rs12774070 compared with those with homozygous C/C in case-control study. Our findings point to the role of variant rs12774070 in the TSR domain near the glycosylation site that may affect the TSR domain and can alter the cell-surface association and sequential processing mechanisms of ADAMTS14. We presented additional evidence in HCC, as elevated ADAMTS14 gene expression was associated with more aggressive cancers and poorer clinical outcomes.
Furthermore, rs61573157 located in the PRR domain of the C-terminal ancillary domain of ADAMTS14 was clinically examined probably because a consensus PxPxP motif had high binding capacity to E1A binding protein p300; the p300-p53 assembly reaction typically influences the alternative TP53 acetylation [46]. Although directly testing this hypothesis was beyond the scope of the current study, evidence suggests that a flexible p300-binding motif exists within the PRR domain and is associated with the susceptibility to HCC in patients with smoking habits, suggesting that ADAMTS14 down-regulation is associated with increased susceptibility to HCC. Hence, the present analyses increased our understanding of naturally occurring ADAMTS14 variants, a lesion category that, although not infrequent, has been relatively neglected in terms of exploring the underlying pathogenic mechanisms. An improved understanding of these variants is a prerequisite for developing therapeutic approaches that can eventually ameliorate the clinical phenotype in patients harboring the corresponding lesions.
Smoking yields toxins with cytotoxic potential which induce the processes of hepatic inflammation and fibrosis. In addition, progressive fibrosis and cirrhosis should be closely monitored in patients with a high risk of developing HCC. Although biomarkers are not widely accepted as important clinical tools [47], they contribute valuable information for the management of patients with HCC with regard to surveillance, diagnosis, evaluation of treatment efficacy, and prediction of outcomes HCC is usually diagnosed in cirrhotic patients (60%-80%). Proteolytic processing occurs in the ECM during the resolution of fibrosis and might predispose to future injury. The non-invasive markers may further improve the prediction to evaluate liver fibrosis status and impaired liver regeneration.
In conclusion, this study comprises a comprehensive effort to supplement medical information regarding HCC pathogenesis and conducting additional bioinformatics analyses of a high number of patients to provide comprehensive evidence of the roles of ADAMTS14 polymorphisms in HCC. Our results suggest that the ADAMTS14 polymorphic variants in the ADAMTS14 ancillary domains contribute to the occurrence and susceptibility to HCC in smoking patients. The joint effect of ADAMTS14 polymorphism in the TSR domain and PRR domain markedly facilitated HCC development. Overall, our analyses provide deeper insights into naturally occurring genetic variants. Characterizing the molecular basis of mutations in cancer cells provides insight into tumorigenesis and accurate biomarkers on such types of variant are required for developing optimal therapeutic approaches that can eventually ameliorate the clinical phenotype in patients harboring the corresponding lesions.
Supporting information
Table A. Association of ADAMTS14 genotypic frequencies with HCC laboratory status. Figure A. ADAMTS14 gene structure and Haploview LD display analysis. (A) ADAMTS14 is large gene and span 90K bp (from Chr.10 72,432,559 to 72,522,197; GRCh37.p.13). There are total 22 exons in this gene (NM_080722.3), the four selected SNPs, rs10823607, rs12774070, rs4747096 and rs61573157, are located on the exons 12, 19, 21 and 22, respectively. In the LD structure of SNPs-pairwise correlation coefficients in East Asian population (HCB+JPT) are shown (B) generated by Haploview version 4.2. A total of 34 haplotype blocks from 716 SNPs in the ADAMTS14 gene region were determined using the define blocks method “Four Gamete Rule” and LD colour scheme “Standard (D’/LOD)”. The four selected SNPs are shown as alleles of that region with recombination rate plotted and haplotypes display, (C) rs10823607 (529th), (D) rs12774070 (621th), (E) rs4747096 (669th) and (F) rs61573157 (689th), located on the LD map of haplotype blocks 22, 26, 31 and 32, respectively. Figure B. Alignment comparison of amino acids sequence. Alignment comparison of amino acids sequence of the amphipathic helix (residues 1098–1125) determined from five proline-rich region in proteins including TPRX1 (NP_940881.2), ARHGAP26 (NP_055886.1), DMRTC2 (NP_001035373.1) and COLEC12 (NP_569057.1) with Crustal Omega software. Amino acids are coloured according to residue type: blue, positive; red, negative; light blue, small; green, hydrophobic; light green, aromatic; brown, cysteine and grey, polar. Numbering is for human ADAMTS14.
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors would like to thank a research grant from the Ministry of Science and Technology, Taiwan, under Contract No. NSC-102-2320-B-040-012-MY2. Additionally, this study was supported by Chung Shan Medical University Hospital and Chi-Mei Foundation. Medical Center (CSMU-CMMC-105-05), Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 2.Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: Worldwide incidence and trends. Gastroenterology. 2004;127(5):S5–S16. [DOI] [PubMed] [Google Scholar]
- 3.Tsai WL, Chung RT. Viral hepatocarcinogenesis. Oncogene. 2010;29(16):2309–24. 10.1038/onc.2010.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6(9):674–87. 10.1038/nrc1934 [DOI] [PubMed] [Google Scholar]
- 5.Wang LY, You SL, Lu SN, Ho HC, Wu MH, Sun CA, et al. Risk of hepatocellular carcinoma and habits of alcohol drinking, betel quid chewing and cigarette smoking: a cohort of 2416 HBsAg-seropositive and 9421 HBsAg-seronegative male residents in Taiwan. Cancer Causes Control. 2003;14(3):241–50. [DOI] [PubMed] [Google Scholar]
- 6.Altamirano J, Bataller R. Cigarette smoking and chronic liver diseases. Gut. 2010;59(9):1159–62. 10.1136/gut.2008.162453 [DOI] [PubMed] [Google Scholar]
- 7.Poonpet T, Honsawek S, Tammachote N, Kanitnate S, Tammachote R. ADAMTS14 gene polymorphism associated with knee osteoarthritis in Thai women. Genet Mol Res. 2013;12(4):5301–9. 10.4238/2013.November.7.5 [DOI] [PubMed] [Google Scholar]
- 8.Colige A, Vandenberghe I, Thiry M, Lambert CA, Van Beeumen J, Li SW, et al. Cloning and characterization of ADAMTS-14, a novel ADAMTS displaying high homology with ADAMTS-2 and ADAMTS-3. J Biol Chem. 2002;277(8):5756–66. 10.1074/jbc.M105601200 [DOI] [PubMed] [Google Scholar]
- 9.Bolz H, Ramirez A, von Brederlow B, Kubisch C. Characterization of ADAMTS14, a novel member of the ADAMTS metalloproteinase family. Biochim Biophys Acta. 2001;1522(3):221–5. [DOI] [PubMed] [Google Scholar]
- 10.Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 2015;16:113 10.1186/s13059-015-0676-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apte SS. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J Biol Chem. 2009;284(46):31493–7. 10.1074/jbc.R109.052340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan K, Duquette M, Liu JH, Dong Y, Zhang R, Joachimiak A, et al. Crystal structure of the TSP-1 type 1 repeats: a novel layered fold and its biological implication. J Cell Biol. 2002;159(2):373–82. 10.1083/jcb.200206062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14(2):231–41. [PubMed] [Google Scholar]
- 14.Opitz R, Muller M, Reuter C, Barone M, Soicke A, Roske Y, et al. A modular toolkit to inhibit proline-rich motif-mediated protein-protein interactions. Proc Natl Acad Sci U S A. 2015;112(16):5011–6. 10.1073/pnas.1422054112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheng S, Carey J, Seftor EA, Dias L, Hendrix MJ, Sager R. Maspin acts at the cell membrane to inhibit invasion and motility of mammary and prostatic cancer cells. Proc Natl Acad Sci U S A. 1996;93(21):11669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverstein RL. The face of TSR revealed: an extracellular signaling domain is exposed. J Cell Biol. 2002;159(2):203–6. 10.1083/jcb.200209138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adzhubei AA, Sternberg MJ, Makarov AA. Polyproline-II helix in proteins: structure and function. J Mol Biol. 2013;425(12):2100–32. 10.1016/j.jmb.2013.03.018 [DOI] [PubMed] [Google Scholar]
- 18.Salah Z, Alian A, Aqeilan RI. WW domain-containing proteins: retrospectives and the future. Front Biosci (Landmark Ed). 2012;17:331–48. [DOI] [PubMed] [Google Scholar]
- 19.Porter S, Scott SD, Sassoon EM, Williams MR, Jones JL, Girling AC, et al. Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin Cancer Res. 2004;10(7):2429–40. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez-Lopez J, Pombo-Suarez M, Loughlin J, Tsezou A, Blanco FJ, Meulenbelt I, et al. Association of a nsSNP in ADAMTS14 to some osteoarthritis phenotypes. Osteoarthritis and cartilage. 2009;17(3):321–7. Epub 2008/09/16. 10.1016/j.joca.2008.07.012 [DOI] [PubMed] [Google Scholar]
- 21.Su SC, Hsieh MJ, Liu YF, Chou YE, Lin CW, Yang SF. ADAMTS14 Gene Polymorphism and Environmental Risk in the Development of Oral Cancer. PLoS One. 2016;11(7):e0159585 10.1371/journal.pone.0159585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruni R, Prosperi M, Marcantonio C, Amadori A, Villano U, Tritarelli E, et al. A computational approach to identify point mutations associated with occult hepatitis B: significant mutations affect coding regions but not regulative elements of HBV. Virol J. 2011;8:394 10.1186/1743-422X-8-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vauthey JN, Lauwers GY, Esnaola NF, Do KA, Belghiti J, Mirza N, et al. Simplified staging for hepatocellular carcinoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2002;20(6):1527–36. Epub 2002/03/16. [DOI] [PubMed] [Google Scholar]
- 24.Yang SF, Yeh CB, Chou YE, Lee HL, Liu YF. Serpin peptidase inhibitor (SERPINB5) haplotypes are associated with susceptibility to hepatocellular carcinoma. Scientific reports. 2016;6:26605 Epub 2016/05/26. 10.1038/srep26605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng HL, Liu YF, Su CW, Su SC, Chen MK, Yang SF, et al. Functional genetic variant in the Kozak sequence of WW domain-containing oxidoreductase (WWOX) gene is associated with oral cancer risk. Oncotarget. 2016;7(43):69384–96. Epub 2016/09/23. 10.18632/oncotarget.12082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zavras AI, Yoon AJ, Chen MK, Lin CW, Yang SF. Metallothionein-1 genotypes in the risk of oral squamous cell carcinoma. Ann Surg Oncol. 2011;18(5):1478–83. 10.1245/s10434-010-1431-3 [DOI] [PubMed] [Google Scholar]
- 27.Lewis TE, Sillitoe I, Andreeva A, Blundell TL, Buchan DW, Chothia C, et al. Genome3D: exploiting structure to help users understand their sequences. Nucleic Acids Res. 2015;43(Database issue):D382–6. 10.1093/nar/gku973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drew K, Winters P, Butterfoss GL, Berstis V, Uplinger K, Armstrong J, et al. The Proteome Folding Project: proteome-scale prediction of structure and function. Genome research. 2011;21(11):1981–94. 10.1101/gr.121475.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DE, Chivian D, Malmstrom L, Baker D. Automated prediction of domain boundaries in CASP6 targets using Ginzu and RosettaDOM. Proteins. 2005;61 Suppl 7:193–200. [DOI] [PubMed] [Google Scholar]
- 31.Breuza L, Poux S, Estreicher A, Famiglietti ML, Magrane M, Tognolli M, et al. The UniProtKB guide to the human proteome. Database (Oxford). 2016;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–6. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 33.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–8. [Google Scholar]
- 34.Reisser S, Strandberg E, Steinbrecher T, Ulrich AS. 3D hydrophobic moment vectors as a tool to characterize the surface polarity of amphiphilic peptides. Biophys J. 2014;106(11):2385–94. 10.1016/j.bpj.2014.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chou K-C, Lin W-Z, Xiao X. Wenxiang: a web-server for drawing wenxiang diagrams. Natural Science. 2011;03(10):862–5. [Google Scholar]
- 36.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–80. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Zhang T, Chen K, Kedarisetti KD, Mizianty MJ, Bao Q, et al. Critical assessment of high-throughput standalone methods for secondary structure prediction. Brief Bioinform. 2011;12(6):672–88. 10.1093/bib/bbq088 [DOI] [PubMed] [Google Scholar]
- 38.Nardi JB, Martos R, Walden KK, Lampe DJ, Robertson HM. Expression of lacunin, a large multidomain extracellular matrix protein, accompanies morphogenesis of epithelial monolayers in Manduca sexta. Insect Biochem Mol Biol. 1999;29(10):883–97. [DOI] [PubMed] [Google Scholar]
- 39.Wang LW, Leonhard-Melief C, Haltiwanger RS, Apte SS. Post-translational modification of thrombospondin type-1 repeats in ADAMTS-like 1/punctin-1 by C-mannosylation of tryptophan. J Biol Chem. 2009;284(44):30004–15. 10.1074/jbc.M109.038059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baert-Desurmont S, Charbonnier F, Houivet E, Ippolito L, Mauillon J, Bougeard M, et al. Clinical relevance of 8q23, 15q13 and 18q21 SNP genotyping to evaluate colorectal cancer risk. European journal of human genetics: EJHG. 2016;24(1):99–105. Epub 2015/04/16. 10.1038/ejhg.2015.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiiba M, Nomura H, Shinozuka K, Saito K, Kouzu Y, Kasamatsu A, et al. Down-regulated expression of SERPIN genes located on chromosome 18q21 in oral squamous cell carcinomas. Oncology reports. 2010;24(1):241–9. Epub 2010/06/02. [DOI] [PubMed] [Google Scholar]
- 42.Zhang B, Jia WH, Matsuda K, Kweon SS, Matsuo K, Xiang YB, et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet. 2014;46(6):533–42. Epub 2014/05/20. 10.1038/ng.2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cal S, Lopez-Otin C. ADAMTS proteases and cancer. Matrix Biol. 2015;44–46: 77–85. [DOI] [PubMed] [Google Scholar]
- 44.Su SC, Hsieh MJ, Yang WE, Chung WH, Reiter RJ, Yang SF. Cancer metastasis: Mechanisms of inhibition by melatonin. Journal of pineal research. 2017;62(1). Epub 2016/10/06. [DOI] [PubMed] [Google Scholar]
- 45.Lee NV, Sato M, Annis DS, Loo JA, Wu L, Mosher DF, et al. ADAMTS1 mediates the release of antiangiogenic polypeptides from TSP1 and 2. EMBO J. 2006;25(22):5270–83. 10.1038/sj.emboj.7601400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dornan D, Shimizu H, Burch L, Smith AJ, Hupp TR. The Proline Repeat Domain of p53 Binds Directly to the Transcriptional Coactivator p300 and Allosterically Controls DNA-Dependent Acetylation of p53. Molecular and Cellular Biology. 2003;23(23):8846–61. 10.1128/MCB.23.23.8846-8861.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu K, Dai Z, Zhou J. Biomarkers for hepatocellular carcinoma: progression in early diagnosis, prognosis, and personalized therapy. Biomark Res. 2013;1(1):10 10.1186/2050-7771-1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A. Association of ADAMTS14 genotypic frequencies with HCC laboratory status. Figure A. ADAMTS14 gene structure and Haploview LD display analysis. (A) ADAMTS14 is large gene and span 90K bp (from Chr.10 72,432,559 to 72,522,197; GRCh37.p.13). There are total 22 exons in this gene (NM_080722.3), the four selected SNPs, rs10823607, rs12774070, rs4747096 and rs61573157, are located on the exons 12, 19, 21 and 22, respectively. In the LD structure of SNPs-pairwise correlation coefficients in East Asian population (HCB+JPT) are shown (B) generated by Haploview version 4.2. A total of 34 haplotype blocks from 716 SNPs in the ADAMTS14 gene region were determined using the define blocks method “Four Gamete Rule” and LD colour scheme “Standard (D’/LOD)”. The four selected SNPs are shown as alleles of that region with recombination rate plotted and haplotypes display, (C) rs10823607 (529th), (D) rs12774070 (621th), (E) rs4747096 (669th) and (F) rs61573157 (689th), located on the LD map of haplotype blocks 22, 26, 31 and 32, respectively. Figure B. Alignment comparison of amino acids sequence. Alignment comparison of amino acids sequence of the amphipathic helix (residues 1098–1125) determined from five proline-rich region in proteins including TPRX1 (NP_940881.2), ARHGAP26 (NP_055886.1), DMRTC2 (NP_001035373.1) and COLEC12 (NP_569057.1) with Crustal Omega software. Amino acids are coloured according to residue type: blue, positive; red, negative; light blue, small; green, hydrophobic; light green, aromatic; brown, cysteine and grey, polar. Numbering is for human ADAMTS14.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.