Abstract
The trans-decalin structure formed as a result of intramolecular Diels–Alder cycloaddition is widely present among bioactive natural products isolated from fungi. In this work, we elucidated the concise, three-enzyme biosynthetic pathway of the cytotoxic myceliothermophin and biochemically characterized the Diels–Alderase (DAase) that catalyzes formation of trans-decalin from an acyclic substrate. Computational studies on the reaction mechanisms rationalizes both the substrate- and stereoselectivity of the enzyme.
Graphical Abstract
Cycloaddition reactions such as intramolecular Diels–Alder (IMDA) reactions are extremely important and versatile synthetic transformations that allow the construction of multicyclic scaffolds with stereochemical control and atom economy.1 Recent findings revealed that nature has evolved enzymes to catalyze [4+2] reactions in natural product biosynthetic pathways.2 Especially from bacterial pathways, a few so-called “Diels-Alderase (DAase)” have been discovered and characterized, including those from the spinosyn,3 solanapyrone,4 tetronate/tetramate containing compounds,5 thiopeptides6 and abyssomycin7 pathways. Structural studies suggest that while there are no conserved sequences for DAases, these enzymes provide an active site environment that accelerates or enables the cycloaddition reactions to occur.7,8 The diversity of DAases found so far inspires the discovery of additional enzymes that can catalyze such reactions, which can enable genome mining of new natural products that contain structural features that are derived from cycloaddition reactions.
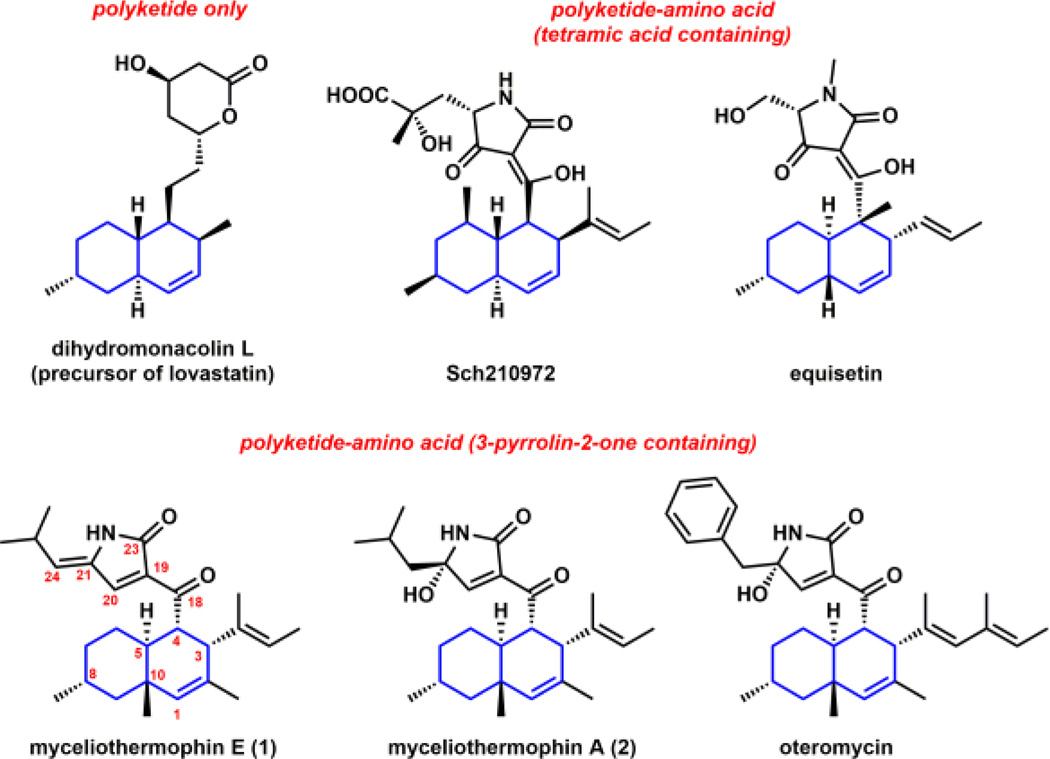
IMDA reactions are also proposed to occur widely among fungal natural product pathways, in particular those of polyketide and polyketide-nonribosomal peptide natural products.9 For example, both the trans-decalin and isoindolone ring systems found in lovastatin10 and cytochalasan,11 respectively, are thought to arrive from cycloaddition reactions from acyclic precursors. Decalin-containing fungal polyketides constitute a large class of natural products with a diverse array of biological activities (Figures 1 and S1). Biosynthesis of the acyclic substrates that contain both the dienophile and the diene are proposed to be catalyzed by the iterative functions of the highly-reducing polyketide synthases (HR-PKSs). While bioinformatic analysis and genetic evidences have suggested a class of lipocalin-like enzymes may be involved in the formation of the decalin ring systems of Sch21097212 and equisetin,13 no direct biochemical evidence of enzyme-catalyzed IMDA reaction has been described. This is in part due to the inability to capture an acyclic substrate required for activity verification. In this work, we provide biochemical confirmation of DAase activity from a fungal polyketide-nonribosomal peptide biosynthetic pathway, using the biosynthesis of myceliothermophin as a model system.
Figure 1.
Structures of decalin containing fungal polyketides and polyketide-amino acid hybrids.
Myceliothermophins, including myceliothermophin E (1) and myceliothermophin A (2), are cytotoxic compounds isolated from the thermophilic fungus Myceliophthora thermophile (Figure 1).14 1 exhibits IC50 values of <100 nM towards a variety of cancer cell lines. Multiple total syntheses of myceliothermophins have been accomplished to establish the absolute stereochemistry as shown in Figure 1.15,16 Both 1 and 2 contain a trans-fused decalin ring system connected to a conjugated 3-pyrrolin-2-one moiety, with 1 containing an exocyclic methylpropylidene unit derived from leucine at C21, while 2 is substituted with a hydroxyl group at the same position. Despite the wide occurrence of the 3-pyrrolin-2-one ring system in natural products (Figure S1),17 the formation of this ring system has not been widely studied. The pyrrolinone is proposed to derive from a Knoevenagel condensation between the α-carbon C19 and a reductively released amino aldehyde C20. This differs from the tetramic acid rings found in equisetin and Sch210972, which arise from Dieckmann cyclization of aminoacylated polyketides.18
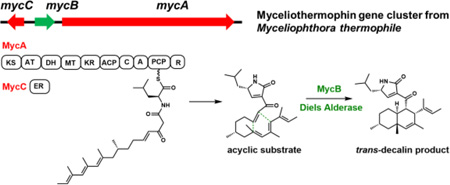
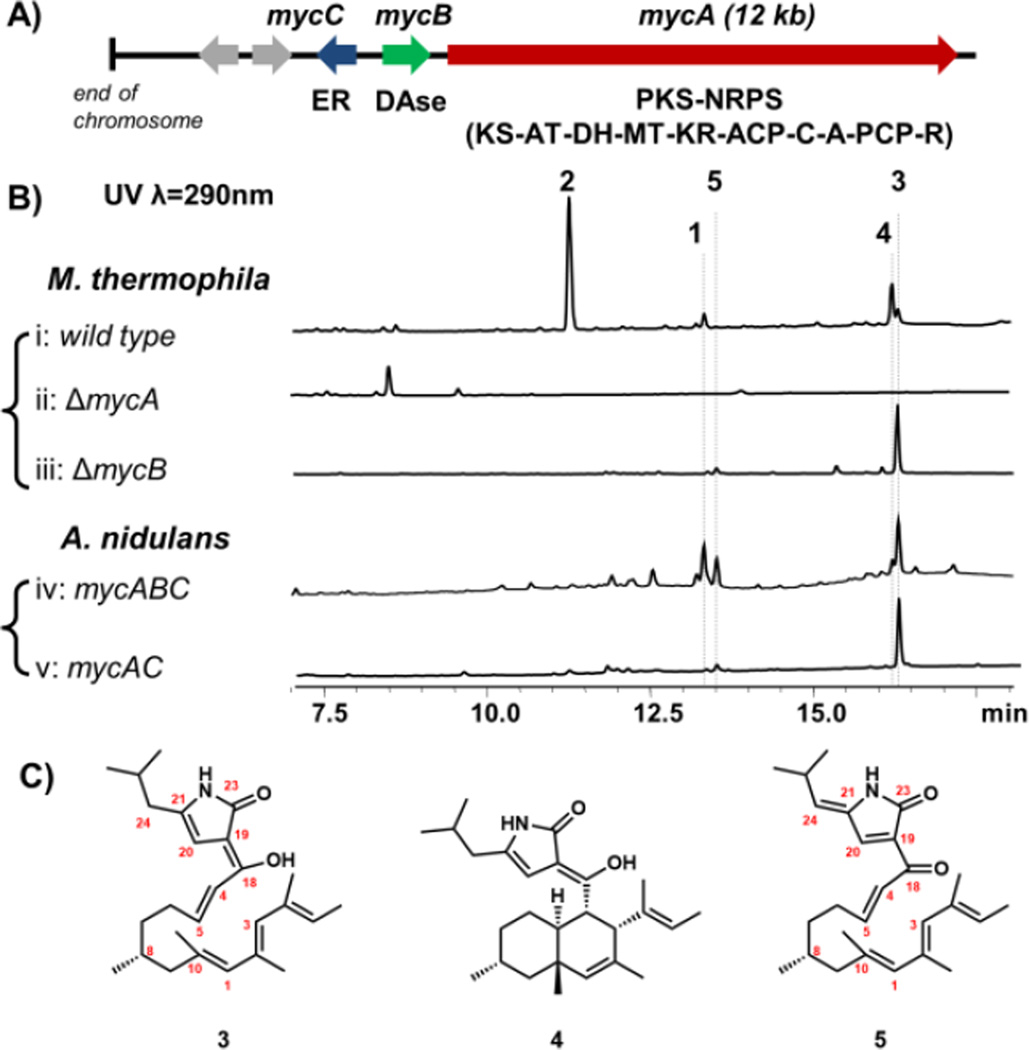
To investigate the biosynthetic pathway of 1 and 2, the genome of the producing organism M. thermophile ATCC 42462 was queried for the presence of PKS-NRPS containing genes. Two PKS-NRPS genes were found, and analysis of the neighboring genes indicated that MYCTH _78013 (named mycA), which is near the end of a chromosome, is a likely candidate (Table S2). Immediately adjacent is mycB encoding a potential DAase that has sequence homology (36% identity) to CghA, which has been implicated to be involved in the cycloaddition during Sch210972 biosynthesis.12 A trans-acting enoylreductase (ER) is encoded in mycC (Figure 2A). This three-gene cassette is found widely among sequenced fungi (Figure S10). To test the link between the gene cluster and production of 1 and 2, which are produced by the wild type strain at yields of 6mg/L and 50 mg/L, respectively (Figure 2B, i) (Figures S11–S12, Table S3–S4), deletion strain of ΔmycA was generated through gene replacement (Figure S2). LC-MS analysis showed that deletion of mycA completely abolished the production of 1 and 2 (Figure 2B, ii), confirming the involvement of MycA in their biosynthesis. Comparison of the LC-MS traces also revealed the abolishment of two additional compounds, 3 and 4, with the same m/z [M+H]+ of 398 that were not previously reported. Both compounds were purified from the wild type strain at yields of 10 mg/L (3) and 16 mg/L (4) as shown in Figure 2C.
Figure 2.
Verification of the myceliothermophin gene cluster. (A) The myc cluster encodes the PKS-NRPS MycA (domain abbreviations: KS: ketosynthase; AT: acyltransferase; DH: dehydratase; MT: methyltransferase; KR: ketoreductase; ACP: acyl carrier protein; C: condensation; A: adenylation; PCP: peptidyl carrier protein; and R: reductase); the in trans enoylreductase (ER) MycC; and the putative DAase MycB; (B) Product profile of wild type and single–gene knockout strains of M. thermophila; and that of A. nidulans transformed with combinations of myc genes; (C) structures of metabolites isolated. The stereochemistries of 3, 4 and 5 are assumed to be similar to 1.
Structural characterization with NMR showed 3 is an acyclic polyolefinic compound that contains a 4-pyrrolin-2-one moiety, which is in conjugation with the enol at C18 via the α-carbon at C19 (Figure 2C and Figures S13, S20–24, Tables S5). This is likely the enolized form of the PKS-NRPS/ER product, which contains eight differentially α- and β-functionalized ketide units aminoacylated with leucine, and has undergone Knoevenagel condensation (Figure 4). To confirm that 3 is synthesized by PKS-NRPS and ER, we introduced both mycA and mycC into the heterologous host Aspergillus nidulans A1145 (Figure S4). Compared to the untransformed host, A. nidulans expressing MycA and MycC showed clear accumulation of a new compound that is identical to 3 in retention time, UV absorbance and mass (Figure 2B, v). This result suggests the myc PKS-NRPS and its ER partner are sufficient to form the pyrrolinone moiety, in contrast to previous work with other R-domain containing fungal PKS-NRPS systems.19
Figure 4.
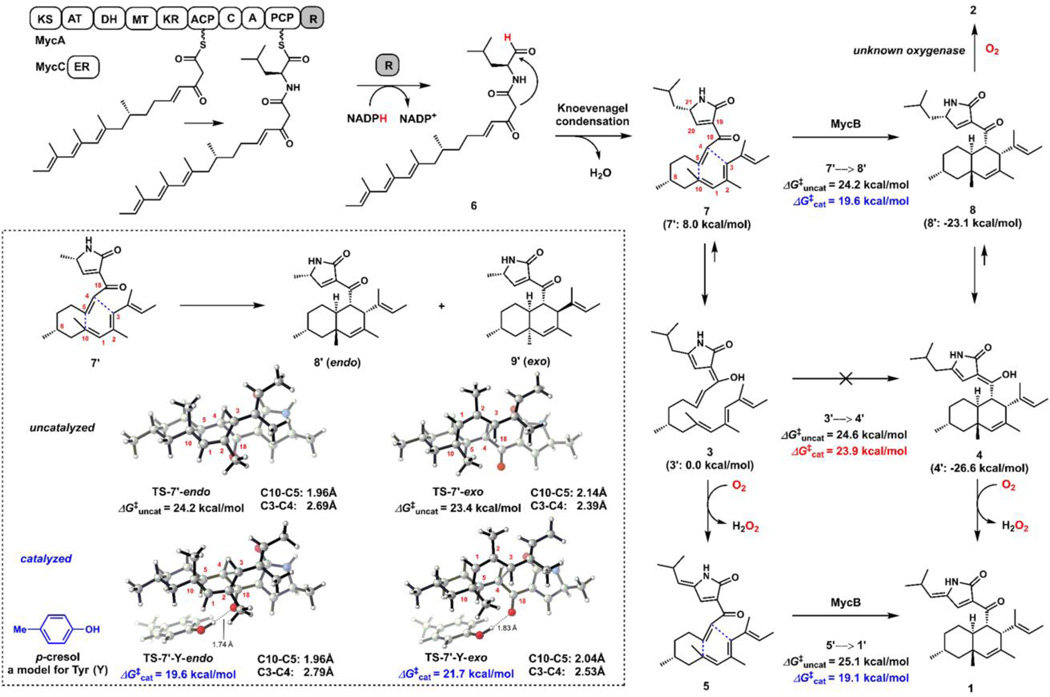
The proposed biosynthetic pathway of 1 based on isolated natural products and biochemical characterization of MycB. DFT analysis were performed on model substrates (1’–8’) of which the isobutyl group in 1–8 is replaced by a methyl group. Calculations were performed at the CPCM (water)/M06-2X/6-311+G(d,p)//M06-2X/6-31G(d) level of theory. The computed free energies of model compounds are shown in parenthesis. Calculations of stereoselectivity under uncatalyzed and catalyzed conditions are shown in the dash box.
On the other hand, 4 is the trans-decalin cyclized form of 3 and contains the enolized dienyl pyrrolinone ring (Figures S14, S25–30, Tables S6). This compound therefore represents the potential intermediate that can be oxidized into both 1 and 2. To examine the role of MycB on the formation of 1, 2, and 4 in M. thermophila, we generated the ΔmycB strain (Figure S3). LC-MS analysis of the metabolites showed that all these cyclized products were abolished, leaving the acyclic 3 as the dominant product (Figure 2B, iii). Hence, MycB is directly involved in the cycloaddition to form the decalin ring. This is further verified when the entire three-gene cassette mycABC was introduced into A. nidulans (Figure 2B, iv). Analysis of the extract showed that compared to mycAC alone, introduction of mycB led to emergence of cyclized product 4 as well as the final metabolite 1. We were not able to detect 2 in this strain, suggesting that the stereoselective hydroxylation of C21 is likely catalyzed by additional enzyme only present in M. thermophila. Analysis of the nearby genes to the myc cluster did not reveal any oxygenases that may perform this reaction (Figure S2), hence the enzyme may be shared with other pathways in the strain.20
In comparing the various extracts analyzed in Figure 2B, we noted a new compound 5 that is consistently found in M. thermophila ΔmycB strain, and the transformed A. nidulans strains. This compound has m/z [M+H]+ of 396 and λmax=327 nm (Figure S15), both of which closely resemble those of 1. Purification of 5 from the ΔmycB strain (3 mg/L) followed by NMR characterization (Figure S31–35) revealed the new compound is the acyclic form of 1, which is consistent with its accumulation in the ΔmycB and in the A. nidulans mycAC strains. We propose that 5 could be an air oxidation product of 3, via likely the same mechanism as oxidation of 1 from 4. We attribute the accumulation of 3 and 5 in the A. nidulans mycABC strain to be insufficient activity of MycB and/or differences in the redox environment compared to the original producer. Indeed, expressing a second copy of the mycB gene does lead to decrease of 5 and increased amount of 4 and 1 (Figure S7).
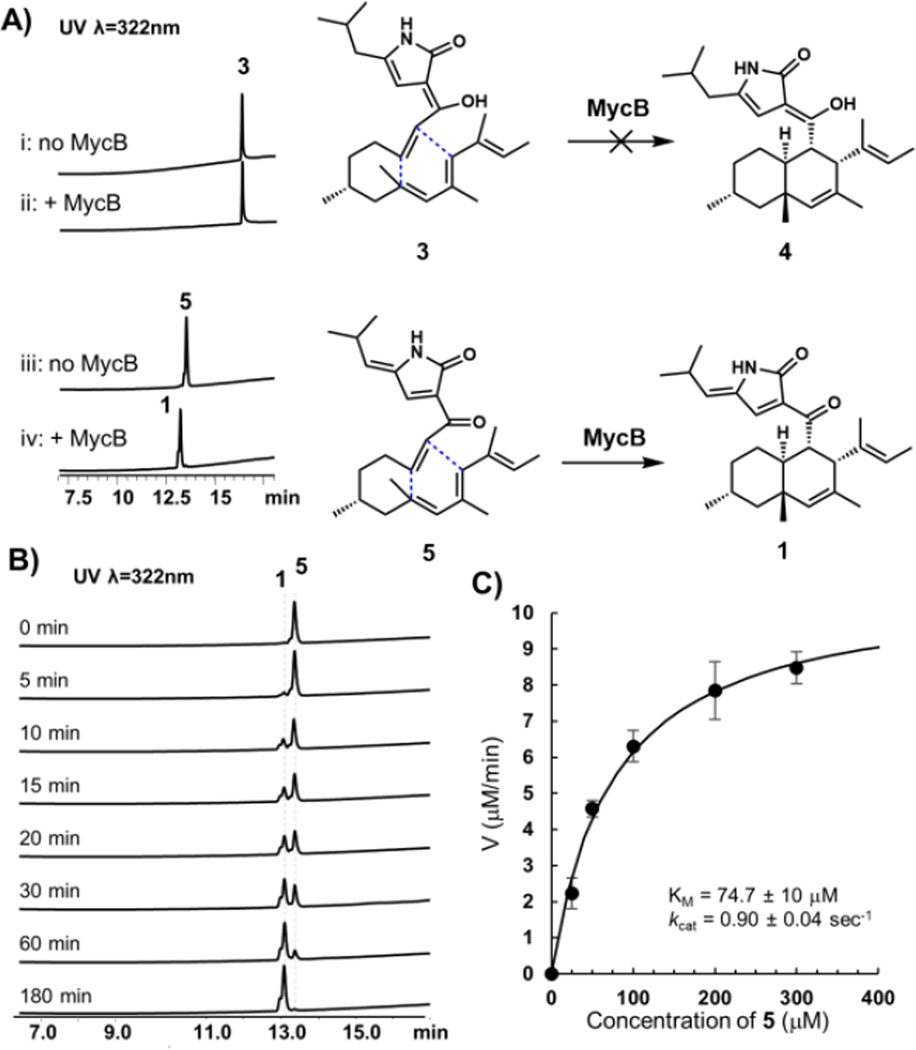
To confirm that MycB is indeed a DAase, N-FLAG-tagged recombinant enzyme was heterologous expressed and purified from Saccharomyces cerevisiae BJ5464-NpgA (0.2 mg/L, Figure S5). Either 0.2 mM 3 or 5 was then incubated with 1 µM MycB in Tris Buffer, pH 7.4 and formation of cyclized products were assayed by LC-MS (Figure 3A). No conversion of 3 to 4 was detected under prolonged incubation (the compound degrades after overnight assay), while complete conversion of 5 to 1 can be observed in three hours (Figure 3A and 3B). Control incubation in the absence of MycB led to degradation of 5 (Figure S6). All chromatographic and spectroscopic properties of 1 from the in vitro assay matches to that purified from in vivo reaction (Figure 2B), illustrating MycB can catalyze the IMDA reaction with regio-and stereospecific control. The kinetic properties of MycB was measured to give a KM of ~ 75 µM towards 5, and a kcat of ~ 0.9 sec−1 (Figure 3C).
Figure 3.
Activity of the DAase MycB. (A) MycB can catalyze the cycloaddition of the ketone 5 to 1, but is not reactive towards the enol 3. (B) Time course analysis of the conversion of 5 to 1. The starting concentrations for 5 and MycB are 200 µM and 1 µM, respectively. (C) Saturation kinetics of MycB. Each data point performed in triplicate.
The in vitro results confirmed that MycB can indeed catalyze the Diels–Alder reaction to convert the C18-keto containing 5 into the cytotoxic natural product 1. The failure of MycB to catalyze the conversion of C18-enol 3 into 4, however, suggests differential reactivity towards the ketone and enol substrates. This difference also leads to a proposal of the myc pathway as shown in Figure 4, to account for the isolation of the metabolites 1−5. Following MycA-catalyzed construction and release of the aminoacyl polyketide aldehyde 6, a Knoevenagel condensation yields the expected ketone product 7. We propose that most likely, this ketone is the natural substrate of MycB, leading to formation of the endo product 8. The keto-3-pyrrolin-2-one can readily enolize to form the observed 4. When MycB is inactivated, the acyclic 7 can also readily enolize to form 3, which is not reactive towards MycB catalyzed IMDA reaction. However, both 3 and 4 may undergo spontaneous air oxidation to yield 5 and 1, respectively. Indeed, when 4 was dissolved in methanol at 37°C for 16 hours, formation of 1 was observed along with an equimolar release of H2O2 from the reaction (Figures S7 and S8). A proposed oxidation mechanism that involves sequential single electron reduction of O2, during which the C18 enol hydrogen is first abstracted, is shown in Figure S9. Significantly (and serendipitously), formation of 5 prevents further tautomerization of the 3-pyrrolin-2-one ring system and results in a stable C18-keto, acyclic substrate for MycB.
To support the proposed pathway and rationalize the reactivity difference between the C18 ketone and enol substrates in the IMDA reaction, we performed density functional theory (DFT) studies on model substrates and products (1’–8’) in which the isobutyl group in 1–8 is replaced by a methyl group. The enols 3’ and 4’ were found to be more stable than the corresponding ketones 7’ and 8’, respectively, with ΔG of −8.0 and −3.5 kcal/mol, respectively (Figure 4), supporting the rapid enolization of 7 into 3, in the absence of MycB.
We then calculated the energetics of the three potential DA reactions shown in Figure 4, starting with either 7’, 3’ or 5’. In each of these cases, the uncatalyzed DA reactions are very slow, with similar ΔG‡uncat values of ~24 – 25 kcal/mol (Figures S36–38). This corresponds to rate constants of ~ 10−5 sec−1 at room temperature according to transition state theory. In contrast, the spontaneous DA reaction in the biosynthesis of Sch210972 or equisetin is very fast.12 The DA reaction of the enol precursor of Sch210972 has a barrier of only 12 kcal/mol, because of the additional electron-withdrawing keto group in the tetramate moiety.12 Interestingly, with 7’, the uncatalyzed reaction is predicted to produce a mixture of diastereomers (Figure 4, dash box), with the unobserved cis-fused decalin 9’ being the major isomer, resulting from the lower energy transition state TS-7’-exo. We propose that a suitable acidic residue in the active site of MycB may accelerate the reaction, by lowering the LUMO of the dienophile and stabilization of the transition state, through hydrogen bonding to the C18-carbonyl group (in 7 and 5, but not 3) next to the dienophile. Such catalytic role for a hydrogen bond donating residue is also proposed based on the structures of SpnF and PyrI4.8 Additionally, because TS-7’-endo is more asynchronous than TS7’-exo, we envisioned that such a catalyst would stabilize the more polarized endo transition state than the exo transition state. To test our hypothesis, we used p-cresol as a model to probe effects of possible catalytic residues such as a tyrosine in the active site of the enzyme (Figure S39). The results showed that the catalyzed DA reaction of either 7’ or 5’ is much faster, with ΔG‡cat of ~19 kcal/mol. This corresponds to a rate constant of ~ 0.1 sec−1 at room temperature, which agrees with the experimentally measured rate (kcat of ~0.9 sec−1, Figure 3C). Also as predicted, the catalyzed DA reaction of 7’ is computed to be endo-selective to give 8’, in agreement with assay results. The hydrogen bond distance in the endo transition state (TS-7’-Y-endo) is shorter than that of the exo transition state (1.74 Å versus 1.83 Å), due to the more polarized nature of the endo TS. In contrast, the hydrogen bond from cresol to the corresponding enol oxygen in 3’ has only a minimal effect on the rate of its DA reaction, with ΔG‡cat = 23.9 kcal/mol. This is in agreement with the experimental observation that MycB could not catalyze the DA reaction of enol 3.
In summary, the nanomolar cytotoxic 1 is synthesized by a concise three-enzyme pathway. Our findings expand the collection of long sought-after DAases from fungi, and may lead to discovery of new decalin-containing natural products using MycB as a signature biosynthetic marker as shown in Figure S10.
Supplementary Material
Acknowledgments
This work was supported by the NIH (1DP1GM106413 and 1R35GM118056). and the NSF (CHE-1361104 to K.N.H.), and JSPS Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers (No. G2604 to K.W.).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Experimental details, spectroscopic and computational data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Takao K-i, Munakata R, Tadano K-i. Chem. Rev. 2005;105:4779. doi: 10.1021/cr040632u. [DOI] [PubMed] [Google Scholar]
- 2.(a) Klas K, Tsukamoto S, Sherman DH, Williams RM. J. Org. Chem. 2015;80:11672. doi: 10.1021/acs.joc.5b01951. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kim HJ, Ruszczycky MW, Liu H-w. Curr. Opin. Chem. Biol. 2012;16:124. doi: 10.1016/j.cbpa.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zheng Q, Tian Z, Liu W. Curr. Opin. Chem. Biol. 2016;31:95. doi: 10.1016/j.cbpa.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 3.Kim HJ, Ruszczycky MW, Choi S-h, Liu Y-n, Liu H-w. Nature. 2011;473:109. doi: 10.1038/nature09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oikawa H, Katayama K, Suzuki Y, Ichihara A. J. Chem. Soc., Chem. Commun. 1995;13:1321. [Google Scholar]
- 5.(a) Tian Z, Sun P, Yan Y, Wu Z, Zheng Q, Zhou S, Zhang H, Yu F, Jia X, Chen D, Mándi A, Kurtán T, Liu W. Nat. Chem. Biol. 2015;11:259. doi: 10.1038/nchembio.1769. [DOI] [PubMed] [Google Scholar]; (b) Hashimoto T, Hashimoto J, Teruya K, Hirano T, Shin-ya K, Ikeda H, Liu H-w, Nishiyama M, Kuzuyama T. J. Am. Chem. Soc. 2015;137:572. doi: 10.1021/ja510711x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Wever WJ, Bogart JW, Baccile JA, Chan AN, Schroeder FC, Bowers AA. J. Am. Chem. Soc. 2015;137:3494. doi: 10.1021/jacs.5b00940. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hudson GA, Zhang Z, Tietz JI, Mitchell DA, van der Donk WA. J. Am. Chem. Soc. 2015;137:16012. doi: 10.1021/jacs.5b10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrne MJ, Lees NR, Han L-C, van der Kamp MW, Mulholland AJ, Stach JEM, Willis CL, Race PR. J. Am. Chem. Soc. 2016;138:6095. doi: 10.1021/jacs.6b00232. [DOI] [PubMed] [Google Scholar]
- 8.(a) Fage CD, Isiorho EA, Liu Y, Wagner DT, Liu H-w, Keatinge-Clay AT. Nat. Chem. Biol. 2015;11:256. doi: 10.1038/nchembio.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zheng Q, Guo Y, Yang L, Zhao Z, Wu Z, Zhang H, Liu J, Cheng X, Wu J, Yang H. Cell Chem. Biol. 2016;23:352. doi: 10.1016/j.chembiol.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Minami A, Oikawa H. J. Antibiot. (Tokyo) 2016;69:500. doi: 10.1038/ja.2016.67. [DOI] [PubMed] [Google Scholar]
- 10.Auclair K, Sutherland A, Kennedy J, Witter DJ, Van den Heever JP, Hutchinson CR, Vederas JC. J. Am. Chem. Soc. 2000;122:11519. [Google Scholar]
- 11.Scherlach K, Boettger D, Remme N, Hertweck C. Nat. Prod. Rep. 2010;27:869. doi: 10.1039/b903913a. [DOI] [PubMed] [Google Scholar]
- 12.Sato M, Yagishita F, Mino T, Uchiyama N, Patel A, Chooi YH, Goda Y, Xu W, Noguchi H, Yamamoto T, Hotta K, Houk KN, Tang Y, Watanabe K. Chem Bio Chem. 2015;16:2294. doi: 10.1002/cbic.201500386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato N, Nogawa T, Hirota H, Jang J-H, Takahashi S, Ahn JS, Osada H. Biochem. Biophys. Res. Commun. 2015;460:210. doi: 10.1016/j.bbrc.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 14.Yang YL, Lu CP, Chen MY, Chen KY, Wu YC, Wu SH. Chem. Eur. J. 2007;13:6985. doi: 10.1002/chem.200700038. [DOI] [PubMed] [Google Scholar]
- 15.Nicolaou K, Shi L, Lu M, Pattanayak MR, Shah AA, Ioannidou HA, Lamani M. Angew. Chem. Int. Ed. 2014;53:10970. doi: 10.1002/anie.201406815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shionozaki N, Yamaguchi T, Kitano H, Tomizawa M, Makino K, Uchiro H. Tetrahedron Lett. 2012;53:5167. [Google Scholar]
- 17.(a) Kontnik R, Clardy J. Org. Lett. 2008;10:4149. doi: 10.1021/ol801726k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Suzuki S, Hosoe T, Nozawa K, Kawai K-i, Yaguchi T, Udagawa S-i. J. Nat. Prod. 2000;63:768. doi: 10.1021/np990371x. [DOI] [PubMed] [Google Scholar]; (c) Osterhage C, Kaminsky R, König GM, Wright AD. J. Org. Chem. 2000;65:6412. doi: 10.1021/jo000307g. [DOI] [PubMed] [Google Scholar]; (d) West RR, Van Ness J, Varming AM, Rassing B, Biggs S, Gasper S, McKernan PA, Piggott J. J. Antibiot. (Tokyo) 1996;49:967. doi: 10.7164/antibiotics.49.967. [DOI] [PubMed] [Google Scholar]; (e) Singh SB, Goetz MA, Jones ET, Bills GF, Giacobbe RA, Herranz L, Stevens-Miles S, Williams DL., Jr J. Org. Chem. 1995;60:7040. [Google Scholar]
- 18.(a) Liu X, Walsh CT. Biochemistry. 2009;48:8746. doi: 10.1021/bi901123r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sims JW, Schmidt EW. J. Am. Chem. Soc. 2008;130:11149. doi: 10.1021/ja803078z. [DOI] [PubMed] [Google Scholar]
- 19.(a) Song Z, Bakeer W, Marshall JW, Yakasai AA, Khalid RM, Collemare J, Skellam E, Tharreau D, Lebrun M-H, Lazarus CM, Bailey AM, Simpson TJ, Cox RJ. Chem. Sci. 2015;6:4837. doi: 10.1039/c4sc03707c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fuji R, Minami A, Gomi K, Oikawa H. Tetrahedron Lett. 2013;54:2999. [Google Scholar]; (c) Niehaus E-M, Kleigrewe K, Weimann P, Studt L, Sieber CMK, Connolly LR, Freitag M, Guldener U, Tudzynski B, Humpt H-U. Chem. Biol. 2013;20:1066. doi: 10.1016/j.chembiol.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Tsunematsu Y, Ishikawa N, Wakana D, Goda Y, Noguchi H, Moriya H, Hotta K, Watanabe K. Nat. Chem. Biol. 2013;9:818. doi: 10.1038/nchembio.1366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.