SUMMARY
Cannabinoid receptor 1 (CB1) is the principal target of Δ9-tetrahydrocannabinol (THC), a psychoactive chemical from Cannabis sativa with a wide range of therapeutic applications and a long history of recreational use. CB1 is activated by endocannabinoids, and is a promising therapeutic target for pain management, inflammation, obesity and substance abuse disorders. Here, we present the 2.8 Å crystal structure of human CB1 in complex with AM6538, a stabilizing antagonist, synthesized and characterized for this structural study. The structure of the CB1-AM6538 complex reveals key features of the receptor and critical interactions for antagonist binding. In combination with functional studies and molecular modeling, the structure provides insight into the binding mode of naturally occurring CB1 ligands, such as THC, and synthetic cannabinoids. This enhances our understanding of the molecular basis for the physiological functions of CB1 and provides new opportunities for the design of next-generation CB1-targeting pharmaceuticals.
Graphical abstract
Introduction
Marijuana from Cannabis sativa L. has been used for both therapeutic and recreational purposes for many centuries (Lemberger, 1980; Li, 1973). In the 1940s, chemistry based on compounds isolated from the plant (Wollner et al., 1942) produced novel biologically active molecules (Adams et al., 1948; Ghosh et al., 1940); however, it was not until the 1960s that the active constituent of marijuana, (−)-Δ9- trans-tetrahydrocannabinol (THC), a terpenoid molecule, was isolated and characterized (Gaoni and Mechoulam, 1964). This provided an early molecular foundation for medicinal chemists to develop related structural analogs and new synthetic ligands (Makriyannis and Rapaka, 1990; Razdan, 1986). Initially, due to their lipophilic nature, it was assumed that cannabinoids exerted their effects by perturbing the physical properties of biological membranes (Makriyannis, 2014; Mavromoustakos et al., 1995). This assumption was challenged with the discovery, cloning, and expression of the first cannabinoid-specific membrane receptor, then designated as the cannabinoid receptor (CB) (Devane et al., 1988; Matsuda et al., 1990). With the subsequent identification of a second receptor, the designation evolved to CB1 and CB2 (Munro et al., 1993). The discovery of the endogenous agonists to the receptors, the endocannabinoids, anandamide (Devane et al., 1992) and 2-arachidonoyl glycerol (2-AG) (Mechoulam et al., 1995) soon followed.
Cannabinoid receptors belong to the class A G protein-coupled receptor (GPCR) family, signal through inhibitory Gαi/o heterotrimeric G proteins (Howlett, 1985) and interact with β-arrestins (Jin et al., 1999). CB1 is the most highly expressed GPCR in the human brain and is expressed throughout the body, with the highest levels found in the central nervous system (Herkenham et al., 1990).
Cannabis has been used for centuries in many cultures to treat a wide range of medical conditions. More recently, therapeutic considerations have moved beyond the plant extract to explore and produce more pharmacologically refined compounds. CB1-selective small molecule agonists have shown therapeutic promise in a wide range of disorders, including pain and inflammation (Cravatt and Lichtman, 2004), multiple sclerosis (Pertwee, 2002; Pryce and Baker, 2015), neurodegenerative disorders (Fernandez-Ruiz et al., 2015). The first CB1-selective antagonist/inverse agonist, rimonabant (SR141716, Acomplia™ (Sanofi-Aventis)) (Rinaldi-Carmona et al., 1994), received approval from the European Medical Agency as an adjunct to diet and exercise for treating obesity (Janero and Makriyannis, 2009). Antagonists of CB1 have been explored as potential therapeutics for obesity-related metabolic disorders (Mazier et al., 2015), mental illness (Black et al., 2011; Rubino et al., 2015), liver fibrosis (Mallat et al., 2013) and nicotine addiction (Schindler et al., 2016). However, rimonabant and other ligands in its class were not approved in the United States due to concerns about adverse events, such as increased anxiety, depression, and suicidal ideation.
Numerous studies have investigated how ligands binding CB1 can mediate downstream signaling. While the variety of compounds exhibiting different pharmacological profiles have provided clues regarding CB1 activation, the molecular details defining the binding modes of both endogenous and exogenous ligands are still largely unknown (Guo et al., 1994; Makriyannis, 2014; Picone et al., 2005). In order to address this deficit in understanding, we have determined the crystal structure of CB1 in complex with a tight binding antagonist AM6538. In conjunction with molecular docking, the structure was used to elucidate the binding modes of a diverse set of antagonists/inverse agonists and agonists of CB1. The structural details of the cannabinoid receptor reported herein improve our understanding of how ligands engage to modulate the cannabinoid system and provide a useful model to facilitate the design of next-generation pharmaceuticals to avoid unwanted side effects. The findings provide insight into mechanisms of slow dissociation of antagonists, which may potentially translate into long acting pharmacological effects.
Results
Synthesis of CB1 Stabilizing Antagonist AM6538 for Structural Studies
One of the key factors facilitating the structure determination of CB1 (Figure 1) is utilization of the antagonist AM6538, the synthesis of which resulted from the strategic modification of rimonabant to enhance its ability to stabilize the ligand-receptor complex and promote CB1 crystal formation. In contrast with rimonabant, the 5-phenyl ring substituent was modified so as to introduce motifs (ex. alkyne unit) that could favor increased affinity for the CB1 receptor (Tam et al., 2010). The rimonabant analog, AM251, (1, Figure 2A) (Lan et al., 1999), a compound that has been used extensively as a pharmacological standard CB1-selective antagonist, was used as the precursor in the AM6538 synthetic process. Synthesis of AM6538 involves the functionalization of the iodo substituent at the para position of the 5-phenyl ring in AM251 with an acetylenic chain system consisting of four carbons and substituted at the omega carbon. To this end, we initially focused on targeting cysteine residues within CB1 by introducing, suitable electrophilic groups (Janero et al., 2015; Li et al., 2005; Mercier et al., 2010; Picone et al., 2005; Szymanski et al., 2011) at the fourth carbon of the alkyne unit, capable of forming a covalent bond with the cysteine thiol group. For AM6538, we introduced at this position, a nitrate group (ONO2) whose role was to serve as a polar group, which may be displaced by a suitable nucleophile (e.g. thiol) (Pattison and Brown, 1956; Yeates et al., 1985) at or near the binding domain or alternatively bind as an intact group so as to obtain a non-covalent, near irreversible attachment by interacting with hydrogen bonding amino acid residues as well as residues capable of π-π interactions. In the present study, affinity mass spectrometry analysis suggests that AM6538 reacts with CB1 as an intact molecule with no evidence of covalent modification of relevant cysteine residues.
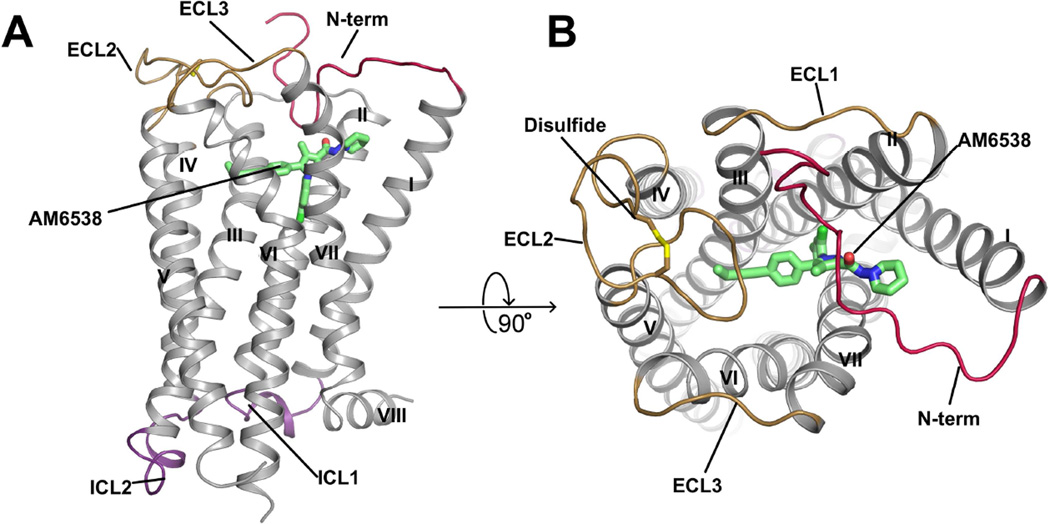
Figure 1. Overall Structure of CB1-AM6538 Complex.
(A) Side view of the CB1-AM6538 complex. The receptor is shown in gray cartoon representation. The ligand AM6538, shown with green sticks, demarcates the binding pocket which is partially occluded by the N-terminal loop (red). The nitrate group is not modeled in the experimental crystal structure as the electron density was not observed. The extracellular loops (ECLs) are shown in brown and the intracellular loops are shown in purple. (B) Top view of the extracellular side. The disulfide bond in ECL2 is shown as yellow sticks. See also Figures S2 and S3.
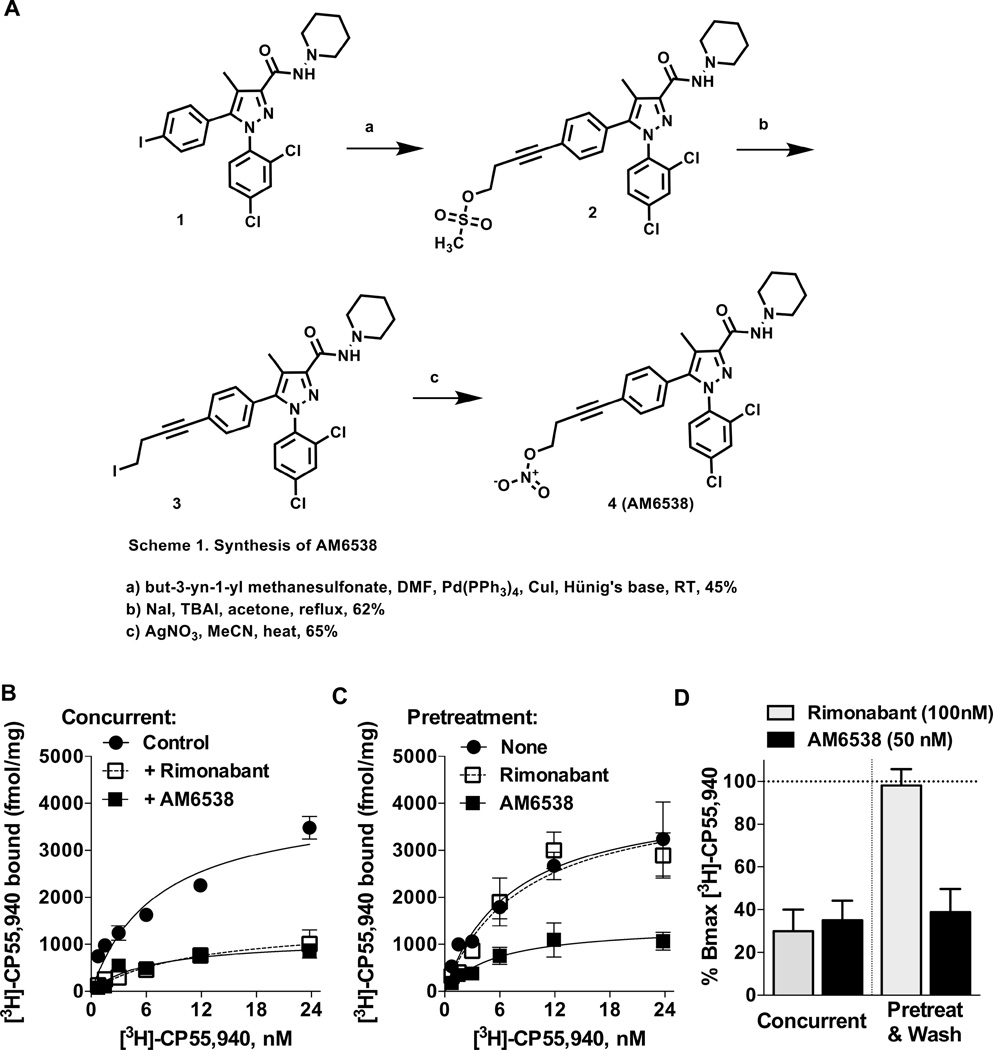
Figure 2. Synthesis and Characterization of AM6538.
(A) Synthetic procedures for compound AM6538. (B) Saturation [3H]-CP55,940 binding assays in the absence (control) or presence of rimonabant (100 nM) or AM6538 (50 nM) demonstrates that both antagonists cause displacement of specific binding of the radioligand when present concurrently in the 1 h binding assay. (C) Following pretreatment of membranes (37 °C, 6 h) with buffer only (None), rimonabant (100 nM) or AM6538 (50 nM); membranes were washed with buffer 3× prior to [3H]-CP55,940 binding as described for B. (D) Percentage of remaining binding (Bmax) detected following the conditions described in (A) (concurrent) and (B) (pretreat & wash). Both antagonists decrease the binding of [3H]-CP55,940 to ~30% when incubated concurrently during the 1 h binding assay. Under pretreatment and washout conditions, rimonabant does not affect subsequent radioligand binding, while AM6538 continues to compete despite washing of the membranes. See also Figure S1 and Table S1.
AM6538 was a strong candidate for crystallographic studies of the receptor based on its high affinity and wash-resistant binding to CB1 as determined by radioligand competition assays against the tritiated agonist, [3H]-CP55,940 (Ki =3.4 ± 1.0 nM) (Figures 2B, C and D). This is in contrast to rimonabant which can be readily washed out of membranes, permitting subsequent radioligand binding (Figures 2B, C and D). Importantly, the crystallized CB1 construct (described below) has comparable affinity for AM6538 as the wild-type receptor (Ki= 5.1 ± 0.9 nM).
In functional assays, AM6538 is a competitive antagonist of the effects of CP55,940 and THC on CB1-mediated inhibition of adenylyl cyclase activity and β-arrestin2 recruitment in overexpression systems (Figures S1A and B, Table S1). Competitive antagonism was confirmed by the [35S]-GTPγS binding assays performed in mouse cerebellum (Figure S1C). For comparison purposes, competitive antagonism was demonstrated for rimonabant in the same systems (Figure S1 and Table S1).
Structure Determination of CB1-AM6538 Complex
To facilitate crystallization it was necessary to modify the wild-type (WT) CB1 sequence. Construct optimization procedures (Lv et al., 2016) identified Flavodoxin (Chun et al., 2012) as a stabilizing fusion partner when inserted within the receptor’s third intracellular loop (ICL3) at Val306 and Pro332. Additionally, the wild-type receptor was truncated on both the N- and C-termini by 98 and 58 residues, respectively. Finally, in order to improve the expression and thermostability of the receptor, four computationally predicted mutations (Thr2103.46Ala (D’Antona et al., 2006), Glu2735.37Lys, Thr2835.47Val and Arg3406.32Glu) were introduced to the CB1 sequence (Figures S2A, B and C) (superscripts denote amino acid position as described by Ballesteros (Ballesteros and Weinstein, 1995)). The modified CB1 construct was inserted into a pTT5 vector for expression in HEK293F cells to generate protein (Figure S2D) that formed crystals in lipidic cubic phase supplemented with cholesterol (Figure S3E); the crystals diffracted to 2.8 Å (Table 1).
Table 1.
Crystallographic Data Collection and Refinement Statistics.
| Data Collection and Refinement Statistics | |
|---|---|
| Ligand | AM6538 |
| Number of Crystals | 29 |
| Data collection | |
| Space Group | C2 |
| Cell Dimensions | |
| a,b,c (Å) | 116.56, 52.63, 143.63 |
| β (°) | 111.14 |
| Number of Reflections Measured | 160,794 |
| Number of Unique Reflections | 19,837 |
| Resolution (Å) | 47.30 – 2.80 (2.90–2.80) |
| Rmerge $ | 0.126 (0.520) |
| Mean I/σ(I) | 10.1 (2.1) |
| Completeness (%) | 97.4 (94.1) |
| Redundancy | 8.1 (4.6) |
| CC1/2 | 0.999 (0.44) |
| Refinement | |
| Resolution (Å) | 47.05 – 2.80 |
| Number of Reflections (test set) | 19,827 (985) |
| Rwork/Rfree | 0.207/0.238 |
| Number of Atoms | |
| CB1 | 2,312 |
| Flavodoxin | 1,103 |
| AM6538& | 33 |
| Lipid and other | 102 |
| Average B Factor (Å2) | |
| Wilson | 73.6 |
| Overall | 87.4 |
| CB1 | 110.6 |
| Flavodoxin | 66.6 |
| AM6538 | 119.5 |
| Lipid and other | 79.7 |
| r.m.s. deviations | |
| Bond Lengths (Å) | 0.004 |
| Bond Angles (°) | 0.638 |
| Ramachandran Plot Statistics (%)* | |
| Favored Regions | 97.7 |
| Allowed Regions | 2.3 |
| Disallowed Regions | 0 |
Data for high resolution shells is shown in parenthesis
Nitrate group is excluded due to the absence of electron density
As defined in MolProbity
Based on affinity mass spectrometry analysis, intact AM6538 is associated with the CB1 protein (Figure S3F). All atoms for AM6538 are observed in the electron density maps except for the terminal nitrate group (Figure 3D). A molecular dynamics (MD) simulation was performed on the CB1-AM6538 complex and the results revealed that the root mean square fluctuation (RMSF) values for the nitrate group and the hinge carbon atom are higher, indicating the nitrate group is more mobile than other atoms in AM6538 (Figure S4B). Further studies on the nitrate group of AM6538 are under investigation.
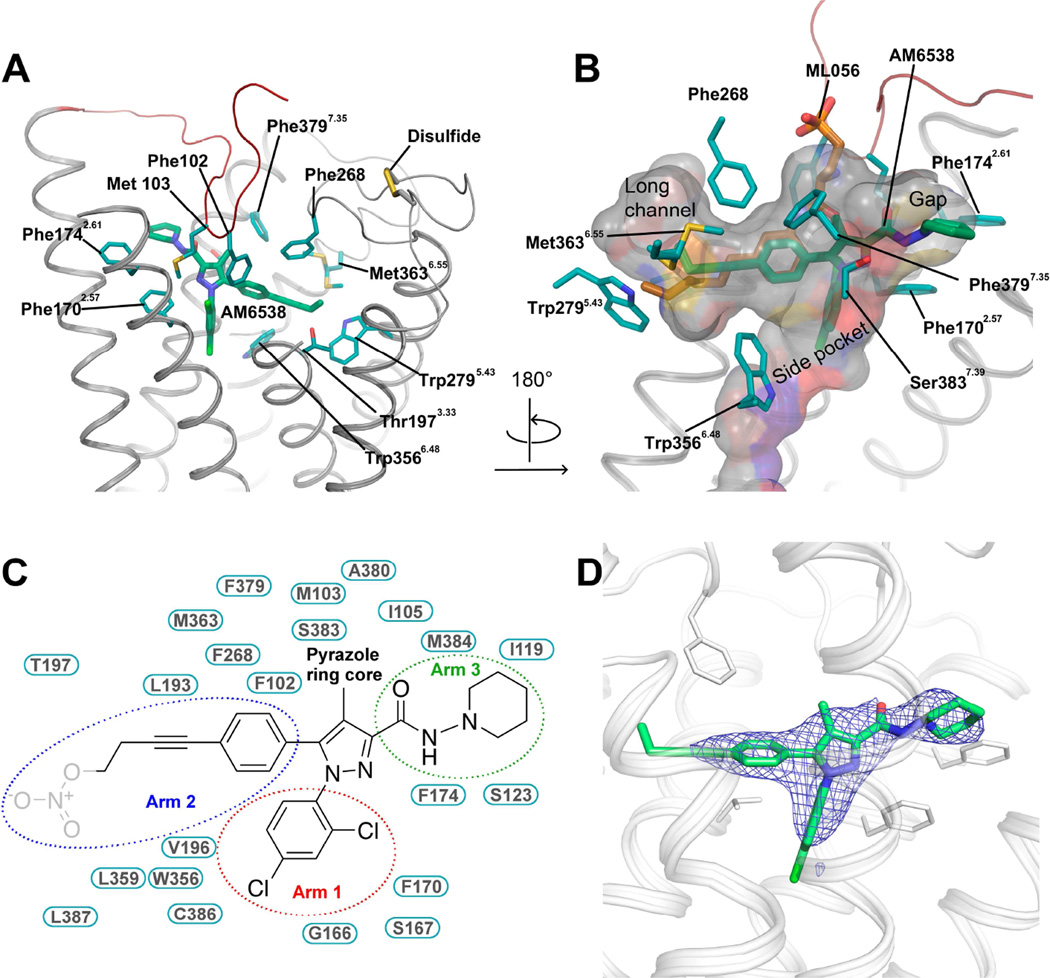
Figure 3. Analysis of the Ligand Binding Pocket of CB1.
(A) Key residues in CB1 for AM6538 binding. AM6538 (green carbons) and CB1 residues (teal carbons) involved in ligand binding are shown in stick representation. The receptor is shown in gray cartoon representation. (B) The shape of the ligand binding pocket. AM6538 (green carbons) and ML056 (brown carbons) are shown in stick representation. (C) Schematic representation of interactions between CB1 and AM6538. The 2,4-dichlorophenyl ring in the red circle is termed as arm 1; The 4-aliphatic chain substituted phenyl ring in the blue circle is termed as arm 2; The piperidin-1-ylcarbamoyl in the green circle is termed as arm 3. The nitrate group, which was not observed in the electron density, is shown in gray. (D) Electron density maps calculated from the refined structure of the CB1-AM6538 complex. |Fo|-|Fc| omit map (blue mesh) of the ligand AM6538 is shown (contoured at 3 σ). See also Figure S3.
Structural Features of CB1 in Complex with AM6538
The overall CB1 structural fold shares a similar architecture with previously solved class A GPCR structures, containing seven transmembrane (7TM) α-helices (I to VII) connected by three extracellular loops (ECL1–3) and three intracellular loops (ICL1–3) and an amphipathic helix VIII (Figure 1A and Figure S3A). The non-truncated part of the N-terminus of CB1, residues 99–112, forms a V-shaped loop, which inserts into the ligand-binding pocket and functions as a plug, restricting access to the pocket from the extracellular side (Figures 1A and B). While the influence of the crystal packing interactions on the conformation of the N-terminus (Figure S3B) cannot be ruled out, it is interesting to note that the N-terminus has been consistently observed in an ordered form in the structures of the related lipid receptors LPA1 (Chrencik et al., 2015) and S1P1 (Hanson et al., 2012), and that the function of CB1 is very sensitive to the presence of the ordered portion of the N-terminus (Andersson et al., 2003; Fay and Farrens, 2013). ECL2 in CB1 consists of 21 residues folding into an intricate structure that projects four residues (268–271) into the binding pocket. Previous work has shown that the four residues are important for mediating interactions with certain classes of ligands (Ahn et al., 2009; Bertalovitz et al., 2010) and that the two cysteines (Cys257 and Cys264) in ECL2 are critical to the function of CB1 (Fay et al., 2005). In the structure, the conformation of ECL2 is constrained by the presence of an intraloop disulfide bond (Cys257-Cys264) (Figure 1B) previously found in the structures of the closely related LPA1 and S1P1 receptors. The highly conserved disulfide bond between ECL2 and helix III (Cys3.25) in most class A GPCRs is lacking in all three lipid receptor structures.
AM6538 Interactions in CB1 Ligand-Binding Pocket
The position of the ligand-binding pocket of CB1 is different from the previously described orthosteric binding sites of other class A GPCRs. AM6538 lies quite low in the binding pocket of CB1, immediately above the conserved Trp3566.48 (Figures 3A and B). The ligand adopts an extended conformation with the ligand strain close to its local minimum as determined by quantum mechanical calculations (Figure S3D).
AM6538 forms mainly hydrophobic interactions with ECL2 and the N-terminus, as well as with all CB1 helices except helix IV (Figures 3 A and B). As described above, the ligand has a pyrazole ring core with three functional groups. For clarity, we have termed the 2,4-dichlorophenyl ring “arm 1,” the 4-aliphatic chain substituted phenyl ring “arm 2,” and the piperidin-1-ylcarbamoyl “arm 3” (Figure 3C). The pyrazole ring core (including the 4-methyl group) is situated between helices II and VII, forming hydrophobic interactions with the side chains of Phe1702.57, Phe3797.35 and Ser3837.39 (Figure 3C), and is capped by the N-terminal loop interactions (Met103N-term). Arm 1 is located in a narrow side pocket (Figure 3B) formed by helices II, III, VI and VII, and forms edge-face π-π interactions with the side chain of Phe1702.57, and with the backbone amide bond between Gly1662.53 and Ser1672.54. This substituted ring moiety forms hydrophobic interactions with Val1963.32, Trp3566.48, Cys3867.42, Leu3877.43 and Met103N-term (Figure 3C). The 2,4-dichlorophenyl ring in arm 1 fits well into the shape of the narrow side pocket (Figure S3C), which explains why 2,4-dichloro or 2-chloro substitutions result in optimal binding (Lange and Kruse, 2005).
Arm 2 of the ligand extends towards a long, narrow channel (Figure 3B) formed by helices III, V, VI and ECL2. The phenyl group in arm 2 establishes π-π interactions with Phe102N-term, Phe268ECL2, and Trp3566.48; the hydrophobic interactions with Leu1933.29, Val1963.32 and Leu3596.51 are observed. A triple bond within the long aliphatic chain in arm 2 forms π-π interactions with Phe268ECL2 and Trp3566.48; and hydrophobic interactions with several residues including Leu1933.29, Val1963.32, Thr1973.33, Leu3596.51, and Met3636.55 (Figure 3C). Interestingly, the binding mode of the long hydrophobic chain is similar to that of ML056 with the S1P1 receptor (Figure 3B), implying this could be a conserved binding pocket for long aliphatic chains in lipid-binding receptors. In regards to the nitrate group that has not been observed in the crystallographic structure, our docking experiments define a domain in which the nitrate group is interacting with residues Thr1973.33, Tyr2755.39 and Trp2795.43 through hydrogen bonding and π-π interactions (Figure S4A).
Finally, arm 3 extends toward a gap constituted by helices I, II, VII, capped by the N-terminal loop (Figure 3B). It forms interactions with hydrophobic residues, Met103N-term, Ile105N-term, Ile1191.35, Ser1231.39, Phe1702.57, Phe1742.61, Ala3807.36, Ser3837.39 and Met3847.40 (Figure 3C). Unlike the π-π interactions formed by the other two arms, the interactions between arm 3 and the receptor are non-specific.
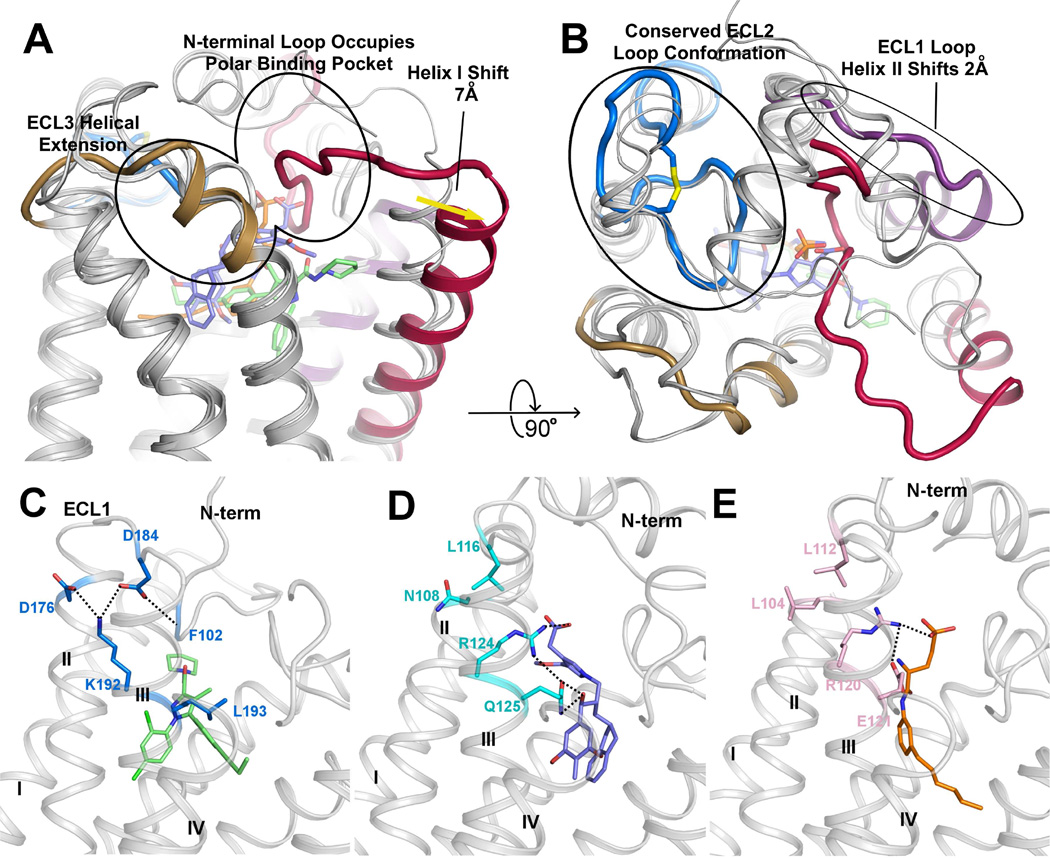
Among the interactions between AM6538 and CB1, Phe1702.57 plays an important role by interacting with the pyrazole ring core as well as rings in arm 1 and arm 3. Moreover, Phe1702.57 is pushed by the ligand to move towards helix I, resulting in a tilt of the last two turns (residues 170–177) of helix II toward helix I, compared to S1P1 and LPA1, the two closest homologs (Figures 4A and B). This tilted helix II, in turn, pushes helix I by about 7 Å, mainly due to interactions with the two bulky residues Phe1702.57 and Phe1742.61.
Figure 4. Comparison of CB1, LPA1 and S1P1 Structural Features.
(A) Side view of CB1 with structurally divergent regions of LPA1 (PDB ID: 4Z34) and S1P1 (PDB ID: 3V2Y) overlaid. LPA1 and S1P1 receptors are shown in gray cartoons. The CB1 N-terminal loop (red) occupies the polar binding pocket, helix I (red helix) is shifted out 7 Å compared with the other two receptors. ECL3 of CB1 shows a three helical turn extension of helix VII (brown helix). (B) 90° rotation of (A) for a top view of CB1 with structurally divergent regions of LPA1 (4Z34) and S1P1 (3V2Y) overlaid. CB1 shows a conserved conformation of ECL2 (blue) with the other two receptors, helix II is shifted out 2 Å (purple helix). (C–E) The interaction network of residues 3.28 of CB1 (K192), LPA1 (R124) and S1P1 (R120). Polar interactions are represented by black dashed lines. (C) CB1 is shown in gray cartoon, AM6538 is shown in green sticks and the key residues are shown in blue sticks; (D) LPA1 is shown in gray cartoon, ONO-9780307 is shown in purple-blue sticks and the key residues are shown in cyan sticks; (E) S1P1 is shown in gray cartoon, ML056 is shown in orange sticks and the key residues are shown in pink sticks. See also Figure S5.
The role of Lys1923.28 in CB1 has been intensively researched. It was reported that Lys192Ala/Lys192Gln/Lys192Glu mutants decreased the affinities of several agonists such as CP55,940, HU-210 and anandamide (Chin et al., 1998; Hurst et al., 2002; Pan et al., 1998; Song and Bonner, 1996). Previously, it was suggested that Lys1923.28 has direct interactions with CB1 ligands. However, in our CB1 structure, Lys1923.28 does not interact directly with AM6538. Instead it forms a salt bridge/hydrogen bond network that stabilizes the conformation of ECL1, the N-terminus and the extracellular parts of helices II and III. The side chain of Lys1923.28 points away from the binding pocket and forms salt bridges with Asp1762.63 and Asp184ECL1, while Asp184ECL1 further stabilizes the N-terminus by forming a hydrogen bond with the backbone of Phe102N-term (Figure 4C).
Structural Comparison of the CB1, LPA1 and S1P1 Receptors
CB1, LPA1 and S1P1 receptors all bind lipid-derived endogenous ligands (anandamide/sn-2-arachidonoylglycerol, sphingosine-1-phosphate, lysophosphatidic acid) (Shimizu, 2009). Early sequence analysis revealed that CB1 has a moderate sequence identity with LPA1 (13% overall, 28% in TM regions) and S1P1 (14% overall, 27% in TM regions) (Bramblett et al., 1995; Isberg et al., 2016) (Figure S5). Crystal structures of LPA1 (Chrencik et al., 2015) and S1P1 (Hanson et al., 2012) have been recently determined. The main structural difference between the three lipid receptors occurs in the extracellular portion, with the most striking being the unique conformation of the N-terminal loop of CB1 (Figures 4A and B). For all three receptors, the N-terminus has a role in ligand recognition. Comparing the ligand binding positions of the three receptors, AM6538 lies more horizontally than the ligands in LPA1 and S1P1, with arm 1 inserted deeper into the side pocket (Figure 4A). Consistently, the N-terminal loop in CB1 is positioned deeper into the binding pocket compared to the N-terminal helices of LPA1 and S1P1, which are both positioned as a cap on their respective ligand binding pockets. As a consequence, helix I of CB1 is pushed outward approximately 7 Å relative to LPA1 and S1P1 by arm 2, opening a wider gap between helices I and VII than what was observed in LPA1 and S1P1 (Figure 4A). Moreover, helix II and ECL1 change their conformation in CB1, with helix II shifting 2 Å further from the binding pocket compared with LPA1 and S1P1, and ECL1 changing from a short helical region in LPA1 and S1P1 to a loop in CB1 (Figure 4B). These conformational changes effectively enlarge the binding pocket of CB1 allowing access to the reentrant N-terminal loop, and contribute to the extensive surface area and multiple sub-pockets associated with CB1. Finally, the ECL3 region of CB1 differs from that of its related receptors by a three helical turn extension of helix VII which increases the rigidity and presumably decreases the flexibility of this loop region in CB1 (Figure 4A).
The arrangement of Lys1923.28 in CB1 is unique when compared with its equivalent residue Arg3.28 in LPA1 and S1P1. In LPA1 and S1P1, Arg3.28 points into the binding pocket forming a strong interaction with the phosphate head group of the ligands (Figures 4D and E); it is stabilized by the negatively charged or polar residue 3.29 (Gln125 in LPA1 and Glu121 in S1P1). However, in CB1 the environment near Leu1933.29 and the ligand is hydrophobic, thus it is energetically favorable for the positively charged Lys1923.28 to point away from the binding pocket (Figure 4C). In fact, Lys1923.28 functions as a stabilization anchor by forming a salt bridge/hydrogen bond network, instead of directly interacting with the ligand as Arg3.28 in LPA1 and S1P1. Another major difference of the endogenous ligands of CB1 (anandamide, 2-AG), LPA1 (LPA) and S1P1 (S1P) is the head group. The heads of LPA1 and S1P1 ligands are negatively charged phosphate groups, while the heads of CB1 ligands are neutral. In fact, phosphorylation of the head group of CB1 ligands anandamide and 2-AG would transform them into ligands of LPA1 (Chrencik et al., 2015).
Binding Modes of Representative Antagonists to CB1
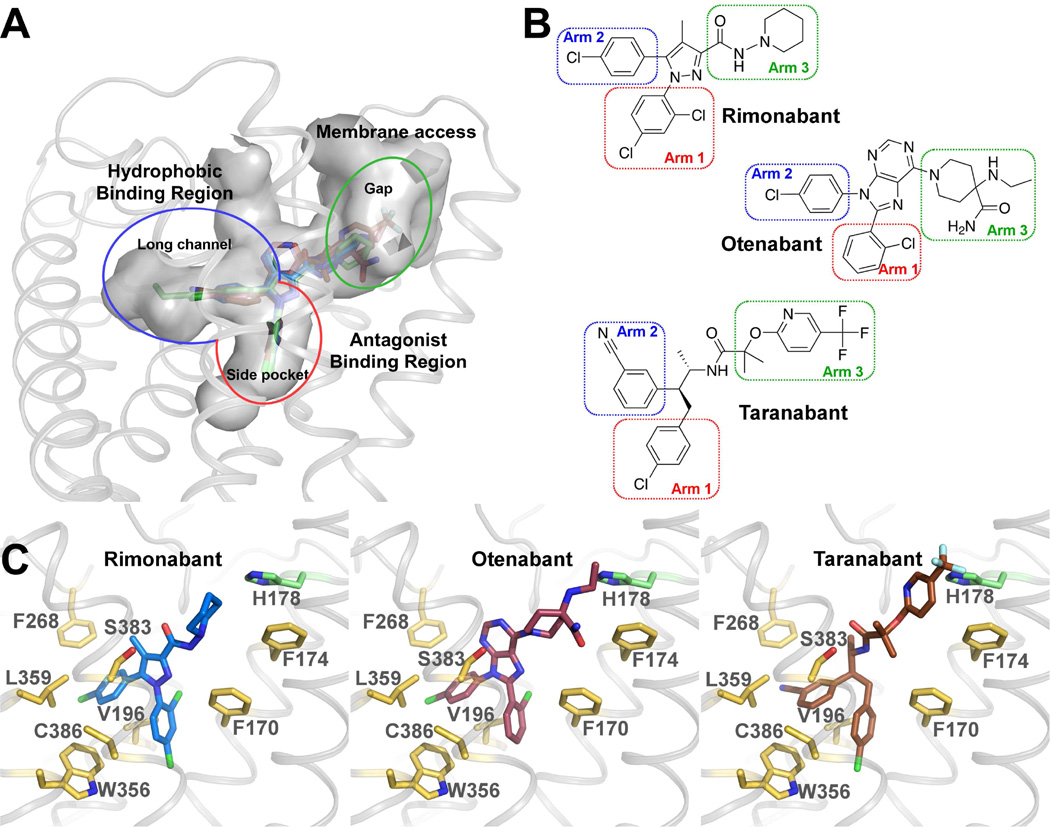
We performed docking of AM6538 and three CB1 antagonists: rimonabant, otenabant and taranabant (Figure 5, Table S2), which represent diverse scaffolds of CB1 antagonists used in clinical trials. For each compound, the top one ranked pose was used for analysis. The docking pose of AM6538 reproduces the crystallographic pose (Figure S4A), with a root mean square deviation (RMSD) of 0.55 Å. For the three antagonists, their 1st-ranked docking poses resemble that of AM6538 in the crystal structure, with three arms that fit into the three branches of the binding pocket, as described in the AM6538 binding mode (Figure 5A). As denoted for AM6538, the arms in the side pocket, long channel and gap are termed arm 1, arm 2 and arm 3 (Figure 5B). The scaffolds of arm 1 and arm 2 of the three antagonists are very similar to each other, and so are the docking poses. The biggest difference is from arm 3, yet they are all quite bulky, which we speculate is the signature for CB1 antagonists. Taranabant has the highest affinity with CB1 among the three ligands (Table S2). It does not have a rigid aromatic ring at the core, allowing its arm 2 and arm 3 to have more freedom to form stronger interactions with surrounding residues (Figure 5C). Mutagenesis of Phe1702.57Ala or Phe1742.61Ala results in dramatically reduced functional affinity for rimonabant and AM6538, while neither mutation alters the potency of the agonist, CP55,940. More conservative mutations to tryptophan (Phe1702.57Trp or Phe1742.61Trp) at either site have no appreciable effect on antagonist binding, further supporting the importance of the hydrophobic interactions at this site (Figures S4G and H). In addition, we performed 50 ns MD simulations to visually assess the predicted ligand-receptor interactions, starting from the docking poses. The RMSD values of AM6538, rimonabant and otenabant are about 1.4 Å. For taranabant, the value is larger (about 3 Å), in accordance with its lacking of the core aromatic ring (Figures S4C–F). The predicted interactions of the ligands are conserved during the short MD simulations. The central structure of AM6538 in MD simulation is closest to the docking pose.
Figure 5. Docking of Different Antagonists in the CB1 Crystal Structure.
(A) CB1 binding pockets with rimonabant (blue sticks), otenabant (raspberry sticks) and taranabant (brown sticks) are shown in gray surface representation. (B) Chemical structures of rimonabant, otenabant and taranabant. The red/blue/green rectangles highlight previously described arms termed as arm 1/arm 2/arm 3 (see Figure 3C). (C) Predicted binding modes of rimonabant (blue sticks), otenabant (raspberry sticks) and taranabant (brown sticks) with CB1. The interacting residues are shown in yellow sticks and H178 is shown in green sticks. See also Figure S4 and Table S2.
Docking Poses of Representative Agonists of CB1
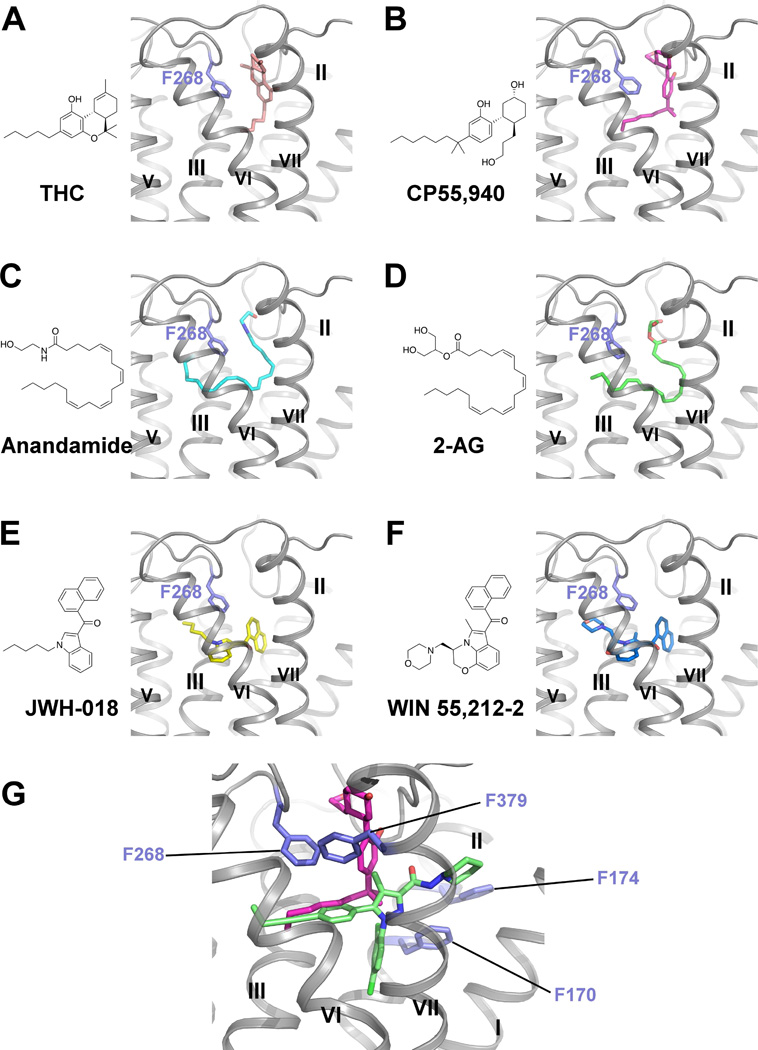
Although the crystal structure we present is in the inactive state, we are able to investigate how representative agonists likely bind to the orthosteric pocket of CB1 by integrating molecular docking, mutagenesis and SAR data (Ahn et al., 2009; Aung et al., 2000; Bertalovitz et al., 2010). Six CB1 agonists (Table S2): the classical cannabinoids, THC and CP55,940; the endogenous agonists, anandamide and 2-AG, and indole derivatives, JWH-018 and WIN 55,212-2 were selected for docking studies. These agonists mainly interact with ECL2, N-terminal loop, helices III, VI and VII, and they do not interact with helices I and II. Their predicted binding modes are shown in Figure 6. For THC and CP55,940, the rings reside between the N-terminal loop and ECL2, forming π-π interactions with Phe268ECL2 (Figures 6A and B), and the carbon chains extend into the long channel and interact with residues from helices III, VI and VII. CP55,940 does not interact with helices I and II, which is supported by our mutagenesis studies: mutations on Phe170 and Phe174 (Phe1702.57Ala/Trp or Phe1742.61Ala/Trp) do not alter the potency of CP55,940 (Figure S4G). Anandamide and 2-AG were predicted to adopt a C-shaped conformation and occupy a similar space as THC (Figures 6C and D). Their hydrophilic heads are sandwiched between the N-terminal loop and ECL2, and their long aliphatic tails extend deeper into the long channel. JWH-018 and WIN 55,212-2, however, are predicted to bind deeper in the pocket than THC (Figures 6E and F). Both the indole rings and naphthalene rings form π-π interactions with Phe268ECL2. The N-substituents reach the end of the long channel and interact with helix V. The binding mode of JWH-018 and WIN55,212-2 is supported by mutations on helix V (McAllister et al., 2003; Song et al., 1999) and SAR study of N-alkyl chain length (Aung et al., 2000). Notably, all of the agonists interact with Phe268ECL2 and Phe3797.35 in our docking poses, which is consistent with mutagenesis studies on ECL2 (Ahn et al., 2009) and Phe3797.35 (Figure S4I). Any of the following individual mutations: Phe268ECL2Trp, Pro269ECL2Ala, His270ECL2Ala, Ile271ECL2Ala, or breaking of the disulfide bond Cys257-Cys264 on ECL2, dramatically decreased the binding of all three different types of agonists, yet had little impact on antagonist binding (Ahn et al., 2009). The prominent role of Phe3797.35 to facilitate CP55,940 functional affinity is supported by the loss of CP55,940 potency with the Phe3797.35 Trp mutation and an even greater loss of agonist activity with the Phe3797.35Ala mutation (Figure S4I). While these mutations dramatically affected CP55,940 agonism, they had no impact on antagonist/inverse agonist (AM6538 or rimonabant ) displacement of agonist (Figure S4J), further supporting the predicted binding pose of CP55,940 (Figure 6G).
Figure 6. Docking Poses of Different Cannabinoid Receptor Agonists.
(A–F) Chemical structures and predicted binding poses of THC (pink sticks) (A), CP55,940 (magenta sticks) (B), Anandamide (cyan sticks) (C), 2-AG (green sticks) (D), JWH-018 (yellow sticks) (E), WIN 55,212-2 (blue sticks) (F). (G) Zoom-in view of predicted CP55,940 binding pose. CP55,940 is shown in magenta sticks, AM6538 is shown in green sticks and the key residues are shown in slate sticks. See also Figure S4 and Table S2.
Discussion
The ligand used in this study, AM6538, was designed with the aim of stabilizing the ligand-CB1 receptor complex and promoting CB1 crystal formation. For this purpose, our approach focused on the use of the substituted biarylpyrazole chemotype based on the structure of rimonabant for obtaining highly utilized proprietary probes. Within this class of compounds, a slight modification of the chemotype led to AM251, a commonly used CB1 inverse agonist/antagonist (Lan et al., 1999) and AM281, a CB1 antagonist whose radiolabeling produced the first in vivo imaging agent for labeling CB1 in nonhuman primates and humans (Berding et al., 2004; Gatley et al., 1998). AM6538 acts as a CB1 stabilizing antagonist and the ligand was effective in allowing structural determination of CB1. AM6538 reacts as an intact molecule with no crystallographic evidence of covalent binding while at the same time not revealing the location of the terminal nitrate group in the X-ray structure. While radioligand binding studies demonstrate that AM6538 binds tightly to the receptor, the precise mode of action for stabilizing the receptor remains to be determined.
To date, there remains considerable controversy with regards to CB1 ligands and their diverse medical applications. This is likely due in part to the wide availability and illicit nature of the most famous CB1 pharmaceutical, marijuana. Marijuana has been widely used across many cultures to treat multiple conditions, with most of the results relayed via oral tradition, anecdote, political position, or with economic interest preventing an objective interpretation of therapeutic in any particular disease state (Whiting et al., 2015). The medicinal marijuana movement continues to gain support, and clinical trials with well-defined endpoints will continue to educate the medical and pharmaceutical communities regarding the relative benefits and drawbacks of targeting this physiological system. The crystal structure of CB1 in complex with AM6538 reveals an expansive and complicated binding pocket network consisting of multiple sub-pockets and channels to various regions of the receptor. The three-arm ligand structure is common to CB1 antagonists and inverse agonists and may be critical for stabilizing the inherent flexibility of the native receptor in a non-signaling conformation. Combining the 3D structure of CB1 and molecular docking of the three representative antagonists, which act as inverse agonists, rimonabant, otenabant and taranabant, the role of each arm is clearly illustrated. Arm 1 is crucial for high affinity binding, while arm 2 extends into the long channel. An aliphatic or aromatic ring on arm 3 pushes on helices I, II, causing them to bend outward, and potentially modulating the pharmacological signaling state of the receptor. Together with structure and modeling data, we speculate that a bulky ring on arm 3 is essential for CB1 antagonism. This observation provides direction for designing more diverse compounds as we have learned that variable chemical groups are tolerated at the core of arm 3, a long carbon chain can be added at the para-position of the phenyl ring in arm 2. For example, introduction of a 4-cyanobut-1-ynyl at arm 2 produces AM6545 (Table S2), a high affinity CB1 neutral antagonist (Tam et al., 2010).
Understanding the nuances of CB1 binding and activation is important, as human use has noted differences between the phytocannabinoid agonist THC and the synthetic cannabinoid constituents of “Spice” or “K-2” such as JWH-018. In general, cannabinoid agonists are routinely abused substances; yet, while overdose of THC/marijuana has not been documented, there have been cases of severe and even deadly responses to the ingestion of such synthetic mixtures resulting in federal restrictions by many countries, including the US. It remains unclear as to why THC can have such a high safety margin, while the synthetic cannabinoid constituents can prove toxic with varying severities of serious side effects (Hermanns-Clausen et al., 2013). Going forward, the study of cannabinoids present in Cannabis sativa will provide clues to its high efficacy and safety margins, and may continue to inspire a rich source of pharmacologically refined compounds and novel therapeutics; the utility of the crystal structure may provide inspiration for drug design toward refining efficacy and avoiding adverse events.
STAR*METHODS
Detailed methods are provided in online version of this paper and include the following:
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
- EXPERIMENTAL MODEL AND SUBJECT DETAILS
- Mice
- METHODS DETAILS
- Synthesis and Characterization of AM6538
- Rational Design of Thermostabilizing Mutations of CB1
- Protein Engineering for Structure Determination
- Protein Expression in Mammalian Expression System
- Protein Purification
- Lipidic Cubic Phase Crystallization of CB1-AM6538 Complex
- Data Collection and Structure Determination
- Quantum Mechanical Optimization of AM6538
- Docking Simulations of CB1 Ligands
- Molecular Dynamics Simulation of CB1 in Complex with AM6538 and Representative Antagonists
- Radioligand Binding Assay
- WT and Mutant hCB1-CHO Cell Line Generation for Functional Studies
- Functional Analysis Studies
- Protein Stability Assays
- Affinity Mass Spectrometry Analysis of AM6538
QUANTIFICATION AND STATISTICAL ANALYSIS
- DATA AND SOFTWARE AVAILABILITY
- Data Resources
METHODS AND RESOURCES
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Raymond C. Stevens (stevens@shanghaitech.edu.cn).
EXPERIMENTAL MODEL AND RESOURCE SHARING
Mice
Experimentally naïve mice (C57BL/6J) were purchased from Jackson Labs at 9 weeks of age and were housed in groups of five per cage in a specific-pathogen free facility, under a 12-h light-dark cycle with food and water ad libitum. At ~16 weeks of age, mice were euthanized by cervical dislocation prior to cerebellum dissection. A total of four mice were used to generate 4 independent experiments for assessing [35S]GTPγS binding. All experiments were performed with the approval of the Institutional Animal Care and Use Committee of The Scripps Research Institute, Jupiter, FL and in accordance to NIH guidelines.
METHOD DETAILS
Synthesis and Characterization of AM6538
All reagents and solvents used for chemical synthesis were purchased from Sigma-Aldrich, TCI Chemicals, Fisher Scientific, Acros or Alfa Aesar. The palladium catalysts were purchased from Sigma-Aldrich or TCI Chemicals. 1H NMR (500 MHz) and 13C spectra (126 MHz) were recorded on a Varian Inova spectrometer. Chemical shifts (δ) are reported in parts per million and are referenced to CDCl3 for 7.26 or 77.7. Multiplicities are indicated as br (broadened), s (singlet), d (doublet), t (triplet) or m (multiplet). Coupling constants (J) are reported in hertz (Hz). Thin layer chromatography (TLC) was performed on Merck-Millipore 210 - 270 µm TLC silica gel plates, (60 Å) and coated with a F254 fluorescent indicator. Flash column chromatography was performed on a Biotage Isolera Spektra system with UV collections at 254 and 280 nm using Luknova flash columns preloaded with normal phase silica gel (50 µm). All moisture sensitive reactions were performed under an atmosphere of high-purity argon while using oven-dried glassware. The intermediates and final compounds were characterized using a combination of 1H NMR, 13C NMR and LC/MS techniques. The LC/MS analysis was performed using a Waters MicroMass ZQ system (electrospray ionization mode) equipped with a Waters 2525 binary gradient module, a Waters 2996 photodiode array detector, a Waters 2424 ELS detector, two Waters 515 HPLC pump, a fluidics organizer and a pump control module II. Compounds were analyzed with gradient elution using acetonitrile/water as the mobile phase and an XTerra MS C18 or an XTerra MS C8, 4.6 mm × 50 mm column (5 µm). Melting-points were recorded on a Fisher Scientific apparatus. IR spectra were obtained on a PerkinElmer Spectrum One FT-IR spectrometer. Elemental analyses were performed using a PerkinElmer Series II 2400 CHNS analyzer.
4-(4-(1-(2,4-dichlorophenyl)-4-methyl-3-(piperidin-1-ylcarbamoyl)-1H-pyrazol-5-yl)phenyl)but-3-yn-1-yl methanesulfonate (2)
To a stirred solution of 1-(2,4-dichlorophenyl)-5-(4-iodophenyl)-4-methyl-N-(piperidin-1-yl)-1H–pyrazole-3-carboxamide (Lan et al., 1999) (1, 1.1 g, 2 mmol) in DMF (30 ml), under argon was added but-3-yn-1-yl methanesulfonate (Tang and Prusov, 2012) (888 mg, 6 mmol), and Hünig’s base (1.9 g, 20 mmol). The reaction mixture was degassed by introducing a steady stream of argon into the solution for 5 min and to this was added tetrakis(triphenylphosphine)palladium (0) (231 mg, 10 mol%) and CuI (76 mg, 20 mol%). The resulting mixture was stirred for 3 h at room temperature. The solvent from the reaction mixture was removed in vacuo at 70 °C and the residue was dissolved in dichloromethane (100 ml) and washed with deionized water (2×~50 mL). The organic layer was separated, dried over anhydrous MgSO4, filtered and the filtrate was removed in vacuo. The residue obtained was purified by flash column chromatography on silica gel (n-hexane/AcOEt = 1/1) to provide 2 as a white solid (511 mg, 45 %); m.p 176–178 °C; 1H NMR (500 MHz, CDCl3-d) δ 7.64 (br. s., 1H, NH), 7.41 (d, J = 1.46 Hz, 1H, ArH), 7.34 (d, J = 8.30 Hz, 2H, ArH), 7.25 - 7.31 (m, 2H, ArH), 7.05 (d, J = 8.30 Hz, 2H, ArH), 4.37 (t, J = 6.84 Hz, 2H, -O-CH2-), 3.07 (s, 3H, -S(O)2-CH3), 2.79 - 2.97 (m, 6H, -CH2-CH2- and -N(CH2CH2)2CH2, 2.37 (s, 3H, HetAr-CH3), 1.75 (m, 4H, -N(CH2CH2)2CH2, 1.43 (br. s., 2H, -N(CH2CH2)2CH2; 13C NMR (126 MHz, CDCl3-d) δ 160.2, 144.7, 143.6, 136.2, 136.1, 133.2, 132.0, 130.8, 130.5, 129.7, 128.8, 128.1, 123.5, 118.5, 85.8, 82.5, 67.4, 57.3, 38.0, 25.7, 23.6, 21.0, 9.6; ES m/z 575.1710 (M++H).
1-(2,4-dichlorophenyl)-5-(4-(4-iodobut-1-ynyl)phenyl)-4-methylN(piperidin-1-yl)-1H-pyrazole-3-carboxamide (3)
To a stirred solution of 2 (500 mg, 0.86 mmol) in anhydrous acetone (50 ml), under argon was added sodium iodide (1.3 g, 8.6 mmol) and tetrabutylammonium iodide (64 g, 0.17 mmol). The resulting reaction mixture was refluxed overnight. The solvent from the reaction mixture was removed in vacuo and the residue was dissolved in dichloromethane (75 ml) and washed with deionized water (2×~50 mL). The organic layer was separated, dried over anhydrous MgSO4, filtered and the filtrate was removed in vacuo. The residue obtained was purified by flash column chromatography on silica gel (n-hexane/AcOEt = 7/3) to provide 3 as a white solid (326 mg, 62 %); m.p 171–173 °C; 1H NMR (500 MHz, CDCl3-d) δ 7.65 (s, 1H, NH), 7.43 (d, J = 1.47 Hz, 1H, ArH), 7.38 (d, J = 7.81 Hz, 2H, ArH), 7.27 - 7.33 (m, 2H, ArH), 7.07 (d, J = 8.30 Hz, 2H, ArH), 3.31 (t, J = 7.32 Hz, 2H, I-CH2), 3.01 (t, J = 7.32 Hz, 2H, -CH2-CH2), 2.88 (m, 4H, -N(CH2CH2)2CH2, 2.39 (s, 3H, HetAr-CH3), 1.68 - 1.87 (m, 4H, -N(CH2CH2)2CH2, 1.45 (m, 2H, -N(CH2CH2)2CH2; 13C NMR (125 MHz, CDCl3-d) δ 160.2, 144.7, 143.7, 136.3, 136.1, 133.2, 132.0, 130.8, 130.6, 129.6, 128.6, 128.1, 123.9, 118.5, 90.3, 81.9, 57.3, 25.7, 24.8, 23.6, 9.6, 1.6; IR (neat) 3308, 2929, 2851, 2787, 2509, 2160, 2031, 1689, 1524, 1486, 1245, 967, 967, 842, 781 cm−1; ES m/z 607.1102 (M++H).
4-(4-(1-(2,4-dichlorophenyl)-4-methyl-3-(piperidin-1-ylcarbamoyl)-1H-pyrazol-5-yl)phenyl)but-3-yn-1-yl nitrate (4, AM6538)
To a stirred solution of 3 (300 mg, 0.5 mmol) taken in anhydrous acetonitrile (30 ml), under argon was added silver nitrate (100 mg, 0.6 mmol). The resulting reaction mixture was heated to 70 °C for 1 h. The contents were cooled to room temperature and the solids were filtered over a celite pad. The filtrate was removed in vacuo and the residue was dissolved in dichloromethane (75 ml) and washed with deionized water (2×~50 mL). The organic layer was separated, dried over anhydrous MgSO4, filtered and the filtrate was removed in vacuo. The residue obtained was purified by flash column chromatography on silica gel (n-hexane/AcOEt = 6/4) to provide 4 (AM6538) (Makriyannis and Vemuri, 2016) as a white solid (173 mg, 65 %); m.p 178–180 °C; 1H NMR (500 MHz, CDCl3-d) δ 7.63 (s, 1H, NH), 7.41 (d, J = 1.95 Hz, 1H, ArH), 7.34 (d, J = 8.30 Hz, 2H, ArH), 7.24 - 7.32 (m, 2H, ArH), 7.05 (d, J = 8.30 Hz, 2H, ArH), 4.62 (t, J = 6.59 Hz, 2H, -O-CH2), 2.81 - 2.91 (m, 6H, -CH2-CH2 and -N(CH2CH2)2CH2), 2.37 (s, 3H, HetAr-CH3), 1.75 (m, 4H, -N(CH2CH2)2CH2, 1.43 (m, 2H, -N(CH2CH2)2CH2; 13C NMR (125 MHz, CDCl3-d) δ 160.2, 144.7, 143.6, 136.2, 136.1, 133.2, 132.0, 130.8, 130.5, 129.7, 128.8, 128.1, 123.5, 118.5, 85.3, 82.4, 70.4, 57.3, 25.7, 23.6, 18.9, 9.6; IR (neat) 3319, 2935, 2857, 2811, 2502, 2160, 2031, 1976, 1682, 1616, 1528, 1488, 1277, 968, 881, 824, 781 cm−1; ES m/z 542.1627 (M++H); Elemental analysis calculated for C26H25Cl2N5O4: C% 57.57, H% 4.65, N% 12.91; found: C% 57.34, H% 4.46, N% 12.74.
Rational Design of Thermostabilizing Mutations of CB1
To improve general stability and homogeneity of the detergent-stabilized CB1, mutations were rationally designed. A 3D homology model of human CB1 was constructed and refined with ICM (Abagyan and Totrov, 1994) using the X-ray structure of sphingosine 1-phosphate receptor 1 (S1P1 receptor, PDB ID: 3V2W) (Hanson et al., 2012) as a template. Best scoring substitutions were visually inspected and evaluated from the evolutionary conservation perspective resulting in a list of proposed substitutions. The predicted substitutions were analyzed for improvement of the receptor monodispersity (as evidenced by SEC traces) and thermal stability (as evidenced by increase in Tm in the CPM assay(Alexandrov et al., 2008).
Protein Engineering for Structure Determination
The sequence of the human CB1 gene was synthesized by GenScript. The Flavodoxin (PDB ID: 1I1O, MW 14.9kDa, with Y98W mutation) fusion protein was fused to the third intracellular loop of the human CB1 gene, using overlapping PCR. The construct has truncations of the CB1 residues 1–98, 307–331 and 415–472. The resulting CB1-Flavodoxin chimera sequence was subcloned into a modified mammalian expression pTT5 vector, which contains a haemagglutinin (HA) signal sequence, a FLAG tag and 10 × His tag, followed by a tobacco etch virus (TEV) protease cleavage site, before the N-terminus of the chimera sequence. The CB1 gene was further modified by introducing four rationally designed mutations, Thr2103.46Ala (D’Antona et al., 2006), Glu2735.37Lys, Thr2835.47Val and Arg3406.32Glu, using standard QuickChange PCR.
Protein Expression in Mammalian Expression System
HEK293F cells (Invitrogen) were grown in suspension starting from the densities at 0.2–0.3 × 106 in a humidified incubator with 5% CO2 at 37 °C with 130rpm. Passage cells when the cell density reaches to 1.6–1.8 × 106 cells/ml, about every 2–3 days. CB1-Flavodoxin construct was transfected and expressed in HEK293F cells (Invitrogen) (Passage number is 12–20) using the FreeStyleTM 293 Expression system (Invitrogen). Briefly, HEK293F cells were seeded on day 0 at 6×105 cells/ml in freeStyle 293 expression medium (Invitrogen). On day 2 the transduction was performed at a cell density of 1.0 to 1.2×106 cells/ml and the cell viability over 95% using PEI-DNA complexes. Approximately 48 hours post-transfection, cells were harvested by centrifugation at 400 g for 20 min at 4 °C.
Protein Purification
Frozen cell pellets were thawed and lysed by repeated washing and centrifugation in the hypotonic buffer of 10 mM HEPES (pH 7.5), 10 mM MgCl2, 20 mM KCl, and the high osmotic buffer of 10 mM HEPES (pH 7.5), 1.0 M NaCl, 10 mM MgCl2, 20 mM KCl, with EDTA-free complete protease inhibitor cocktail tablets (Roche). The washed membranes were suspended in hypotonic buffer with 30% glycerol and flash-frozen with liquid nitrogen and stored at −80 °C until further use. Purified membranes were thawed at room temperature and incubated with 20 µM AM6538 and inhibitor cocktail at 4 °C for 3 h. The membranes were further incubated with 1.0mg/ml iodoacetamide (Sigma) for 1 h. The membranes were solubilized in the buffer containing 50 mM HEPES (pH 7.5), 500 mM NaCl, 1% (w/v) n-dodecyl-beta-D-maltopyranoside (DDM, Anatrace) and 0.2% (w/v) cholesterol hemisucinate (CHS, Sigma-Aldrich) at 4 °C for 2.5–3 h. The supernatants containing the solubilized CB1 proteins were isolated by high-speed centrifugation, and then incubated with TALON IMAC resin (Clontech) and 20 mM imidazole, at 4 °C overnight. The resin was washed with 15 column volumes of washing buffer I containing 25 mM HEPES (pH 7.5), 500 mM NaCl, 10% (v/v) glycerol, 0.05% (w/v) DDM, 0.01% (w/v) CHS, 30 mM imidazole and 20 µM AM6538, and 5 column volumes of washing buffer II containing 25 mM HEPES (pH 7.5), 500 mM NaCl, 10% (v/v) glycerol, 0.05% (w/v) DDM, 0.01% (w/v) CHS, 50 mM imidazole and 20 µM AM6538. The proteins were eluted by 2.5 column volumes of eluting buffer containing 25 mM HEPES (pH 7.5), 500 mM NaCl, 10% (v/v) glycerol, 0.02% (w/v) DDM, 0.004% (w/v) CHS, 250 mM imidazole and 10 µM AM6538. PD MiniTrap G-25 column (GE Healthcare) was used to remove imidazole. The protein was treated overnight with TEV protease to cleave the N-terminal FLAG/His tags from the proteins. Finally, the purified protein was concentrated to about 50mg/ml with a 100 kDa cutoff concentrator (Sartorius) and used in crystallization trials. The protein yield and monodispersity were tested by analytical size exclusion chromatography (aSEC).
Lipidic Cubic Phase Crystallization of CB1-AM6538 Complex
For the initial crystallization setup, purified CB1-Flavodoxin in complex with AM6538 was reconstituted into lipidic cubic phase (LCP) by mixing with molten lipid (90% (w/v) monoolein and 10% (w/v) cholesterol) at a protein/lipid ratio of 2:3 (v/v) using a mechanical syringe mixer (Caffrey and Cherezov, 2009). LCP crystallization trials were performed using an NT8-LCP crystallization robot (Formulatrix). 96-well glass sandwich plates were incubated and imaged at 20 °C using an automatic incubator/imager (RockImager 1000, Formulatrix). The crystals grew in the condition of 0.1M HEPES pH 7.0–7.4, 100mM (NH4)2HPO4, 25%-32% PEG400, 2–20 mM Ethylenediaminetetraacetic acid disodium salt dehydrate (EDTA) and grew to the full size after 2 wk. The crystals were harvested using micromounts (MiTeGen) and flash-frozen in liquid nitrogen.
Data Collection and Structure Determination
X-ray diffraction data were collected at the Spring-8 beam line 41XU, Hyogo, Japan, using a Pilatus3 6M detector (X-ray wavelength 1.0000 Å). A rastering and data-collection strategy was followed as previously described (Cherezov et al., 2009; Hanson et al., 2012). Data were integrated and scaled using XDS (Kabsch, 2010) and merged using SCALA (Collaborative Computational Project, 1994). Initial phase information was obtained by molecular replacement (MR) with Phaser (McCoy et al., 2007) using the receptor portion of LPA1 (PDB ID: 4Z34) and Flavodoxin structure (PDB ID: 1I1O) as search models. Refinement was performed with Phenix (Adams et al., 2010) and Buster (Smart et al., 2012) followed by manual examination and rebuilding of the refined coordinates in the program COOT (Emsley et al., 2010) using both |2Fo|-|Fc| and |Fo|-|Fc| maps.
Quantum Mechanical Optimization of AM6538
Optimizing the geometry of AM6538 by quantum-mechanical energy minimization was performed using Jaguar 9.0 (Schrödinger, 2015a). Density functional theory (DFT) with B3LYP functional and basis set 6-31G**++ was used. The conformation in the crystal structure was used as starting point.
Docking Simulations of CB1 Ligands
Prediction of ligand binding to CB1 was done with Schrodinger Suite 2015-4. Processing of the protein structure was performed with the Protein Preparation Wizard. Converting of ligands from 2D to 3D structures was performed using LigPrep. Molecular docking was performed by two different methods: for antagonists, rigid protein docking in extra precision was used with Glide 6.9 (Friesner et al., 2004; Friesner et al., 2006; Halgren et al., 2004; Schrödinger, 2015a); for agonists, Induced Fit Docking (Felder and Schrott-Fischer, 1995; Schrödinger, 2015b; Sherman et al., 2006), allowing optimization of residues within 5.0Å, in extra precision, was used.
Molecular Dynamics Simulation of CB1 in Complex with AM6538 and Representative antagonists
Molecular dynamics simulation was performed using GROMACS 5.1.2 (Abraham et al., 2015), using force field CHARMM27 (Feller and MacKerell, 2000; MacKerell et al., 1998; Mackerell et al., 2004). Crystal structures of CB1 with AM6538 or inverse agonists in the binding pocket (binding mode predicted by molecular docking) was embedded into a pre-equilibrated POPC (1-palmytoil-2-oleoyl-sn-glycero-3-phosphatidylcholine) lipid bilayer following an online protocol (Kandasamy and Larson, 2006). The topology files of ligands and POPC molecules were generated using online server SwissParam (Zoete et al., 2011). The systems were solvated with water, and chloride ions were added to neutralize the system. Molecular Dynamics simulations were performed in the NPT ensemble, at temperature of 310K and pressure of 1 atm using semi-isotropic coupling. First, each system was balanced position-restrained MD for 10 ns (total energy was stable). Then 50 ns MD simulations with no position restraints were performed to each system for three independent runs, and these trajectories are used for analysis. Ligand RMSD and RMSF values were calculated with non-hydrogen protein atoms superimposed to the starting structure. The central structure was the structure with the smallest RMSD of all protein and ligand atoms from all three trajectories, followed by energy minimization.
Radioligand Binding Assay
Stably transfected, hCB1 HEK-293F cell lines expressing the WT receptor were used for saturation and competition binding assays using [3H]-CP55,940 (0.79 nM, specific activity: 88.3 Ci/mmol, Perkin Elmer) with unlabeled CP55,940 (30 µM) used for nonspecific binding determination. Binding assays were performed at 37 °C for 1 hour in the presence of 25 mg protein per well prior to collection of membranes by rapid filtration, washing and scintillation facilitated detection of tritium retained on the membranes according to standard procedures (Janero et al., 2015).Saturation binding assays and subsequent nonlinear hyperbolic curve fitting analysis (Graphpad Prism 6.0) revealed a Bmax of 10.2 ± 2.6 pmol/mg and Kd = 5.6 ± 2.3 nM. Assays were performed in the same manner for determination of affinity for the crystallization CB1 construct wherein [3H]-CP55,940 (revealed a Bmax of 47.2 ±13.1 pmol/mg and Kd = 37.8 ± 13.9 nM). For wash out experiments at the WT CB1, membranes were incubated at 37 °C for 1 h in the presence of vehicle (buffer with 1% DMSO), 100 nM rimonabant or 50 nM AM6538. The membranes were then resuspended in assay buffer containing 1% BSA followed by centrifugation 2 times and then washed and collected a third time in assay buffer alone. These membranes were then subject to the saturation binding assay as described above in this section.
WT and Mutant hCB1-CHO Cell Line Generation for Functional Studies
The 3×HA (haemagglutinin)-N-terminus tagged hCB1 receptor cDNA was obtained from cDNA.org and subcloned into a murine stem cell virus for cell line transduction (pMSCV-puro, Clontech). Point mutations were introduced to the N terminus-3xHA-tagged human CB1 receptor cDNA in MSCV retroviral vector by using Q5® Site-Directed Mutagenesis kit (New England Biolabs) to create mutant CB1 plasmids (F170A, F170W, F174A, F174W, F379A and F379W). The primers used to make the mutations are given in the Key Resources table. The WT and mutant CB1 constructs were then packaged into retrovirus via Phoenix package system, and the produced retroviruses were applied to CHO-K1 cells for gene transduction. Stable cell lines were generated following selection with antibiotic (puromycin) selection. Cells were passaged onto 384-well plates for pharmacological characterization. Cells were maintained in DMEM/F-12 media supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 5 µg/mL puromycin (Invitrogen, Waltham, MA).
FUNCTIONAL ANALYSIS STUDIES
CISBIO® cAMP HTRF Assay
Inhibition of forskolin-stimulated cAMP was determined using the CISBIO® cAMP Homogenous Time-Resolved Fluorescence resonance energy transfer (FRET) (HTRF) HiRange assay according to the manufacturer’s instructions (Cisbio Assays, Bedford, MA). Cells (5,000 cells/well in low-volume 384 well plates) were cotreated at room temperature for 30 min with 25 µM RO-20-1724 and 20 µM forskolin (Sigma-Aldrich), and hCB1R ligands at concentrations ranging from 0.03 – 10,000 nM. Cells were then incubated with cAMP-d2 antibody in assay diluent and cryptate solution in lysis buffer for 60 min at room temperature (Cisbio Assays). Fluorescence was measured at 620/665 nm using a Perkin-Elmer EnVision™ plate reader (Waltham, MA). FRET was calculated as fluorescence at 665 nm/620 nm. Basal cAMP levels were determined from cells incubated in the absence of RO-20-1724, forskolin, and ligand. cAMP levels were determined in cells incubated with RO-20-1724 and forskolin. Compounds were dissolved in DMSO in PBS and diluted to final solvent concentrations of 1% at the times and concentrations indicated. Agonist properties in the assays used for the WT cells used competitive analysis were as follows: EC50= 7.9 ± 2.0 nM, Emax= 3.21 ± 0.39 (fold over baseline, set as 100% CP55,940); THC: EC50= 160.1 ± 26.0 nM; Emax= 1.36 ± 0.05 (fold over baseline, set as 100% THC). N≥3 individual experiments performed in duplicate presented as mean ± SEM.
DiscoveRx® βArrestin2 Assay
βArrestin2 recruitment was determined using the CHO-hCB1R PathHunter assay (DiscoveRx®) according to the manufacturer’s instructions following a 1.5 h incubation at 37°C with the indicated concentrations of ligands. All experiments included a vehicle control. Agonist properties in the cell lines were as follows: CP55,940: EC50= 23.8 ± 4.6 nM, Emax= 4.03 ± 0.19 (fold over baseline, set as 100% CP55,940); THC: EC50= 89.2 ± 38.9 nM; Emax= 2.24 ± 0.16 (fold over baseline, set as 100% THC). N≥3 individual experiments performed in duplicate presented as mean ± S. E. M.
[35S]GTPγS Binding to CB1 in C57 Mouse Cerebellum Membranes
G protein-coupling in mouse brain was measured using a previously published protocol (Bohn et al., 2015; Janero et al., 2015). The cerebellum from C57BL/6 mice (4–7 months old) were homogenized (glass on glass) in homogenization buffer (10 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM DTT). The homogenate was passed through a 26-gauge needle, centrifuged twice at 20,000 x g for 30 min at 4 °C, and resuspended in ice-cold assay buffer (50 mM Tris-HCl pH 7.4, 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA and 2 µM GDP, 1 mM DTT). For each reaction, 2.5 µg of membrane protein was incubated in assay buffer containing ~ 0.1 nM [35S]GTPγS and increasing concentrations of test compound in a total volume of 200 µL for 2 h at room temperature. Test compounds (CP55,904, rimonabant, AM6538, and THC) were first diluted through serial dilutions in DMSO, then with assay buffer to a final DMSO concentration of 1%. Reactions were terminated by filtering membrane-bound and free [35S]GTPγS through GF/B filters using a 96-well plate harvester (Brandel Inc., Gaithersburg, MD) and rinsing with ice-cold dH2O. Filters were dried overnight, and radioactivity was determined with a microplate scintillation counter. For experiments used to analyze drug competition, agonists and antagonists were incubated with the membranes simultaneously. Agonist properties in the mouse cerebellum: CP55,940: EC50= 63.0 ± 5.5 nM, Emax= 2.40 ± 0.04 (fold over baseline, set as 100% CP55,940); N=4 individual mouse cerebella preparations performed in duplicate presented as mean ± S. E. M.
Protein Stability Assays
Protein thermostability was tested by a microscale fluorescent thermal stability assay using the thiol-specific fluorochrome N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM), which reacts with the native cysteines embedded in the protein interior as a sensor for the overall integrity of the folded state. The CPM dye (Invitrogen) was dissolved in DMSO at 4 mg/ml and stored at −80 °C. Prior to use the dye stock was diluted 1:20 in buffer 25 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 0.05% (w/v) DDM and 0.01% (w/v) CHS. The tested protein (~5 µg) was diluted in the same buffer to a final volume of 120 µl. 1 µl of the diluted dye was added and thoroughly mixed with protein. The reaction mixture was incubated at room temperature for 15 min, and subsequently transferred to a sub-micro quartz fluorimeter cuvette (Starna Cells, Inc.) and heated in a controlled way with a ramp rate of 1 °C/min over a temperature range from 20–90 °C in a Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies). The excitation wavelength was set at 387 nm, while the emission wavelength was 463 nm. Protein homogeneity was also tested by analytical size-exclusion chromatography (aSEC) using a 1260 Infinity HPLC system (Agilent Technologies).
Affinity Mass Spectrometry Analysis of AM6538
The CB1 protein co-purified with AM6538 was first desalted using PD MiniTrap G-25 column cartridge to deplete free ligands. Then the protein complex sample (~1 µg) was filtered through 30 kDa MW cutoff ultrafiltration membrane (Sartorius, Germany) by centrifugation at 13,000 g for 10 min at 4 °C in the buffer containing 150 mM ammonium acetate, 0.02% (w/v) DDM and 0.004% (w/v) CHS. The protein complexes retained on the ultrafiltration membrane was transferred to a new centrifugal tube. The ligands were dissociated from the complexes with 90% methanol and separated from the denatured protein by centrifugation at 13,000 g for 20 min at 25 °C. The supernatant was dried out in speed vacuum, reconstituted in 50% methanol, diluted by 50-fold prior to LC-MS analysis using Agilent 6230 TOF equipped with an Agilent 1260 HPLC system. The compound was eluted with 95% methanol/0.1% formic acid from a Hypersil GOLD C18 column (2.1 mm × 100 mm, 3 µm, Thermo Fisher Scientific, USA) at a flow rate of 0.4 mL/min. Full-scan mass spectra were acquired in the range of 100–1000 m/z on Agilent 6230 TOF with major ESI source settings: voltage 3000 V, gas temperature 350 °C, fragmentor 100 V.
QUANTIFICATION AND STATISTICAL ANALYSIS
Concentration-response curves for cAMP and β-arrestin2 are presented as % of CP55,940 or THC at 1 µM, as indicated. Concentration-response curves were fit to a non-linear regression (three parameter) model to determine EC50 and Emax, or a Gaddum/Schild EC50 shift global nonlinear regression model in Prism (v. 6.0, GraphPad Software Inc., San Diego, CA), as indicated. In order to best fit data to the Gaddum/Schild EC50 shift global non-linear regression, pEC50, pA2, Hill slope, Emax, and Emin were shared for all data sets. Schild slope was constrained to unity after determining a competitive antagonism model (i.e. Schild slope = 1) was the preferred model for these data.
DATA AND SOFTWARE AVAILABILITY
Data Resources
Coordinates and structures factors have been deposited in the Protein Data Bank for CB1-AM6538 (PDB: 5TGZ).
Supplementary Material
Rimonabant and AM6538 are Competitive Antagonists of CP55,940 and THC. Related to Figure 2. Rimonabant (left) and AM6538 (right) competitively antagonize CP55,940 (top) and THC (bottom)-induced (A) Inhibition of forskolin-stimulated cAMP accumulation in hCB1-CHO cells and (B) β-arrestin2-recruitment in hCB1-CHO DiscoveRx PathHunter cells. (C) Rimonabant (left) and AM6538 (right) competitively antagonize CP55,940-stimulated [35S]-GTPγS binding in mouse cerebellum membrane preparations. Data are presented as the mean ± S. E. M. of 3 or more assays run in duplicate or triplicate. Competitive nonlinear analysis was performed in GraphPad Prism 6.0 and parameters are included in Table S1. Characterization of agonist effects in the cells and cerebellum are provided in the supplemental methods.
Stability Assay of CB1 Mutants. Related to Figure 1. (A, B) Analytical size exclusion chromatography (aSEC) of CB1 mutants testing the effects of mutations and mutation combination on protein homogeneity. (A) The raw data show that the CB1 mutant containing all four mutations (orange trace) has the highest protein yield. (B) The normalized data show that the combination (orange trace) improves protein homogeneity. (C) CPM ramping assay of CB1 mutants testing the effects of mutations and mutation combinations on protein thermostability. The Tm value of the CB1 mutant containing all four mutations (orange trace) is higher than other mutants, indicating that this mutation combination improves protein thermostability. (D) Coommassie-stained polyacrylamide gel electrophoresis (PAGE) of the purified crystallization-grade CB1. CB1-1: Coommassie-stained PAGE of sample purified by cobalt immobilized-metal affinity chromatography (TALON-IMAC); CB1-2: Coommassie-stained PAGE of sample after TEV protease digestion.
Fusion Protein Engineering and Crystal Packing of CB1-AM6538 Complex. Related to Figures 1 and 3. (A) Overall structure of CB1-AM6538 complex. CB1 and Flavodoxin are colored in orange and slate, respectively. AM6538 is shown in green stick representation and FMN (the substrate of Flavodoxin) is shown in magenta stick representation. (B) Crystal packing of the CB1-AM6538 complex, N-terminus involved in the crystal packing. (C) The 2,4-dichorophenyl ring of AM6538 fits well into the shape of the side-pocket. The receptor is shown in gray cartoon and gray surface, AM6538 is shown in cyan carbons. (D) Superposition of AM6538 in crystal structure (green) and local minimum calculated by quantum mechanics (magenta). (E) Crystal images for CB1 in complex with AM6538 (scale bar is 90 mm). (F) The high-resolution mass spectrometry spectra of the pure standard AM6538 (top) and the ligand dissociated from CB1 complex (bottom). The theoretical m/z for the monoisotopic peak of AM6538 is 542.1356.
MD Simulations of the Predicted Antagonists and Mutagenesis Validation of the Docking Predictions. Related to Figure 5 and Figure 6. (A) Docking pose of AM6538 (magenta sticks) and predicted hydrogen bond (orange dashed lines) and π-π interaction (blue dashed lines) in comparison with AM6538 in crystal structure (green sticks). (B) RMSF of AM6538 in MD simulation. (C-F) AM6538 (C), rimonabant (D), otenabant (E) and taranabant (F) Central structures in MD (protein is shown in light cyan cartoon; ligands are shown in blue sticks) versus docking poses (protein is shown in white cartoon and ligands are shown in yellow sticks), and ligand RMSD (three independent runs). (G) Mutations of hCB1 at Phe170Ala or Phe174Ala disrupts the ability of AM6538 and rimonabant to displace CP55,940 while preserving its functional activity. The corresponding tryptophan mutations have no effect on agonist or antagonist actions. (H) A loss of antagonist affinity is further demonstrated in competition response curves agonist 100nM CP55,940. (I) Mutation of Phe379Trp and Phe379Ala progressively worsen the functional affinity of CP55,940. (J) When tested against concentrations of CP55,940 that maximally stimulate the receptor, AM6538 and rimonabant maintain their potency as antagonists (100 nM CP55,940 for WT and Phe379Trp and 10 µM CP55,940 for Phe379Ala; normalization is made per mutant). Data are presented as mean ± S. E. M. of 3 or more assays performed in duplicate or triplicate.
Multiple Sequence Alignment of Human CB1 with CB2, LPA1 and S1P1 Receptors. Related to Figure 4. Positions that are identical between the receptors are highlighted with a red background. Alignment was done by MAFFT v7.299b (Katoh and Standley, 2013) and the graphics were prepared using ESPript 3.0 server (Robert and Gouet, 2014).
Highlights.
AM6538 is presented as a stabilizing, tight binding antagonist of CB1
Crystal structure of human CB1 in complex with AM6538 is determined
Molecular docking predicts CB1 binding modes of THC and synthetic cannabinoids
Resolution of the binding pocket provides path for rational CB1 drug design
Acknowledgments
This work was supported by: the Ministry of Science and Technology of China grants 2014CB910400 & 2015CB910104, and The National Nature Science Foundation of China grant 31330019 to (Z.J.L); National Institutes of Health grants P01DA009158 (A.M., L.M.B.), R37DA023142 (A.M.), R01AI118985 (I.K.) and NSF grants. We thank the Shanghai Municipal Government, ShanghaiTech University and GPCR Consortium for financial support. The synchrotron radiation experiments were performed at the BL41XU of Spring-8 with approval of the Japan Synchrotron Radiation Research Institute (JASRI) (proposal no. 2015B1031 and 2016A2731), GM/CA@APS of Argonne National Lab which is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under contract number No. DE-AC02-06CH11357 and beamline BL17U1 (Shanghai Synchrotron Radiation Facility [SSRF], China). We thank the Cloning, Cell Expression and Protein Purification Core Facilities of iHuman Institute for their support. We thank A. Walker for assistance with the manuscript; F. Xu, V. Cherezov, V. Katritch, A. Ishchenko, H. Tao, J. Cheng, D. Liu, W. Zhong and W. Liu for helpful discussions. The NIDA Drug supply program provided standards used in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Coordinates and structures factors have been deposited in the Protein Data Bank for CB1-AM6538 (PDB: 5TGZ).
Author Contributions
Conceptualization, L.M.B., A.M., R.C.S., Z.J.L.; Methodology, K.V., A.M. J-H. H., R.B.L. E.L.S., B.W., Q.Z.; Validation, G.W.H.; Formal Analysis, T.H., M.P., L.Q., G.W.H., Y.W., S.Z., W.S., S.L., R.B.L., E.L.S., N.Z., B.W., Q.Z., M.A.H., L.M.B., A.M., Z.J.L.; Investigation, T.H., K.V., L.Q., G.W.H., W.S., S.L., A.K., R.B.L., E.L.S., J-H.H., N.Z., H.Z., I.K.; Writing – Original Draft, T.H.; Writing, Reviewing & Editing, T.H., K.V., Y.W., S.Z., I.K., M.A.H., L.M.B., A.M., R.C.S., Z.J.L.; Visualization, T.H., M.P., M.A.H.; Supervision, L.M.B., A.M., R.C.S., Z.J.L.
References
- Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R, Mac KS, Jr, Loewe S. Tetrahydrocannabinol homologs with double branched alkyl groups in the 3-position. J Am Chem Soc. 1948;70:664–668. doi: 10.1021/ja01182a068. [DOI] [PubMed] [Google Scholar]
- Ahn KH, Bertalovitz AC, Mierke DF, Kendall DA. Dual role of the second extracellular loop of the cannabinoid receptor 1: ligand binding and receptor localization. Mol Pharmacol. 2009;76:833–842. doi: 10.1124/mol.109.057356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov AI, Mileni M, Chien EY, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Andersson H, D’Antona AM, Kendall DA, Von Heijne G, Chin CN. Membrane assembly of the cannabinoid receptor 1: impact of a long N-terminal tail. Mol Pharmacol. 2003;64:570–577. doi: 10.1124/mol.64.3.570. [DOI] [PubMed] [Google Scholar]
- Aung MM, Griffin G, Huffman JW, Wu M, Keel C, Yang B, Showalter VM, Abood ME, Martin BR. Influence of the N-1 alkyl chain length of cannabimimetic indoles upon CB(1) and CB(2) receptor binding. Drug Alcohol Depend. 2000;60:133–140. doi: 10.1016/s0376-8716(99)00152-0. [DOI] [PubMed] [Google Scholar]
- Ballesteros JA, Weinstein H. [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. In: Stuart CS, editor. Methods in Neurosciences. Academic Press; 1995. pp. 366–428. [Google Scholar]
- Ben-Shabat S, Fride E, Sheskin T, Tamiri T, Rhee MH, Vogel Z, Bisogno T, De Petrocellis L, Di Marzo V, Mechoulam R. An entourage effect: inactive endogenous fatty acid glycerol esters enhance 2-arachidonoyl-glycerol cannabinoid activity. Eur J Pharmacol. 1998;353:23–31. doi: 10.1016/s0014-2999(98)00392-6. [DOI] [PubMed] [Google Scholar]
- Berding G, Muller-Vahl K, Schneider U, Gielow P, Fitschen J, Stuhrmann M, Harke H, Buchert R, Donnerstag F, Hofmann M, et al. [123I]AM281 single-photon emission computed tomography imaging of central cannabinoid CB1 receptors before and after Delta9-tetrahydrocannabinol therapy and whole-body scanning for assessment of radiation dose in tourette patients. Biol Psychiatry. 2004;55:904–915. doi: 10.1016/j.biopsych.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Bertalovitz AC, Ahn KH, Kendall DA. Ligand Binding Sensitivity of the Extracellular Loop Two of the Cannabinoid Receptor 1. Drug Dev Res. 2010;71:404–411. doi: 10.1002/ddr.20388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black MD, Stevens RJ, Rogacki N, Featherstone RE, Senyah Y, Giardino O, Borowsky B, Stemmelin J, Cohen C, Pichat P, et al. AVE1625, a cannabinoid CB1 receptor antagonist, as a co-treatment with antipsychotics for schizophrenia: improvement in cognitive function and reduction of antipsychotic-side effects in rodents. Psychopharmacology (Berl) 2011;215:149–163. doi: 10.1007/s00213-010-2124-0. [DOI] [PubMed] [Google Scholar]
- Bohn LM, Zhou L, Ho JH. Approaches to Assess Functional Selectivity in GPCRs: Evaluating G Protein Signaling in an Endogenous Environment. Methods Mol Biol. 2015;1335:177–189. doi: 10.1007/978-1-4939-2914-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramblett RD, Panu AM, Ballesteros JA, Reggio PH. Construction of a 3D model of the cannabinoid CB1 receptor: determination of helix ends and helix orientation. Life Sci. 1995;56:1971–1982. doi: 10.1016/0024-3205(95)00178-9. [DOI] [PubMed] [Google Scholar]
- Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V, Hanson MA, Griffith MT, Hilgart MC, Sanishvili R, Nagarajan V, Stepanov S, Fischetti RF, Kuhn P, Stevens RC. Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 microm size X-ray synchrotron beam. J R Soc Interface. (6 Suppl 5):S587–S597. doi: 10.1098/rsif.2009.0142.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CN, Lucas-Lenard J, Abadji V, Kendall DA. Ligand binding and modulation of cyclic AMP levels depend on the chemical nature of residue 192 of the human cannabinoid receptor 1. J Neurochem. 1998;70:366–373. doi: 10.1046/j.1471-4159.1998.70010366.x. [DOI] [PubMed] [Google Scholar]
- Chrencik JE, Roth CB, Terakado M, Kurata H, Omi R, Kihara Y, Warshaviak D, Nakade S, Asmar-Rovira G, Mileni M, et al. Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1. Cell. 2015;161:1633–1643. doi: 10.1016/j.cell.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun E, Thompson AA, Liu W, Roth CB, Griffith MT, Katritch V, Kunken J, Xu F, Cherezov V, Hanson MA, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cravatt BF, Lichtman AH. The endogenous cannabinoid system and its role in nociceptive behavior. J Neurobiol. 2004;61:149–160. doi: 10.1002/neu.20080. [DOI] [PubMed] [Google Scholar]
- D’Antona AM, Ahn KH, Kendall DA. Mutations of CB1 T210 produce active and inactive receptor forms: correlations with ligand affinity, receptor stability, and cellular localization. Biochemistry. 2006;45:5606–5617. doi: 10.1021/bi060067k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devane WA, Dysarz FA, 3rd, Johnson MR, Melvin LS, Howlett AC. Determination and characterization of a cannabinoid receptor in rat brain. Mol Pharmacol. 1988;34:605–613. [PubMed] [Google Scholar]
- Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JF, Dunham TD, Farrens DL. Cysteine residues in the human cannabinoid receptor: only C257 and C264 are required for a functional receptor, and steric bulk at C386 impairs antagonist SR141716A binding. Biochemistry. 2005;44:8757–8769. doi: 10.1021/bi0472651. [DOI] [PubMed] [Google Scholar]
- Fay JF, Farrens DL. The membrane proximal region of the cannabinoid receptor CB1 N-terminus can allosterically modulate ligand affinity. Biochemistry. 2013;52:8286–8294. doi: 10.1021/bi400842k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder CC, Joyce KE, Briley EM, Mansouri J, Mackie K, Blond O, Lai Y, Ma AL, Mitchell RL. Comparison of the pharmacology and signal transduction of the human cannabinoid CB1 and CB2 receptors. Mol Pharmacol. 1995;48:443–450. [PubMed] [Google Scholar]
- Felder E, Schrott-Fischer A. Quantitative evaluation of myelinated nerve fibres and hair cells in cochleae of humans with age-related high-tone hearing loss. Hear Res. 1995;91:19–32. doi: 10.1016/0378-5955(95)00158-1. [DOI] [PubMed] [Google Scholar]
- Feller SE, MacKerell AD. An improved empirical potential energy function for molecular simulations of phospholipids. J Phys Chem B. 2000;104:7510–7515. [Google Scholar]
- Fernandez-Ruiz J, Romero J, Ramos JA. Endocannabinoids and Neurodegenerative Disorders: Parkinson’s Disease, Huntington’s Chorea, Alzheimer’s Disease, and Others. Handb Exp Pharmacol. 2015;231:233–259. doi: 10.1007/978-3-319-20825-1_8. [DOI] [PubMed] [Google Scholar]
- Fong TM, Guan XM, Marsh DJ, Shen CP, Stribling DS, Rosko KM, Lao J, Yu H, Feng Y, Xiao JC, et al. Antiobesity efficacy of a novel cannabinoid-1 receptor inverse agonist, N-[(1S,2S)-3-(4-chlorophenyl)-2-(3-cyanophenyl)-1-methylpropyl]-2-methyl-2-[[5-(t rifluoromethyl)pyridin-2-yl]oxy]propanamide (MK-0364), in rodents. J Pharmacol Exp Ther. 2007;321:1013–1022. doi: 10.1124/jpet.106.118737. [DOI] [PubMed] [Google Scholar]
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Gaoni Y, Mechoulam R. Isolation, Structure, and Partial Synthesis of an Active Constituent of Hashish. J Am Chem Soc. 1964;86:1646–1647. [Google Scholar]
- Gatley SJ, Lan R, Volkow ND, Pappas N, King P, Wong CT, Gifford AN, Pyatt B, Dewey SL, Makriyannis A. Imaging the brain marijuana receptor: development of a radioligand that binds to cannabinoid CB1 receptors in vivo. J Neurochem. 1998;70:417–423. doi: 10.1046/j.1471-4159.1998.70010417.x. [DOI] [PubMed] [Google Scholar]
- Ghosh R, Todd AR, Wilkinson S. 206. Cannabis indica. Part IV. The synthesis of some tetrahydrodibenzopyran derivatives. J Am Chem Soc. 1940:1121–1125. [Google Scholar]
- Griffith DA, Hadcock JR, Black SC, Iredale PA, Carpino PA, DaSilva-Jardine P, Day R, DiBrino J, Dow RL, Landis MS, et al. Discovery of 1-[9-(4-chlorophenyl)-8-(2-chlorophenyl)-9H–purin-6-yl]-4-ethylaminopiperidine-4- carboxylic acid amide hydrochloride (CP-945,598), a novel, potent, and selective cannabinoid type 1 receptor antagonist. J Med Chem. 2009;52:234–237. doi: 10.1021/jm8012932. [DOI] [PubMed] [Google Scholar]
- Guo Y, Abadji V, Morse KL, Fournier DJ, Li X, Makriyannis A. (−)-11-Hydroxy-7’-isothiocyanato-1’,1’-dimethylheptyl-delta 8-THC: a novel, high-affinity irreversible probe for the cannabinoid receptor in the brain. J Med Chem. 1994;37:3867–3870. doi: 10.1021/jm00049a002. [DOI] [PubMed] [Google Scholar]
- Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Hanson MA, Roth CB, Jo E, Griffith MT, Scott FL, Reinhart G, Desale H, Clemons B, Cahalan SM, Schuerer SC, et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkenham M, Lynn AB, Little MD, Johnson MR, Melvin LS, de Costa BR, Rice KC. Cannabinoid receptor localization in brain. Proc Natl Acad Sci U S A. 1990;87:1932–1936. doi: 10.1073/pnas.87.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanns-Clausen M, Kneisel S, Szabo B, Auwarter V. Acute toxicity due to the confirmed consumption of synthetic cannabinoids: clinical and laboratory findings. Addiction. 2013;108:534–544. doi: 10.1111/j.1360-0443.2012.04078.x. [DOI] [PubMed] [Google Scholar]
- Howlett AC. Cannabinoid inhibition of adenylate cyclase. Biochemistry of the response in neuroblastoma cell membranes. Mol Pharmacol. 1985;27:429–436. [PubMed] [Google Scholar]
- Hurst DP, Lynch DL, Barnett-Norris J, Hyatt SM, Seltzman HH, Zhong M, Song ZH, Nie J, Lewis D, Reggio PH. N-(piperidin-1-yl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H–pyrazole −3-carboxamide (SR141716A) interaction with LYS 3.28(192) is crucial for its inverse agonism at the cannabinoid CB1 receptor. Mol Pharmacol. 2002;62:1274–1287. doi: 10.1124/mol.62.6.1274. [DOI] [PubMed] [Google Scholar]
- Isberg V, Mordalski S, Munk C, Rataj K, Harpsoe K, Hauser AS, Vroling B, Bojarski AJ, Vriend G, Gloriam DE. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 2016;44:D356–D364. doi: 10.1093/nar/gkv1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janero DR, Lindsley L, Vemuri VK, Makriyannis A. Cannabinoid 1 G protein-coupled receptor (periphero-)neutral antagonists: emerging therapeutics for treating obesity-driven metabolic disease and reducing cardiovascular risk. Expert Opin Drug Discovery. 2011;6:995–1025. doi: 10.1517/17460441.2011.608063. [DOI] [PubMed] [Google Scholar]
- Janero DR, Makriyannis A. Cannabinoid receptor antagonists: pharmacological opportunities, clinical experience, and translational prognosis. Expert Opin Emerg Drugs. 2009;14:43–65. doi: 10.1517/14728210902736568. [DOI] [PubMed] [Google Scholar]
- Janero DR, Yaddanapudi S, Zvonok N, Subramanian KV, Shukla VG, Stahl E, Zhou L, Hurst D, Wager-Miller J, Bohn LM, et al. Molecular-interaction and signaling profiles of AM3677, a novel covalent agonist selective for the cannabinoid 1 receptor. ACS Chem Neurosci. 2015;6:1400–1410. doi: 10.1021/acschemneuro.5b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Brown S, Roche JP, Hsieh C, Celver JP, Kovoor A, Chavkin C, Mackie K. Distinct domains of the CB1 cannabinoid receptor mediate desensitization and internalization. J Neurosci. 1999;19:3773–3780. doi: 10.1523/JNEUROSCI.19-10-03773.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy SK, Larson RG. Molecular dynamics simulations of model trans-membrane peptides in lipid bilayers: a systematic investigation of hydrophobic mismatch. Biophys J. 2006;90:2326–2343. doi: 10.1529/biophysj.105.073395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuster JE, Stevenson JI, Ward SJ, D’Ambra TE, Haycock DA. Aminoalkylindole binding in rat cerebellum: selective displacement by natural and synthetic cannabinoids. J Pharmacol Exp Ther. 1993;264:1352–1363. [PubMed] [Google Scholar]
- Lan R, Liu Q, Fan P, Lin S, Fernando SR, McCallion D, Pertwee R, Makriyannis A. Structure-activity relationships of pyrazole derivatives as cannabinoid receptor antagonists. J Med Chem. 1999;42:769–776. doi: 10.1021/jm980363y. [DOI] [PubMed] [Google Scholar]
- Lange JH, Kruse CG. Keynote review: Medicinal chemistry strategies to CB1 cannabinoid receptor antagonists. Drug Discov Today. 2005;10:693–702. doi: 10.1016/S1359-6446(05)03427-6. [DOI] [PubMed] [Google Scholar]
- Lemberger L. Potential therapeutic usefulness of marijuana. Annu Rev Pharmacol Toxicol. 1980;20:151–172. doi: 10.1146/annurev.pa.20.040180.001055. [DOI] [PubMed] [Google Scholar]
- Li C, Xu W, Vadivel SK, Fan P, Makriyannis A. High affinity electrophilic and photoactivatable covalent endocannabinoid probes for the CB1 receptor. J Med Chem. 2005;48:6423–6429. doi: 10.1021/jm050272i. [DOI] [PubMed] [Google Scholar]
- Li H-L. An archaeological and historical account of cannabis in China. Economic Botany. 1973;28:437–448. [Google Scholar]
- Lv X, Liu J, Shi Q, Tan Q, Wu D, Skinner JJ, Walker AL, Zhao L, Gu X, Chen N, et al. In vitro expression and analysis of the 826 human G protein-coupled receptors. Protein Cell. 2016;7:325–337. doi: 10.1007/s13238-016-0263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Mackerell AD, Jr, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. (3rd) 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Makriyannis A. 2012 Division of medicinal chemistry award address. Trekking the cannabinoid road: a personal perspective. J Med Chem. 2014;57:3891–3911. doi: 10.1021/jm500220s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makriyannis A, Rapaka RS. The molecular basis of cannabinoid activity. Life Sci. 1990;47:2173–2184. doi: 10.1016/0024-3205(90)90147-j. [DOI] [PubMed] [Google Scholar]
- Makriyannis A, Vemuri VK, inventors. Cannabinergic nitrate esters and related analogs. US 2016096822A1 Patent Application. 2016
- Mallat A, Teixeira-Clerc F, Lotersztajn S. Cannabinoid signaling and liver therapeutics. J Hepatol. 2013;59:891–896. doi: 10.1016/j.jhep.2013.03.032. [DOI] [PubMed] [Google Scholar]
- Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- Mavromoustakos T, Yang DP, Makriyannis A. Small angle X-ray diffraction and differential scanning calorimetric studies on O-methyl-(−)-delta 8-tetrahydrocannabinol and its 5’ iodinated derivative in membrane bilayers. Biochim Biophys Acta. 1995;1237:183–188. doi: 10.1016/0005-2736(95)00101-8. [DOI] [PubMed] [Google Scholar]
- Mazier W, Saucisse N, Gatta-Cherifi B, Cota D. The Endocannabinoid System: Pivotal Orchestrator of Obesity and Metabolic Disease. Trends Endocrinol Metab. 2015;26:524–537. doi: 10.1016/j.tem.2015.07.007. [DOI] [PubMed] [Google Scholar]
- McAllister SD, Rizvi G, Anavi-Goffer S, Hurst DP, Barnett-Norris J, Lynch DL, Reggio PH, Abood ME. An aromatic microdomain at the cannabinoid CB(1) receptor constitutes an agonist/inverse agonist binding region. J Med Chem. 2003;46:5139–5152. doi: 10.1021/jm0302647. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechoulam R, Ben-Shabat S, Hanus L, Ligumsky M, Kaminski NE, Schatz AR, Gopher A, Almog S, Martin BR, Compton DR, et al. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem Pharmacol. 1995;50:83–90. doi: 10.1016/0006-2952(95)00109-d. [DOI] [PubMed] [Google Scholar]
- Mercier RW, Pei Y, Pandarinathan L, Janero DR, Zhang J, Makriyannis A. hCB2 ligand-interaction landscape: cysteine residues critical to biarylpyrazole antagonist binding motif and receptor modulation. Chem Biol. 2010;17:1132–1142. doi: 10.1016/j.chembiol.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- Pan X, Ikeda SR, Lewis DL. SR 141716A acts as an inverse agonist to increase neuronal voltage-dependent Ca2+ currents by reversal of tonic CB1 cannabinoid receptor activity. Mol Pharmacol. 1998;54:1064–1072. doi: 10.1124/mol.54.6.1064. [DOI] [PubMed] [Google Scholar]
- Pattison FLM, Brown GM. Organic Nitrates As Synthetic Intermediates: Preparations Of Nitrates And Some Representative Reactions. Can J Chem. 1956;34:879–884. [Google Scholar]
- Pertwee RG. Cannabinoids and multiple sclerosis. Pharmacol Ther. 2002;95:165–174. doi: 10.1016/s0163-7258(02)00255-3. [DOI] [PubMed] [Google Scholar]
- Picone RP, Khanolkar AD, Xu W, Ayotte LA, Thakur GA, Hurst DP, Abood ME, Reggio PH, Fournier DJ, Makriyannis A. (−)-7’-Isothiocyanato-11-hydroxy-1’,1’-dimethylheptylhexahydrocannabinol (AM841), a high-affinity electrophilic ligand, interacts covalently with a cysteine in helix six and activates the CB1 cannabinoid receptor. Mol Pharmacol. 2005;68:1623–1635. doi: 10.1124/mol.105.014407. [DOI] [PubMed] [Google Scholar]
- Pryce G, Baker D. Endocannabinoids in Multiple Sclerosis and Amyotrophic Lateral Sclerosis. Handb Exp Pharmacol. 2015;231:213–231. doi: 10.1007/978-3-319-20825-1_7. [DOI] [PubMed] [Google Scholar]
- Razdan RK. Structure-activity relationships in cannabinoids. Pharmacol Rev. 1986;38:75–149. [PubMed] [Google Scholar]
- Rinaldi-Carmona M, Barth F, Heaulme M, Shire D, Calandra B, Congy C, Martinez S, Maruani J, Neliat G, Caput D, et al. SR141716A, a potent and selective antagonist of the brain cannabinoid receptor. FEBS Lett. 1994;350:240–244. doi: 10.1016/0014-5793(94)00773-x. [DOI] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino T, Zamberletti E, Parolaro D. Endocannabinoids and Mental Disorders. Handb Exp Pharmacol. 2015;231:261–283. doi: 10.1007/978-3-319-20825-1_9. [DOI] [PubMed] [Google Scholar]
- Schindler CW, Redhi GH, Vemuri K, Makriyannis A, Le Foll B, Bergman J, Goldberg SR, Justinova Z. Blockade of Nicotine and Cannabinoid Reinforcement and Relapse by a Cannabinoid CB1-Receptor Neutral Antagonist AM4113 and Inverse Agonist Rimonabant in Squirrel Monkeys. Neuropsychopharmacology. 2016;41:2283–2293. doi: 10.1038/npp.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger . Glide, version 6.9, Schrödinger, LLC. New York, NY: 2015a. [Google Scholar]
- Schrödinger . Induced Fit Docking protocol 2015–4, Glide version 6.4, Prime version 3.7, Schrödinger, LLC. New York, NY: 2015b. [Google Scholar]
- Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- Shimizu T. Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu Rev Pharmacol Toxicol. 2009;49:123–150. doi: 10.1146/annurev.pharmtox.011008.145616. [DOI] [PubMed] [Google Scholar]
- Showalter VM, Compton DR, Martin BR, Abood ME. Evaluation of binding in a transfected cell line expressing a peripheral cannabinoid receptor (CB2): identification of cannabinoid receptor subtype selective ligands. J Pharmacol Exp Ther. 1996;278:989–999. [PubMed] [Google Scholar]
- Smart OS, Womack TO, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song ZH, Bonner TI. A lysine residue of the cannabinoid receptor is critical for receptor recognition by several agonists but not WIN55212-2. Mol Pharmacol. 1996;49:891–896. [PubMed] [Google Scholar]
- Song ZH, Slowey CA, Hurst DP, Reggio PH. The difference between the CB(1) and CB(2) cannabinoid receptors at position 5.46 is crucial for the selectivity of WIN55212-2 for CB(2) Mol Pharmacol. 1999;56:834–840. [PubMed] [Google Scholar]
- Szymanski DW, Papanastasiou M, Melchior K, Zvonok N, Mercier RW, Janero DR, Thakur GA, Cha S, Wu B, Karger B, et al. Mass spectrometry-based proteomics of human cannabinoid receptor 2: covalent cysteine 6.47(257)-ligand interaction affording megagonist receptor activation. J Proteome Res. 2011;10:4789–4798. doi: 10.1021/pr2005583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J, Vemuri VK, Liu J, Batkai S, Mukhopadhyay B, Godlewski G, Osei-Hyiaman D, Ohnuma S, Ambudkar SV, Pickel J, et al. Peripheral CB1 cannabinoid receptor blockade improves cardiometabolic risk in mouse models of obesity. J Clin Invest. 2010;120:2953–2966. doi: 10.1172/JCI42551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Prusov EV. Total synthesis of RNA-polymerase inhibitor ripostatin B and 15-deoxyripostatin A. Angew Chem Int Ed Engl. 2012;51:3401–3404. doi: 10.1002/anie.201108749. [DOI] [PubMed] [Google Scholar]
- Totrov M, Abagyan R. Protein-ligand docking as an energy optimization problem. In Drug-receptor thermodynamics: introduction and applications. New York: John Wiley & Sons; 2001. pp. 603–624. [Google Scholar]
- Whiting PF, Wolff RF, Deshpande S, Di Nisio M, Duffy S, Hernandez AV, Keurentjes JC, Lang S, Misso K, Ryder S, et al. Cannabinoids for Medical Use: A Systematic Review and Meta-analysis. JAMA. 2015;313:2456–2473. doi: 10.1001/jama.2015.6358. [DOI] [PubMed] [Google Scholar]
- Wiley JL, Compton DR, Dai D, Lainton JA, Phillips M, Huffman JW, Martin BR. Structure-activity relationships of indole- and pyrrole-derived cannabinoids. J Pharmacol Exp Ther. 1998;285:995–1004. [PubMed] [Google Scholar]
- Wollner HJ, Matchett JR, Levine J, Loewe S. Isolation of a Physiologically Active Tetrahydrocannabinol from Cannabis Sativa Resin. J Am Chem Soc. 1942;64:26–29. [Google Scholar]
- Yeates RA, Laufen H, Leitold M. The reaction between organic nitrates and sulfhydryl compounds. A possible model system for the activation of organic nitrates. Mol Pharmacol. 1985;28:555–559. [PubMed] [Google Scholar]
- Zoete V, Cuendet MA, Grosdidier A, Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rimonabant and AM6538 are Competitive Antagonists of CP55,940 and THC. Related to Figure 2. Rimonabant (left) and AM6538 (right) competitively antagonize CP55,940 (top) and THC (bottom)-induced (A) Inhibition of forskolin-stimulated cAMP accumulation in hCB1-CHO cells and (B) β-arrestin2-recruitment in hCB1-CHO DiscoveRx PathHunter cells. (C) Rimonabant (left) and AM6538 (right) competitively antagonize CP55,940-stimulated [35S]-GTPγS binding in mouse cerebellum membrane preparations. Data are presented as the mean ± S. E. M. of 3 or more assays run in duplicate or triplicate. Competitive nonlinear analysis was performed in GraphPad Prism 6.0 and parameters are included in Table S1. Characterization of agonist effects in the cells and cerebellum are provided in the supplemental methods.
Stability Assay of CB1 Mutants. Related to Figure 1. (A, B) Analytical size exclusion chromatography (aSEC) of CB1 mutants testing the effects of mutations and mutation combination on protein homogeneity. (A) The raw data show that the CB1 mutant containing all four mutations (orange trace) has the highest protein yield. (B) The normalized data show that the combination (orange trace) improves protein homogeneity. (C) CPM ramping assay of CB1 mutants testing the effects of mutations and mutation combinations on protein thermostability. The Tm value of the CB1 mutant containing all four mutations (orange trace) is higher than other mutants, indicating that this mutation combination improves protein thermostability. (D) Coommassie-stained polyacrylamide gel electrophoresis (PAGE) of the purified crystallization-grade CB1. CB1-1: Coommassie-stained PAGE of sample purified by cobalt immobilized-metal affinity chromatography (TALON-IMAC); CB1-2: Coommassie-stained PAGE of sample after TEV protease digestion.
Fusion Protein Engineering and Crystal Packing of CB1-AM6538 Complex. Related to Figures 1 and 3. (A) Overall structure of CB1-AM6538 complex. CB1 and Flavodoxin are colored in orange and slate, respectively. AM6538 is shown in green stick representation and FMN (the substrate of Flavodoxin) is shown in magenta stick representation. (B) Crystal packing of the CB1-AM6538 complex, N-terminus involved in the crystal packing. (C) The 2,4-dichorophenyl ring of AM6538 fits well into the shape of the side-pocket. The receptor is shown in gray cartoon and gray surface, AM6538 is shown in cyan carbons. (D) Superposition of AM6538 in crystal structure (green) and local minimum calculated by quantum mechanics (magenta). (E) Crystal images for CB1 in complex with AM6538 (scale bar is 90 mm). (F) The high-resolution mass spectrometry spectra of the pure standard AM6538 (top) and the ligand dissociated from CB1 complex (bottom). The theoretical m/z for the monoisotopic peak of AM6538 is 542.1356.
MD Simulations of the Predicted Antagonists and Mutagenesis Validation of the Docking Predictions. Related to Figure 5 and Figure 6. (A) Docking pose of AM6538 (magenta sticks) and predicted hydrogen bond (orange dashed lines) and π-π interaction (blue dashed lines) in comparison with AM6538 in crystal structure (green sticks). (B) RMSF of AM6538 in MD simulation. (C-F) AM6538 (C), rimonabant (D), otenabant (E) and taranabant (F) Central structures in MD (protein is shown in light cyan cartoon; ligands are shown in blue sticks) versus docking poses (protein is shown in white cartoon and ligands are shown in yellow sticks), and ligand RMSD (three independent runs). (G) Mutations of hCB1 at Phe170Ala or Phe174Ala disrupts the ability of AM6538 and rimonabant to displace CP55,940 while preserving its functional activity. The corresponding tryptophan mutations have no effect on agonist or antagonist actions. (H) A loss of antagonist affinity is further demonstrated in competition response curves agonist 100nM CP55,940. (I) Mutation of Phe379Trp and Phe379Ala progressively worsen the functional affinity of CP55,940. (J) When tested against concentrations of CP55,940 that maximally stimulate the receptor, AM6538 and rimonabant maintain their potency as antagonists (100 nM CP55,940 for WT and Phe379Trp and 10 µM CP55,940 for Phe379Ala; normalization is made per mutant). Data are presented as mean ± S. E. M. of 3 or more assays performed in duplicate or triplicate.
Multiple Sequence Alignment of Human CB1 with CB2, LPA1 and S1P1 Receptors. Related to Figure 4. Positions that are identical between the receptors are highlighted with a red background. Alignment was done by MAFFT v7.299b (Katoh and Standley, 2013) and the graphics were prepared using ESPript 3.0 server (Robert and Gouet, 2014).