Abstract
Since its discovery 75 years ago, a wealth of knowledge has accumulated on the role of cardiolipin, the hallmark phospholipid of mitochondria, in bioenergetics and particularly on the structural organization of the inner mitochondrial membrane. A surge of interest in this anionic doubly-charged tetra-acylated lipid found in both prokaryotes and mitochondria has emerged based on its newly discovered signaling functions. Cardiolipin displays organ, tissue, cellular and transmembrane distribution asymmetries. A collapse of the membrane asymmetry represents a pro-mitophageal mechanism whereby externalized cardiolipin acts as an “eat-me” signal. Oxidation of cardiolipin's polyunsaturated acyl chains - catalyzed by cardiolipin complexes with cytochrome c. - is a pro-apoptotic signal. The messaging functions of myriads of cardiolipin species and their oxidation products are now being recognized as important intracellular and extracellular signals for innate and adaptive immune systems. This newly developing field of research exploring cardiolipin signaling is the main subject of this review.
Keywords: Cardiolipin signaling, peroxidase, cardiolipin oxidation, mitophagy, innate and adaptive immunity, apoptosis
If you are out to describe truth, leave elegance to the tailor
– Albert Einstein
Introduction
Eukaryotic cells are believed to have evolved from a chimeric combination of an archaeon and bacterium [1-3]. Their subsequent evolution to multicellular organisms changed the biosphere and geology of our planet [4, 5]. This review will focus on the ancient, conserved and intriguing lipid, cardiolipin (CL) and its signaling functions. Understanding the context of how eukaryotic cells and subsequently multicellular organisms became so systemically prominent provides clues to eukaryotic cellular diversity, adaptation and survival. Evidence based on genomic studies suggests that a bacterial ancestor of the eukaryotic cell evolved into mitochondria (and chloroplasts) [3, 6]. Mitochondria allow aerobic eukaryotes to generate vastly more ATP per cell than their prokaryotic ancestors. Eukaryotes have up to 4-5 orders of magnitude more energy per gene than do bacteria and thus have a huge evolutionary edge over prokaryotes in adapting to changing environments [3]. This energetic and metabolic complexity, however, requires immensely more sophisticated communications between the cellular and mitochondrial components, thus demanding and stimulating new signaling and control mechanisms and pathways.
In both mitochondria and bacteria, in addition to ionic gradients, substrate oxidation-linked electron transport through the proton-extruding respiratory complexes I, III and IV forms a trans-membrane proton gradient [7]. Transduction of this proton gradient energy to ATP synthesis via complex V is a core function of mitochondria in aerobic eukaryotic cells. Respiratory complexes I, II and III also reduce O2 to O2•−. Disproportionation of O2•− catalyzed by superoxide dismutase forms H2O2, a potent source of oxidizing equivalents [8], is utilized in many signaling reactions [9-12]. The importance of H2O2 dependent reactions catalyzed by many metalloproteins, particularly heme-peroxidases, is so high that the functional significance of superoxide dismutase enzymatic activity may be attributed not to the elimination of O2•− but to the synthesis of H2O2 [13]. Notably, not only electron transport complexes but also several mitochondrial dehydrogenase complexes, such as pyruvate and 2-oxoglutarate dehydrogenases, are major potential sources of O2•−. The formation of O2•− is dependent in part on the electrochemical gradient and the redox state of electron transport components which is also dependent on oxygen and the redox balance of the cell[14]. Collectively mitochondrial and cellular oxidants are often referred to as reactive oxygen species (ROS) [15, 16].
Eukaryotic cells capable of replication have mitochondria (with one known exception, [17]) and this includes anaerobic eukaryotes that do not generate ATP using a “bioenergetic” membrane. The functions of the mitochondria clearly extend beyond ATP production. Mitochondria have a bacterial type circular genome in multiple copies that is separate from but is coordinated with the nuclear genome [18, 19] but the number of mitochondrial genes is less than 1% of their proteome. Significant gene loss also occurs in obligate intracellular pathogens and symbionts and even descriptions of the minimum gene size needed by engineered bacteria are now being defined[20, 21]. Why the mitochondrial genome is even preserved at all is not clear, but the mitochondrial genome codes for 13 mRNA's that translate to membrane protein constituents of the respiratory chain as well as for the tRNA for a 12 S and 16S ribozyme and for 22 tRNAs [18]. As mitochondria are present in most anaerobic eukaryotes, non-ATP generating functions of mitochondria, notably signaling and control mechanisms, may have dictated the evolutionary preservation of the mitochondrial genome. In spite of the likely relevance of the mitochondrial genome to non-bioenergetic functions of the organelle, all the 13 mitochondrial genes encode subunits of electron transport complexes. The enigma of the essentiality of mitochondria and mtDNA remains unresolved.
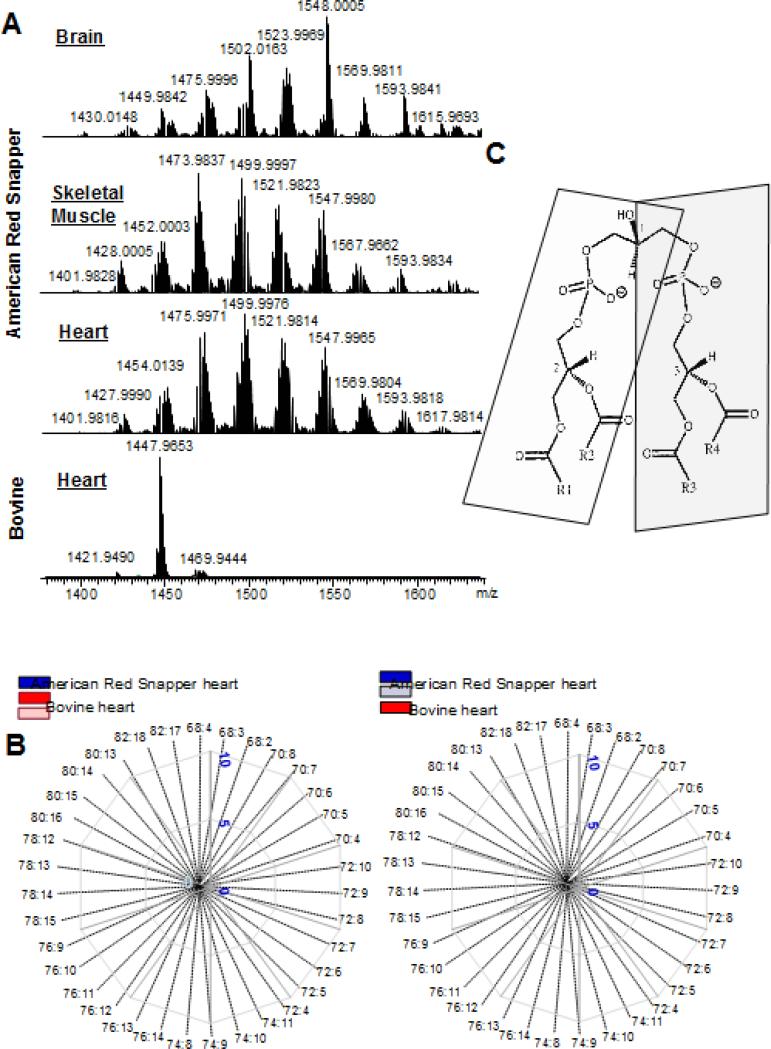
While maintaining their ATP generating functions mitochondria must undergo biogenesis, mitophagy and turnover of their complex membrane systems to optimize their many functions. The proteins comprising the respiratory chain, mitochondrial transport systems and protein synthesis along with a cohort of mitochondrial specific lipids are ancient and highly conserved mitochondrial constituents. A hallmark mitochondrial lipid present in all “bioenergetic” prokaryotic and mitochondrial membranes is CL. Cardiolipin is a unique double-charged tetra-acyl chained phospholipid that is conserved in bacteria and mitochondria. Functions ascribed to CL include maintaining a non-spheroidal membrane structure, binding specifically to membrane proteins, respiratory and super-complex stabilization and signaling [22-31]. The signaling features of CL are the focus of this review. The mitochondrial CL differs from its prokaryotic precursor by having diverse polyunsaturated acyl side chains. In addition to structurally altering the membrane bilayer, polyunsaturated acyls are also oxidation targets giving rise to myriads of signaling lipid mediators [32] The initially synthesized nascent mitochondrial CLs may include a variety of acyls (32 fatty acid residues in Lipid Maps (www.lipidmaps.org) generating a highly diversified molecular speciation of CLs. In several tissues, however, the nascent CLs with different acyls (hetero-acylated CLs) undergo subsequent synthetic transformations - a process also called remodeling - to form CLs with identical acyls, homo-acylated CLs [33-36]. This stage of the biosynthesis requires remarkable specificity of the engaged enzymatic catalysts as it has to overcome the entropy-driven diversification of randomly integrated acyls in favor of stochastically almost impossible selection of only few homo-acylated CL species. The predominant presence of tetra-linoleoyl CLs in the heart, skeletal muscles and liver of mammals [35, 37, 38] illustrates the extraordinary transacylase selectivity of the process. While it is evident that high levels of selectivity is presumably associated with some function of homo-acylated polyunsaturated fatty acid (PUFA) CLs, their actual unique role remains unidentified. It is noteworthy, however, that this exceptional and “energetically expensive” selectivity may be a mammalian evolutionary achievement as phylogenetically more ancient eukaryotic organisms, like cold blooded fish, do not display the selective accumulation of homo-acylated CLs in heart and liver. In Fig. 1A-B the hetero-acylation of CLs detected in the brain, heart and skeletal muscle of Northern Red Snapper (Lutjanus campechanus) is contrasted with bovine heart which is mostly homo-acylated. Figure 1 compares the diversification and the preponderance of different molecular species of CL in the fish and bovine heart. The fish has a CL hetero-acylation that more closely resembles the diverse range of CL seen in the mammalian brain – which has the highest tissue diversification in mammalian tissues [39-41]. The enormous diversity of Cls in the heart of a fish (Northern Red Snapper) contrasts with extremely high dominance of homo-acylated tetralinoleoyl-CL in the bovine heart.
Fig. 1. Highly diversified molecular speciation of cardiolipin are typical of fish tissues compared to the high uniformity of CLs in bovine heart.
A. LC-MS spectra of CLs from brain, skeletal muscle and heart from a Northern Red Snapper (Lutjanus campechanus) and bovine heart CL. The m/z shown are for the most abundant species in each cluster of CL. The CL species with m/z of 1447.9653 (72:8) is the major species in bovine heart but not in fish heart. B. Spider graph plots of the number (left panel) and amounts (right panel) CL molecular species in fish and bovine hearts. Species are presented as a ratio of carbon to double bonds number (C:DB). For clarity, the scales for the number of CL species in bovine heart (pale pink, left panel) and for the amount of CL species in fish heart (pale blue, right panel) were magnified in 4 and 5 times respectively. C. Cardiolipin structure. A pro-chiral carbon (1) is located on the central glycerol. The 4 acyl chains (R1, R2, R3 and R4) are linked to 2 glycerols each with a chiral carbon with the same (R) chirality on the left and right side of the molecule (2,3), highlighted by the shading. The phosphates are charged at physiological pH.
While CLs and oxidized products derived from them are now recognized as a rich and diverse source of cell signals, the significance of the diversification vs uniformity of CL is still enigmatic and represents an exciting field of future research [35, 42-44]. In this context, CLs and their oxidation products should be recognized as a novel cellular signaling language that is part of and integral to, many cellular functions [22, 24, 26, 45]. The complexity required for a harmonized spatiotemporal organization of the myriads of reactions in a multi-cellular organism as well as interactions with extra- and intracellular bacterial pathogens, involving CL, is apparent. This includes signaling during cell death, mitophagy, innate immunity and potentially many hereto unknown signaling functions. With increasing signaling complexity, the potential for defects derived from CL biosynthesis resulting in pathology is already apparent in Barth Syndrome [33, 35, 46-48] and it is likely other diseases linked to CL structure, location and stability may subsequently be defined.
This review is not at all intended to restate or rework the many fine reviews about CL, which nicely summarize and review the explosion of new literature and knowledge about CLs [26, 35, 42-44, 49-51]. Instead, our focus will be on issues of CL as a signal that are not well understood and require wisdom and experimental precision to unravel. We will provide a brief overview of CL structure and synthesis pertaining to its role in cellular signaling. A more detailed discussion of CL binding to proteins, especially cytochrome c (cyt c), its asymmetric distribution in the mitochondria, the scope of CL diversity and the role of CL in mitophagy, apoptosis and innate and adaptive immunity will provide opportunities for the reader to understand that CLs function as sophisticated cell signals. While our understanding of the complex differential tissue distribution of unsaturated and oxidized CL and its role in cell signaling is just now becoming apparent, we trust that the readers of this review will grasp the depth and complexity that CLs play in biology.
Cardiolipin, Its Molecular Structure, Homo- and Hetero-acylated Species, Asymmetry – Links with the Biosynthesis
Structure and ionization state
Cardiolipin, (1,3-bis(sn-3’-phosphatidyl)-sn-glycerol) demonstrates a molecular asymmetry, an acyl chain asymmetry and a membrane distribution asymmetry which are integral features of its functions in mitochondria. Cardiolipin is a dimeric phosphatidic acid with a “backbone” glycerol containing a prochiral central carbon, two phosphates and 4 acyl chains (Fig. 1C) [52]. The two phosphates are ionized at physiological pH resulting in CL being a doubly-negatively charged amphiphilic lipid. While the pKa of the phosphates of CL had originally been described as strong acids and were doubly negatively charged [53], a pH titration study comparing a native and 2'-deoxy-glycerol backbone suggested that there was both a low and high pKa[54]. Subsequently membrane measurements of CL using external proton adsorption, electrophoretic mobility of membranes and magic angle spinning P31 NMR have clearly shown that the pKa of CL as well as lyso-CL is ~2-3 indicating that both phosphates are ionized at physiological pH in lipid bilayers [55-57]. Thus it seems logical or almost certain that at physiological pH, CLs in lipid bilayers adopt the states with two fully ionized phosphate groups. It remains to be determined whether CL molecules interacting with a wide variety of proteins may encounter microenvironments leading to incomplete ionization of one or both phosphate groups? It is likely that binding initiated by predominantly electrostatic interactions - as happens during CL-induced peroxidase activation of cytochrome c with its positive surface charge of ~8+ [58] - is realized with fully ionized CLs, as evidenced by the iso-electric behavior of the complexes at a ratio of 1:4 during electrophoresis [59]. Similarly, 1H-NMR titrations of cyt c/CL complexes (at a 1:4 ratio) in bilayer membranes display saturable broadening of the characteristic resolved signals (Yanamala, et al, personal communications). Whether this arrangement of CL is universally characteristic of its interactions with numerous other proteins or rather engages singly ionized CL molecules, similar to the predominant formation of its singly-charged species in gas phase by electro-spray ionization during mass–spectrometric analysis (see below), - is an interesting subject for future studies.
Biosynthesis and Remodeling
Of the three known prokaryotic CL synthetic pathways, mitochondria and actinobacteria share a CL synthetic schema utilizing two CDP-diacyl glycerol (CDP-DAG) precursors while most prokaryotes use phosphatidyl glycerols. Many more details of these differential syntheses have been described elegantly including the stepwise synthesis of CL, has been the subject of numerous good recent reports and reviews [51, 60-63]. There are some aspects of the initial synthesis that have ramifications for the structure and potentially to the function of constitutive and remodeled CLs and this we highlight here.
In mitochondria, in order to attach the two phosphatidyl groups to the 1 and 3 positions of the “backbone” glycerol, two molecules of CDP-DAG are required and two separate enzymes are used in the synthesis, phosphatidyl-glycerol-phosphate synthetase, and CL synthetase. We note that the CDP-DAG has a chiral (R) carbon in position 2 of the glycerol and it is not altered during CL synthesis [64]. Accordingly the constitutive CL is synthesized with an asymmetry such that any binding across one of the two peripheral chiral centers in CL (note shading in Fig 1C), we could call it “trans-chiral binding”, confers a distinct binding motif that will differ on the “left” versus “right” sides of CL. This asymmetry is independent of the composition of the CL acyl chains. Any imputed function for this structural determinant of CL has not been addressed experimentally but may play a role in subtle tissue specific CL binding and acyl-chain remodeling.
There is a divergence between prokaryotic and eukaryotic CL synthesis after its initial synthesis. The bacterial CLs undergo limited acyl chain modifications, including (limited) formation of plasmalogen acyl chains that contain an alk-1'-enyl ether linkage as demonstrated in Clostridium difficile [65]. The mitochondrial CL however undergoes massive cell and tissue-dependent enzyme linked acyl-chain remodeling. Following its initial synthesis, CL is believed to be de-acylated (possibly by a Ca2+ independent phospholipases A2) resulting in formation of monolyso-CL (MLCL) [66-68].
Three separate enzymes have been identified in the remodeling of CL, two are acyl-transferases requiring an acyl-CoA, and the third is a trans-acylase that can rapidly “re-sort” the acyl chains by trans-acylation. While one can envision the acyl-specificity of CLs remodeled through acyl-CoA-driven pathways, it is more difficult to ascribe the production of acyl-specific, rather than random, CL products formed through entirely stochastic trans-acylation mechanism. These enzymes are i) MLCL acyltransferase-1 (MLCAT1) which is located on the matrix side of the inner mitochondrial membrane (IMM) and has also been described as a splice variant of trifunctional protein (TFP). Both TFP and its splice variant MLCLAT1 have a role in re-acylating lyso-CL using acyl-CoA [69]. ii) Acyl-CoA:lyso-CL-acyltransferase-1 (ALCAT1) is located on the outer side of the outer mitochondrial membrane (OMM) and also requires an acyl-CoA to acylate MLCL (or di-Lyso CL) [70, 71]. As either CL or MLCL would need to move from the inner side of the IMM to the outer side of the OMM for this enzyme to play a substantive role in CL remodeling, its role in remodeling may be limited to a small fraction of the CL pool. iii) Taffazin is a trans-acylase located in the inter-membrane space (IMS) in yeast and mammals. [72-76]. It is believed to be the major enzyme involved in CL remodeling. As has been mentioned above, the enzyme lacks acyl chain specificity and it is unclear how tafazzin-linked CL remodeling can result in large fractions of homo-acylated CL species such as for example, tetralinoleoyl-CL (TLCL) which constitutes up to 80% of mammalian cardiac and skeletal muscle CL. In contrast to muscle tissue, mammalian neural tissue is rich in diverse hetero-acylated CLs [77, 78]. While the heart's main function is pumping blood, this requires both a variable and uninterrupted energy supply, TLCL may be a CL structure that is speculated to have a special role in responding to the high energetic demands needed for the cardiac contractile function [79]. Acyl chain substrate availability and nutritional pressure, and/or the removal of selected CL-acyl products could drive this enzyme linked trans-acylation to concentrate some forms of homo-acylated CL, but details of these enzyme-linked synthetic/remodeling activities are lacking. In the absence of tafazzin in yeast mitochondria, homo-acyl-CL is lost but the mitochondria still function normally but with a far more hetero-acylated CL composition [36, 42, 64]. As the role of remodeled CL is not exclusively to maintain the integrity of the mitochondrial inner membrane, alternative functions of CL including signaling are likely [80].
Cardiolipin is located asymmetrically within the IMM. Structurally, the four acyl chains of CL coupled with the double-negatively charged polar end provide CL with a “conical” shape, with the polar region being the cone cusp, and the flexible and variable acyl chains being the base of the “cone”. The length, saturation and oxidation of these side chains, provide differential influences on the shape, binding, function and stability of CL [35, 48, 81, 82]. This “conical” shaped and double negatively charged CL is asymmetrically distributed in the IMM with a higher concentration of CL on the inner side of the IMM. The respiratory complexes and the adenine nucleotide carrier have an absolute requirement for CL and these complexes constitute much of the IMM protein [83-86]. With a high protein/lipid ratio ~3:1 in the IMM [87] and with CL being about ~20% of the (cardiac) mitochondrial phospholipids, the weight ratio of protein/CL phospholipids is ~15:1. It is likely much of the CL is utilized to facilitate tight membrane folding [27, 30] and as a “lipid stabilizer” of the respiratory complexes and supercomplexes [38, 88-91]. A cryo-EM map of one super-complex consisting of a dimer of complex III and two monomers of complex IV was reported to contain 50 CLs [92]. Clearly a significant portion of the CL in the IMM is bound to respiratory-linked proteins. Thus CL has many obligate protein binding sites in the IMM as well as opportunities to relay information about the structural integrity of the respiratory complexes and supercomplexes as it tethers lipophilic domains of membrane proteins.
The process of synthesizing homo-acylated TLCL may seem cumbersome but why the mitochondria in select tissues “remake” CL as mostly homo-acylated CL is open to speculation and experimentation. In terms of signaling and even more intriguing are the CL modifications that occur to “the rest” of the CL. Hetero-acylated CLs are synthesized with varying acyl chain lengths, desaturation and tissue specificity [77, 80]. Each CL species (the diversity range and synthetic locations are outlined later in this review) will have subtle structural differences and the polyunsaturated acyl chains are each candidates for lipid peroxidation. Like their unsaturated phospholipid counterparts, many CL oxidation products have the potential to convey information to receptors of lipid oxidation products. Oxidized CL acyl chain fragments can dissociate from CL to convey information. With multiple binding sites of CL to proteins, its synthesis as a molecule with a chiral asymmetry, its subsequent acyl exchange and re-modeling, then its requisite movement to non-mitochondrial sites invite questions about understanding more about the remodeling process, why it occurs and how information from CL molecules is used to convey signals within and beyond the cell.
Diversity of Cardiolipins – Theoretically Possible vs Experimentally Detectable CL Species in Cells and Tissues Detectable by LC-MS
The structure of CLs with their four acyl chains (and 32 fatty acids) makes them not only unique but also one of the more structurally diverse lipid molecules with a potential diversity exceeding 106 molecular species across different types and classes of life. Considering possible CL metabolites, particularly oxidative metabolites with several oxygen-containing functionalities such as hydroperoxy-, hydroxy-, oxo-, epoxy- that may be formed in different positions of the polyunsaturated acyls and adding to this oxidatively-truncated CL derivatives – the total number of CLs and their metabolites may exceed 107 individual species. This huge diversity is not found experimentally as only some of the polyunsaturated fatty acids are present (Fig. 2), but tissue and cell specific CL diversity is broad and seems to be enzymatically and dynamically controlled. What is clear is that nutritional pressure as well as disease conditions associated with enhanced phospholipid oxidation may significantly increase the diversity of CLs. Assuming that CLs are used for signaling purposes where every molecular species or a combination of several of them acts as a regulatory signal or a word in the signaling language, then the richness of this communication language is extraordinary. Notably, one can trace the evolutionary trends in the development of CL language. For example, 5 major fatty acids in prokaryotes (without oxidizable polyunsaturated species) can produce up to 625 species. The 14 major fatty acids, including polyunsaturated forms, in eukaryotes can produce 38,416 distinct CL species [93]. These numbers may increase an order of magnitude if CL oxidation products are included. Although these calculated levels of diversification are theoretically possible, most CLs reported in biological systems are enriched with a select number of fatty acids reducing the actual number of species observed [64].
Fig. 2. Expansion of the diversity of CLs.
The initially synthesized 4 CL acyl chains can be saturated, mono- unsaturated or poly-unsaturated. Remodeling is initiated by deacylation of one or more acyl chains and with enzymatic precision a wide variety of fatty acids can re-acylate the lyso-CLs forming organ and tissue specific variations of CL content.
There is currently no single comprehensive analytical method for identification of all possible CL species. Recent developments in soft ionization techniques and mass spectrometry (MS) have provided better insights into the identification and characterization of diverse CL species in biological systems [32]. Most protocols currently used for MS analysis of CL consist of two stages. A schematic outline of sample preparation extraction, lipid analysis, data and statistical analysis are presented in Supplemental Fig.1.
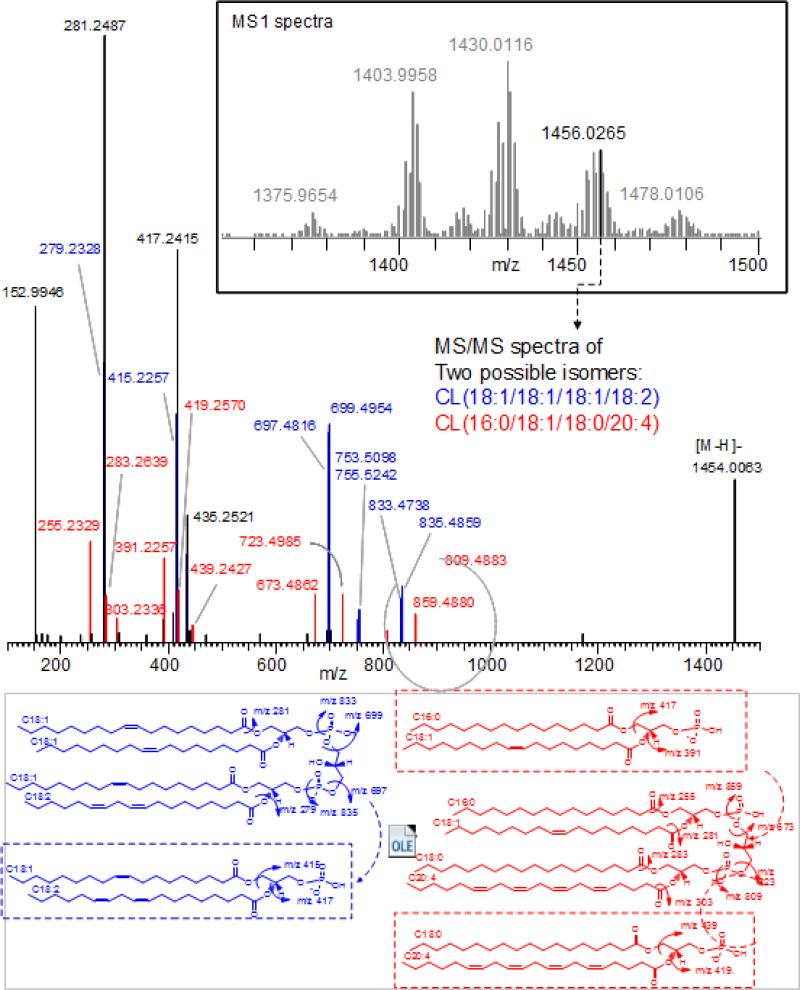
The first stage or MS1 distinguishes the species based on their molecular weight. Since many of the CL species are isomers this stage usually identifies groups of species that have the same molecular mass. For example 12 different molecular species of CL could account for the molecular weight of 1451 (m/z 1450.0) [80]. The second stage of mass spectrometric analysis or MS2 identifies the fragments generated from a particular CL molecule with a specific mass to charge ratio. Mass spectra obtained in MS2 provide the identities of acyl chains present in a particular CL molecule (Fig. 3) [94]. However the exact location of the acyl chains (regioisomers) cannot be identified.
Fig. 3. Detailed structural information can be obtained by contemporary high resolution MS equipment: The Orbitrap™ high energy collision dissociation (HCD) MS2-spectra of cardiolipins with the [M-H]- ion at m/z 1371.9359.
Two isomers CL(18:1/18:1/18:1/18:2) and CL(16:0/18:1/18:0/20:4) were identified. CL(18:1/18:1/18:1/18:2) (in blue): The characteristic structural fragments includes: [PG(18:1/18:1)+Phosphate-H]− and [PG(18:1/18:2)+Phosphate-H]− ions of m/z 835 and m/z 833, respectively; neutral loss (NL) of phosphate group from above moieties generating [PG-H]− ions with m/z 755 and m/z 753, respectively. [PA(18:1/18:1)-H]− and [PA(18:1/18:2)-H]− ions of m/z 699 and m/z 697, respectively; NL of C18:1 from above PA moieties generating m/z 417 and m/z 415 (box), respectively. The individual fatty acid carboxylate anion was also observed with m/z 281 and m/z 279, corresponding to C18:1 and C18:2, respectively. CL(16:0/18:1/18:0/20:4) (in red): The characteristics structural fragments includes: [PG(16:0/18:1)+Phosphate-H]− and [PG(18:0/20:4)+Phosphate-H]− ions of m/z 809 and m/z 859, respectively; [PA(16:0/18:1)-H]− and [PA(18:0/20:4)-H]− ions of m/z 673 and m/z 723, respectively; neutral loss (NL) of fatty acids from above PA moieties generating m/z 417, 391, 439 and 419 (box). The individual fatty acid carboxylate anion was also observed with m/z 255, 281, 283 and 303, corresponding to C16:0, C18:1, C18:0 and C20:4, respectively. m/z 153 was ion of glycerol-3-phosphoate with NL of H2O, the characteristic ion of phospholipids. Shared fragments were marked in black in MS/MS spectra. Abbreviation: NL, neutral loss; PA, phosphatidic acid; PG, phosphatidylglycerol
The two most commonly utilized MS approaches for the analysis of CL include: shotgun lipidomics and chromatographic separation followed by tandem MS (LC-MS/MS). Shotgun lipidomics employs direct infusion of lipid extracts into the mass spectrometer. Using a shotgun lipidomics strategy, Han and colleagues identified 56 isomeric and ~100 individual molecular species (excluding regioisomers) of CL in mouse brain tissue [95]. Because CLs comprises a small percentage of total phospholipids it is often difficult to analyze them in sufficient detail without separation of lipid extracts by different chromatographic techniques. Different liquid chromatography (LC) techniques thus have been developed to separate phospholipids and enhance detection of CL [95-97]. Using a two-dimensional LC (2D-LC) MS, we have identified 178 individual molecular species of CL in rat brain [77]. Notably mammalian brain tissue has the most diversified CL assortment compared to other tissues [98, 99]. For example heart, liver and skeletal muscle display 39, 25 and 40, respectively, individual species of CL in mice; laboratory yeast cells have 17 and E. coli has 56 different CL species [95].
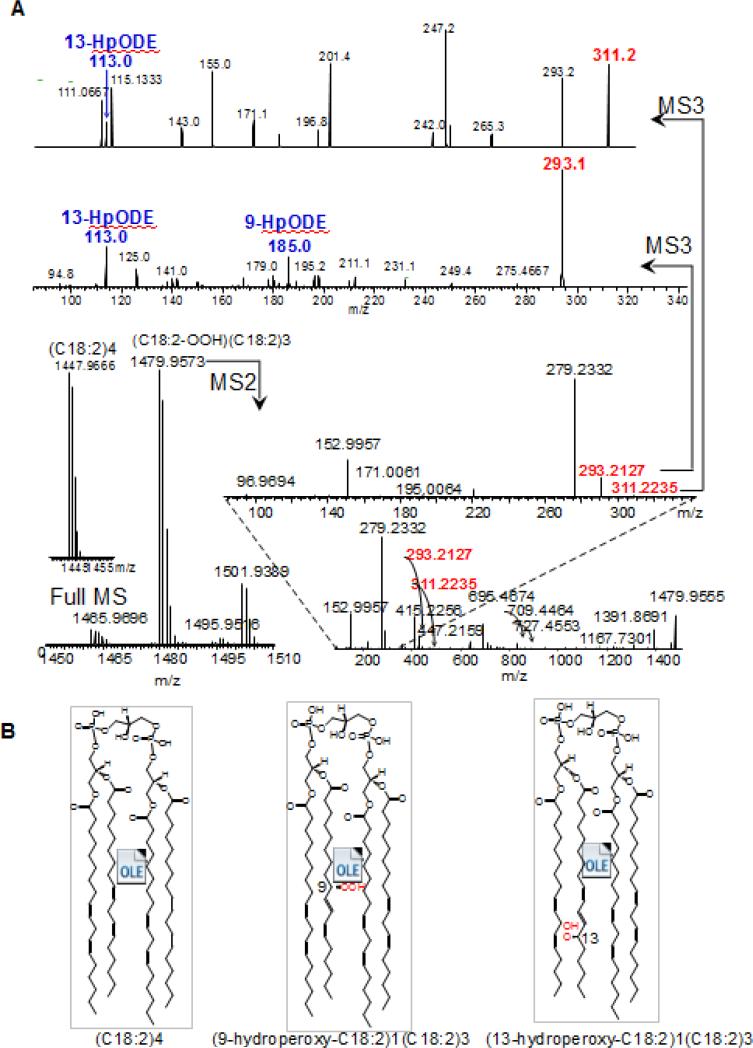
Recent developments in accurate mass and high sensitivity MS further expands the limits for identification and quantification of low abundance CL species in plasma and oxidized CL species in tissues [32, 41]. As an example, a newly developed Tribrid™ Ion-trap/Orbitrap™ technology (Fusion-Lumos, ThermoFisher Scientific) allows for unequivocal structural characterization of very low abundance oxidation products of CL with identification of the positions of oxygen-containing group in the fatty acyls in MS3 spectra (Fig. 4). While these enhanced CL analytical methods improve the identification of diverse and low abundance CLs, the emerging challenges of lipidomics and redox lipidomics will be to discover the links between the low abundance CL species, including oxygenated CLs, and their functional roles in normal physiology and disease conditions.
Fig. 4. MS2/MS3 fragmentation analysis leads to detailed structural characterization of oxidized CLs.
An example showing the increased resolution of the structural characterization of CL-(C18:2)4 hydroperoxy molecular species formed in an AAPH (2,2'-azobis-2-methyl-propanimidamide, dihydrochloride) driven reaction using a Tribrid™ MS Fusion Lumos (Thermo Fisher Scientific, San Jose, CA). A. Full, MS2 and MS3 spectra of (hydroperoxy-C18:2)1(C18:2)3. Daughter molecular ions formed during MS2 analysis corresponding to the oxygenated C18:2 are shown in red. Characteristic fragments of 9-hydroperoxy-C18:2 and 13-hydroperoxy-C18:2 formed during MS3 analysis are shown in blue. B. The structures of the detected nascent CL (C18:2)4 and two of its oxidized products, (9-hydroperoxy-C18:2)1(C18:2)3-CL and (13-hydroperoxy-C18:2)1(C18:2)3-CL as identified and characterized using MS2/MS3.
Imaging mass spectrometry: CL distribution in neural tissues
As much as the power of contemporary LC-MS technologies allows for complete structural identification and in many cases quantitative analysis of different diversified CLs in organelles and cell bio-fluids the spatial distribution of CLs in tissues cannot easily be assessed by these protocols. Recent successes in MALDI-MS based imaging have resulted in the development of MS-imaging or “biochemical microscopy” [100], that has been applied to a number of different phospholipids [101]. Imaging mass spectrometry has the unique advantage of allowing the correlation of molecular mass to charge (m/z) information with the topographical (regional/spatial) orientation of a broad spectrum of analytes including peptides, proteins, lipids, small molecules/cellular metabolites, and drugs in a label-free manner [40, 101-103]. The low abundance and high diversification of CL in some tissues such as brain made it difficult to perform MS imaging of CLs on frozen tissue sections because of CL diversity and low abundant species. However, by using protocols that minimize interfering signals from the major membranes phospholipids – such as phosphatidylcholines and phosphatidylethanolamines – the task of imaging of CLs in brain has been resolved (Fig. 5). These techniques use a combination of chemical and enzymatic treatments of brain tissue sections combined with a modified matrix application device that allows for the visualization, mapping and fragmentation of CL species [101]. Employment of contemporary MALDI-MS equipment (e.g, RapiFlex™, Bruker-Daltonics) permits acquisition of excellent images of CLs in tissues with their utmost diversification, such as measured in brain.
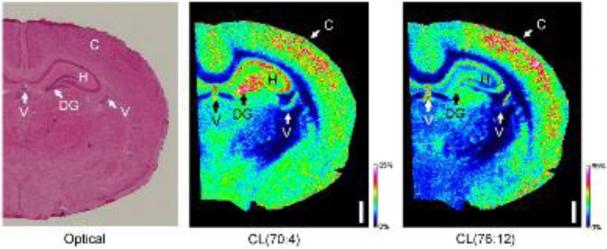
Fig. 5. Biochemical microscopy by MALDI-MS imaging reveals the differential distribution of CL species in various anatomical areas of the brain.
Serial rat brain coronal sections were prepared. The left panel shows an optical image from a hematoxylin and eosin stained section using a Nikon-90i™ microscope. MALDI-MS imaging detects 23 species of CL at a lateral resolution (pixel size) of 50 microns, two of these species are shown in the middle and right panels. Predominantly saturated species of CL such as CL(70:4) (middle panel) are strongest in the hippocampal region (H), especially the dentate gyrus (DG), and moderately less intense in the cortex (C) and ventricles ( V). Highly unsaturated species of CL such as CL(76:12) (right panel) are strongest in the cortex and ventricles (C, V) and less intense in the hippocampus (H). The images were acquired on a Bruker Daltonics-RapiFlex™ MALDI, using the protocol from [101]. Ions detected as [M-H]1- and intensities are relative with respect to the given ion. The white bar = 1 mm.
Principles of Cardiolipin Binding to Proteins – Emphasis on Mitochondrial Proteins
Interactions of CL with proteins are pivotal for understanding many CL functions. Numerous proteins have been documented to interact with CL, and the presence of CL is essential for many of them structurally and functionally [104]. For instance, CLs bound to respiratory complexes III and IV are crucial for electron transport. The CL binding sites of cytochrome bc1 are located on the membrane-exposed surface of the respiratory chain complex III [105]. In addition to maintaining the structural integrity of complex III, CLs spontaneously diffuse to the interface of the cytochrome bc1 dimer to the higher potential heme b groups of the complex's catalytic Qi-site (the quinone reduction site), which suggests that CLs have a role in proton uptake [106]. In the same manner, CL molecules are tightly bound to cytochrome c oxidase, the terminal complex of the respiratory chain and are essential for its function [107]. Two CL binding sites of cytochrome c oxidase are involved in pathways linking CLs to the entrance of the D and H proton channels across the enzyme complex, which highlights the role of CLs in the proton delivery machinery to complex IV [108].
Not only membrane proteins, but also several peripheral membrane proteins and even soluble proteins are known to interact with CL, stressing its role in diverse cellular pathways. For example, microtubule-associated protein 1A/1B-light chain 3 (LC3), which is involved in mitophagy, recognizes the externalized CL on the damaged mitochondria [109]. The double point mutation of two arginines in the N-terminus of LC3, an area rich in hydrophobic residues, diminished the degradation of defective mitochondria [109]. It has also been documented that nucleoside diphosphate kinase Nm23-H4/NDPK-D, binds to CL on the IMS side of IMM and transports CL to OMM through a proposed rotary machinery system [110]. Interestingly, the analysis of the CL-binding sites of these proteins revealed the presence of both, basic and hydrophobic residues, confirming the need for both, Coulomb and van der Waals forces in CL interactions with proteins.
To investigate the generality of this statement, a recent systematic analysis of known CL binding proteins with particular emphasis on those where structural information was available has been carried out [104]. An in-depth analysis of the amino acids, secondary structure elements and structural motifs in CL-binding sites in co-crystal structures allowed deriving of several common features. Most strikingly, CL-binding sites are highly flexible. This was suggested by a higher content of Gly, a lower content of Pro and predominance of coiled-coil secondary structure. Furthermore, as expected, overrepresentation of Arg and Lys in phosphate binding patches and of Leu, Ile, and Val in acyl chain binding patches confirmed the previously known involvement of electrostatic and hydrophobic interactions in binding. Further supporting the conclusion of the importance of positive charges is the fact that negatively charged residues (Asp and Glu) are either underrepresented or disallowed in head group binding sites. There was also an overrepresentation of Tyr in the head group binding region, which is a widely used residue in signaling due to its ability to be phosphorylated. Phosphorylation and the introduction of negative charge at a Tyr site would be expected to drastically alter CL binding properties. Specific super-secondary structural motifs also known as arches or iLoops [111] were also identified in binding patches (Fig. 6) and in particular loop MCL.GH.2.19.1, (Fig. 6A) which is characteristic for the mitochondrial ATP/ADP carrier. Paralleling previous studies in Protein-protein interactions [112, 113], it is tempting to speculate that this flexible loop containing a positively charged residue in the phosphate groups binding region, and a helix facilitating an anchor for acyl chain binding, may be considered a footprint for CL binding.
Fig. 6. Computer modeling illustrates the major modes of CL interactions with proteins.
Structural details of the interaction between CL and: (A) the ATP/ADP carrier (PDB 1okc); (B) the cytochrome C oxidase (PDB 3w7g) and cytochrome C1 heme protein (PDB 1P84). The CL molecule is shown in sticks, oxygen atoms in red, phosphorus atoms in orange and carbon atoms in pale gray. When super-secondary structural motifs (loops) were identified near the CL molecule, these are highlighted in cyan (helix) and light-pink (coiled coil) colors (A and B) the closest protein region is shown in light blue (C). Over the cartoon representation of secondary structures, a mesh network displays physico-chemical properties of amino-acids neighboring the CL molecule: in orange, hydrophobic; in blue: positively charged; in red, negatively charged. The key CL-binding motif [KR]-(X)-G is highlighted using sticks representations and lime-green color for Gly and blue-purple color for Arg/Lys.
There are two areas that require further work, (1) the specificity of these features for CL, and (2) the classification of CL-binding modes according to protein/enzyme functions. Thus, future work needs to explore the generality of the findings for CL. In particular, other anionic phospholipids or other doubly negatively charged small molecules such as GDP or ADP may show overlap in the structural and dynamic features in their respective binding pockets. Furthermore, the CL binding mode and affinity to respiratory complexes, in which CL mainly is in bound state, may vary from proteins proposed to be involved in CL translocation —alternating bound/unbound states are essential for this functionality— either between IMM and OMM (such as NDPK-D and mitochondrial creatine kinase) or across the membrane (like the adenine nucleotide translocator) [114, 115].
Interactions of Cytochrome c with Cardiolipins – Role in Catalysis of Cardiolipin Oxidation
Redistribution of CL between the IMM and OMM in depolarized mitochondria facilitates its interactions with unusual positively charged protein partners spatially separated in intact organelles. One of such partner is cyt c - a highly positively charged hemoprotein located in the IMS and shuttling electrons from respiratory complex III (cytochrome c reductase) to complex IV (cytochrome c oxidase) [116]. The disruption in the asymmetric distribution of CL, which occurs at the onset of apoptosis, leads to tight membrane binding of cyt c. The complex of cyt c/CL gains peroxidase activity realized in the presence of oxidizing equivalents such as H2O2 generated by disrupted electron transport activity or organic (lipid) hydroperoxides [59]. As CL is a highly selective substrate of the peroxidase reaction (see below), CL peroxidation is a characteristic feature of apoptosis required for the completion of this death program [117]. Not surprisingly, substantial efforts have been directed towards design and development of new inhibitors of cyt c /CL peroxidase activity as anti-apoptotic modalities [118, 119]. In spite of numerous studies [120-125] the universal structural characterization of CL-driven activation of cyt c into a peroxidase still has not been achieved.
Clearly, the peroxidase function of cyt c may require a subtle “loosening” or more substantial reorganization of its hexa-coordinated heme-iron environment [126]. The degree of the structural re-arrangements defines the potency of peroxidase activation. Several models based on detailed solid state NMR studies as well as crystallographic work suggest that small scale structural changes are sufficient for inducing the peroxidase competence of cyt c/CL complexes [127, 128]. These data complement the kinetic studies [129] in which a very early peroxidase activity was detectable before any substantial changes in the coordination of heme Fe with one of the axial ligands, (Met80), were detectable.
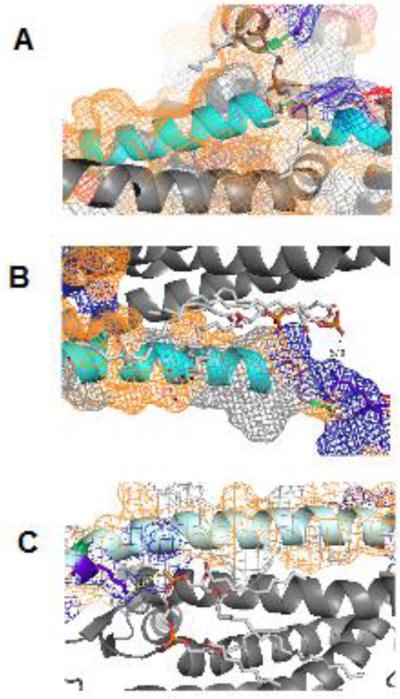
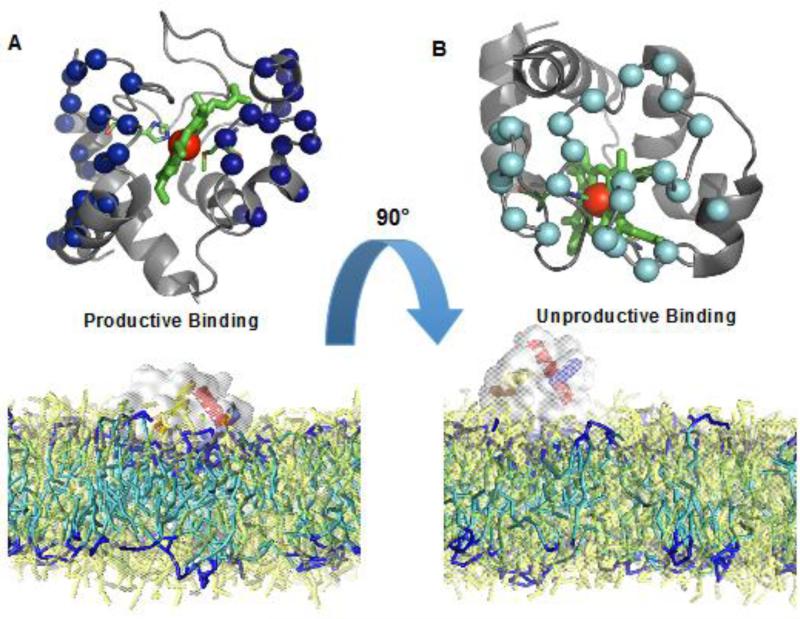
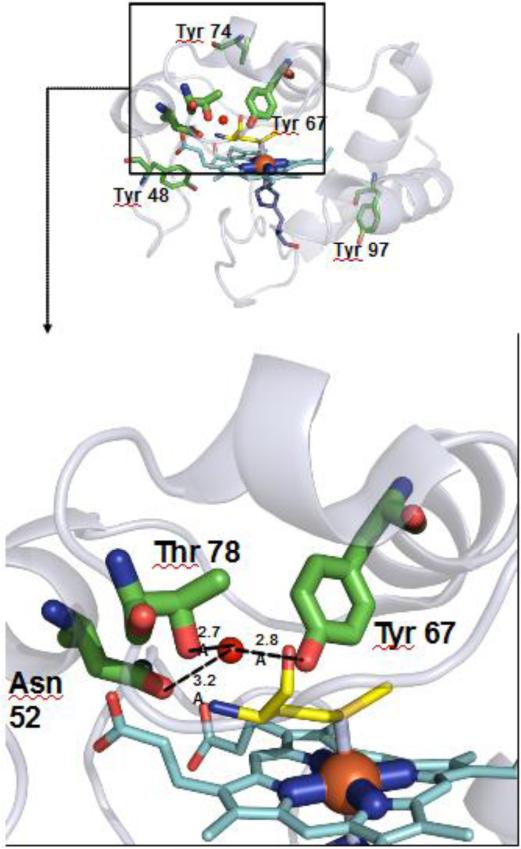
More profound structural changes realized via interactions with three CL-binding sites for cyt c were revealed by studies using solution NMR and molecular modeling (Fig. 7). Involvement of two binding sites located on both sides of a heme crevice introduces a membrane deformation and tight interactions. The ability to bind CLs simultaneously at the proximal and distal sides of the heme and the consequent induced strain due to this binding, opens up the heme crevice providing a path to the formation of the peroxidase. Thus simultaneous binding of the distal and proximal sites can be considered as “productive” binding mode leading to a robust peroxidase activation. Time-resolved FRET measurements of fluorescently-labeled cyt c revealed a conformational diversity of the CL-bound protein with distinct populations of the polypeptide structures that varied in their degree of protein unfolding [130]. Notably, a fraction of the complexes were strongly unfolded similarly to the fully denatured state of the hemoprotein induced by treatment with guanidine hydrochloride. Based on several studies of the peroxidase activation of cyt c by denaturing agents [131] it is likely that these “robustly” denatured forms of cyt c/CL complexes are mostly accountable for the peroxidase activity. CL peroxidation requires the formation of a protein-immobilized radical, specifically tyrosyl radicals (Tyr•). The latter act as immediate reactive intermediates of the peroxidase cycle [132]. Among four tyrosines (Y48, Y67, Y74, and Y94), mutation of Tyr67 (Tyr67Phe), the closest to the heme-iron caused the most significant loss of the protein-immobilized radical EPR signal and loss of the peroxidase activity of the cyt c/CL complex [132]. This suggests the highly conserved Tyr67 and its radical are likely redox intermediates of the cyt c/CL peroxidase complex and formation of the radical is facilitated by the protein structural rearrangements that are necessary for CL peroxidation [132] [133] (Fig. 8). An interesting aspect of cyt c's involvement in CL peroxidation is its phosphorylation [134]. Notably, two tyrosines, Tyr97 in bovine heart and Tyr48 in bovine liver – but not Tyr67 - have been identified so far as phosphorylation sites in cyt c [132, 135].
Fig. 7. Molecular dynamic simulations illustrate different modes of cyt c interactions with CL-containing lipid bilayers.
Two separate binding orientations are shown. A) Productive binding involves tighter interactions and deeper penetration into the membrane facilitated by CL sequestration leading to peroxidase activation. B) Unproductive binding engages smaller amounts of sequestered CL and does not lead to peroxidase activation.
Fig. 8. Partial x-ray structure of cyt c illustrates the proximity of a candidate Tyr residue involved in peroxidase activation by CL.
The upper panel shows the positions of the cyt c tyrosines (Y48, Y67, Y74, and Y94). Y67 is located in the closest proximity to the heme-iron (lower panel). Y67 acts as a donor of an electron for a porphyrin cation-radical generated as a reactive peroxidase intermediate. In the oxygenase half-reaction of the cyt c /CL peroxidase complex, Y67 radical acts as an electron acceptor from oxidizable CL.
According to the results of computational modeling, one of the characteristic features of the peroxidase gain of function is that increasing the CL concentration enhances the affinity of cyt c for the membrane. Remarkably, cyt c recruits CL to form a cluster in the membrane, which demonstrates that multiple CL molecules (not individual molecules in isolation) stabilize the cyt c /CL complex. On the basis of coarse-grained molecular dynamics studies a model has been proposed [136] in which cyt c is partially and stably embedded in a locally curved and CL-rich membrane patch. In the context of patho-physiologically relevant events, the structural data clearly indicate that the “deepness” of cyt c rearrangements are dictated by the amounts of available CL in the membrane microenvironment. Translocation of CLs from the IMM to the OMM may be the major factor controlling peroxidase activation of the hemoprotein enhancing CL oxidation.
Mitophagy
Being central to the bioenergetics and regulation of metabolism, mitochondria also represent the major threat to the existence of cells, tissues and the entire organism. This teetering between the remarkable benefits offered by and dangers associated with mitochondrial functions has been known for more than five decades since the recognition of the substantial production of O2•− radicals and H2O2 by altered electron transport through the respiratory complexes [137-140]. Recently, it has become evident that several redox enzymes of the Krebs cycle (e.g. 2-oxoglutarate dehydrogenase (OGDH), branched-chain 2-oxoacid dehydrogenase (BCKDH), and pyruvate dehydrogenase (PDH) complexes are also capable of considerable superoxide/H2O2 production. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I [141] and represent an additional source of ROS with the rates of their generation exceeding that of electron transport [142]. This redox-associated imbalance (also called oxidative stress) is believed to potentially turn into a completely uncontrolled pro-oxidant reactions catalyzed by high concentrations of a variety of catalysts containing transition metals. Indeed, mitochondria are the site of heme synthesis and they are enriched with hemoproteins, in addition to abundant non-heme iron-sulfur clusters, and catalytic copper [143, 144]. Aside from the most potent inducers of redox stress, mitochondria's calamitous potential has become particularly obvious with the appreciation of the pro-apoptotic propensity of mitochondria-compartmentalized cyt c to facilitate the formation of apoptosomes thus triggering the programmed apoptotic death pathway [145]. Moreover, activation of the intracellular pro-inflammatory machinery and extra-cellular immunogenic responses to mitochondrial DNA and formyl-peptides – the attributes of their association with the prokaryotic ancestors – add-up to the potential perilous consequences of mitochondrial dis-organization. Finally, because of the strict dependence of mitochondria on the trafficking of most of their proteins encoded by the nuclear genome, defective protein import may occur that results in the accumulation of mistargeted proteins as well as misfolded and aggregated proteins. These metabolic misfortunes cause stress responses the mitochondrial unfolded protein response (UPRmt) that aims to increase the capacity of protein quality control mechanisms inside mitochondria [146].
Not surprisingly, cells developed highly effective systems of mitochondrial quality control in which the prominent roles are played by the mitochondrial unfolded protein response and mitophagy - a specialized mitochondria-targeted process of autophagy, or piecemeal autophagy of mitochondrial subdomains. These specialized mechanisms appear to mitigate mitochondrial impairment [147] by removing dysfunctional mitochondria under normal physiological conditions and in response to pathological stresses.
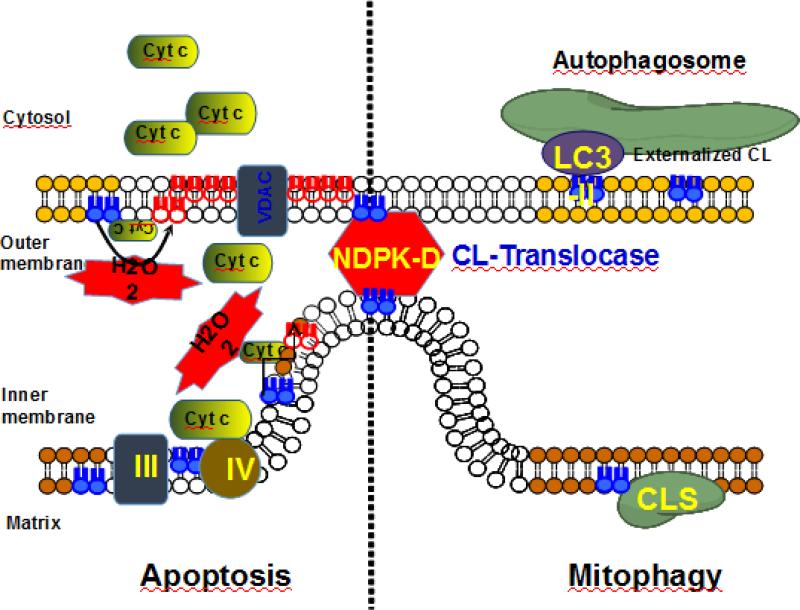
Autophageal machinery delivers damaged mitochondria into double-membrane vesicles, autophagosomes, that eventually fuse with late endosomes or lysosomes where lysosomal enzymes degrade the autophagosome contents including the inner autophagosomal membrane. Selective autophageal cargo receptors (p62 and its paralog NBR1, NDP52 and its paralog T6BP, and optineurin) link their bound mitochondrial cargo to the autophagosomal membrane by interacting with lipidated (by phosphatidylethanolamine) ATG8 proteins, microtubule-associatedprotein-1 light chain 3 (LC3)/Gamma-aminobutyric acid receptor-associated protein (GABARAP) that are intimately associated with the autophagosome membrane cargo receptors [148]. A separate and important issue is the nature of signals on the surface of damaged mitochondria that are recognized by ATG8 proteins. It is likely that both protein-based signals as well as lipid signals may be involved in the recognition process. Among the former the mitochondrial kinase PINK1 and subsequently recruited RBR E3 ubiquitin ligase, Parkin are believed to be involved in the detection of damaged mitochondria [149]. Among the latter, CLs have recently attracted significant attention as signaling molecules appearing on the surface of damaged mitochondria [26, 109]. The autophagy protein (LC3), which mediates both autophagosome formation and cargo recognition, contains CL-binding sites important for the engulfment of mitochondria by the autophagic system (Fig. 9).
Fig. 9. Externalization of CLs to the mitochondrial surface occurs during mitophagy whereas oxidation of CLs is necessary for the execution of apoptosis.
In mitophagy CL is translocated by NDPK-D which binds to both the IMM and OMM, facilitating CL movement to the OM where it can bind and activate LC3-II which initiates autophagosome formation leading to mitophagy. In the figure the CL is colored blue, other lipids are brown and oxidized CL is colored red. In apoptosis CL forms a complex with cyt c converting it to a peroxidase, H2O2 is used as the oxidant of CL and the net result is the release of cyt c through the OMM and into the cytoplasm, as well as externalization of CL to the OMM resulting in apoptotic death.
In normally functioning mitochondria, CL is confined to the IMM, predominantly to its matrix-oriented leaflet in the proximity of the site of its synthesis (see above). Recognition by the cytosolic LC3 requires the presence of CL on the mitochondrial surface suggesting that trans-membrane re-distribution of CL has to take place in damaged mitochondria undergoing mitophagy. In mitochondria maintaining their membrane potential, the majority of the IMM CL is engaged in the interactions with different proteins, including ETC super-complexes and only negligible amounts of CL are present in protein-free form [89]. This is well illustrated by recent 31P-magic angle NMR data demonstrating essentially lack of discernable CL-derived signals from normal mitochondria [150]. This organization, including the tight binding of CL, is dramatically changed in depolarized mitochondria treated by protonophoric uncouplers and other mitochondria damaging agents. Indeed, direct LC-MS measurements established significant enrichment of the OMM with CL [151] after exposure to the uncouplers, rotenone, staurosporine, 6-hydroxydopamine. Because molecular speciation of the IMM and OMM CL is very similar, it is most likely that redistribution - rather than activation of biosynthetic pathways - is mostly accountable for the externalized on the mitochondrial surface CL [80].
The information on the mechanisms of CL-translocation from the IMM to the OMM is scarce. Assuming that most of the CLs are present on the matrix side of the IMM, three steps are required for CL to reach the mitochondrial surface; i) through the IMM, ii) through the intermembrane space, and iii) through the OMM. In spite of the significant progress in characterization of the machinery for protein trafficking into mitochondria, the mechanisms of CL translocations remain largely unknown. Recent studies indicate that a hexameric intermembrane space protein, NDPK-D (or NM23-H4), binds CL and facilitates its redistribution to the OMM thus identifying one of the candidate proteins involved in the CL transfer through the intermembrane space [151]. The putative binding site of the protein is in the proximity of R90 as established by the loss of activity in R90D mutants of NDPKD. Interestingly, the affinity of NDPKD for CL is not high suggesting that there may be an important threshold effect preventing excessive sensitivity of the transfer mechanisms to stochastic fluctuations of CL concentrations. This specific feature is also important for the prevention of withdrawal of CL from its associations with many mitochondrial proteins. The regulation of the transfer process may include several control mechanisms. Mitophagy-inducing CL-transfer activity of NDPK-D is closely associated with the dynamin-like GTPase OPA1, implicating fission-fusion dynamics in mitophagy regulation. In agreement with this regulatory mechanism, CL binding suppresses kinase activity of NDPKD. While the overall dependence of mitophagy and CL externalization on the total content of CL and its integrity have been established, the translocation mechanisms of CLs within IMM and OMM are yet to be deciphered although several proteins have been proposed as suspects. For example, uncoupling proteins and anionic transporters in the IMM as well as phospholipid scramblase-3 in the OMM may participate in the respective translocation pathways [26]. The necessity of focused research in this area is emphasized by the discovered associations between disturbances in CL metabolism and mitophagy in disease (eg, Barth syndrome, Parkinson disease [33, 46, 152, 153].
Selective Cardiolipin Oxidation: Role in Apoptosis
There is a belief that excessive production of superoxide radicals in mitochondria by ETC and Krebs cycle dehydrogenases may cause poorly controlled random reactions of chemical oxidation of different biomolecules, including (phospho) lipids [143, 154]. This concept has brought to life the idea of antioxidants - acting predominantly via mechanisms of free radical scavenging – as protectors against oxidative stress and its exuberant manifestations as oxidative injury. While the concept may still retain its value as an interesting chemical excursion into the land of metabolic biochemistry under extreme conditions, experimental tests and clinical trials have not provided any significant support to the dominant stochastic chemistry of oxidation reactions in cells, tissues and the body [155]. In contrast, alternative ideas of enzymatically regulated oxidative reactions, including reactions of phospholipid peroxidation, have been put forward as the leading cause of oxidative signaling whereby dis-regulation of these reactions may lead to the accumulation of excessive amounts of lipid oxidation products, albeit in specific rather than random molecular targets [156].
One of the most informative approaches to experimental testing of these ideas is via the use of redox lipidomics based on quantitative and structural characterization of phospholipid oxidation products. Because the rate of lipid oxidation strongly depends on the number of double bonds in the molecule, phospholipids are expected to be oxidized dependently on the levels of their poly-unsaturation rather than on the nature of their polar head-group [157]. In contrast, enzymatically catalyzed oxidation reactions may affect specific classes of phospholipids. With this in mind, it is interesting to perform analysis of the specificity of phospholipid peroxidation in several experimental models of tissue injury leading to a specific type of programmed cell death, apoptosis. Brain trauma, neuronal damage after cardiac arrest, substantia nigra after exposure of animals to rotenone, hyperoxia and nanoparticle exposure of the lung, gamma-radiation induced damage to small intestine have been long recognized to initiate tissue damage and mitochondria-mediated intrinsic apoptotic cell death accompanied by characteristic and significant oxidative damage. Intriguingly, a very high level of phospholipid specificity has been discovered as one of the most characteristic features of oxidation reactions triggered by these very different stimuli in different tissues with the common denominator – massive apoptosis. Mitochondrial CLs were found universally and most abundantly oxidized during injury associated with these insults [25, 41, 77, 80, 158]. Strikingly, CL oxidation turned to be non-random as well but had a pronounced specificity unrelated to the pattern of CL polyunsaturation. Accumulation of hydroxyl- and hydroperoxy- derivatives of CL was observed in tissues of the affected animals whereby C18:2 acyls – rather than more polyunsaturated C20:4, C22:5 or C22:6 acyls – were predominantly involved in the oxidation process. This suggests that a specific enzymatic mechanism, such as cyt c/CL peroxidase, might be predominantly responsible for the production of pro-apoptotic death signals [80].
A recently developed optolipidomics approach based on a combination of high affinity light-sensitive ligands for CL with LC-MS based lipidomics demonstrated selective accumulation of hydroxy- and hydroperoxy-C18:2-containing CL species in mouse embryonic fibroblasts and HeLa cells triggered to apoptosis [159]. In this case, selectivity of intrinsic apoptotic death has been achieved by photo-oxidation of mitochondria-accumulated and CL-selective dye, nonyl-acridine orange and confirmed by a bioinformatics n-gram analysis. Similar oxygenated CL-derived death signals were detected in cells during apoptosis triggered by different stimuli such as actinomycin D [160], staurosporine [39], mechanical stretch of neurons [77], rotenone [97], hyperoxia [161] and ionizing radiation [162]. Notably CL oxidation is highly specific for apoptotic but not other types of programmed cell death. In fact CL oxidation is selectively excluded from ferroptotic cell death, where phosphatidyl-ethanolamine (PE) plays a signaling role [163].
Intra- and Extra-Cellular Cardiolipin Regulation of Immune Responses
In addition to acting as a regulatory platform for intra-mitochondrial and intra-cellular metabolism, CLs can also participate in defining and fine-tuning immune responses. As descendants of protobacteria that entered into an endosymbiotic relationship with ancestral anaerobes, mitochondria bear unique microbial molecular motifs - pathogen associated molecular patterns (PAMPs) recognized by the innate immune system [164]. Hidden from the mitochondrial surface CLs can, upon externalization to the surface and release into extracellular environments, serve as a source of PAMP like danger-associated molecular patterns (DAMPs) [164, 165]. Thus, the process of CL translocation may be not the end rather the beginning of the CL-driven journey in which the major roles are played by different components of the immune system.
Intracellular Signaling via Inflammasome Activation
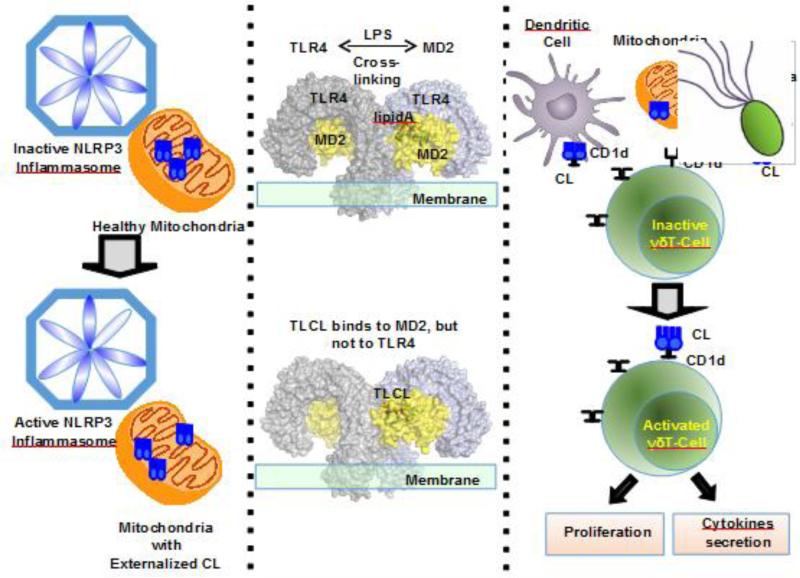
Damaged (depolarized) mitochondria with externalized CL represent one of the common intracellular activating stimuli for the responses of innate immune cells realized via the formation of inflammasomes. One of them, NLRP3 inflammasome, activates caspase-1 [166] ultimately leading to the proteolytic activation of the pro-inflammatory cytokines, interleukin-1β (IL-1β) and interleukin-18 (IL-18) [167]. Several features indicate that mitochondria with externalized CLs are tightly involved in the activation of NLRP3 inflammasomes: i) relocation from endoplasmic reticulum to mitochondria [168] ii) negative regulation by mitophagy/autophagy [169, 170] iii) dependence of inflammasome activation on CL contents (e.g. via knockdown of cardiolipin synthase); iv) CL binding to the leucine rich repeat (LRR) region of Nlrp3; v) direct activation of the inflammasome by CLs [171]. While the nature of the inflammasome activation signal produced by damaged mitochondria still is not fully identified, it is likely that CL externalized to the surface can serve as a mitochondrial docking site for the inflammasome assembly and subsequent activation or alternatively as the direct activating ligand for NLRP3 (Fig. 10), [171, 172].
Fig. 10. Intra- and Extracellular CLs are involved in the regulation of the immune responses.
The left panel shows that mitochondrial externalized CLs are involved in the activation of NLRP3 inflammasomes, which in turn activates caspase-1; The middle panel shows that LPS (which contains lipid A) cross-links two TLR4-MD2 complexes to activate an inflammatory response. However, CL only binds to MD2, but cannot cross-links TLR4's; In the right panel, mitochondria with externalized CL have characteristics of bacteria. CD1d protein is able to bind and present mammalian or bacterial CL to CL-responsive γδ T cells that exist in the spleen and liver of healthy mice. In response to CL these cells proliferate in a dose-dependent manner, and secrete the cytokines IFN-γ and RANTES.
Extracellular signaling: phagocytosis and immune-suppression
Dying or injured cells can release mitochondria and mitochondrial fragments with externalized CLs [173-175] which – similarly to phosphatidylserine (PS) – can be recognized by professional phagocytes as an “eatme” signal for uptake and degradation. Indeed, it has been demonstrated that mitochondria or liposomes with CLs exposed on their surface attract phagocytes and stimulate engulfment mediated through the cell surface scavenger receptor CD36 [22]. Of note, the effectiveness of the uptake is not dependent on CL oxidation – similar to autophagy of mitochondrial with externalized CL [24]. Independently of the uptake process, membranes with integrated CL can be effectively bound by the phagocyte Toll-like receptor 4 (TLR4)/Md2. The significance of the latter interaction becomes particularly obvious in the context of the inflammatory response elicited by macrophages challenged and activated by the bacterial agonist lipopolysaccharide (LPS). The antigenicity and inflammatory potency of LPS is defined by hexa-acylated Lipid A [176, 177]. By acting as a structural homologue of antigenically inactive immature Lipid A, CL potently suppresses TLR4/Md2-driven cytokine production and release thus causing immune-paralysis [22]. This dichotomous role for extracellular CL - as an “eat-me” signal and inhibitor of the innate immune response – should be considered in the context of host-pathogen relationships and recognition of (PAMPs).
Adaptive immune response and anti-CL antibodies
The continuous presence of CL externalized on damaged mitochondria – due to failure of its effective removal by the innate immune system or constant production as a result of cell death - can activate antigen-presenting cells (APCs) such as dendritic cells (DC) and macrophages and, through interactions with T cells and B cells, stimulate the adaptive immune response resulting in the production of anti-CL antibodies [178]. These effects of CLs may be stimulated by beta-2-glycoprotein 1 (β2GPI), a glycoprotein present in high concentrations in plasma and capable of effective binding to anionic phospholipids, including CLs [179, 180]. Excessive formation and accumulation of antiphospholipid antibodies, particularly antibodies against CLs, may lead to the development of a systemic autoimmune disease - the anti-phospholipid syndrome (APS). Recent studies indicate that viral infection provoked damage of mitochondria, CL externalization and impaired mitophagy may be essential in the development of Lupus erythematosus and other systemic autoimmune diseases [178].
Concluding remarks and future directions
Mitochondria have retained a number of their ancestral bacterial attributes including retention of the bioenergetic molecular machinery for making ATP, a bacterial type genome (albeit diminutive) and the conservation of the ancient prokaryotic phospholipid CL. In multicellular eukaryotic organisms, mitochondria have evolved to do so much more than their prokaryotic ancestors. The role of mitochondria in regulating cellular energy balance, oxidant production, mitochondrial biogenesis, mitophagy, cell death pathway(s) and innate immunity, now define their central role in sensing metabolic and invasive perturbations that are essential for controlling cellular stability and homeostasis. Cardiolipin has a role in all of these functions. Over the 75 years since its discovery, remarkable progress in the understanding of the roles that CLs and their metabolites play in mitochondrial functions has been attained. One of the latest achievements is the discovery of multiple signaling functions of CLs not just within mitochondria but also in cells and the entire body.
Perhaps the single most important tool used to measure CLs and their oxidized products is MS which has proven invaluable in most modern studies of lipids including CLs. Increasing sensitivity and mass accuracy, sample handling and sophisticated lipidomic analysis have continued to improve, providing better structural characterization and more reliable quantitative measurements of CLs and their products. The identification of the myriad of CLs and their oxidized products may be compared with the discovery of a CL-based language for effective signaling. However, the structure of this language, its grammar, words and even its “alphabet” are still unknown. Significantly more work in which not only improved technologies, including MS-based technologies, but also more advanced lipidomic and bioinformatics analysis combined with focused molecular and cell biology experiments will be necessary.
Still unexplained, several mysteries of CL should be mentioned. The synthesis of CLs is lacking specificity in regards to a detailed explanation of the formation of tissue specific homo-acylated and hetero-acylated CL as well as their functions. Cells go to an energetically costly effort to produce homo-acylated CLs. In mammalian cardiac cells ~80% of the CL has 4 identical tetra-linoleoyl chains, how and why this is done is unclear. The responsible biosynthetic pathways involved still wait for their accurate identification; so do specific functions of tetra-linoleoyl-CL in the heart and skeletal muscles. Hetero-acylated CL present in all mammalian tissues and notably in brain are the likely precursors of many CL signals, particularly lipid mediators whose identification and physiological roles are still obscure.
While the differences in diversity, molecular speciation and susceptibility to oxidation between bacterial and mitochondrial CLs have been overall known, surprisingly, one of the central issues - the cross-communications of bacterial and mitochondrial CLs during infection still remain enigmatic. One may also wonder whether the role of mitochondrial symbionts in host cells is always beneficial or the evil “call of the wild” can be awakened in mitochondria in the face of the invasion by extra- and intracellular bacterial pathogens. Many CL-dependent features of host/pathogen relationships are yet to be explored. As one of the examples, mitochondria and bacteria share the common evolutionary conserved mechanisms for CL externalization that are used for signaling purposes – elimination of dysfunctional mitochondria via mitophagy and virulence/survival for the host cells and bacterial pathogens, respectively. Molecular speciation of externalized CLs defines the specific meaning of the CL-encoded signals. Given the recently established redistribution of CLs from the inner to the outer membrane of intracellular bacterial pathogens upon the encounter with a host cell, it is tempting to speculate that these externalized pathogen CLs act as a signal by mimicking mitochondrial CLs. This facilitates effective uptake and delivery of bacteria into the endo-phagosomal compartment of host cells thus facilitating the growth and subsequent proliferation of bacteria. Based on this new concept, new anti-bacterial peptides with a high affinity towards externalized bacterial CLs can be designed. Moreover, the protein machinery involved in the CL externalization process may represent new targets for drug discovery. By mimicking mitochondrial CLs – bacterial non-oxidizable CL species may interfere with the critical signaling by oxygenated species thus hijacking essential mammalian metabolic networks. Understanding this may lead to targeted employment of PUFA-containing CLs as antibacterial agents.
Mitochondria are known to act as epigenomic regulators, however the nature of the signals causing chromatin-modifying agents remains largely unknown. Similar to ancient ‘quorum sensing’ mechanisms of bacterial communities, mitochondria are believed to act as networks generating integrated signals essential for cell adaptation and plasticity [181]. It is possible that CLs represent important signals integrating intracellular and extracellular signaling systems. Mitochondria with externalized CLs released from dying cells (via necrosis, necroptosis or ferroptosis) may confuse professional phagocytes resulting in “false” uptake and mishandled innate immune responses. Mitochondria-targeted stabilizers may attenuate these undesirable effects.
Polyunsaturated CLs of eukaryotes evolved with aerobic life and are essential for coordination of multicellular organization. Tracing of the evolutionary development of the “cardiolipin-ome” will contribute to understanding the fundamental principles of mitochondrial CL signaling.
Supplemental Material
Highlights.
Cardiolipin (CL) is a unique ancient lipid common to bacteria and mitochondria
Cardiolipin is essential for mitochondrial bioenergetic functions
While located in the inner membrane, CL can externalize to signal mitophagy
Enzymatic oxidation of mitochondrial cardiolipins is required for apoptosis
Oxidized CL are precursors of lipid mediators
Diversification of mitochondrial CLs is tissue specific
Homo-acylated CLs are characteristic of the heart
Acknowledgments
This study was supported by NIH: PO1HL114453, CA165065, ES020693, U19AIO68021, NS076511, NS061817; NIOSH: OH008282, Human Frontier Science Program HFSP-RGP00132014, the Barth Syndrome Foundation, Inc. and the Barth Syndrome Foundation of Canada.
Abbreviations
- AAPH
(2,2'-azobis-2-methyl-propanimidamide, dihydrochloride)
- CL
cardiolipin
- cyt c
cytochrome c
- DAMPs
danger-associated molecular patterns
- IMM
inner mitochondria membrane
- IMS
mitochondrial inter membrane space
- LC
liquid chromatography
- MLCL
monolyso-cardiolipin
- MS
mass spectrometry
- NL
neutral loss
- OMM
outer mitochondrial membrane
- PA
phosphatidic acid
- PAMP
pathogen-associated molecular patterns
- PG
phosphatidyl glycerol
- PS
phosphatidylserine
- PUFA
polyunsaturated fatty acids
- ROS
reactive oxygen species
- TLCL
tetralinoleoyl-cardiolipin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest None
References
- 1.Lane N. Bioenergetic constraints on the evolution of complex life. Cold Spring Harbor perspectives in biology. 2014;6:a015982. doi: 10.1101/cshperspect.a015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 3.Martin WF, Garg S, Zimorski V. Endosymbiotic theories for eukaryote origin, Philosophical transactions of the Royal Society of London. Series B. Biological sciences. 2015;370:20140330. doi: 10.1098/rstb.2014.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyons TW, Reinhard CT, Planavsky NJ. The rise of oxygen in Earth's early ocean and atmosphere. Nature. 2014;506:307–315. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 5.Westbroek P. Life as a geological force — dynamics of the earth. W. W. Norton; New York, London: 1991. [Google Scholar]
- 6.Lake JA. Eukaryotic origins, Philosophical transactions of the Royal Society of London. Series B. Biological sciences. 2015;370:20140321. doi: 10.1098/rstb.2014.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell P, Moyle J. Chemiosmotic hypothesis of oxidative phosphorylation. Nature. 1967;213:137–139. doi: 10.1038/213137a0. [DOI] [PubMed] [Google Scholar]
- 8.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) The Journal of biological chemistry. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 9.Bartz RR, Suliman HB, Piantadosi CA. Redox mechanisms of cardiomyocyte mitochondrial protection. Frontiers in physiology. 2015;6:291. doi: 10.3389/fphys.2015.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breton-Romero R, Lamas S. Hydrogen peroxide signaling in vascular endothelial cells. Redox biology. 2014;2:529–534. doi: 10.1016/j.redox.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reczek CR, Chandel NS. ROS-dependent signal transduction. Current opinion in cell biology. 2015;33:8–13. doi: 10.1016/j.ceb.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sies H. Role of metabolic H2O2 generation: redox signaling and oxidative stress. The Journal of biological chemistry. 2014;289:8735–8741. doi: 10.1074/jbc.R113.544635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halliwell B, Clement MV, Long LH. Hydrogen peroxide in the human body. FEBS letters. 2000;486:10–13. doi: 10.1016/s0014-5793(00)02197-9. [DOI] [PubMed] [Google Scholar]
- 14.Brand MD. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free radical biology & medicine. 2016 doi: 10.1016/j.freeradbiomed.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Fridovich I. Oxygen: how do we stand it? Medical principles and practice : international journal of the Kuwait University, Health Science Centre. 2013;22:131–137. doi: 10.1159/000339212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halliwell B. Reactive oxygen species and the central nervous system. Journal of neurochemistry. 1992;59:1609–1623. doi: 10.1111/j.1471-4159.1992.tb10990.x. [DOI] [PubMed] [Google Scholar]
- 17.Karnkowska A, Vacek V, Zubacova Z, Treitli SC, Petrzelkova R, Eme L, Novak L, Zarsky V, Barlow LD, Herman EK, Soukal P, Hroudova M, Dolezal P, Stairs CW, Roger AJ, Elias M, Dacks JB, Vlcek C, Hampl V. A Eukaryote without a Mitochondrial Organelle. Current biology : CB. 2016;26:1274–1284. doi: 10.1016/j.cub.2016.03.053. [DOI] [PubMed] [Google Scholar]
- 18.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 19.Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchison CA, 3rd, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, Pelletier JF, Qi ZQ, Richter RA, Strychalski EA, Sun L, Suzuki Y, Tsvetanova B, Wise KS, Smith HO, Glass JI, Merryman C, Gibson DG, Venter JC. Design and synthesis of a minimal bacterial genome. Science (New York, N.Y.) 2016;351:aad6253. doi: 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- 21.Keeling PJ, Corradi N. Shrink it or lose it: balancing loss of function with shrinking genomes in the microsporidia. Virulence. 2011;2:67–70. doi: 10.4161/viru.2.1.14606. [DOI] [PubMed] [Google Scholar]
- 22.Balasubramanian K, Maeda A, Lee JS, Mohammadyani D, Dar HH, Jiang JF, St Croix CM, Watkins S, Tyurin VA, Tyurina YY, Kloditz K, Polimova A, Kapralova VI, Xiong Z, Ray P, Klein-Seetharaman J, Mallampalli RK, Bayir H, Fadeel B, Kagan VE. Dichotomous roles for externalized cardiolipin in extracellular signaling: Promotion of phagocytosis and attenuation of innate immunity. Science signaling. 2015;8:ra95. doi: 10.1126/scisignal.aaa6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogdanov M, Mileykovskaya E, Dowhan W. Lipids in the assembly of membrane proteins and organization of protein supercomplexes: implications for lipid-linked disorders. Sub-cellular biochemistry. 2008;49:197–239. doi: 10.1007/978-1-4020-8831-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu CT, Bayir H, Kagan VE. LC3 binds externalized cardiolipin on injured mitochondria to signal mitophagy in neurons: implications for Parkinson disease. Autophagy. 2014;10:376–378. doi: 10.4161/auto.27191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji J, Baart S, Vikulina AS, Clark RS, Anthonymuthu TS, Tyurin VA, Du L, St Croix CM, Tyurina YY, Lewis J, Skoda EM, Kline AE, Kochanek PM, Wipf P, Kagan VE, Bayir H. Deciphering of mitochondrial cardiolipin oxidative signaling in cerebral ischemia-reperfusion. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2015;35:319–328. doi: 10.1038/jcbfm.2014.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kagan VE, Tyurina YY, Tyurin VA, Mohammadyani D, Angeli JP, Baranov SV, Klein-Seetharaman J, Friedlander RM, Mallampalli RK, Conrad M, Bayir H. Cardiolipin signaling mechanisms: collapse of asymmetry and oxidation. Antioxidants & redox signaling. 2015;22:1667–1680. doi: 10.1089/ars.2014.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchenkova MA, Dyakova YA, Tereschenko EY, Kovalchuk MV, Vladimirov YA. Cytochrome c Complexes with Cardiolipin Monolayer Formed under Different Surface Pressure. Langmuir : the ACS journal of surfaces and colloids. 2015;31:12426–12436. doi: 10.1021/acs.langmuir.5b03155. [DOI] [PubMed] [Google Scholar]
- 29.Paul SS, Sil P, Haldar S, Mitra S, Chattopadhyay K. Subtle Change in the Charge Distribution of Surface Residues May Affect the Secondary Functions of Cytochrome c. The Journal of biological chemistry. 2015;290:14476–14490. doi: 10.1074/jbc.M114.607010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phan MD, Shin K. Effects of cardiolipin on membrane morphology: a Langmuir monolayer study. Biophysical journal. 2015;108:1977–1986. doi: 10.1016/j.bpj.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlattner U, Tokarska-Schlattner M, Epand RM, Boissan M, Lacombe ML, Klein-Seetharaman J, Kagan VE. Mitochondrial NM23-H4/NDPK-D: a bifunctional nanoswitch for bioenergetics and lipid signaling. Naunyn-Schmiedeberg's archives of pharmacology. 2015;388:271–278. doi: 10.1007/s00210-014-1047-4. [DOI] [PubMed] [Google Scholar]
- 32.Tyurina YY, Domingues RM, Tyurin VA, Maciel E, Domingues P, Amoscato AA, Bayir H, Kagan VE. Characterization of cardiolipins and their oxidation products by LC-MS analysis. Chemistry and physics of lipids. 2014;179:3–10. doi: 10.1016/j.chemphyslip.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu YW, Claypool SM. Disorders of phospholipid metabolism: an emerging class of mitochondrial disease due to defects in nuclear genes. Frontiers in genetics. 2015;6:3. doi: 10.3389/fgene.2015.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renne MF, Bao X, De Smet CH, de Kroon AI. Lipid Acyl Chain Remodeling in Yeast. Lipid insights. 2015;8:33–40. doi: 10.4137/LPI.S31780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saric A, Andreau K, Armand AS, Moller IM, Petit PX. Barth Syndrome: From Mitochondrial Dysfunctions Associated with Aberrant Production of Reactive Oxygen Species to Pluripotent Stem Cell Studies. Frontiers in genetics. 2015;6:359. doi: 10.3389/fgene.2015.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye C, Shen Z, Greenberg ML. Cardiolipin remodeling: a regulatory hub for modulating cardiolipin metabolism and function. Journal of bioenergetics and biomembranes. 2016;48:113–123. doi: 10.1007/s10863-014-9591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fajardo VA, McMeekin L, Saint C, LeBlanc PJ. Cardiolipin linoleic acid content and mitochondrial cytochrome c oxidase activity are associated in rat skeletal muscle. Chemistry and physics of lipids. 2015;187:50–55. doi: 10.1016/j.chemphyslip.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Mejia EM, Cole LK, Hatch GM. Cardiolipin metabolism and the role it plays in heart failure and mitochondrial supercomplex formation. Cardiovascular & hematological disorders drug targets. 2014;14:98–106. doi: 10.2174/1871529x14666140505123753. [DOI] [PubMed] [Google Scholar]
- 39.Tyurin VA, Tyurina YY, Feng W, Mnuskin A, Jiang J, Tang M, Zhang X, Zhao Q, Kochanek PM, Clark RS, Bayir H, Kagan VE. Mass-spectrometric characterization of phospholipids and their primary peroxidation products in rat cortical neurons during staurosporine-induced apoptosis. Journal of neurochemistry. 2008;107:1614–1633. doi: 10.1111/j.1471-4159.2008.05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sparvero LJ, Amoscato AA, Kochanek PM, Pitt BR, Kagan VE, Bayir H. Mass-spectrometry based oxidative lipidomics and lipid imaging: applications in traumatic brain injury. Journal of neurochemistry. 2010;115:1322–1336. doi: 10.1111/j.1471-4159.2010.07055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tyurina YY, Polimova AM, Maciel E, Tyurin VA, Kapralova VI, Winnica DE, Vikulina AS, Domingues MR, McCoy J, Sanders LH, Bayir H, Greenamyre JT, Kagan VE. LC/MS analysis of cardiolipins in substantia nigra and plasma of rotenone-treated rats: Implication for mitochondrial dysfunction in Parkinson's disease. Free radical research. 2015;49:681–691. doi: 10.3109/10715762.2015.1005085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baile MG, Lu YW, Claypool SM. The topology and regulation of cardiolipin biosynthesis and remodeling in yeast. Chemistry and physics of lipids. 2014;179:25–31. doi: 10.1016/j.chemphyslip.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Claypool SM, Koehler CM. The complexity of cardiolipin in health and disease. Trends in biochemical sciences. 2012;37:32–41. doi: 10.1016/j.tibs.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlame M. Cardiolipin remodeling and the function of tafazzin. Biochimica et biophysica acta. 2013;1831:582–588. doi: 10.1016/j.bbalip.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 45.Kagan VE, Chu CT, Tyurina YY, Cheikhi A, Bayir H. Cardiolipin asymmetry, oxidation and signaling. Chemistry and physics of lipids. 2014;179:64–69. doi: 10.1016/j.chemphyslip.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaspard GJ, McMaster CR. Cardiolipin metabolism and its causal role in the etiology of the inherited cardiomyopathy Barth syndrome. Chemistry and physics of lipids. 2015;193:1–10. doi: 10.1016/j.chemphyslip.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 47.Saini-Chohan HK, Mitchell RW, Vaz FM, Zelinski T, Hatch GM. Delineating the role of alterations in lipid metabolism to the pathogenesis of inherited skeletal and cardiac muscle disorders: Thematic Review Series: Genetics of Human Lipid Diseases. Journal of lipid research. 2012;53:4–27. doi: 10.1194/jlr.R012120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Z, Ye C, McCain K, Greenberg ML. The Role of Cardiolipin in Cardiovascular Health. BioMed research international. 2015;2015:891707. doi: 10.1155/2015/891707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolinsky VW, Cole LK, Sparagna GC, Hatch GM. Cardiac mitochondrial energy metabolism in heart failure: Role of cardiolipin and sirtuins. Biochimica et biophysica acta. 2016 doi: 10.1016/j.bbalip.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 50.Horvath SE, Daum G. Lipids of mitochondria. Progress in lipid research. 2013;52:590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 51.Ren M, Phoon CK, Schlame M. Metabolism and function of mitochondrial cardiolipin. Progress in lipid research. 2014;55:1–16. doi: 10.1016/j.plipres.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 52.Lecocq J, Ballou CE. On the Structure of Cardiolipin. Biochemistry. 1964;3:976–980. doi: 10.1021/bi00895a023. [DOI] [PubMed] [Google Scholar]
- 53.Few AV, Gilby AR, Seaman GV. An electrophoretic study on structural components of Micrococcus lysodeikticus. Biochimica et biophysica acta. 1960;38:130–136. doi: 10.1016/0006-3002(60)91202-6. [DOI] [PubMed] [Google Scholar]
- 54.Kates M, Syz JY, Gosser D, Haines TH. pH-dissociation characteristics of cardiolipin and its 2'-deoxy analogue. Lipids. 1993;28:877–882. doi: 10.1007/BF02537494. [DOI] [PubMed] [Google Scholar]
- 55.Kooijman S, E. E. L, Graber ZT, Tyurina YY, Bayir H, Kagan VE. Magic angle spinning (31)P NMR spectroscopy reveals two identical ionization states for the cardiolipin phosphates in phospholipid liposomes. 2016 doi: 10.1016/j.bbamem.2016.10.013. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olofsson G, Sparr E. Ionization constants pKa of cardiolipin. PloS one. 2013;8:e73040. doi: 10.1371/journal.pone.0073040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sathappa M, Alder NN. The ionization properties of cardiolipin and its variants in model bilayers. Biochimica et biophysica acta. 2016;1858:1362–1372. doi: 10.1016/j.bbamem.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barlow GH, Margoliash E. Electrophoretic behavior of mammalian-type cytochromes c. The Journal of biological chemistry. 1966;241:1473–1477. [PubMed] [Google Scholar]
- 59.Belikova NA, Vladimirov YA, Osipov AN, Kapralov AA, Tyurin VA, Potapovich MV, Basova LV, Peterson J, Kurnikov IV, Kagan VE. Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry. 2006;45:4998–5009. doi: 10.1021/bi0525573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koprivnjak T, Zhang D, Ernst CM, Peschel A, Nauseef WM, Weiss JP. Characterization of Staphylococcus aureus cardiolipin synthases 1 and 2 and their contribution to accumulation of cardiolipin in stationary phase and within phagocytes. Journal of bacteriology. 2011;193:4134–4142. doi: 10.1128/JB.00288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin TY, Weibel DB. Organization and function of anionic phospholipids in bacteria. Applied microbiology and biotechnology. 2016;100:4255–4267. doi: 10.1007/s00253-016-7468-x. [DOI] [PubMed] [Google Scholar]
- 62.Sandoval-Calderon M, Geiger O, Guan Z, Barona-Gomez F, Sohlenkamp C. A eukaryote-like cardiolipin synthase is present in Streptomyces coelicolor and in most actinobacteria. The Journal of biological chemistry. 2009;284:17383–17390. doi: 10.1074/jbc.M109.006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan BK, Bogdanov M, Zhao J, Dowhan W, Raetz CR, Guan Z. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16504–16509. doi: 10.1073/pnas.1212797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schlame M, Ren M, Xu Y, Greenberg ML, Haller I. Molecular symmetry in mitochondrial cardiolipins. Chemistry and physics of lipids. 2005;138:38–49. doi: 10.1016/j.chemphyslip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 65.Guan Z, Katzianer D, Zhu J, Goldfine H. Clostridium difficile contains plasmalogen species of phospholipids and glycolipids. Biochimica et biophysica acta. 2014;1842:1353–1359. doi: 10.1016/j.bbalip.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baile MG, Whited K, Claypool SM. Deacylation on the matrix side of the mitochondrial inner membrane regulates cardiolipin remodeling. Molecular Biology of the Cell. 2013;24:2008–2020. doi: 10.1091/mbc.E13-03-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beranek A, Rechberger G, Knauer H, Wolinski H, Kohlwein SD, Leber R. Identification of a Cardiolipin-specific Phospholipase Encoded by the Gene CLD1 (YGR110W) in Yeast. The Journal of biological chemistry. 2009;284:11572–11578. doi: 10.1074/jbc.M805511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsu YH, Dumlao DS, Cao J, Dennis EA. Assessing phospholipase A2 activity toward cardiolipin by mass spectrometry. PloS one. 2013;8:e59267. doi: 10.1371/journal.pone.0059267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor WA, Mejia EM, Mitchell RW, Choy PC, Sparagna GC, Hatch GM. Human Trifunctional Protein Alpha Links Cardiolipin Remodeling to Beta-Oxidation. PloS one. 2012;7:e48628. doi: 10.1371/journal.pone.0048628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao J, Liu Y, Lockwood J, Burn P, Shi Y. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. The Journal of biological chemistry. 2004;279:31727–31734. doi: 10.1074/jbc.M402930200. [DOI] [PubMed] [Google Scholar]
- 71.Zhao Y, Chen YQ, Li S, Konrad RJ, Cao G. The microsomal cardiolipin remodeling enzyme acyl-CoA lysocardiolipin acyltransferase is an acyltransferase of multiple anionic lysophospholipids. Journal of lipid research. 2009;50:945–956. doi: 10.1194/jlr.M800567-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Claypool SM, McCaffery JM, Koehler CM. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. The Journal of Cell Biology. 2006;174:379–390. doi: 10.1083/jcb.200605043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gu Z, Valianpour F, Chen S, Vaz FM, Hakkaart GA, Wanders RJ, Greenberg ML. Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Molecular microbiology. 2004;51:149–158. doi: 10.1046/j.1365-2958.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- 74.Lu YW, Galbraith L, Herndon JD, Lu YL, Pras-Raves M, Vervaart M, Van Kampen A, Luyf A, Koehler CM, McCaffery JM, Gottlieb E, Vaz FM, Claypool SM. Defining functional classes of Barth syndrome mutation in humans. Human molecular genetics. 2016 doi: 10.1093/hmg/ddw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma L, Vaz FM, Gu Z, Wanders RJ, Greenberg ML. The human TAZ gene complements mitochondrial dysfunction in the yeast taz1Delta mutant. Implications for Barth syndrome. The Journal of biological chemistry. 2004;279:44394–44399. doi: 10.1074/jbc.M405479200. [DOI] [PubMed] [Google Scholar]
- 76.Schlame M, Rustow B. Lysocardiolipin formation and reacylation in isolated rat liver mitochondria. The Biochemical journal. 1990;272:589–595. doi: 10.1042/bj2720589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ji J, Kline AE, Amoscato A, Samhan-Arias AK, Sparvero LJ, Tyurin VA, Tyurina YY, Fink B, Manole MD, Puccio AM, Okonkwo DO, Cheng JP, Alexander H, Clark RS, Kochanek PM, Wipf P, Kagan VE, Bayir H. Lipidomics identifies cardiolipin oxidation as a mitochondrial target for redox therapy of brain injury. Nature neuroscience. 2012;15:1407–1413. doi: 10.1038/nn.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kiebish MA, Han X, Cheng H, Lunceford A, Clarke CF, Moon H, Chuang JH, Seyfried TN. Lipidomic analysis and electron transport chain activities in C57BL/6J mouse brain mitochondria. Journal of neurochemistry. 2008;106:299–312. doi: 10.1111/j.1471-4159.2008.05383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hatch GM. Cardiolipin biosynthesis in the isolated heart. The Biochemical journal. 1994;297(Pt 1):201–208. doi: 10.1042/bj2970201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tyurina YY, Poloyac SM, Tyurin VA, Kapralov AA, Jiang J, Anthonymuthu TS, Kapralova VI, Vikulina AS, Jung MY, Epperly MW, Mohammadyani D, Klein-Seetharaman J, Jackson TC, Kochanek PM, Pitt BR, Greenberger JS, Vladimirov YA, Bayir H, Kagan VE. A mitochondrial pathway for biosynthesis of lipid mediators. Nature chemistry. 2014;6:542–552. doi: 10.1038/nchem.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frohman MA. Role of mitochondrial lipids in guiding fission and fusion. Journal of molecular medicine (Berlin, Germany) 2015;93:263–269. doi: 10.1007/s00109-014-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mejia EM, Hatch GM. Mitochondrial phospholipids: role in mitochondrial function. Journal of bioenergetics and biomembranes. 2016;48:99–112. doi: 10.1007/s10863-015-9601-4. [DOI] [PubMed] [Google Scholar]
- 83.Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol. 2008;182:937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fry M, Green DE. Cardiolipin requirement for electron transfer in complex I and III of the mitochondrial respiratory chain. The Journal of biological chemistry. 1981;256:1874–1880. [PubMed] [Google Scholar]
- 85.Hoffmann B, Stockl A, Schlame M, Beyer K, Klingenberg M. The reconstituted ADP/ATP carrier activity has an absolute requirement for cardiolipin as shown in cysteine mutants. The Journal of biological chemistry. 1994;269:1940–1944. [PubMed] [Google Scholar]
- 86.Vergeade A, Bertram CC, Bikineyeva AT, Zackert WE, Zinkel SS, May JM, Dikalov SI, Roberts LJ, 2nd, Boutaud O. Cardiolipin fatty acid remodeling regulates mitochondrial function by modifying the electron entry point in the respiratory chain. Mitochondrion. 2016;28:88–95. doi: 10.1016/j.mito.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Daum G. Lipids of mitochondria. Biochimica et biophysica acta. 1985;822:1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 88.Peyta L, Jarnouen K, Pinault M, Guimaraes C, Pais de Barros JP, Chevalier S, Dumas JF, Maillot F, Hatch GM, Loyer P, Servais S. Reduced cardiolipin content decreases respiratory chain capacities and increases ATP synthesis yield in the human HepaRG cells. Biochimica et biophysica acta. 2016;1857:443–453. doi: 10.1016/j.bbabio.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 89.Mileykovskaya E, Dowhan W. Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chemistry and physics of lipids. 2014;179:42–48. doi: 10.1016/j.chemphyslip.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vartak R, Porras CA, Bai Y. Respiratory supercomplexes: structure, function and assembly. Protein & cell. 2013;4:582–590. doi: 10.1007/s13238-013-3032-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bazan S, Mileykovskaya E, Mallampalli VK, Heacock P, Sparagna GC, Dowhan W. Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. The Journal of biological chemistry. 2013;288:401–411. doi: 10.1074/jbc.M112.425876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mileykovskaya E, Penczek PA, Fang J, Mallampalli VK, Sparagna GC, Dowhan W. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. The Journal of biological chemistry. 2012;287:23095–23103. doi: 10.1074/jbc.M112.367888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schlame M. Assays of cardiolipin levels. Methods in cell biology. 2007;80:223–240. doi: 10.1016/S0091-679X(06)80011-7. [DOI] [PubMed] [Google Scholar]
- 94.Hsu FF, Turk J, Rhoades ER, Russell DG, Shi Y, Groisman EA. Structural characterization of cardiolipin by tandem quadrupole and multiple-stage quadrupole ion-trap mass spectrometry with electrospray ionization. Journal of the American Society for Mass Spectrometry. 2005;16:491–504. doi: 10.1016/j.jasms.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 95.Han X, Yang K, Yang J, Cheng H, Gross RW. Shotgun lipidomics of cardiolipin molecular species in lipid extracts of biological samples. Journal of lipid research. 2006;47:864–879. doi: 10.1194/jlr.D500044-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Minkler PE, Hoppel CL. Separation and characterization of cardiolipin molecular species by reverse-phase ion pair high-performance liquid chromatography-mass spectrometry. Journal of lipid research. 2010;51:856–865. doi: 10.1194/jlr.D002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tyurina YY, Winnica DE, Kapralova VI, Kapralov AA, Tyurin VA, Kagan VE. LC/MS characterization of rotenone induced cardiolipin oxidation in human lymphocytes: Implications for mitochondrial dysfunction associated with Parkinson's disease. Molecular nutrition & food research. 2013;57:1410–1422. doi: 10.1002/mnfr.201200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cheng H, Mancuso DJ, Jiang X, Guan S, Yang J, Yang K, Sun G, Gross RW, Han X. Shotgun lipidomics reveals the temporally dependent, highly diversified cardiolipin profile in the mammalian brain: temporally coordinated postnatal diversification of cardiolipin molecular species with neuronal remodeling. Biochemistry. 2008;47:5869–5880. doi: 10.1021/bi7023282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kagan VE, Chu CT, Tyurina YY, Cheikhi A, Bayir H. Cardiolipin asymmetry, oxidation and signaling. Chemistry and physics of lipids. 2014;179:64–69. doi: 10.1016/j.chemphyslip.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Norris JL, Caprioli RM. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chemical reviews. 2013;113:2309–2342. doi: 10.1021/cr3004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Amoscato AA, Sparvero LJ, He RR, Watkins S, Bayir H, Kagan VE. Imaging Mass Spectrometry of Diversified Cardiolipin Molecular Species in the Brain. Analytical Chemistry. 2014;86:6587–6595. doi: 10.1021/ac5011876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aichler M, Walch A. MALDI Imaging mass spectrometry: current frontiers and perspectives in pathology research and practice. Laboratory investigation; a journal of technical methods and pathology. 2015;95:422–431. doi: 10.1038/labinvest.2014.156. [DOI] [PubMed] [Google Scholar]
- 103.Sparvero LJ, Amoscato AA, Dixon CE, Long JB, Kochanek PM, Pitt BR, Bayir H, Kagan VE. Mapping of phospholipids by MALDI imaging (MALDI-MSI): realities and expectations. Chemistry and physics of lipids. 2012;165:545–562. doi: 10.1016/j.chemphyslip.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Planas-Iglesias J, Dwarakanath H, Mohammadyani D, Yanamala N, Kagan VE, Klein-Seetharaman J. Cardiolipin Interactions with Proteins. Biophysical journal. 2015;109:1282–1294. doi: 10.1016/j.bpj.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arnarez C, Mazat JP, Elezgaray J, Marrink SJ, Periole X. Evidence for cardiolipin binding sites on the membrane-exposed surface of the cytochrome bc1. Journal of the American Chemical Society. 2013;135:3112–3120. doi: 10.1021/ja310577u. [DOI] [PubMed] [Google Scholar]
- 106.Poyry S, Cramariuc O, Postila PA, Kaszuba K, Sarewicz M, Osyczka A, Vattulainen I, Rog T. Atomistic simulations indicate cardiolipin to have an integral role in the structure of the cytochrome bc1 complex. Biochimica et biophysica acta. 2013;1827:769–778. doi: 10.1016/j.bbabio.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 107.Paradies G, Petrosillo G, Pistolese M, Ruggiero FM. The effect of reactive oxygen species generated from the mitochondrial electron transport chain on the cytochrome c oxidase activity and on the cardiolipin content in bovine heart submitochondrial particles. FEBS letters. 2000;466:323–326. doi: 10.1016/s0014-5793(00)01082-6. [DOI] [PubMed] [Google Scholar]
- 108.Arnarez C, Marrink SJ, Periole X. Identification of cardiolipin binding sites on cytochrome c oxidase at the entrance of proton channels. Scientific reports. 2013;3:1263. doi: 10.1038/srep01263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, Qiang Wang KZ, Zhu J, Klein-Seetharaman J, Balasubramanian K, Amoscato AA, Borisenko G, Huang Z, Gusdon AM, Cheikhi A, Steer EK, Wang R, Baty C, Watkins S, Bahar I, Bayir H, Kagan VE. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nature cell biology. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schlattner U, Tokarska-Schlattner M, Ramirez S, Tyurina YY, Amoscato AA, Mohammadyani D, Huang Z, Jiang J, Yanamala N, Seffouh A, Boissan M, Epand RF, Epand RM, Klein-Seetharaman J, Lacombe ML, Kagan VE. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. The Journal of biological chemistry. 2013;288:111–121. doi: 10.1074/jbc.M112.408633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bonet J, Planas-Iglesias J, Garcia-Garcia J, Marin-Lopez MA, Fernandez-Fuentes N, Oliva B. ArchDB 2014: structural classification of loops in proteins. Nucleic acids research. 2014;42:D315–319. doi: 10.1093/nar/gkt1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Planas-Iglesias J, Bonet J, Garcia-Garcia J, Marin-Lopez MA, Feliu E, Oliva B. Understanding protein-protein interactions using local structural features. Journal of molecular biology. 2013;425:1210–1224. doi: 10.1016/j.jmb.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 113.Planas-Iglesias J, Marin-Lopez MA, Bonet J, Garcia-Garcia J, Oliva B. iLoops: a protein-protein interaction prediction server based on structural features. Bioinformatics (Oxford, England) 2013;29:2360–2362. doi: 10.1093/bioinformatics/btt401. [DOI] [PubMed] [Google Scholar]
- 114.Schlattner U, Tokarska-Schlattner M, Ramirez S, Bruckner A, Kay L, Polge C, Epand RF, Lee RM, Lacombe ML, Epand RM. Mitochondrial kinases and their molecular interaction with cardiolipin. Biochimica et biophysica acta. 2009;1788:2032–2047. doi: 10.1016/j.bbamem.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 115.Schlattner U, Tokarska-Schlattner M, Rousseau D, Boissan M, Mannella C, Epand R, Lacombe ML. Mitochondrial cardiolipin/phospholipid trafficking: the role of membrane contact site complexes and lipid transfer proteins. Chemistry and physics of lipids. 2014;179:32–41. doi: 10.1016/j.chemphyslip.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 116.Skulachev VP. Cytochrome c in the apoptotic and antioxidant cascades. FEBS letters. 1998;423:275–280. doi: 10.1016/s0014-5793(98)00061-1. [DOI] [PubMed] [Google Scholar]
- 117.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 118.Bakan A, Kapralov AA, Bayir H, Hu F, Kagan VE, Bahar I. Inhibition of Peroxidase Activity of Cytochrome c: De Novo Compound Discovery and Validation. Molecular pharmacology. 2015;88:421–427. doi: 10.1124/mol.115.097816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jiang J, Bakan A, Kapralov AA, Silva KI, Huang Z, Amoscato AA, Peterson J, Garapati VK, Saxena S, Bayir H, Atkinson J, Bahar I, Kagan VE. Designing inhibitors of cytochrome c/cardiolipin peroxidase complexes: mitochondria-targeted imidazole-substituted fatty acids. Free radical biology & medicine. 2014;71:221–230. doi: 10.1016/j.freeradbiomed.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rytomaa M, Kinnunen PK. Evidence for two distinct acidic phospholipid-binding sites in cytochrome c. The Journal of biological chemistry. 1994;269:1770–1774. [PubMed] [Google Scholar]
- 121.Rytomaa M, Kinnunen PK. Reversibility of the binding of cytochrome c to liposomes. Implications for lipid-protein interactions. The Journal of biological chemistry. 1995;270:3197–3202. doi: 10.1074/jbc.270.7.3197. [DOI] [PubMed] [Google Scholar]
- 122.Salamon Z, Tollin G. Surface plasmon resonance studies of complex formation between cytochrome c and bovine cytochrome c oxidase incorporated into a supported planar lipid bilayer. II. Binding of cytochrome c to oxidase-containing cardiolipin/phosphatidylcholine membranes. Biophysical journal. 1996;71:858–867. doi: 10.1016/S0006-3495(96)79287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stepanov G, Gnedenko O, Mol'nar A, Ivanov A, Vladimirov Y, Osipov A. Evaluation of cytochrome c affinity to anionic phospholipids by means of surface plasmon resonance. FEBS letters. 2009;583:97–100. doi: 10.1016/j.febslet.2008.11.029. [DOI] [PubMed] [Google Scholar]
- 124.Tuominen EK, Wallace CJ, Kinnunen PK. Phospholipid-cytochrome c interaction: evidence for the extended lipid anchorage. The Journal of biological chemistry. 2002;277:8822–8826. doi: 10.1074/jbc.M200056200. [DOI] [PubMed] [Google Scholar]
- 125.Zucchi MR, Nascimento OR, Faljoni-Alario A, Prieto T, Nantes IL. Modulation of cytochrome c spin states by lipid acyl chains: a continuous-wave electron paramagnetic resonance (CW-EPR) study of haem iron. The Biochemical journal. 2003;370:671–678. doi: 10.1042/BJ20021521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Atkinson J, Kapralov AA, Yanamala N, Tyurina YY, Amoscato AA, Pearce L, Peterson J, Huang Z, Jiang J, Samhan-Arias AK, Maeda A, Feng W, Wasserloos K, Belikova NA, Tyurin VA, Wang H, Fletcher J, Wang Y, Vlasova II, Klein-Seetharaman J, Stoyanovsky DA, Bayir H, Pitt BR, Epperly MW, Greenberger JS, Kagan VE. A mitochondria-targeted inhibitor of cytochrome c peroxidase mitigates radiation-induced death. Nature communications. 2011;2:497. doi: 10.1038/ncomms1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mandal A, Hoop CL, DeLucia M, Kodali R, Kagan VE, Ahn J, van der Wel PC. Structural Changes and Proapoptotic Peroxidase Activity of Cardiolipin-Bound Mitochondrial Cytochrome c. Biophysical journal. 2015;109:1873–1884. doi: 10.1016/j.bpj.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McClelland LJ, Mou TC, Jeakins-Cooley ME, Sprang SR, Bowler BE. Structure of a mitochondrial cytochrome c conformer competent for peroxidase activity. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:6648–6653. doi: 10.1073/pnas.1323828111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kapralov AA, Kurnikov IV, Vlasova II, Belikova NA, Tyurin VA, Basova LV, Zhao Q, Tyurina YY, Jiang J, Bayir H, Vladimirov YA, Kagan VE. The hierarchy of structural transitions induced in cytochrome c by anionic phospholipids determines its peroxidase activation and selective peroxidation during apoptosis in cells. Biochemistry. 2007;46:14232–14244. doi: 10.1021/bi701237b. [DOI] [PubMed] [Google Scholar]
- 130.Hanske J, Toffey JR, Morenz AM, Bonilla AJ, Schiavoni KH, Pletneva EV. Conformational properties of cardiolipin-bound cytochrome c. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:125–130. doi: 10.1073/pnas.1112312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hong Y, Muenzner J, Grimm SK, Pletneva EV. Origin of the conformational heterogeneity of cardiolipin-bound cytochrome C. Journal of the American Chemical Society. 2012;134:18713–18723. doi: 10.1021/ja307426k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kapralov AA, Yanamala N, Tyurina YY, Castro L, Samhan-Arias A, Vladimirov YA, Maeda A, Weitz AA, Peterson J, Mylnikov D, Demicheli V, Tortora V, Klein-Seetharaman J, Radi R, Kagan VE. Topography of tyrosine residues and their involvement in peroxidation of polyunsaturated cardiolipin in cytochrome c/cardiolipin peroxidase complexes. Biochimica et biophysica acta. 2011;1808:2147–2155. doi: 10.1016/j.bbamem.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ying T, Wang ZH, Lin YW, Xie J, Tan X, Huang ZX. Tyrosine-67 in cytochrome c is a possible apoptotic trigger controlled by hydrogen bonds via a conformational transition. Chemical communications (Cambridge, England) 2009:4512–4514. doi: 10.1039/b904347k. [DOI] [PubMed] [Google Scholar]
- 134.Yu H, Lee I, Salomon AR, Yu K, Huttemann M. Mammalian liver cytochrome c is tyrosine-48 phosphorylated in vivo, inhibiting mitochondrial respiration. Biochimica et biophysica acta. 2008;1777:1066–1071. doi: 10.1016/j.bbabio.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pecina P, Borisenko GG, Belikova NA, Tyurina YY, Pecinova A, Lee I, Samhan-Arias AK, Przyklenk K, Kagan VE, Huttemann M. Phosphomimetic substitution of cytochrome C tyrosine 48 decreases respiration and binding to cardiolipin and abolishes ability to trigger downstream caspase activation. Biochemistry. 2010;49:6705–6714. doi: 10.1021/bi100486s. [DOI] [PubMed] [Google Scholar]
- 136.Mohammadyani D, Tyurin VA, O'Brien M, Sadovsky Y, Gabrilovich DI, Klein-Seetharaman J, Kagan VE. Molecular speciation and dynamics of oxidized triacylglycerols in lipid droplets: Mass spectrometry and coarse-grained simulations. Free radical biology & medicine. 2014;76:53–60. doi: 10.1016/j.freeradbiomed.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Boveris A, Chance B. The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. The Biochemical journal. 1973;134:707–716. doi: 10.1042/bj1340707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiological reviews. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 139.Jensen PK. Antimycin-insensitive oxidation of succinate and reduced nicotinamide-adenine dinucleotide in electron-transport particles. I. pH dependency and hydrogen peroxide formation. Biochimica et biophysica acta. 1966;122:157–166. doi: 10.1016/0926-6593(66)90057-9. [DOI] [PubMed] [Google Scholar]
- 140.Loschen G, Flohe L, Chance B. Respiratory chain linked H(2)O(2) production in pigeon heart mitochondria. FEBS letters. 1971;18:261–264. doi: 10.1016/0014-5793(71)80459-3. [DOI] [PubMed] [Google Scholar]
- 141.Quinlan CL, Goncalves RL, Hey-Mogensen M, Yadava N, Bunik VI, Brand MD. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. The Journal of biological chemistry. 2014;289:8312–8325. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bunik VI, Artiukhov A, Kazantsev A, Goncalves R, Daloso D, Oppermann H, Kulakovskaya E, Lukashev N, Fernie A, Brand M, Gaunitz F. Specific inhibition by synthetic analogs of pyruvate reveals that the pyruvate dehydrogenase reaction is essential for metabolism and viability of glioblastoma cells. Oncotarget. 2015;6:40036–40052. doi: 10.18632/oncotarget.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free radical biology & medicine. 2000;29:222–230. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- 144.Sies H. Oxidative stress: a concept in redox biology and medicine. Redox biology. 2015;4:180–183. doi: 10.1016/j.redox.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wu CC, Bratton SB. Regulation of the intrinsic apoptosis pathway by reactive oxygen species. Antioxidants & redox signaling. 2013;19:546–558. doi: 10.1089/ars.2012.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lin YF, Haynes CM. Metabolism and the UPR(mt) Molecular cell. 2016;61:677–682. doi: 10.1016/j.molcel.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sun N, Youle RJ, Finkel T. The Mitochondrial Basis of Aging. Molecular cell. 2016;61:654–666. doi: 10.1016/j.molcel.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mancias JD, Kimmelman AC. Mechanisms of Selective Autophagy in Normal Physiology and Cancer. Journal of molecular biology. 2016;428:1659–1680. doi: 10.1016/j.jmb.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Randow F, Youle RJ. Self and nonself: how autophagy targets mitochondria and bacteria. Cell host & microbe. 2014;15:403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sani MA, Keech O, Gardestrom P, Dufourc EJ, Grobner G. Magic-angle phosphorus NMR of functional mitochondria: in situ monitoring of lipid response under apoptotic-like stress. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:2872–2878. doi: 10.1096/fj.09-134114. [DOI] [PubMed] [Google Scholar]
- 151.Kagan VE, Jiang J, Huang Z, Tyurina YY, Desbourdes C, Cottet-Rousselle C, Dar HH, Verma M, Tyurin VA, Kapralov AA, Cheikhi A, Mao G, Stolz D, St Croix CM, Watkins S, Shen Z, Li Y, Greenberg ML, Tokarska-Schlattner M, Boissan M, Lacombe ML, Epand RM, Chu CT, Mallampalli RK, Bayir H, Schlattner U. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell death and differentiation. 2016;23:1140–1151. doi: 10.1038/cdd.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ghio S, Kamp F, Cauchi R, Giese A, Vassallo N. Interaction of alpha-synuclein with biomembranes in Parkinson's disease -role of cardiolipin. Progress in lipid research. 2016;61:73–82. doi: 10.1016/j.plipres.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 153.Schlame M, Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS letters. 2006;580:5450–5455. doi: 10.1016/j.febslet.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 154.Forman HJ, Davies KJ, Ursini F. How do nutritional antioxidants really work: nucleophilic tone and para-hormesis versus free radical scavenging in vivo. Free radical biology & medicine. 2014;66:24–35. doi: 10.1016/j.freeradbiomed.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kagan VE, Wipf P, Stoyanovsky D, Greenberger JS, Borisenko G, Belikova NA, Yanamala N, Samhan Arias AK, Tungekar MA, Jiang J, Tyurina YY, Ji J, Klein-Seetharaman J, Pitt BR, Shvedova AA, Bayir H. Mitochondrial targeting of electron scavenging antioxidants: Regulation of selective oxidation vs random chain reactions. Advanced drug delivery reviews. 2009;61:1375–1385. doi: 10.1016/j.addr.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Niki E, Yoshida Y, Saito Y, Noguchi N. Lipid peroxidation: mechanisms, inhibition, and biological effects. Biochemical and biophysical research communications. 2005;338:668–676. doi: 10.1016/j.bbrc.2005.08.072. [DOI] [PubMed] [Google Scholar]
- 157.Halliwell B, Chirico S. Lipid peroxidation: its mechanism, measurement, and significance. The American journal of clinical nutrition. 1993;57:715S–724S. doi: 10.1093/ajcn/57.5.715S. discussion 724S-725S. [DOI] [PubMed] [Google Scholar]
- 158.Kagan VE, Kapralov AA, St Croix CM, Watkins SC, Kisin ER, Kotchey GP, Balasubramanian K, Vlasova II, Yu J, Kim K, Seo W, Mallampalli RK, Star A, Shvedova AA. Lung macrophages “digest” carbon nanotubes using a superoxide/peroxynitrite oxidative pathway. ACS nano. 2014;8:5610–5621. doi: 10.1021/nn406484b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mao G, Qu F, St Croix CM, Tyurina YY, Planas-Iglesias J, Jiang J, Huang Z, Amoscato AA, Tyurin VA, Kapralov AA, Cheikhi A, Maguire J, Klein-Seetharaman J, Bayir H, Kagan VE. Mitochondrial Redox Opto-Lipidomics Reveals Mono-Oxygenated Cardiolipins as Pro-Apoptotic Death Signals. ACS chemical biology. 2016;11:530–540. doi: 10.1021/acschembio.5b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jiang J, Huang Z, Zhao Q, Feng W, Belikova NA, Kagan VE. Interplay between bax, reactive oxygen species production, and cardiolipin oxidation during apoptosis. Biochemical and biophysical research communications. 2008;368:145–150. doi: 10.1016/j.bbrc.2008.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tyurina YY, Tyurin VA, Kaynar AM, Kapralova VI, Wasserloos K, Li J, Mosher M, Wright L, Wipf P, Watkins S, Pitt BR, Kagan VE. Oxidative lipidomics of hyperoxic acute lung injury: mass spectrometric characterization of cardiolipin and phosphatidylserine peroxidation. American journal of physiology. Lung cellular and molecular physiology. 2010;299:L73–85. doi: 10.1152/ajplung.00035.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Belikova NA, Jiang J, Tyurina YY, Zhao Q, Epperly MW, Greenberger J, Kagan VE. Cardiolipin-specific peroxidase reactions of cytochrome C in mitochondria during irradiation-induced apoptosis. International journal of radiation oncology, biology, physics. 2007;69:176–186. doi: 10.1016/j.ijrobp.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 163.Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch A, Eggenhofer E, Basavarajappa D, Radmark O, Kobayashi S, Seibt T, Beck H, Neff F, Esposito I, Wanke R, Forster H, Yefremova O, Heinrichmeyer M, Bornkamm GW, Geissler EK, Thomas SB, Stockwell BR, O'Donnell VB, Kagan VE, Schick JA, Conrad M. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nature cell biology. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, Vandenabeele P. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends in immunology. 2011;32:157–164. doi: 10.1016/j.it.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 165.Zitvogel L, Kepp O, Kroemer G. Decoding cell death signals in inflammation and immunity. Cell. 2010;140:798–804. doi: 10.1016/j.cell.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 166.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Molecular cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 167.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 168.Sutterwala FS, Haasken S, Cassel SL. Mechanism of NLRP3 inflammasome activation. Annals of the New York Academy of Sciences. 2014;1319:82–95. doi: 10.1111/nyas.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature immunology. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 171.Iyer SS, He Q, Janczy JR, Elliott EI, Zhong Z, Olivier AK, Sadler JJ, Knepper-Adrian V, Han R, Qiao L, Eisenbarth SC, Nauseef WM, Cassel SL, Sutterwala FS. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39:311–323. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.O'Neill LA. Cardiolipin and the Nlrp3 inflammasome. Cell metabolism. 2013;18:610–612. doi: 10.1016/j.cmet.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 173.Dorio A, Cerella C, De Nicola M, D'Alessio M, Gualandi G, Ghibelli L. Non-apoptogenic Ca2+-related extrusion of mitochondria in anoxia/reoxygenation stress. Annals of the New York Academy of Sciences. 2007;1099:512–515. doi: 10.1196/annals.1387.067. [DOI] [PubMed] [Google Scholar]
- 174.Goldmann O, Medina E. The expanding world of extracellular traps: not only neutrophils but much more. Frontiers in immunology. 2012;3:420. doi: 10.3389/fimmu.2012.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nakajima A, Kurihara H, Yagita H, Okumura K, Nakano H. Mitochondrial Extrusion through the cytoplasmic vacuoles during cell death. The Journal of biological chemistry. 2008;283:24128–24135. doi: 10.1074/jbc.M802996200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bryant CE, Spring DR, Gangloff M, Gay NJ. The molecular basis of the host response to lipopolysaccharide. Nature reviews. Microbiology. 2010;8:8–14. doi: 10.1038/nrmicro2266. [DOI] [PubMed] [Google Scholar]
- 177.Steimle A, Autenrieth IB, Frick JS. Structure and function: Lipid A modifications in commensals and pathogens. International journal of medical microbiology : IJMM. 2016 doi: 10.1016/j.ijmm.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 178.Broder A, Chan JJ, Putterman C. Dendritic cells: an important link between antiphospholipid antibodies, endothelial dysfunction, and atherosclerosis in autoimmune and non-autoimmune diseases. Clinical immunology (Orlando, Fla.) 2013;146:197–206. doi: 10.1016/j.clim.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Alessandri C, Conti F, Pendolino M, Mancini R, Valesini G. New autoantigens in the antiphospholipid syndrome. Autoimmunity reviews. 2011;10:609–616. doi: 10.1016/j.autrev.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 180.van den Hoogen LL, van Roon JA, Radstake TR, Fritsch-Stork RD, Derksen RH. Delineating the deranged immune system in the antiphospholipid syndrome. Autoimmunity reviews. 2016;15:50–60. doi: 10.1016/j.autrev.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 181.Picard M. Mitochondrial synapses: intracellular communication and signal integration. Trends in neurosciences. 2015;38:468–474. doi: 10.1016/j.tins.2015.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.