Abstract
Although 1p/19q codeletion is the genetic hallmark defining oligodendrogliomas, approximately 30-40% of oligodendroglial tumors have intact 1p/19q in the literature and they demonstrate a worse prognosis. This group of 1p/19q intact oligodendroglial tumors is frequently suggested to be astrocytic in nature with TP53 and ATRX mutations but actually remains under-investigated. In the present study, we provided evidence that not all 1p/19q intact oligodendroglial tumors are astrocytic through histologic and molecular approaches. We examined 1p/19q status by FISH in a large cohort of 337 oligodendroglial tumors and identified 39.8% lacking 1p/19q codeletion which was independently associated with poor prognosis. Among this 1p/19q intact oligodendroglial tumor cohort, 58 cases demonstrated classic oligodendroglial histology which showed older patient age, better prognosis, association with grade III histology, PDGFRA expression, TERTp mutation, as well as frequent IDH mutation. More than half of the 1p/19q intact oligodendroglial tumors showed lack of astrocytic defining markers, p53 expression and ATRX loss. TP53 mutational analysis was additionally conducted in 45 cases of the 1p/19q intact oligodendroglial tumors. Wild-type TP53 was detected in 71.1% of cases which was associated with classic oligodendroglial histology. Importantly, IDH and TERTp co-occurred in 75% of 1p/19q intact, TP53 wild-type oligodendrogliomas, highlighting the potential of the co-mutations in assisting diagnosis of oligodendrogliomas in tumors with clear cell morphology and non-codeleted 1p/19q status. In summary, our study demonstrated that not all 1p/19q intact oligodendroglial tumors are astrocytic and co-evaluation of IDH and TERTp mutation could potentially serve as an adjunct for diagnosing 1p/19q intact oligodendrogliomas.
Keywords: oligodendroglial tumors, intact 1p/19q, TERT, IDH, Pathology Section
INTRODUCTION
Oligodendroglial tumors comprise of oligodendrogliomas and oligoastrocytomas and are associated with favorable clinical behaviors compared with astrocytomas and some can be chemosensitive [1]. Codeletion of 1p and 19q is a characteristic genetic signature which is observed in 39-70% of oligodendrogliomas and 21-59% of oligoastrocytomas in most series [2-4]. Codeletion is associated with longer survival and is also a predictive marker for response to chemotherapy with procarbazine, lomustine and vincristine [1, 5].
Aside from codeletion of 1p/19q, oligodendroglial tumors are strongly associated with mutations of IDH1/2, TERT promoter (TERTp) and they are also associated with methylation of MGMT and upregulation of PDGFRA [6-8]. TERTp mutations are regarded as a common mechanism of upregulation of telomerase in primary glioblastoma and oligodendroglial tumors [9]. 1p/19q codeleted and TERTp mutated tumors are mutually exclusive with tumors exhibiting mutations in ATRX (alpha thalassemia mental retardation syndrome X linked) and TP53. The latter two are usually regarded as markers of astrocytic lineage [7, 10-12].
The fact remains that in most series with many coming from important international trials, 30-40% of oligodendroglial tumors are 1p/19q non-codeleted and these tumors exhibit a worse prognosis [1, 13-15]. It has been proposed that most, if not all, of these non-codeleted tumors are actually astrocytic in nature with either TP53 or ATRX mutations [11]. However, there have been few large-scale studies on molecular characterization of oligodendroglial tumors without 1p/19q codeletion. They make up a significant proportion of “oligodendroglial tumors” diagnosed in international series and daily practice and there is no standard-of-care treatment strategy. In this study, we aim to characterize the molecular and clinical features of this neglected group, interrogating them with standard biomarkers and correlating with overall survival.
RESULTS
Cohort characteristics
The 337 cases of oligodendroglial tumors included 125 oligodendrogliomas (WHO grade II), 105 oligoastrocytomas (WHO grade II), 72 anaplastic oligodendrogliomas (WHO grade III) and 33 anaplastic oligoastrocytomas (WHO grade III). The mean and median ages of the cohort were 43.1 and 43.0 years, respectively (range 5 to 70 years). The male to female ratio was 1:0.77. Operation data was available in 80.9% (271/335) of patients, with 72.7% (197/271) of patients received total resection and 27.3% (74/271) of patients received non-total resection. Adjuvant treatment data was available in 75.8% (254/335) of patients, with 76% (193/254) of patients receiving radiotherapy and 57.1% (145/254) of patients receiving chemotherapy. Survival data was available in 74.3% (249/335) of patients, with median follow-up and median overall survival being 8.2 years and 10.6 years, respectively.
Clinical and molecular differences between 1p/19q codeleted and non-codeleted oligodendroglial tumors
Chromosomal 1p and 19q status were examined in all 337 samples of oligodendroglial tumors, with 1p loss detected in 63.8% (215/337) and 19q loss detected in 62.0% (209/337) of the cohort. Combined 1p/19q codeletion was observed in 60.2% (203/337) of cases including 95 oligodendrogliomas, 44 anaplastic oligodendrogliomas, 51 oligoastrocytomas and 13 anaplastic oligoastrocytomas. 1p loss only and 19q loss only were found in 8.2% and 4.5% among the 134 1p/19q non-codeleted oligodendroglial tumors, respectively. Clinical and molecular characteristics were summarized according to 1p/19q codeletion status in Table 1. Comparing the variables between 1p/19q codeleted and non-codeleted oligodendroglial tumors, tumors with codeletion showed significant associations with classic oligodendroglial histology (p < 0.00001), frontal lobe localization (p = 0.012), and were more amenable to total surgical resection (p = 0.008). Molecularly, 1p/19q codeleted tumors also exhibited co-occurring associations with IDH mutation (p < 0.000001), TERTp mutation (p < 0.000001), and MGMT promoter methylation (p = 0.015) (Figure 1a to 1f).
Table 1. Clinical and molecular characteristics of oligodendroglial tumors according to 1p/19q status.
| n | 1p/19q non-codeleted OTs | 1p/19q codeleted OTs | All OTs | ||
| Gender (male / female) | 337 | 74 / 60 | 115 / 88 | 189 / 148 | |
| Age (mean / median / range) | 337 | 40.8 / 42 / 5 - 72 years | 44.6 / 44 / 21 - 75 years | 43.1 /43 / 5 - 75 years | |
| Histologic type | 337 | ||||
| Oligodendroglioma | 58 (43.3%) | 139 (68.5%) | 197 (58.5%) | ||
| Oligoastrocytoma | 76 (56.7%) | 64 (31.5%) | 140 (41.5%) | ||
| Histologic grade | 337 | ||||
| II | 86 (64.2%) | 146 (71.9%) | 232 (68.8%) | ||
| III | 48 (35.8%) | 57 (28.1%) | 105 (31.2%) | ||
| Tumor location | 334 | ||||
| Frontal | 67 (50.8%) | 126 (62.4%) | 193 (57.8%) | ||
| Temporal | 20 (15.2%) | 23 (11.4%) | 43 (12.9%) | ||
| Parietal | 5 (3.8%) | 11 (5.4%) | 16 (4.8%) | ||
| Occipital | 5 (3.8%) | 3 (1.5%) | 8 (2.4%) | ||
| More than one cerebral lobe | 21 (15.9%) | 27 (13.4%) | 48 (14.4%) | ||
| Other locations | 14 (10.6%) | 12 (5.9%) | 26 (7.8%) | ||
| Operation | 273 | ||||
| Total resection | 74 (63.8%) | 123 (78.3%) | 197 (72.2%) | ||
| Non-total resection | 42 (36.2%) | 34 (21.7%) | 76 (27.8%) | ||
| Adjuvant therapy | 256 | ||||
| Concomitant chemo-radiotherapy | 47 (45.2%) | 84 (55.3%) | 131 (51.2%) | ||
| Radiotherapy only | 23 (22.1%) | 41 (27.0%) | 64 (25.0%) | ||
| Chemotherapy only | 4 (3.8%) | 9 (5.9%) | 13 (5.1%) | ||
| No adjuvant therapy | 30 (28.8%) | 18 (11.8%) | 48 (18.8%) | ||
| IDH | 305 | ||||
| IDH1 mut | 70 (58.3%) | 172 (93.0%) | 242 (79.3%) | ||
| IDH2 mut | 7 (5.8%) | 8 (4.3%) | 15 (4.9%) | ||
| wt | 43 (35.8%) | 5 (2.7%) | 48 (15.7%) | ||
| TERTp | 248 | ||||
| C228T | 21 (23.3%) | 91 (57.6%) | 112 (45.2%) | ||
| C250T | 12 (13.3%) | 37 (23.4%) | 49 (19.8%) | ||
| wt | 57 (63.3%) | 30 (19.0%) | 87 (35.1%) | ||
| mut, mutant; wt, wild-type; n, number of cases with data available. | |||||
Figure 1. Clinical and molecular characteristics of oligodendroglial tumors based on 1p/19q codeletion status.
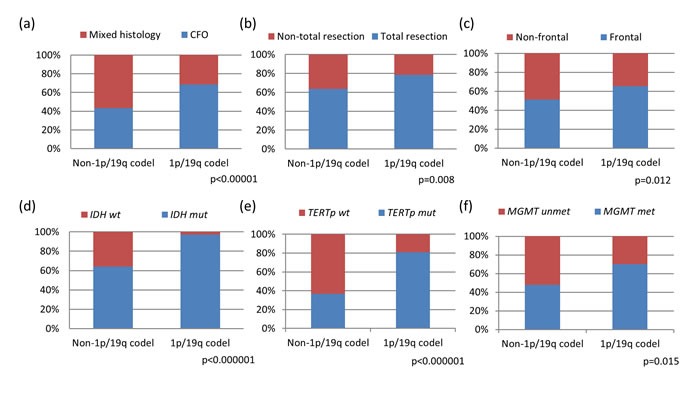
Tumors with 1p/19q codeletion showed significant associations with classical oligodendroglial histology (p < 0.00001) a. frontal lobe localization (p = 0.012) c. and were more amenable to total surgical resection (p = 0.008) b. 1p/19q codeleted tumors also exhibited co-occurring associations with IDH mutation (p < 0.000001) d. TERTp mutation (p < 0.000001) e. and MGMT promoter methylation (p = 0.015) f. CFO, classic for oligodendroglial morphology; mixed, mixed oligoastrocytic histology; mut, mutant; wt, wild-type; met, methylated; unmet, unmethylated.
Univariate survival analysis was conducted in the cohort according to the clinical and molecular variables (Table 2). Oligodendroglial tumors with 1p/19q codeletion exhibited significantly longer overall survival than 1p/19q non-codeleted tumors (median OS 11.8 years vs 7.0 years, p < 0.00001). Other favorable prognostic factors in univariate analysis included age ≤ 50 years (p < 0.000001), histologic grade II (p > 0.000001), and IDH mutation (p = 0.001). Trend of better prognosis was observed in tumors with classic oligodendroglial histology (p = 0.059) and TERTp mutation (p = 0.053). Independent favorable prognostic value of 1p/19q codeletion was demonstrated in multivariate analysis by adjusting with significant factors in univariate analysis (Table 3). Favorable prognostic value of 1p/19q codeletion (HR = 0.51, p = 0.015) was independent of age (p < 0.000001), histologic grade (p < 0.00001), histologic type (p = 0.003), IDH status (p = 0.014), and TERTp status (p = 0.415).
Table 2. Univariate analysis of clinical and molecular variables in oligodendroglial tumors.
| n | HR | [95%CI] | Median OS (years) | p | ||
| Gender | Male | 137 | 1 | 10.1 | 0.18 | |
| Female | 106 | 0.755 | [0.499 - 1.141] | 11.8 | ||
| Age | ≤ 50 years | 181 | 1 | 11.8 | <0.000001 | |
| > 50 years | 62 | 3.289 | [2.173 - 4.98] | 5.9 | ||
| Histologic grade | Grade II | 169 | 1 | 11.8 | <0.000001 | |
| Grade III | 74 | 3.84 | [2.517-5.859] | 6.7 | ||
| Histologic type | Oligodendroglial | 132 | 1 | 11 | 0.059 | |
| Mixed oligoastrocytic | 111 | 1.481 | [0.982-2.234] | 10 | ||
| Frontal involvement | Yes | 173 | 1 | 10.6 | 0.238 | |
| No | 69 | 1.302 | [0.839 - 2.02] | 7.8 | ||
| Operation | Total resection | 173 | 1 | 11.3 | 0.172 | |
| Non-total resection | 67 | 1.345 | [0.878 - 2.059] | 9.7 | ||
| Adjuvant therapy | Concomitant chemo-radiotherapy | 127 | 1 | 10.6 | 0.361 | |
| Radiotherapy only | 57 | 1.512 | [0.867 - 2.639] | 8.8 | ||
| Chemotherapy only | 9 | 1.464 | [0.869 - 2.464] | NR | ||
| No adjuvant therapy | 38 | 0.872 | [0.21 - 3.63] | 10.1 | ||
| 1p/19q | codeleted | 143 | 1 | 11.8 | <0.00001 | |
| non-codeleted | 100 | 2.563 | [1.689 - 3.889] | 7 | ||
| IDH1/2 | mut | 195 | 1 | 10.6 | 0.001 | |
| wt | 25 | 2.553 | [1.454 - 4.483] | 3.8 | ||
| TERTp | mut | 121 | 1 | 11.1 | 0.053 | |
| wt | 58 | 1.638 | [0.988 - 2.715] | 10.1 | ||
| mut, mutant; wt, wild-type; n, number of cases with data available. | ||||||
Table 3. Multivariate analysis of clinical and molecular variables in oligodendroglial tumors.
| HR | [95%CI] | p | ||
| Age | 1.074 | [1.046-1.103] | <0.000001 | |
| Histologic grade | Grade II | 1 | <0.00001 | |
| Grade III | 3.736 | [2.087-6.688] | ||
| Histologic type | Oligodendroglial | 1 | 0.003 | |
| Mixed oligoastrocytic | 2.413 | [1.359-4.284] | ||
| 1p/19q | codeleted | 0.51 | [0.296-0.879] | 0.015 |
| non-codeleted | 1 | |||
| IDH1/2 | mut | 0.364 | [0.162-0.817] | 0.014 |
| wt | 1 | |||
| TERTp | mut | 0.786 | [0.441-1.402] | 0.415 |
| wt | 1 | |||
| mut, mutant; wt, wild-type; n, number of cases with data available. | ||||
Presence of classic oligodendroglial morphology in 1p/19q non-codeleted gliomas with distinct features
Among the 134 cases of 1p/19q non-codeleted oligodendroglial tumors, 43.3% (58/134) of cases demonstrated classic oligodendroglial histology, including 30 oligodendrogliomas and 28 anaplastic oligodendrogliomas. Interestingly, patients with classic oligodendroglial histology were significantly older than those with mixed oligoastrocytic histology (mean age 44.5 years vs 38.0 years, p = 0.01) (Figure 2a). The difference in age was still observed after excluding 11 paediatric (18 years or below) cases (2 oligodendrogliomas, 1 anaplastic oligodendroglioma and 8 oligoastrocytomas) (46.2 years vs 41.4 years, p = 0.023) (Supplementary Figure S1a). Prognostically, classic oligodendroglial histology was also associated with better prognosis across the 1p/19q non-codeleted cohort (p < 0.000001) (Figure 2e). Correlating with histologic grade, 48.3% (28/58) of 1p/19q non-codeleted oligodendrogliomas showed grade III histology, compared to 26.3% (20/76) in 1p/19q non-codeleted oligoastrocytomas (p = 0.009) (Figure 2b). Molecularly, classic oligodendroglial histology in a background of non-codeleted 1p/19q was also associated with TERTp mutation (p = 0.007) (Figure 2c) and showed frequent IDH mutation (67.4%) (Supplementary Figure S1b). In a subset of the 1p/19q non-codeleted tumors (N = 80), PDGFRA immunohistochemical expression was also evaluated and classic oligodendroglial histology was associated with positive PDGFRA expression (p = 0.019) (Figure 2d).
Figure 2. Correlations between clinicopathological factors and molecular variables among 1p/19q intact oligodendroglial tumors.

Patients with classic oligodendroglial histology were significantly older than those with mixed oligoastrocytic histology (p = 0.01) a. Grade III histology was more common in oligodendrogliomas (48.3%) than oligoastrocytomas (26.3%) among 1p/19q non-codeleted tumors b. Classic oligodendroglial histology in a background of non-codeleted 1p/19q was associated with p mutation (p = 0.007) c. and positive PDGFRA expression (p = 0.019) d. Kaplan-Meier curves for overall survival (OS) of tumor histology in 1p/19q intact oligodendroglial tumors e. CFO, classic for oligodendroglial morphology; mixed, mixed oligoastrocytic histology; mut, mutant; wt, wild-type; -ve, negative; +ve, positive; OS, overall survival.
Infrequent astrocytic markers identified in 1p/19q non-codeleted oligodendroglial tumors
To further interrogate lineage of the oligodendrogliomas lacking 1p/19q codeletion, p53 and ATRX immunohistochemical expression were also evaluated. p53 immunohistochemistry positivity was detectable in only 30% of 1p/19q non-codeleted oligodendroglial tumors among 90 cases examined, including seven anaplastic oligodendrogliomas, 13 oligoastrocytomas and seven anaplastic oligoastrocytomas (Supplementary Figure S1c). Among 70 cases of 1p/19q non-codeleted gliomas examined for ATRX expression, internal positivity was detected in 61 cases and only 26.2% (16/61) of cases demonstrated ATRX loss. 83.3% (15/18) of oligodendrogliomas and 69.8% (30/43) of oligoastrocytomas showed ATRX positivity even in a background of intact 1p/19q (Supplementary Figure S1d). Co-evaluation of p53 and ATRX immunohistochemistry identified 52.6% (30/57) of 1p/19q non-codeleted oligodendroglial tumors lacking the important astrocytic markers (Figure 3).
Figure 3. Co-evaluation of p53 and ATRX immunohistochemistry in 1p/19q non-codeleted oligodendroglial tumors.
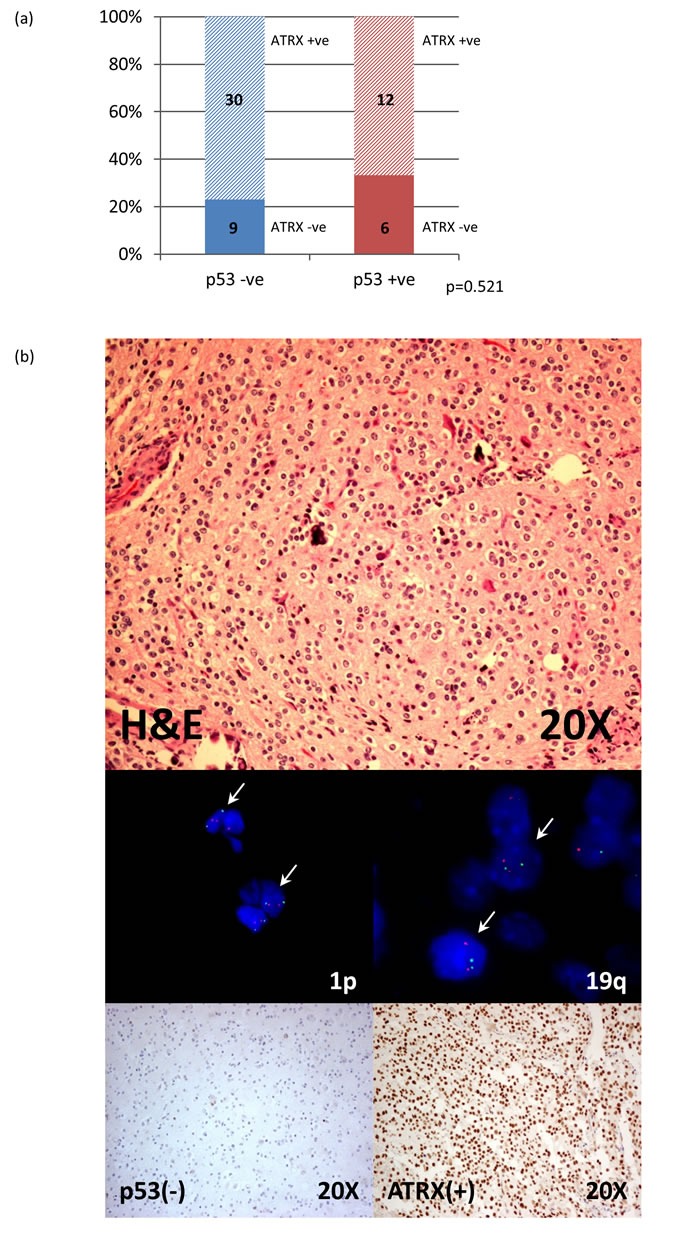
52.6% (30/57) of 1p/19q non-codeleted oligodendroglial tumors lacking p53 positivity and loss of ATRX expression a. Photos of 1p/19q intact oligodendroglioma with negative p53 expression and positive ATRX staining b. -ve, negative; +ve, positive.
1p/19q non-codeleted oligodendroglial tumors with wild-type TP53 are frequently IDH/TERTp mutated
We additionally conducted mutational analysis of TP53 (exon 4 to 9) in 45 cases of oligodendroglial tumors lacking 1p/19q codeletion. TP53 mutations detected were summarized in Supplementary Table S2. Wild-type TP53 was observed in 71.1% (32/45) of 1p/19q non-codeleted oligodendroglial tumors and trended to better survival (p = 0.058) (Figure 4c). Importantly, 50% (16/32) of TP53 wild-type tumors showed classic oligodendroglial histology (p = 0.045) (Figure 4a). Among this subset of 1p/19q non-codeleted, TP53 wild-type oligodendroglial tumors, IDH and TERTp mutations were present in 87.5% (28/32) and 50% (16/32) of cases, respectively. The mutations co-occurred in 75% (12/16) of oligodendrogliomas without 1p/19q codeletion and TP53 mutation (p = 0.012) (Figure 4b).
Figure 4. Correlations between TP53 mutational status and clinicopathological and molecular variables.

Half of TP53 wild-type tumors showed classic oligodendroglial histology (p = 0.045) a. Mutations of IDH and TERTp co-occurred in 75% of oligodendrogliomas without 1p/19q codeletion and TP53 mutation (p = 0.012) b. Patients with Wild-type TP53 tend to have better survival (p = 0.058) c. CFO, classic for oligodendroglial morphology; mixed, mixed oligoastrocytic histology; mut, mutant; wt, wild-type; OS, overall survival.
DISCUSSION
In the present study, we examined a large cohort of oligodendroglial tumors (N = 337) and identified 60.2% of cases harbored 1p/19q codeletion. Since first reported by Reifenberger and colleagues in 1994 [16], combined chromosomal whole arm deletion in 1p and 19q has been referred as genetic hallmark of oligodendrogliomas [17]. Mediated by an unbalanced translocation between chromosome 1 and 19, with formation of derivative chromosomes der(1;19)(q10;p10) and der(1;19)(p10;q10) and the latter one being subsequently deleted during gliomagenesis [13, 18], 1p/19q codeletion was one of the most representative and remarkable molecular markers in diffuse gliomas with diagnostic, prognostic and predictive utilities in clinical practice [7, 19]. The marker served as a stratifier in clinical trials and was shown to predict therapeutic response to PCV chemotherapy (procarbazine, lomustine and vincristine) and radiotherapy in oligodendroglial tumors [1, 5]. 1p/19q codeletion will be one of the molecular markers being incorporated into the integrated morphological and molecular diagnosis of CNS tumors as proposed by the International Society of Neuropathology-Haarlem Consensus Guidelines [20].
While classical oligodendrogliomas would be defined by 1p/19q codeletion [7], there were clearly oligodendroglial tumors lacking the molecular hallmark in various retrospective cohorts and important clinical trials, some of which with histology having been vigorously interrogated by a central review panel of experienced neuropathologists [1, 5, 6, 10, 11, 14, 15, 21-37] (Table 4). In the EORTC 26951 study, 75% of 316 oligodendroglial tumors had non-codeleted 1p/19q and 175 cases were oligodendroglioma [5]. In the RTOG 9402 trial, 29% of 150 oligodendroglial tumors had intact 1p or 19q alleles [1], when the histology of the same cohort was subject to central panel review, 20% (19/97) of the CFO (classic for oligodendroglial morphology) tumors had intact 1p/19q [38]. So clearly, “oligodendroglial tumors” without 1p/19q codeletion are a distinct, sizeable group which cannot be put aside. In our study, we identified 24% (30/125) of oligodendrogliomas and 38.9% (28/72) of anaplastic oligodendrogliomas had intact 1p or 19q, yielding an overall frequency of 1p/19q non-codeleted oligodendrogliomas of 29.4% (58/197). Our results corroborated the observations of the existence of 1p/19q intact oligodendrogliomas from previous studies and suggested the need of additional biomarker(s) to characterize this subset of oligodendrogliomas.
Table 4. Review of various retrospective cohorts and important clinical trials.
| Study | No. of O examined | No. of AO examined | No. of non-1p/19q codeleted O identified | No. of non-1p/19q codeleted AO identified | Method used |
|---|---|---|---|---|---|
| Ducray F and Sanson M et al. 2008 [25] | 22 | 24 | 9 (40.9%) | 12 (50%) | array-CGH |
| Durand KS and Labrousse FJ et al. 2010 [30] | 2 | 5 | 0 | 0 | LOH |
| Labussiere M and Sanson M et al. 2010 [27] | 90 | 119 | 43 (47.8%) | 55 (46.2%) | array-CGH |
| Buckley PG and Farrell MA et al. 2011 [33] | 15 | 12 | 3 (20%) | 3 (25%) | FISH and array-CGH |
| Ducray F and Sanson M et al. 2011 [35] | 73 | 133 | 30 (51%) | 42 (45%) | array-CGH |
| Eigenbrod S and Kretzschmar HA et al. 2011 [37] | 10 | 10 | 1 (10%) | 4 (40%) | LOH |
| Goze C and Duffau H et al. 2012 [29] | 21 | - | 6 (29%) | - | LOH |
| Jiao Y and Yan H et al. 2012 [36] | 21 | 29 | 8 (38.1%) | 3 (10.3%) | LOH |
| Li S and Jiang T et al. 2012 [14] | - | 36 | - | 22 (61.1%) | DHPLC |
| Sahm F and Hartmann C et al. 2012 [34] | 9 | 9 | 2 (22.2%) | 1 (11.1%) | LOH and MLPA |
| Arita H and Ichimura K et al. 2013 [10] | 34 | 31 | 7 (23%) | 7 (27%) | MLPA |
| Cairncross G and Mehta M et al. 2013 [1] | - | 150 | - | 43 (29%) | FISH |
| Frenel JS and Campone M et al. 2013 [32] | 0 | 43 | 0 | 17 (42.5%) | FISH |
| Jiang H and Lin S et al. 2013 [28] | - | 24 | - | 7 (29.2%) | FISH |
| Mur P and Melendez B et al. 2013 [31] | 19 | 14 | 14 (42.4%) | FISH | |
| van den Bent MJ and Hoang-Xuan K et al.2013 [5] | - | 316 (AO and AOA) | - | 236 (75%, AO and AOA) | FISH |
| Chan AK and Ng HK et al.2014 [15] | 19 | 11 | 7 (36.8%) | 4 (36.4%) | FISH |
| Gillet E and Idbaih A et al. 2014 [23] | 32 | - | 16 (50%) | - | LOH |
| Sahm F and von Deimling A et al. 2014 [11] | 43 (OA) | 11 (25.6%, OA) | FISH | ||
| Brat DJ and Zhang J et al. 2015 [51] | 65 | 44 | 27 (41.5%) | 13 (29.5%) | Genomic Analysis |
| Chan AK and Ng HK et al.2015 [6] | 18 | 3 | 7 (41.2%) | 1 (33.3%) | FISH |
| Gleize V and Sanson M et al. 2015 [24] | 60 | 43 | 4 (6.7%) | 9 (20.9%) | LOH |
| Weller M and Reifenberger G et al. 2015 [26] | 3 | 3 | 0 | 1 (33.3%) | array-CGH |
| Dubbink HJ and van den Bent MJ et al. 2016 [22] | - | 93 | - | 40 (47.1%) | LOH, FISH and NGS |
O, oligodendroglioma; AO, anaplastic oligodendroglioma; OA, Oligoastrocytoma; FISH, fluorescence in situ hybridization; LOH, loss of heterozygosity; MLPA, multiplex ligation-dependent probe amplification; DHPLC, denaturing high-performance liquid chromatography; NGS, next-generation sequencing; array-CGH, array-based comparative genomic hybridization
Genomic landscape of grades II and III diffuse gliomas was comprehensively interrogated in the landmark study reported by the TCGA Research Network using multi-omic platforms [39]. The genomewide analyses identified concordant classification of three prognostically relevant molecular subgroups of lower grade gliomas based on unsupervised clustering of multi-omic data, which can be robustly identified by IDH mutation and 1p/19q codeletion. Importantly, IDH mutated, 1p/19q codeleted gliomas were characterized by mutations in CIC, FUBP1, NOTCH1, and TERT promoter, suggesting these mutations could serve as oligodendroglial lineage markers. IDH mutated gliomas lacking 1p/19q codeletion showed high frequency of TP53 mutation (94%) and ATRX inactivation (86%), compatible with their role as astrocytic markers. These substantial genomic data provided the basis of our choice of astrocytic and oligodendroglial markers in our current study (Supplementary Table S3). It was worthwhile to note that in the TCGA report, OncoSign analysis based on recurrent copy-number variations, mutations, and gene fusions, was performed and the group identified four dominant OncoSign classes, namely OSC1 to OSC4, which were correlated with the IDH-1p/19q defined subgroups. 1p/19q non-codeleted oligodendrogliomas were enriched in OSC3 and were associated with IDH mutation and TERTp mutation, validating the observations in our study. Another remarkable study from Weller et al. interrogated grades II and III diffuse gliomas using genome- and transcriptome-wide profiling and reported molecular subgroups harboring distinct genomic aberrations and expression patterns which provided prognostically significant information on top of IDH mutation and 1p/19q codeletion [26]. Genomic profiling revealed five molecular subgroups with Group I tightly correlating with IDH mutation and 1p/19q codeletion and exhibiting the best clinical outcome, and Group V harboring glioblastoma-like copy number changes including 7q gain and 10q loss, and showing the poorest survival. Importantly, the genomic profiling provided clinically relevant information with biological implication for prognostication of diffuse gliomas beyond histologic classification and grading.
1p/19q non-codeleted oligodendrogliomas in our study were associated with TERTp mutation (p = 0.007) and demonstrated frequent IDH mutation (67.4%). Co-mutations of IDH and TERTp occurred in 75% of 1p/19q intact, TP53 wild-type oligodendrogliomas in our cohort. Notably, IDH mutation was also frequently detected in the 1p/19q intact oligodendroglial tumors and conferred chemo-radiation sensitivity in the absence of 1p/19q codeletion as shown by Cairncross and colleagues [21]. In terms of classification and diagnostic utility, presence of IDH mutation, even without 1p/19q codeletion, has been suggested to indicate the diagnosis of oligodendrogliomas over other tumors with similar histo-morphological features [40]. Multiple studies have shown the association between TERTp mutation and 1p/19q codeletion as well as classic oligodendroglial morphology [6, 9, 10, 41, 42]. Our current study validated previous observations and additionally demonstrated the association between TERTp mutation and classic oligodendroglial morphology in a molecular background of intact 1p/19q. Notably, in the study by Killela and colleagues examining more than 470 diffuse gliomas, co-mutations of IDH and TERTp were identified in 79.3% (69/87) of oligodendrogliomas while 1p/19q codeletion was only observed in 54% (47/87) of the cases [41]. Our study and previous observations suggested the potential of IDH mutation plus TERTp mutation in assisting the diagnosis of oligodendroglioma for a tumor with clear cell morphology but lacking 1p/19q codeletion.
It was crucial to avoid misclassification of true 1p/19q codeleted oligodendrogliomas as non-deleted tumors (ie false negativity) in our study. We employed FISH and used commercial probe targeting the “minimally deleted regions” in order to maximize the sensitivity in detecting 1p/19q codeletion [2]. The technique had been well standardized and adopted in various studiesand could be applied robustly on formalin-fixed paraffin-embedded tissues in routine diagnostic neuropathology practice. Importantly, FISH probes targeting at the same loci were also adopted by the EORTC and RTOG clinical trials [1, 5]. Since 1p/19q loss classically involved deletion of the entire 1p and 19q arms, tumors with retained minimally deleted regions in our study were extremely unlikely to be false negative cases.
We further examined astrocytic markers, p53 expression and ATRX loss, in the 1p/19q non-codeleted oligodendroglial tumors. Co-evaluation of the two astrocytic immuno-markers revealed more than 50% of the 1p/19q non-codeleted oligodendroglial tumors did not show p53 positivity and ATRX loss, i.e. they were not molecularly astrocytic. Among 38 cases of mixed oligoastrocytomas with analyzable data of ATRX expression and TERTp mutation, the two biomarkers showed mutual exclusivity in all but one case [Supplementary Figure S1e]. TERTp and ATRX mutations represented independent genetic mechanisms in maintaining telomere lengths in tumorigenesis of diffuse gliomas [42]. In the study by Sahm and colleagues investigating 43 mixed oligoastrocytomas, 1p/19q codeletion and ATRX loss were mutually exclusive in all but one case, suggesting parting with the diagnosis of oligoastrocytoma. According to these authors, all non-codeleted oligodendroglial tumors are in fact astrocytic [11]. With the morphological and molecular evidence of several cases of “true” oligoastrocytoma being reported [46-48], complete farewell to oligoastrocytoma can be argued. Notably, in our series, we also did not find any true cases of “oligoastrocytoma”.
Clinical trials by Cairncross et al. and van den Bent et al. have set the standard therapeutic protocol for patients with 1p/19q codeleted anaplastic oligodendroglial tumors [1, 5]. The former research group has additionally demonstrated that the survival benefit of PCV (procarbazine, lomustine, and vincristine) in 1p/19q non-codeleted anaplastic oligodendroglial tumors was associated with IDH mutation [21]. Clinical benefits of radiation plus PCV have been extended to treatment of low grade diffuse gliomas [49]. The chemoradiotherapy was associated with prolonged overall survival and progression-free survival in subsets of oligodendroglioma, oligoastrocytoma, and IDH mutated low grade gliomas. Our group has previously reported the potential clinical value of TERTp mutation in predicting differential responses to genotoxic therapies in grades II and III diffuse gliomas, on top on IDH status [50]. Our current study identified a major portion of oligodendrogliomas lacking 1p/19q codeletion was characterized by IDH and TERTp co-mutation, with a background of ATRX nuclear positivity wild-type TP53 (Figure 5), indicating that these patients could potentially benefit from chemoradiotherapy.
Figure 5. Overview of the sequential work-flow based on The 2016 WHO classification of CNS tumors and Haarlem scheme [20, 56].
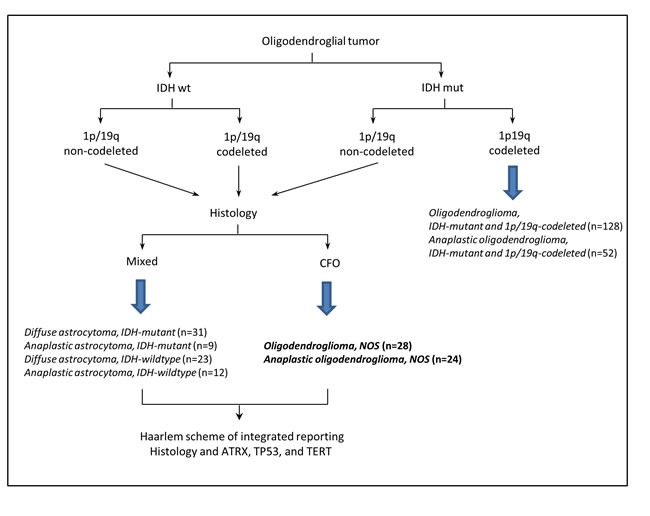
mixed, mixed oligoastrocytic histology; CFO, classic for oligodendroglial morphology.
In summary, this study examined a large cohort of oligodendroglial tumors and demonstrated that astrocytic tumors could not account for all 1p/19q non-codeleted oligodendroglial tumors and oligodendroglioma lacking 1p/19q codeletion may form a distinct subgroup of diffuse glioma. Co-evaluation of IDH and TERTp mutation had diagnostic value for oligodendroglioma and could serve as an adjunct in oligodendroglial tumors after evaluation of the 1p/19q status.
MATERIALS AND METHODS
Patients and tissue samples
Oligodendroglial tumors diagnosed from our service archives at the Prince of Wales Hospital (The Chinese University of Hong Kong) and Huashan Hospital (Fudan University, Shanghai) were included in the present study. Totally 337 oligodendroglial tumors were included in our study and 71 of them were from Hong Kong and 266 from Shanghai. Only cases which were reviewed by two neuropathologists (HK N and H C) as oligodendroglial were included based on The 2007 WHO classification of tumors of the central nervous system [51]. The present study used only formalin-fixed, paraffin-embedded material. Clinical and survival data of the patients were retrieved from the respective institutional medical record systems. This study was approved by the Ethics Committee of Shanghai Huashan Hospital and the New Territories East Cluster-Chinese University of Hong Kong Ethics Committee. The cohort in this study partially overlapped with previous studies [6, 15, 52].
Fluorescence In Situ hybridization for chromosome 1p and 19q
Fluorescent in situ hybridization (FISH) was performed on formalin-fixed, paraffin-embedded tumor tissue to detect deletion of chromosome 1p and 19q [15]. 4-μm-thick sections were deparaffinized, treated with sodium thiocyanate and followed by digestion with pepsin solution at 37°C. Dual-color-probe hybridization was then performed using Vysis 1p36/1q25 and 19q13/19p13 FISH Probe Kit (Abbott Molecular) and the spectrum-green-probe was labeled on chromosome 1q and 19p, respectively. Both probes and target tumor DNA were denatured in an 80°C oven for 30 minutes and followed by an overnight incubation at 37°C. Nuclei were counterstained with Vectashield mounting medium containing 4′, 6-diamidino-2- phenylindole (Vector Laboratories, Burlingame, CA, USA) and the number of FISH signals was assessed under a Zeiss Axioplan fluorescence microscope (Carl Zeiss Microscopy LLC, NY, USA) equipped with a triple-pass filter (DAPI/Green/Orange). Hybridizing signals of at least 100 non-overlapping nuclei were enumerated and a sample will be considered as 1p or 19q deleted when more than 25% of counted nuclei presented one target (orange) signal and two reference (green) signals [2].
Mutation analysis of IDH1/2, TERT promoter and TP53
Mutation status of IDH1/2, TERT promoter and TP53 were studied with direct sequencing [6, 52, 53]. Tissues from representative areas with tumor content > 70% were collected from 5 to 7 dewaxed formalin-fixed, paraffin-embedded sections and re-suspended in 10mM Tris-HCl buffer (pH 8.5) which also contained proteinase K with a final concentration of 2 mg/ml. After that, the mixture was incubated at 55°C for 2-18 h and then at 98°C for 10 min. After the above incubation, the buffer comprising the cell lysate was used for polymerase chain reaction (PCR) analysis of IDH1, IDH2, TP53 and TERTp. We performed direct sequencing for IDH1, IDH2, TERT promoter region and exon 4-9 of TP53 as per previous protocols [6, 15, 52-54]. All mutational status was validated by sequencing of a newly amplified DNA fragment. Sequences of primers used for PCR analysis of IDH1/2, TERT promoter and TP53 were listed in Supplementary Table S1. For sequencing of IDH1/2 and TP53, target DNA fragments were amplified in a total 10μl reaction volume which containing 1μl of cell lysate, 1μl of MgCl2 (25mM), 1μl of 10*PCR buffer II, 0.2μl of each deoxyribonucleoside triphosphate (10mM), 0.3μl of each forward and reverse primers of target DNA fragment (10nmol), 0.075μl of AmpliTaq Gold DNA polymerase (Life Technologies Corporation, Hong Kong, China). PCR for TERT promoter region was performed in 10μl reaction mixture containing 1μl of cell lysate, 0.3μl of each forward and reverse primers of target DNA fragment (10nmol), and 5μl of KAPA HiFi HotStart ReadyMix DNA Polymerase (Kapa Biosystems Wilmington, DE, USA). PCR reaction started with a denature procedure of 95°C for 10 min, then followed by 45 cycles of 95°C for 20 sec, annealing temperature (Supplementary Table 1) for 20 sec and 72°C for 30 sec, and a final extension step of 72°C for 3 min. Products were then treated with Exonuclease I (TakaRa Biotechnology Limited, Dalian, China) of 2μl (0.25U/μl) per 5μl PCR product at 37°C for 15min followed by 80°C for 15min. Sequencing of target DNA fragment was performed using BigDye Terminator Cycle Sequencing kit v.1.1 (Life Technologies). The Genetic Analyzer 3130xl was used for the following sequencing and the results were analyzed by Sequencing Analysis Software. Hotspot mutations of IDH1/2 and TERTp, all missense mutations of TP53 could be detected. All detected missense mutations of TP53 were listed in Supplementary Table S2.
Immunohistochemistry of ATRX, p53 and PDGFRA
Immunohistochemistry was performed for detecting the expression of p53, PDGFRA, and ATRX. 4μm thick formalin-fixed, paraffin-embedded (FFPE) sections of each sample were de-waxed by xylene and rehydrated in graded alcohols. Sections were then treated with citrate buffer (pH 6.0) by heating for antigen retrieval. After antigen retrieval, they were subjected to immuno-staining by BenchMark XT automated tissue staining systems (Ventana Medical Systems, Tucson, AZ, USA) using validated protocols for detecting the expression of p53 and ATRX. Sections were incubated with antibodies of anti-p53 (Dako DO-7, 1:100) and anti-ATRX (SIGMA CAT: HPA001906, 1:400) at 37°C for 32 min and followed by an incubation with UltraView HRP-conjugated multimer antibody reagent (Ventana Medical Systems). Subsequent antigen detection was performed using UltraView diamino benzidine chromogen step (Ventana Medical Systems). Expression of PDGFRA was detected using the automated Bond-max system (Leica Bond-Max) with anti-PDGFRA antibody (Santa Cruz, C-20, 1:200) and subsequent antigen detection was performed using Ultra View diamino benzidine chromogen step (Bond-Max). At last, slides were counterstained with Mayer's hematoxylin. A tumor was scored p53 positive if > 10% of tumor cells showed strong nuclear staining [53]. Samples with more than 10% of tumor cells showing positive nuclear staining of ATRX were scored as positive. Endothelial cells, cortical neurons and infiltrating inflammatory cells were generally positive and served as internal positive controls [12]. Cytoplasmic and membrane staining were considered for evaluation of PDGFRA expression. Both the distribution and intensity of staining were semi-quantitatively scored as previously reported [55].
Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics Version 20 (IBM Corporation, NY, USA). Univariate analysis was conducted using Chi-square or Fisher's test to compare categorical variables. Independent-Samples T Test was used to compare mean age between 2 populations. Kaplan-Meier estimator and log rank test were performed for univariate survival analysis. Whereas a Cox proportional hazards model was employed for multivariate survival analysis. Statistical significance was considered when p < 0.05 (two-side).
SUPPLEMENTARY MATERIAL FIGURES AND TABLES
Acknowledgments
This study was supported by Health and Medical Research Fund of Hong Kong (grant no. 02133146), 973 Program (grant no: 2015CB755500), and Shanghai Sailing Program (grant no:16YF1415200) .
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Cairncross G, Wang M, Shaw E, Jenkins R, Brachman D, Buckner J, Fink K, Souhami L, Laperriere N, Curran W, Mehta M. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. Journal of clinical oncology. 2013;31(3):337–343. doi: 10.1200/JCO.2012.43.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horbinski C, Miller CR, Perry A. Gone FISHing: clinical lessons learned in brain tumor molecular diagnostics over the last decade. Brain pathology. 2011;21(1):57–73. doi: 10.1111/j.1750-3639.2010.00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourne TD, Schiff D. Update on molecular findings, management and outcome in low-grade gliomas. Nature reviews Neurology. 2010;6(12):695–701. doi: 10.1038/nrneurol.2010.159. [DOI] [PubMed] [Google Scholar]
- 4.Appin CL, Brat DJ. Molecular Genetics of Gliomas. The Cancer Journal. 2014;20(NO.1):66–72. doi: 10.1097/PPO.0000000000000020. [DOI] [PubMed] [Google Scholar]
- 5.van den Bent MJ, Brandes AA, Taphoorn MJ, Kros JM, Kouwenhoven MC, Delattre JY, Bernsen HJ, Frenay M, Tijssen CC, Grisold W, Sipos L, Enting RH, French PJ, Dinjens WN, Vecht CJ, Allgeier A, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. Journal of clinical oncology. 2013;31(3):344–350. doi: 10.1200/JCO.2012.43.2229. [DOI] [PubMed] [Google Scholar]
- 6.Chan AK, Yao Y, Zhang Z, Chung NY, Liu JS, Li KK, Shi Z, Chan DT, Poon WS, Zhou L, Ng HK. TERT promoter mutations contribute to subset prognostication of lower-grade gliomas. Modern pathology. 2015;28(2):177–186. doi: 10.1038/modpathol.2014.94. [DOI] [PubMed] [Google Scholar]
- 7.Wesseling P, van den Bent M, Perry A. Oligodendroglioma: pathology, molecular mechanisms and markers. Acta neuropathologica. 2015;129(6):809–827. doi: 10.1007/s00401-015-1424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majumdar K, Radotra BD, Vasishta RK, Pathak A. Platelet-derived growth factor expression correlates with tumor grade and proliferative activity in human oligodendrogliomas. Surgical neurology. 2009;72(1):54–60. doi: 10.1016/j.surneu.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Koelsche C, Sahm F, Capper D, Reuss D, Sturm D, Jones DT, Kool M, Northcott PA, Wiestler B, Bohmer K, Meyer J, Mawrin C, Hartmann C, Mittelbronn M, Platten M, Brokinkel B, et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta neuropathologica. 2013;126(6):907–915. doi: 10.1007/s00401-013-1195-5. [DOI] [PubMed] [Google Scholar]
- 10.Arita H, Narita Y, Fukushima S, Tateishi K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP, Kawahara N, Shibui S, Ichimura K. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta neuropathologica. 2013;126(2):267–276. doi: 10.1007/s00401-013-1141-6. [DOI] [PubMed] [Google Scholar]
- 11.Sahm F, Reuss D, Koelsche C, Capper D, Schittenhelm J, Heim S, Jones DT, Pfister SM, Herold-Mende C, Wick W, Mueller W, Hartmann C, Paulus W, von Deimling A. Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta neuropathologica. 2014;128(4):551–559. doi: 10.1007/s00401-014-1326-7. [DOI] [PubMed] [Google Scholar]
- 12.Wiestler B, Capper D, Holland-Letz T, Korshunov A, von Deimling A, Pfister SM, Platten M, Weller M, Wick W. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta neuropathologica. 2013;126(3):443–451. doi: 10.1007/s00401-013-1156-z. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, Flynn H, Passe S, Felten S, Brown PD, Shaw EG, Buckner JC. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer research. 2006;66(20):9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Yan C, Huang L, Qiu X, Wang Z, Jiang T. Molecular prognostic factors of anaplastic oligodendroglial tumors and its relationship: a single institutional review of 77 patients from China. Neuro-oncology. 2012;14(1):109–116. doi: 10.1093/neuonc/nor185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan AK, Pang JC, Chung NY, Li KK, Poon WS, Chan DT, Shi Z, Chen L, Zhou L, Ng HK. Loss of CIC and FUBP1 expressions are potential markers of shorter time to recurrence in oligodendroglial tumors. Modern pathology. 2014;27(3):332–342. doi: 10.1038/modpathol.2013.165. [DOI] [PubMed] [Google Scholar]
- 16.Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W, Collins VP. Molecular Genetic Analysis of Oligodendroglial Tumors Shows Preferential Allelic Deletions on 19q and 1p. American Journal of Pathology. 1994;145(No.5):1175–1190. [PMC free article] [PubMed] [Google Scholar]
- 17.Cairncross G, Jenkins R. Gliomas With 1p/19q Codeletion: a.k.a. Oligodendroglioma. The Cancer Journal. 2008;14(6):352–357. doi: 10.1097/PPO.0b013e31818d8178. [DOI] [PubMed] [Google Scholar]
- 18.Griffin CA, Burger P, Morsberger L, Yonescu R, Swierczynski S, Weingart JD, Murphy KM. Identification of der(1;19)(q10;p10) in Five Oligodendrogliomas Suggests Mechanism of Concurrent 1p and 19q Loss. Journal of neuropathology and experimental neurology. 2006;65(No.10):988–994. doi: 10.1097/01.jnen.0000235122.98052.8f. [DOI] [PubMed] [Google Scholar]
- 19.Brandner S, von Deimling A. Diagnostic, prognostic and predictive relevance of molecular markers in gliomas. Neuropathology and applied neurobiology. 2015;41(6):694–720. doi: 10.1111/nan.12246. [DOI] [PubMed] [Google Scholar]
- 20.Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A, Aldape K, Brat D, Collins VP, Eberhart C, Figarella-Branger D, Fuller GN, Giangaspero F, Giannini C, Hawkins C, Kleihues P, et al. International Society Of Neuropathology—Haarlem consensus guidelines for nervous system tumor classification and grading. Brain pathology. 2014;24(5):429–435. doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cairncross JG, Wang M, Jenkins RB, Shaw EG, Giannini C, Brachman DG, Buckner JC, Fink KL, Souhami L, Laperriere NJ, Huse JT, Mehta MP, Curran WJ., Jr Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. Journal of clinical oncology. 2014;32(8):783–790. doi: 10.1200/JCO.2013.49.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dubbink HJ, Atmodimedjo PN, Kros JM, French PJ, Sanson M, Idbaih A, Wesseling P, Enting R, Spliet W, Tijssen C, Dinjens WN, Gorlia T, van den Bent MJ. Molecular classification of anaplastic oligodendroglioma using next-generation sequencing: a report of the prospective randomized EORTC Brain Tumor Group 26951 phase III trial. Neuro-oncology. 2016;18(3):388–400. doi: 10.1093/neuonc/nov182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillet E, Alentorn A, Doukoure B, Mundwiller E, van Thuijl HF, Reijneveld JC, Medina JA, Liou A, Marie Y, Mokhtari K, Hoang-Xuan K, Sanson M, Delattre JY, Idbaih A. TP53 and p53 statuses and their clinical impact in diffuse low grade gliomas. Journal of neuro-oncology. 2014;118(1):131–139. doi: 10.1007/s11060-014-1407-4. [DOI] [PubMed] [Google Scholar]
- 24.Gleize V, Alentorn A, Connen de Kerillis L, Labussiere M, Nadaradjane AA, Mundwiller E, Ottolenghi C, Mangesius S, Rahimian A, Ducray F, network P, Mokhtari K, Villa C, Sanson M. CIC inactivating mutations identify aggressive subset of 1p19q codeleted gliomas. Annals of neurology. 2015;78(3):355–374. doi: 10.1002/ana.24443. [DOI] [PubMed] [Google Scholar]
- 25.Ducray F, Idbaih A, de Reynies A, Bieche I, Thillet J, Mokhtari K, Lair S, Marie Y, Paris S, Vidaud M, Hoang-Xuan K, Delattre O, Delattre JY, Sanson M. Anaplastic oligodendrogliomas with 1p19q codeletion have a proneural gene expression profile. Molecular cancer. 2008;7:41. doi: 10.1186/1476-4598-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weller M, Weber RG, Willscher E, Riehmer V, Hentschel B, Kreuz M, Felsberg J, Beyer U, Loffler-Wirth H, Kaulich K, Steinbach JP, Hartmann C, Gramatzki D, Schramm J, Westphal M, Schackert G, et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta neuropathologica. 2015;129(5):679–693. doi: 10.1007/s00401-015-1409-0. [DOI] [PubMed] [Google Scholar]
- 27.Labussiere M, Idbaih A, Wang XW, Marie Y, Boisselier B, Falet C, Paris S, Laffaire J, Carpentier C, Criniere E, Ducray F, Hallani SE, Mokhtari K, Hoang-Xuan K, Delattre JY, Sanson M. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74:1886–1890. doi: 10.1212/WNL.0b013e3181e1cf3a. [DOI] [PubMed] [Google Scholar]
- 28.Jiang H, Ren X, Cui X, Wang J, Jia W, Zhou Z, Lin S. 1p/19q codeletion and IDH1/2 mutation identified a subtype of anaplastic oligoastrocytomas with prognosis as favorable as anaplastic oligodendrogliomas. Neuro-oncology. 2013;15(6):775–782. doi: 10.1093/neuonc/not027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goze C, Bezzina C, Goze E, Rigau V, Maudelonde T, Bauchet L, Duffau H. 1P19Q loss but not IDH1 mutations influences WHO grade II gliomas spontaneous growth. Journal of neuro-oncology. 2012;108(1):69–75. doi: 10.1007/s11060-012-0831-6. [DOI] [PubMed] [Google Scholar]
- 30.Durand KS, Guillaudeau A, Weinbreck N, DeArmas R, Robert S, Chaunavel A, Pommepuy I, Bourthoumieu S, Caire F, Sturtz FG, Labrousse FJ. 1p19q LOH patterns and expression of p53 and Olig2 in gliomas: relation with histological types and prognosis. Modern pathology. 2010;23(4):619–628. doi: 10.1038/modpathol.2009.185. [DOI] [PubMed] [Google Scholar]
- 31.Mur P, Mollejo M, Ruano Y, de Lope AR, Fiano C, Garcia JF, Castresana JS, Hernandez-Lain A, Rey JA, Melendez B. Codeletion of 1p and 19q determines distinct gene methylation and expression profiles in IDH-mutated oligodendroglial tumors. Acta neuropathologica. 2013;126(2):277–289. doi: 10.1007/s00401-013-1130-9. [DOI] [PubMed] [Google Scholar]
- 32.Frenel JS, Leux C, Loussouarn D, Le Loupp AG, Leclair F, Aumont M, Mervoyer A, Martin S, Denis MG, Campone M. Combining two biomarkers, IDH1/2 mutations and 1p/19q codeletion, to stratify anaplastic oligodendroglioma in three groups: a single-center experience. Journal of neuro-oncology. 2013;114(1):85–91. doi: 10.1007/s11060-013-1152-0. [DOI] [PubMed] [Google Scholar]
- 33.Buckley PG, Alcock L, Heffernan J, Woods J, Brett F, Stallings RL, Farrell MA. Loss of Chromosome 1p/19q in Oligodendroglial Tumors: Refinement of Chromosomal Critical Regions and Evaluation of Internexin Immunostaining as a Surrogate Marker. Journal of neuropathology and experimental neurology. 2011;70(No.3):177–182. doi: 10.1097/NEN.0b013e31820c765b. [DOI] [PubMed] [Google Scholar]
- 34.Sahm F, Koelsche C, Meyer J, Pusch S, Lindenberg K, Mueller W, Herold-Mende C, von Deimling A, Hartmann C. CIC and FUBP1 mutations in oligodendrogliomas, oligoastrocytomas and astrocytomas. Acta neuropathologica. 2012;123(6):853–860. doi: 10.1007/s00401-012-0993-5. [DOI] [PubMed] [Google Scholar]
- 35.Ducray F, Mokhtari K, Criniere E, Idbaih A, Marie Y, Dehais C, Paris S, Carpentier C, Dieme MJ, Adam C, Hoang-Xuan K, Duyckaerts C, Delattre JY, Sanson M. Diagnostic and prognostic value of alpha internexin expression in a series of 409 gliomas. European journal of cancer. 2011;47(5):802–808. doi: 10.1016/j.ejca.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 36.Jiao Y, Killela PJ, Reitman ZJ, Rasheed BA, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Nagahashi Marie SK, Bettegowda C, Agrawal N, Lipp E, Pirozzi CJ, Lopez GY, He Y, et al. Frequent ATRX, CIC, and FUBP1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eigenbrod S, Roeber S, Thon N, Giese A, Krieger A, Grasbon-Frodl E, Egensperger R, Tonn JC, Kreth F-W, Kretzschmar HA. α-Internexin in the Diagnosis of Oligodendroglial Tumors and Association With 1p/19q Status. Journal of neuropathology and experimental neurology. 2011;70(11):970–978. doi: 10.1097/NEN.0b013e3182333ef5. [DOI] [PubMed] [Google Scholar]
- 38.Giannini C, Burger PC, Berkey BA, Cairncross JG, Jenkins RB, Mehta M, Curran WJ, Aldape K. Anaplastic oligodendroglial tumors: refining the correlation among histopathology, 1p 19q deletion and clinical outcome in Intergroup Radiation Therapy Oncology Group Trial 9402. Brain pathology. 2008;18(3):360–369. doi: 10.1111/j.1750-3639.2008.00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cancer Genome Atlas Research N, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O, Robertson AG, Noushmehr H, Laird PW, Cherniack AD, Akbani R, et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. The New England journal of medicine. 2015;372(26):2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanboon J, Williams EA, Louis DN. The Diagnostic Use of Immunohistochemical Surrogates for Signature Molecular Genetic Alterations in Gliomas. Journal of neuropathology and experimental neurology. 2015 doi: 10.1093/jnen/nlv009. [DOI] [PubMed] [Google Scholar]
- 41.Killela PJ, Pirozzi CJ, Healy P, Reitman ZJ, Lipp E, Rasheed BA, Yang R, Diplas BH, Wang Z, Greer PK, Zhu H, Wang CY, Carpenter AB, Friedman H, Friedman AH, Keir ST, et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget. 2014;5(6):1515–1525. doi: 10.18632/oncotarget.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, Grollman AP, He TC, He Y, Hruban RH, Jallo GI, Mandahl N, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proceedings of the National Academy of Sciences of the United States of America; 2013; pp. 6021–6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbashina V, Salazar P, Holland EC, Rosenblum MK, Ladanyi M. Allelic Losses at 1p36 and 19q13 in Gliomas: Correlation with Histologic Classification, Definition of a 150-kb Minimal Deleted Region on 1p36, and Evaluation of CAMTA1 as a Candidate Tumor Suppressor Gene. Clinical Cancer Research. 2005;11:1119–1128. [PubMed] [Google Scholar]
- 44.Smith JS, Alderete B, Minn Y, Borell TJ, Perry A, Mohapatra G, Hosek SM, Kimmel D, O’Fallon J, Yates A, Feuerstein BG, Burger PC, Scheithauer BW, Jenkins RB. Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene. 1999;18:4144–4152. doi: 10.1038/sj.onc.1202759. [DOI] [PubMed] [Google Scholar]
- 45.Tews B, Felsberg J, Hartmann C, Kunitz A, Hahn M, Toedt G, Neben K, Hummerich L, von Deimling A, Reifenberger G, Lichter P. Identification of novel oligodendroglioma-associated candidate tumor suppressor genes in 1p36 and 19q13 using microarray-based expression profiling. International journal of cancer. 2006;119(4):792–800. doi: 10.1002/ijc.21901. [DOI] [PubMed] [Google Scholar]
- 46.Qu M, Olofsson T, Sigurdardottir S, You C, Kalimo H, Nister M, Smits A, Ren ZP. Genetically distinct astrocytic and oligodendroglial components in oligoastrocytomas. Acta neuropathologica. 2007;113(2):129–136. doi: 10.1007/s00401-006-0142-0. [DOI] [PubMed] [Google Scholar]
- 47.Wilcox P, Li CC, Lee M, Shivalingam B, Brennan J, Suter CM, Kaufman K, Lum T, Buckland ME. Oligoastrocytomas: throwing the baby out with the bathwater? Acta neuropathologica. 2015;129(1):147–149. doi: 10.1007/s00401-014-1353-4. [DOI] [PubMed] [Google Scholar]
- 48.Huse JT, Diamond EL, Wang L, Rosenblum MK. Mixed glioma with molecular features of composite oligodendroglioma and astrocytoma: a true “oligoastrocytoma”? Acta neuropathologica. 2015;129(1):151–153. doi: 10.1007/s00401-014-1359-y. [DOI] [PubMed] [Google Scholar]
- 49.Buckner JC, Shaw EG, Pugh SL, Chakravarti A, Gilbert MR, Barger GR, Coons S, Ricci P, Bullard D, Brown PD, Stelzer K, Brachman D, Suh JH, Schultz CJ, Bahary JP, Fisher BJ, et al. Radiation plus Procarbazine, CCNU, and Vincristine in Low-Grade Glioma. The New England journal of medicine. 2016;374(14):1344–1355. doi: 10.1056/NEJMoa1500925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang ZY, Chan AK, Ding XJ, Qin ZY, Hong C.S, Chen LC, Zhang X, Zhao FP, Wang Y, Wang Y, Zhou LF, Zhuang ZP, Ng HK, Yan H, Yao Y, Mao Y. TERT promoter mutations contribute to IDH mutations in predicting differential responses to adjuvant therapies in WHO grade II and III diffuse gliomas. Oncotarget. 2015;6(28):24871–24883. doi: 10.18632/oncotarget.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta neuropathologica. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan AK, Yao Y, Zhang Z, Shi Z, Chen L, Chung NY, Liu JS, Li KK, Chan DT, Poon WS, Wang Y, Zhou L, Ng HK. Combination genetic signature stratifies lower-grade gliomas better than histological grade. Oncotarget. 2015;6(25):20885–20901. doi: 10.18632/oncotarget.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takami H, Yoshida A, Fukushima S, Arita H, Matsushita Y, Nakamura T, Ohno M, Miyakita Y, Shibui S, Narita Y, Ichimura K. Revisiting TP53 Mutations and Immunohistochemistry—A Comparative Study in 157 Diffuse Gliomas. Brain pathology. 2015;25(3):256–265. doi: 10.1111/bpa.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang RQ, Shi Z, Chen H, Chung NY, Yin Z, Li KK, Chan DT, Poon WS, Wu J, Zhou L, Chan AK, Mao Y, Ng HK. Biomarker-based prognostic stratification of young adult glioblastoma. Oncotarget. 2015;7(4):5030–5041. doi: 10.18632/oncotarget.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinho O, Longatto-Filho A, Lambros MB, Martins A, Pinheiro C, Silva A, Pardal F, Amorim J, Mackay A, Milanezi F, Tamber N, Fenwick K, Ashworth A, Reis-Filho JS, Lopes JM, Reis RM. Expression, mutation and copy number analysis of platelet-derived growth factor receptor A (PDGFRA) and its ligand PDGFA in gliomas. British journal of cancer. 2009;101(6):973–982. doi: 10.1038/sj.bjc.6605225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta neuropathologica. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


