Abstract
Missing covariate data hampers variable selection in multilevel regression settings. Current variable selection techniques for multiply-imputed data commonly address missingness in the predictors through list-wise deletion and stepwise-selection methods which are problematic. Moreover, most variable selection methods are developed for independent linear regression models and do not accommodate multilevel mixed effects regression models with incomplete covariate data. We develop a novel methodology that is able to perform covariate selection across multiply-imputed data for multilevel random effects models when missing data is present. Specifically, we propose to stack the multiply-imputed data sets from a multiple imputation procedure and to apply a group variable selection procedure through group lasso regularization to assess the overall impact of each predictor on the outcome across the imputed data sets. Simulations confirm the advantageous performance of the proposed method compared with the competing methods. We applied the method to reanalyze the Healthy Directions-Small Business cancer prevention study, which evaluated a behavioral intervention program targeting multiple risk-related behaviors in a working-class, multi-ethnic population.
Keywords: BIC, cancer prevention, group lasso, intervention studies, multilevel, multiple imputation, regularization, Rubin’s rules
1. Introduction
Multilevel models are commonly used in large-scale, community-based intervention or medical trial studies to describe the relationship of the predictors on mean response through fixed effects while also describing the clustering of data (e.g. workers within worksites, students within schools) through random effects. It is becoming standard practice to collect as many predictors as possible to study the impact of contextual, social or comorbidity conditions on the scientific outcome of interest. When performing multilevel models with a high number of predictors, variable selection is useful for discovering and understanding important underlying associations. A demonstrative example is the Healthy-Directions Small-Business (HD-SB) study which studied workers clustered within worksites that were randomized to an intervention or control group. The aim of the HD-SB study was to identify relevant factors that relate to increased consumption of fruits and vegetables. As often encountered in multilevel and longitudinal studies, the selection of important variables is hindered by missing data in the covariates and by the introduction of random effects.
There has been considerable amount of work on the topic of variable selection for mixed effects models (Fan & Li, 2004; Qu & Li, 2006; Johnson et al., 2008; Ni et al., 2010; Chen & Dunson, 2003; Zhu & Zhang, 2006; Crainiceanu, 2008; Zhang & Lin, 2008; Kinney & Dunson, 2008; Wang et al., 2010; Bondell et al., 2010; Ibrahim et al., 2011). These methods require the data to be fully observed (i.e. no missing data). The need to adequately handle missing data is being recognized as a very important aspect of statistical practice with implications for main analyses and sensitivity analyses. Often, researchers resort to complete case analyses where subjects are only included if there are no missing values for all the variables included in the analysis. This strategy is widely known to give rise to bias in model parameters, except for the very special setting where the missing values are missing completely at random (MCAR) (Little & Rubin, 2002).
To address these issues, methods have been developed to perform variable selection with missing data. Garcia et al. (2010) proposed an expectation-maximization (EM) algorithm to simultaneously optimize the penalized likelihood function and estimate the tuning parameter in the presence of missing data. Johnson et al. (2008) considered a penalized estimating function approach to variable selection when missing data is present. Variants of the Akaike information criterion (AIC) to select models from partially observed data have been proposed by Shimodaira (1994), Hens et al. (2006), and Claeskens & Consentino (2008). A criterion for model selection in the presence of incomplete data based on Kullback’s symmetric divergence was also proposed by Seghouane et al. (2005). Similarly, Ibrahim et al. (2008) developed a class of information-based model selection criteria dependent only on output from the EM algorithm to address the missing data problem. A similar EM approach for model selection is taken by Bueso et al. (1999). While these methods are important, there is a gap in the literature for performing variable selection when the method of dealing with missing data is through multiple imputation.
Multiple imputation is a statistical technique that maintains the observed relationship of the data while reflecting the uncertainty present in the missing data through multiple datasets. Performing multiple imputations in lieu of EM and GEE approaches for statistical inference is appealing since it is easy to communicate with collaborators and it tends to be robust against departures from the complete-data model (Schafer, 1997). Multiple imputation methods for linear mixed-effects models are a recent development (Schafer & Yucel, 2002; Demirtas, 2004; Stuart et al., 2009; Demirtas & Hedeker, 2008; Schafer, 1997; Schafer & Yucel, 2002; Liu et al., 2000; Yucel, 2008; Goldstein et al., 2009; Van Buuren & Groothuis-Oudshoorn, 2011). Whichever imputation method is chosen, a total of m complete data sets each with p predictors will be produced. The appealing quality of these m imputed data sets is that complete-data methods can be used.
Although much attention has been given to constructing parameter estimates with an appropriate measure of uncertainty for multiply imputed data, there is no clear guidance on how to perform variable selection on the multiply imputed data sets. Practically, variable selection can be performed on each imputed dataset. However, it is unclear how to combine the selection results as each data set will presumably select different variables in the model. Brand (1999) proposed an ad-hoc procedure that constructs a final model with variables that are deemed significant in at least 60% of the imputed models. Yang et al. (2005) proposed two Bayesian alternative strategies for variable selection in classical linear regression models with missing covariates. Heymans et al. (2007) and Wood et al. (2008) developed methodology that performs automatic backward selection of multiply-imputed data sets. Moreover, none of these methods tackle missing data in the context of mixed models. To fill this serious knowledge gap, we propose a new framework for performing variable selection for multilevel models when multiply-imputed data are considered.
We make several contributions in this paper. First, we describe a penalized likelihood approach for multilevel models that simultaneously uses every multiply-imputed data set to select relevant predictors. Secondly, to overcome the challenges of combining across multiply-imputed datasets, we propose a novel approach that stacks the multiply-imputed data sets which can allow the use of group variable selection via group lasso regularization to assess the overall significance of each predictor on the outcome across all the imputed data sets. Finally, as the selection of an appropriate tuning parameter poses additional problems for multiply imputed data sets, we provide a Bayesian information criterion (BIC) for tuning parameter selection.
The format of this paper is as follows. In section 2, we present the multilevel model and develop a penalized procedure to perform variable selection for multiply-imputed multilevel data. Section 3 provides simulation studies of the proposed methodology. Section 4 applies the developed methodology to the analysis of the Healthy Directions-Small Business cancer prevention study, followed by our concluding remarks in Section 5.
2. Penalized Multiply-Imputed Likelihood
2.1. Model Representation
Suppose that there are n clusters indexed by i = 1, 2, . . . , n and the nth cluster has a total of ki subjects indexed by j = 1, 2, . . . , ki. Let Yi j denote the response on the jth subject within the ith cluster. For example, Yi j can denote the outcome for the jth worker in the ith worksite. Associated with each Yi j is a p × 1 vector of covariates, Xi j. The vector Xi j can include covariates defined at each of the two levels and can also include covariates formed by aggregating values over lower-level units. We consider a two-level linear mixed-effects model, though the proposal can be extended to a more general mixed-effects setting. In particular, it can be adapted to longitudinal multilevel data because longitudinal data is a special case of multilevel data with only a single level of clustering and a specific ordering of observations within the cluster.
The two-level linear mixed model (i.e. multilevel model) is given by
| (1) |
where β is the p × 1 vector of regression coefficients, Zi j is a q × 1 design matrix for the random effects which is typically formed from a subset of the covariates, bi is a q × 1 vector of latent random effects and is distributed MV N(0, σ2Φ) and εi j are assumed to be i .i .d N(0, σ2).
For the ith cluster, let Yi = (Yi1, . . . , Yiki)T denote the ki × 1 vector of outcomes and εi = (εi1, . . . , εiki)T the residual vector. Similarly, let denote the ki × p design matrix of covariates and Zi the appropriate subset of Xi . Model (1) can be expressed as
| (2) |
where εi ~ N(0, Σ). We assume independence among the different (Yi ,Xi ). We also assume that portions of Xi are ignorably missing (i.e. MCAR or MAR). Let denote the missing parts of Xi and denote the observed parts.
2.2. Multiply-Imputed Likelihood
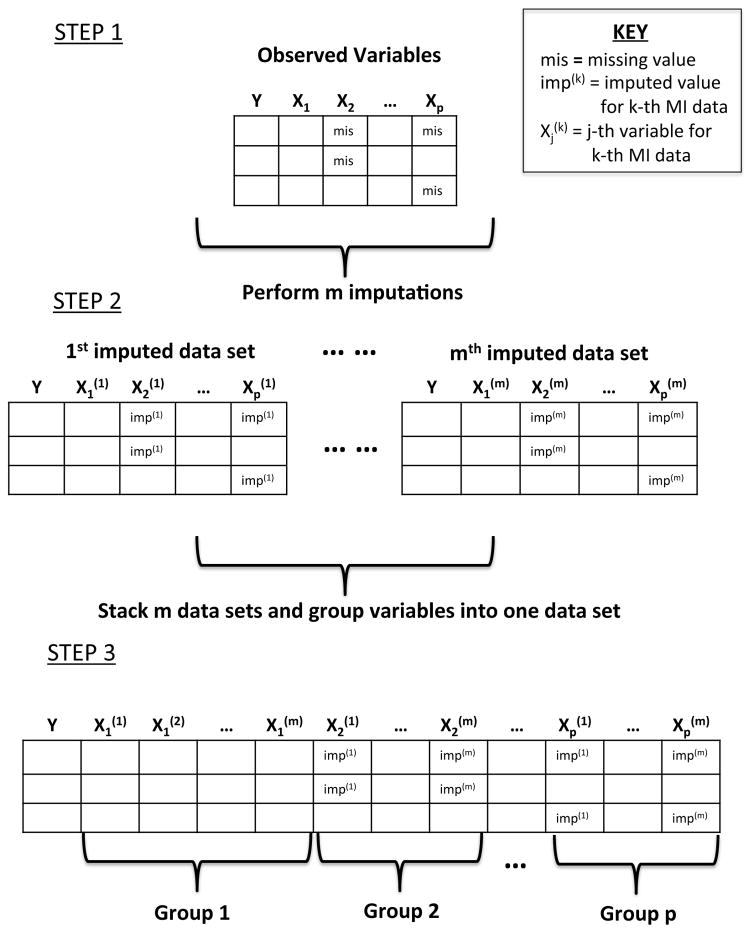
We propose a penalized likelihood method that performs variable selection simultaneously on the multiply imputed data sets for multilevel models via the group lasso. The group lasso was first introduced by Yuan & Lin (2006) as a means of selecting grouped factors for accurate prediction in regression. The procedure begins by stacking the m complete data sets into one wide complete data set. We transform m different multilevel models into one multilevel model with up to p × m covariates where each predictor will be represented by up to m imputed variables. The scheme of the data stacking procedure is found in figure 1.
Figure 1.
Data stacking scheme for proposed variable selection procedure. STEP 1: Identify covariates to be included for selection and their corresponding missing values. STEP 2: Perform m imputations to produce m complete data sets. STEP 3: Stack the m complete data sets into a single wide complete data to be analyzed. Group relevant variables from the m imputed data sets.
We propose the following multilevel model to identify relevant variables across multiply-imputed data sets is
| (3) |
where denotes the gth predictor for the ith cluster from the ℓth imputed data set (g = 1, . . . , p and ℓ = 1, . . . , m) and denotes its corresponding regression coefficient. For simplicity, we rewrite model (3) as
| (4) |
where and . Under model (4), we have that the marginal distribution of outcome where . For multilevel model (4), we build our variable selection procedure on the restricted maximum likelihood (REML) method of estimation in linear mixed models. The REML log-likelihood for the data under model (4) is
| (5) |
The maximum likelihood estimate of β(·) is obtained by maximizing (5) with respect to β(·).
To perform variable selection and to identify non-zero components of β(·), we maximize the profile penalized log-REML function
| (6) |
where λ is a nonnegative tuning parameter, ug is the group size for the gth group (ug = 1 when there is no missing data on covariate g and ug = m when missing data is present on covariate g and m imputations were performed), and || · || is the L2 norm on the Euclidean space. The penalty term in (6) encourages sparsity at the group level since the Euclidean norm of a vector is zero if all the components of are zero. The innovation of the data stacking scheme and group lasso penalization formulation is that by treating ( ) as a group and by summing the Euclidean norms of the loadings in each group, we can shrink all the regression estimates in one group to zero simultaneously, leading to overall variable selection across imputed data sets. For some values of λ, an entire predictor (across the m imputations) can be removed entirely out of the model across imputations, leading to overall variable selection.
There are a few subtleties regarding the proposed procedure that merit attention. If there is no missing data present in the design matrix and no imputations are performed, then (6) reduces to the typical lasso by considering each covariate as their own individual group. The same is true if there exists missingness in the design matrix and only one imputation is performed. If one imputed data set is generated then by treating each variable in the imputed data as its own group, (6) reduces to
which is equivalent to the traditional lasso penalized likelihood.
Another subtlety that needs to be addressed is the number of predictors to be used in the final stacked data set. When performing the m imputations, the columns without missing data will be exactly the same across the m imputed data sets (i.e. ) for all Xg with completely observed data. It would be inappropriate to treat the these variables as a group with m members, as they are perfectly collinear. In the case of a fully observed variable, we simply construct a stacked data set where the m imputed columns of a fully observed covariate is represented by only one of the columns of the complete variable. For example, consider the situation where X = (X1,X2). Of the two predictors of Y, suppose X1 contains some missing values and X2 is fully observed. For sake of illustration, suppose that to address the missingness in X1, only two imputations (m = 2) are performed. The new stacked data structure will contain three predictors, ( , X2). The two imputed variables for X1 (i.e. and ) will be represented as one group and X2 as its own group because no missing data was present in X2 and thus no imputation was constructed for that predictor. The proposed conditional penalized likelihood function for this illustration will take on the following form
| (7) |
which is an intermediate between the ℓ2 penalty used in ridge regression for the imputed variable and an ℓ1 penalty for the completely observed variable.
2.3. Algorithm
Maximizing the penalized profile log-REML function (6), with respect to β(·), presents some computational challenges. To maximize (6) with respect to β(·), we consider an approach similar to Lin & Zhang (2006) and Wang et al. (2010) that transforms the optimization problem into a simpler, but equivalent optimization function.
Proposition 1
Consider the following two optimization procedures
| (8) |
| (9) |
Denote the maximizer of (8) as and the maximizer of (9) as ( , τ̃g) for g = 1, . . . , p and ℓ = 1, . . . , m. Then it follows that
| (10) |
| (11) |
The proof of Proposition 1 is provided in the appendix. The relevance of Proposition 1 is that instead of maximizing (8) directly, we can maximize (9) and obtain equivalent results for . Computationally, we prefer objective function (9) over (8) because (9) resembles a generalized ridge regression, which can be solved through a Newton-Raphson algorithm when τg is fixed.
We propose to maximize (9) by iteratively cycling between and τg. The algorithm is as follows:
Initialize and with conceivable values. Unless other information is known, initial values can be set to 0 and can be set to 1.
- For iteration k, update through
- For iteration k, update through
Continue steps 2 and 3 until is sufficiently small.
2.4. Penalty Selection Procedure
A fundamental issue with the proposed penalty procedure is how to choose the best approximating model among a class of competing models with varying number of parameters. This is equivalent to deciding how to choose the tuning parameter λ. Widely used variable selection criterion for selecting λ include the BIC and the general cross-validation (GCV) method. It is widely known that GCV and BIC are not easily computed in the presence of missing data because they are functions of the missing data, which lead to intractable integrals (Garcia et al., 2010). However, one of the advantages of our proposed methodology is that because we generate m complete multiply-imputed datasets, our procedure avoids the limitation of GCV and BIC under missing data. It has been shown that GCV significantly over fits in most problems, and the BIC has been shown to provide consistent variable selection (Wang et al., 2007). We propose a BIC-type criterion to choose the appropriate tuning parameter. Given any two estimated models, we choose the tuning parameter, λ, that minimizes the following BIC criterion
| (12) |
where , the total sample size. Although N is not the effective sample size (Jiang et al., 2008), this BIC criterion has performed well in our simulation studies and data analysis and has some precedence in this form (Pu & Niu, 2006; Bondell et al., 2010). Additionally, the degrees of freedom q is the total number of nonzero estimates of β̂(·).
2.5. Post-Procedure Estimation
Once the m multiple imputations have been constructed and the proposed procedure has performed variable selection, it is of interest to obtain parameter estimates of the final model. For each of the m unstacked imputed data sets, a linear mixed-effects model with only the selected variables from the proposed procedure can be performed. To obtain an overall estimate of the regression coefficients and standard errors, we can combine the results from each of the m data sets using Rubin’s Rules (Rubin, 1987). Rubin’s Rules proceed as follows: let denote the estimated regression coefficient for the jth predictor and the ℓth imputation and its corresponding estimated variance. The overall regression parameter estimate can be obtained through
and its variance estimate as
where the first component of the addition takes into account the variability within each imputed data set and the second component accounts for the between-imputation variance. A 95% confidence interval for βj can be obtained using the approximation where
It has been shown by Rubin (1987) that a small number of imputations can lead to high-quality inference.
3. Numerical Studies
We performed simulation studies to compare the merits and finite sample performance of the proposed methodology with standard statistical practices. We compare our proposed penalized likelihood procedure to multiple competitors. First, we compare to the regularized lasso on full data without any missingness. This will be considered the gold standard as variable selection will be performed on the complete data. Second, we compare the proposed methodology to the regularized lasso on complete-cases only data. This will assess how missing data under a MAR mechanism affects variable selection and whether the proposed model improves variable selection performance. We also compared our approach to the Brand ad-hoc procedure of selecting covariates that are significant in at least 60% of imputed models.
We simulated complete data from a two-level linear mixed effects model with a random intercept. We consider three scenarios:
-
SCENARIO 1. Data is generated from n = 40 independent clusters with 5 observations in each cluster where
where β = (3, 1.5, 0, 0, 2, 0, 0, 0)T, εi j ~ N(0, 1), Xi j = (Xi j,1, . . . , Xi j,8) and Xi j,1, . . . , Xi j,8 are N(0, 1) variables and Cor r (Xi j,g,Xi j,g′) = ρ|g−g′| with ρ set to 0.3.
SCENARIO 2. The setting is similar to scenario 1, except we increase the number of clusters to n = 60 and increase the number of observations per cluster to be 25 to assess a larger sample performance.
SCENARIO 3. The setting is similar to scenario 2, except we increase the number of clusters to n = 150 to correspond to large cluster studies.
We induced missing values from an MAR mechanism. Let ri j indicate the missingness of , where ri j = 1 when Xi j is observed and ri j = 0 when Xi j is missing. We select ri j from Bernoulli sampling with success probability given by logit(π(Xobs ;α0,α)) = α0 + αXobs, thus imposing MAR. The values of α0 and α were selected to induce 25% missing data (missing data was only induced on X1, X2 and X3).
For each of the scenarios above, 500 data sets were produced. For the full data and complete cases lasso regularization, the traditional BIC was used to select predictors. For the proposed methods, the BIC in (12) was used to select relevant predictors. Multiple imputations were performed on the missing data using the MICE methodology (White et al., 2011).
We considered changes in the values of m (number of imputations) in our simulation study. The results of the numerical studies are presented in table 1. In particular, we present the model selection frequency (the percentage of times the true model was selected), the average model size, the percentage of false negatives, and the percentage of false positives.
Table 1.
Simulation study results for lasso on complete data (Lasso-Full), lasso on complete cases only (Lasso-CCO) and proposed methodology. “Model Size”, “F+”, “F−” indicate the mean model size, false positive rate and false negative rate over the 500 simulated data sets, respectively. “Correct Model” denotes the percentage of times the correct model was selected.
| Method | Model Size | Correct Model | F+ | F− | X1 | X2 | X3 | X4 | X5 | X6 | X7 | X8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SCENARIO 1: Small Study (n = 40, ki = 5) | ||||||||||||
|
| ||||||||||||
| Lasso-Full | 3.50 | 66.6 | 0.11 | 0.01 | 1.00 | 1.00 | 0.21 | 0.19 | 0.96 | 0.13 | 0.01 | 0.02 |
| Lasso-CCO | 3.46 | 62.6 | 0.11 | 0.02 | 1.00 | 0.98 | 0.22 | 0.18 | 0.92 | 0.10 | 0.03 | 0.02 |
| Brand-m1 | 3.43 | 64.4 | 0.09 | 0.00 | 1.00 | 1.00 | 0.15 | 0.08 | 1.00 | 0.06 | 0.09 | 0.06 |
| Proposed-m1 | 2.69 | 62.8 | 0.02 | 0.08 | 1.00 | 0.78 | 0.03 | 0.04 | 0.81 | 0.02 | 0.00 | 0.00 |
| Brand-m3 | 3.34 | 68.2 | 0.07 | 0.00 | 1.00 | 1.00 | 0.17 | 0.06 | 1.00 | 0.03 | 0.06 | 0.03 |
| Proposed-m3 | 3.60 | 63.4 | 0.13 | 0.01 | 1.00 | 0.97 | 0.20 | 0.26 | 1.00 | 0.14 | 0.02 | 0.01 |
| Brand-m5 | 3.32 | 72.6 | 0.06 | 0.00 | 1.00 | 1.00 | 0.14 | 0.05 | 1.00 | 0.04 | 0.05 | 0.03 |
| Proposed-m5 | 3.07 | 66.2 | 0.05 | 0.04 | 0.99 | 0.83 | 0.07 | 0.12 | 0.96 | 0.05 | 0.00 | 0.00 |
|
| ||||||||||||
| SCENARIO 2: Medium Study (n = 60, ki = 25) | ||||||||||||
|
| ||||||||||||
| Lasso-Full | 3.29 | 87.0 | 0.06 | 0.00 | 1.00 | 1.00 | 0.12 | 0.10 | 1.00 | 0.06 | 0.00 | 0.00 |
| Lasso-CCO | 3.61 | 63.6 | 0.12 | 0.00 | 1.00 | 1.00 | 0.32 | 0.21 | 1.00 | 0.08 | 0.00 | 0.00 |
| Brand-m1 | 4.08 | 15.8 | 0.22 | 0.00 | 1.00 | 1.00 | 0.79 | 0.08 | 1.00 | 0.07 | 0.09 | 0.06 |
| Proposed-m1 | 3.58 | 72.4 | 0.12 | 0.00 | 1.00 | 1.00 | 0.24 | 0.22 | 1.00 | 0.12 | 0.00 | 0.00 |
| Brand-m3 | 4.01 | 15.6 | 0.20 | 0.00 | 1.00 | 1.00 | 0.82 | 0.06 | 1.00 | 0.05 | 0.06 | 0.03 |
| Proposed-m3 | 3.35 | 74.6 | 0.08 | 0.00 | 1.00 | 0.98 | 0.09 | 0.23 | 1.00 | 0.06 | 0.00 | 0.00 |
| Brand-m5 | 4.03 | 14.0 | 0.21 | 0.00 | 1.00 | 1.00 | 0.82 | 0.05 | 1.00 | 0.06 | 0.08 | 0.03 |
| Proposed-m5 | 3.05 | 78.8 | 0.03 | 0.02 | 0.99 | 0.89 | 0.04 | 0.10 | 0.97 | 0.04 | 0.00 | 0.00 |
|
| ||||||||||||
| SCENARIO 3: Large Study (n = 150, ki = 25) | ||||||||||||
|
| ||||||||||||
| Lasso-Full | 3.00 | 91.6 | 0.01 | 0.01 | 1.00 | 1.00 | 0.02 | 0.02 | 0.94 | 0.02 | 0.00 | 0.00 |
| Lasso-CCO | 3.44 | 77.0 | 0.09 | 0.00 | 1.00 | 1.00 | 0.22 | 0.12 | 1.00 | 0.10 | 0.00 | 0.00 |
| Brand-m1 | 4.25 | 0.60 | 0.25 | 0.00 | 1.00 | 1.00 | 0.99 | 0.08 | 1.00 | 0.05 | 0.08 | 0.05 |
| Proposed-m1 | 0.82 | 3.45 | 0.09 | 0.00 | 1.00 | 1.00 | 0.17 | 0.16 | 1.00 | 0.13 | 0.00 | 0.00 |
| Brand-m3 | 4.20 | 0.60 | 0.24 | 0.00 | 1.00 | 1.00 | 0.99 | 0.07 | 1.00 | 0.04 | 0.06 | 0.03 |
| Proposed-m3 | 3.22 | 84.4 | 0.04 | 0.00 | 1.00 | 1.00 | 0.04 | 0.15 | 1.00 | 0.03 | 0.00 | 0.00 |
| Brand-m5 | 4.17 | 0.60 | 0.23 | 0.00 | 1.00 | 1.00 | 0.99 | 0.04 | 1.00 | 0.04 | 0.06 | 0.03 |
| Proposed-m5 | 3.15 | 88.0 | 0.03 | 0.00 | 1.00 | 1.00 | 0.02 | 0.12 | 1.00 | 0.02 | 0.0 | 0.0 |
Using the full data and performing lasso to select variables as our benchmark method, the results from table 1 indicate that having missing observations in the covariates present in any data set lowers the ability for any method to select the correct model. Overall, when missing data is present, the method that performs the best at recovering the correct model is our proposed method (with m = 5 imputations). We note that performing lasso on the complete cases only data set performs adequate variable selection. We also note that having more subjects within a cluster (ki = 25), results in higher model selection frequency for all methods expect for the Brand approach. As cluster and sample size increases, the Brand approach does not select the correct model as frequently as our proposed method.
Simulation results suggest that one imputation is not sufficient to obtain reliable variable selection. For instance, the CCO lasso and the proposed methodology have very similar model selection frequencies (CCO: 63.6%, proposed: 72.4%; scenario 2) when m = 1, well below the full data selection frequency of 87.0%. This should be expected because one imputation does not account for the uncertainty in the imputation method. However, when more imputations are considered (m = 3), the proposed method outperforms the complete case method (proposed: 74.6%, CCO: 63.6%; scenario 2). The proposed methodology is almost as good at identifying the correct model as having the full data (full: 91.6%, proposed: 88.0%; scenario 3) after 5 imputations.
In terms of model size, the full model lasso selects models that are closest to the true model size on average. Both the CCO and the Brand procedure tend to select larger models on average. The proposed methodology produces smaller models than the CCO and Brand methods as the number of imputations increase.
4. Data Analysis
This statistical work was motivated by the Healthy-Directions Small-Business study (Sorensen et al. 2005). Current epidemiological studies have shown the relationship between dietary patterns and physical inactivity with multiple cancers and chronic diseases. One of the primary goals of the Healthy-Directions Small-Business study is to investigate whether or not the cancer prevention that incorporates occupational health and health promotion can lead to significant improvements in the mean consumption of fruits and vegetables, levels of physical activity, smoking cessation and reduction of occupational carcinogens. The HD-SB study was a randomized, controlled intervention study conducted between 1999 and 2003 as part of the Harvard Center Prevention Program Project. The study population of the HD-SB study were small manufacturing worksites that employed multi-ethnic, low-wage workers. Details of worksite eligibility and recruitment can found in Sorensen et al. (2005). Participating worksites were randomized to either the 18-month intervention group or minimal intervention control group.
For the purpose of this data analysis, we focus on predictors that are hypothesized to relate to mean consumption of fruits and vegetables at followup. Along with intervention status, a substantial number of covariates were collected to determine their impact on the primary outcome: consumption of red meat per week, levels of leisure physical activity, smoking status (1 current and 0 otherwise), educational level (1 if college degree or more and 0 otherwise), gender (1 if female and 0 otherwise), body mass index, at least one child in household less than eighteen years of age (1 if true and 0 otherwise), marital status (1 if married and 0 otherwise), race (1 if nonwhite and 0 otherwise), age, multivitamin use (1 if takes ≥ 6 days/week and 0 otherwise), poor (1 if ≥ 185% of poverty threshold and 0 otherwise) and nonimmigrant (1 if participant was born in the United States and 0 otherwise). The study had 974 respondents of which only complete information on all the variables of interest was obtained for 793 respondents (i.e. 18.5% missing data present).
The linear mixed model to answer the primary goal takes on the following form:
where these 15 predictors were considered for selection. A random intercept model was used to model the clustering of workers within worksites. We constructed a multilevel model for the complete cases data (all missing observations removed) and also using the proposed methodology for m = 1, m = 3 and m = 5 imputations. Selection of the tuning parameter was based on the BIC in (12). Regression estimates selected by the m = 5 proposed model are provided using Rubin’s Rules. The results of the data analysis are in table 2.
Table 2.
Data analysis results for the Healthy Directions - Small Business study. The stars represent the variables selected from the given variable selection procedure. CCO denotes the complete cases only results, where all observations with missing data were removed and the remaining observations were used in estimation and variable selection. The final column presents estimates of the regression coefficients based on m =5 imputations using the proposed method.
| Variable Selection Method | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Variable | % | CCO | CCO | CCO | Proposed | Proposed | ||
| miss | β̂ | P-value | LASSO | m=1 | m=3 | m=5 | β̂ (95% CI) | |
| Baseline Fruit | 0.6 | 0.56 | * | * | * | * | * | 0.51 (0.45,0.57) |
| Intervention | 0.0 | 0.34 | * | * | * | * | * | 0.31 (0.07,0.54) |
| Meat | 0.7 | −0.01 | * | * | * | * | −0.03 (−0.05, 0.00) | |
| Phys. Act. | 6.4 | 0.02 | * | |||||
| Smoking | 0.1 | −0.18 | * | * | * | * | −0.23 (−0.46,0.00) | |
| Education | 1.3 | 0.23 | * | |||||
| Female | 0.0 | 0.40 | * | * | * | * | * | 0.24 (0.02,0.45) |
| BMI | 4.8 | 0.03 | * | * | ||||
| Kids≤ 18 | 0.9 | 0.12 | ||||||
| Married | 0.4 | 0.03 | ||||||
| NonWhite | 0.0 | −0.02 | ||||||
| Age | 1.1 | 0.01 | * | * | ||||
| Multivit | 0.7 | 0.17 | * | * | * | * | 0.10 (−0.11,0.32) | |
| Poor | 1.1 | 0.14 | ||||||
| Immigrant | 0.4 | −0.04 | * | * | * | * | −0.16 (−0.37,0.04) | |
The complete-cases only multilevel model does not perform variable selection; the estimated regression coefficients are non-zero for the 15 predictors. What is commonly done in practice is to select significant variables to be those with p – value < 0.05. Based on the p-value criterion, the significant predictors of fruits and vegetables under the CCO scenario are baseline fruit, intervention, gender and BMI. Performing lasso on the complete case data reveals a model with 11 relevant predictors. The proposed methodology with m = 1, m = 3 and m = 5 imputations selected baseline fruit, intervention, meat consumption, smoking, gender, multivitamin and nonimmigrant, with m = 3 additionally including age.
The proposed methodology in this data analysis produces smaller models than the complete cases only lasso, a pattern which was observed in the simulations section. Compared to choosing significant variables via p-values, the proposed methodology additionally identifies meat consumption, smoking status, immigrant status and multivitamin use to be relevant predictors of follow-up fruit/vegetable intake. The parameter estimates for the final model, as selected by the proposed method with five imputations, are presented in the last column of table 2. Overall, there seems to be a strong positive intervention effect on follow-up fruit and vegetable intake. We note the gender gap where females tend to consume more fruits and vegetables at followup on average than men, which has been shown in previous studies (Sorensen et al., 2007).
5. Discussion
We describe methodology that can perform variable selection for multilevel models with missing covariate data. When the method of handling missingness in the covariates is through multiple imputation, we describe a penalized likelihood approach that performs variable selection across the m imputed data sets simultaneously through group lasso regularization. Numerical studies demonstrate the benefits of imputing and then performing variable selection instead of doing a complete-cases only analysis, which is typically done in practice. Ignoring missing data through methods like complete-cases analyses potentially undermine scientific credibility of causal conclusions from intervention studies (Little et al., 2012).
The proposed methodology may be extended to generalized linear-mixed models (GLMMs) as our approach is likelihood based. An additional aspect of the analysis of mixed models is the selection of random effects. There are two types of variable selection approaches for multilevel models: the first is selecting significant fixed-effect variables (i.e. columns from Xi when the random effects are not considered in the selection) and the second is selection of both fixed and random effects (i.e. columns from Zi ). The mean and variance of Yi based on model (2) are given by
The fixed-effects selection through Xi affects the mean structure of Yi and the selection through Zi affects the covariance structure of Yi. We focus our methodology on fixed effects variable selection, though extensions of this work could be developed to identify both significant fixed and random effects.
This study does have limitations and indicates areas for future research. First, after completing the post-estimation procedure described in this paper, it remains necessary to account for modeling bias. Performing variable selection and then using the selected model to perform estimation is commonly done in practice, but is likely to yield overly optimistic inferences. This is due to the underestimation of variability of the estimated parameters. Shen et al. (2004), Hu & Dong (2007), Wang & Lagakos (2009) and Minnier et al. (2011) have used data perturbation methods to account for the variable selection process to make approximately unbiased inferences. Extensions of data perturbation methods to multiply-imputed variable selection is needed. Second, the proposed procedure requires a substantial amount of observations and clusters to successfully perform these models. As the number of covariates with missing data increases, the model increases in size (by multiple of m) which could potentially lead to model instability. Third, more works needs to be developed to provide rules of thumb for how many imputations are needed. In general, more imputations are preferred (Bodner, 2008), but this has the potential to introduce very large, potentially unstable proposed models. Lastly, model (3) is a working model and not the true model. Other selection criterion such as prediction error or likelihood could have been entertained for the selection of the final model. We hope the proposed developments will make it possible for researchers to maximize the use of available information in their data and uncover important underlying associations.
Acknowledgments
This research was supported by a grant from the Robert Wood Johnson Foundation Health & Society Scholars program at Harvard University (Grant # 69248) and from the National Heart, Lung, and Blood Institute (R01HL107240).
Appendix
PROOF OF PROPOSITION 1
We begin by showing (11). Denote the objective function and corresponding to the two optimization equations respectively
The result in (11) falls after rewriting as
The expression inside the square brackets can be rewritten as
We know that a2 + b2 ≥ 2ab, thus
Equality holds if and only if . To show (10), we first show .
Therefore, . Now, let . After some algebra similar to above, we get observe that . Thus . As a result, , which leads to the unique maximizer because is convex.
References
- Bodner TE. What improves with increased missing data imputations? Structural Equation Modeling. 2008;(15):651–675. [Google Scholar]
- Bondell HD, Krishna A, Ghosh SK. Joint variable selection for fixed and random effects in linear mixedeffects models. Biometrics. 2010;66(4):1069–1077. doi: 10.1111/j.1541-0420.2010.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso M, Qian G, Angulo J. Stochastic complexity and model selection from incomplete data. Journal of Statistical Planning and Inference. 1999;76(1):273–284. [Google Scholar]
- Chen Z, Dunson D. Random effects selection in linear mixed models. Biometrics. 2003;59(4):762–769. doi: 10.1111/j.0006-341x.2003.00089.x. [DOI] [PubMed] [Google Scholar]
- Claeskens G, Consentino F. Variable selection with incomplete covariate data. Biometrics. 2008;64(4):1062–1069. doi: 10.1111/j.1541-0420.2008.01003.x. [DOI] [PubMed] [Google Scholar]
- Crainiceanu C. Likelihood ratio testing for zero variance components in linear mixed models. Random Effect and Latent Variable Model Selection. 2008;192:3–17. [Google Scholar]
- Demirtas H. Simulation driven inferences for multiply imputed longitudinal datasets. Statistica Neerlandica. 2004;58(4):466–482. [Google Scholar]
- Demirtas H, Hedeker D. An imputation strategy for incomplete longitudinal ordinal data. Statistics in medicine. 2008;27(20):4086–4093. doi: 10.1002/sim.3239. [DOI] [PubMed] [Google Scholar]
- Fan J, Li R. New estimation and model selection procedures for semiparametric modeling in longitudinal data analysis. Journal of the American Statistical Association. 2004;99(467):710–723. [Google Scholar]
- Garcia R, Ibrahim J, Zhu H. Variable Selection for regression models with missing data. Statistica Sinica. 2010;20(1):149–165. [PMC free article] [PubMed] [Google Scholar]
- Goldstein H, Carpenter J, Kenward M, Levin K. Multilevel models with multivariate mixed response types. Statistical Modelling. 2009;9(3):173–197. [Google Scholar]
- Hens N, Aerts M, Molenberghs G. Model selection for incomplete and design-based samples. Statistics in medicine. 2006;25(14):2502–2520. doi: 10.1002/sim.2559. [DOI] [PubMed] [Google Scholar]
- Heymans M, Van Buuren S, Knol D, Van Mechelen W, De Vet H. Variable selection under multiple imputation using the bootstrap in a prognostic study. BMC medical research methodology. 2007;7(1):33–43. doi: 10.1186/1471-2288-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Dong Y. Estimating the predictive quality of dose–response after model selection. Statistics in medicine. 2007;26(16):3114–3139. doi: 10.1002/sim.2786. [DOI] [PubMed] [Google Scholar]
- Ibrahim J, Zhu H, Garcia R, Guo R. Fixed and Random Effects Selection in Mixed Effects Models. Biometrics. 2011;67(2):495–503. doi: 10.1111/j.1541-0420.2010.01463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim J, Zhu H, Tang N. Model Selection Criteria for Missing-Data Problems Using the EM Algorithm. Journal of the American Statistical Association. 2008;103(484):1648–1658. doi: 10.1198/016214508000001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Rao JS, Gu Z, Nguyen T. Fence methods for mixed model selection. The Annals of Statistics. 2008:1669–1692. [Google Scholar]
- Johnson B, Lin D, Zeng D. Penalized estimating functions and variable selection in semiparametric regression models. Journal of the American Statistical Association. 2008;103(482):672–680. doi: 10.1198/016214508000000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney S, Dunson D. Bayesian Model Uncertainty in Mixed Effects Models. Random effect and latent variable model selection. 2008;192:37–62. [Google Scholar]
- Lin Y, Zhang H. Component selection and smoothing in multivariate nonparametric regression. The Annals of Statistics. 2006;34(5):2272–2297. [Google Scholar]
- Little R, Rubin D. Statistical analysis with missing data. J. Wiley & Sons; 2002. [Google Scholar]
- Little RJ, D’Agostino R, Cohen ML, Dickersin K, Emerson SS, Farrar JT, Frangakis C, Hogan JW, Molenberghs G, Murphy SA, et al. The prevention and treatment of missing data in clinical trials. New England Journal of Medicine. 2012;367(14):1355–1360. doi: 10.1056/NEJMsr1203730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Taylor J, Belin T. Multiple imputation and posterior simulation for multivariate missing data in longitudinal studies. Biometrics. 2000;56(4):1157–1163. doi: 10.1111/j.0006-341x.2000.01157.x. [DOI] [PubMed] [Google Scholar]
- Minnier J, Tian L, Cai T. A perturbation method for inference on regularized regression estimates. Journal of the American Statistical Association. 2011;106(496):1371–1382. doi: 10.1198/jasa.2011.tm10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni X, Zhang D, Zhang H. Variable selection for semiparametric mixed models in longitudinal studies. Biometrics. 2010;66(1):79–88. doi: 10.1111/j.1541-0420.2009.01240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu W, Niu XF. Selecting mixed-effects models based on a generalized information criterion. Journal of multivariate analysis. 2006;97(3):733–758. [Google Scholar]
- Qu A, Li R. Quadratic Inference Functions for Varying-Coefficient Models with Longitudinal Data. Biometrics. 2006;62(2):379–391. doi: 10.1111/j.1541-0420.2005.00490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin D. Multiple imputation for nonresponse in surveys. J. Wiley & Sons; 1987. [Google Scholar]
- Schafer J. Analysis of incomplete multivariate data. Chapman & Hall/CRC; 1997. [Google Scholar]
- Schafer J, Yucel R. Computational strategies for multivariate linear mixed-effects models with missing values. Journal of Computational and Graphical Statistics. 2002;11(2):437–457. [Google Scholar]
- Seghouane A, Bekara M, Fleury G. A criterion for model selection in the presence of incomplete data based on Kullback’s symmetric divergence. Signal Processing. 2005;85(7):1405–1417. [Google Scholar]
- Shen X, Huang H, Ye J. Inference after model selection. Journal of the American Statistical Association. 2004;99(467):751–762. [Google Scholar]
- Shimodaira H. A new criterion for selecting models from partially observed data. Selecting Models from Data: Artificial Intelligence and Statistics IV, Lecture Notes in Statistics. 1994;89:21–29. [Google Scholar]
- Sorensen G, Barbeau E, Stoddard A, Hunt M, Kaphingst K, Wallace L. Promoting behavior change among working-class, multiethnic workers: results of the healthy directions–small business study. American Journal of Public Health. 2005;95(8):1389–1395. doi: 10.2105/AJPH.2004.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen G, Stoddard A, Dubowitz T, Barbeau E, Bigby J, Emmons K, Berkman L, Peterson K. The influence of social context on changes in fruit and vegetable consumption: results of the healthy directions studies. American Journal of Public Health. 2007;97(7):1216–1227. doi: 10.2105/AJPH.2006.088120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart E, Azur M, Frangakis C, Leaf P. Multiple imputation with large data sets: a case study of the children’s mental health initiative. American journal of epidemiology. 2009;169(9):1133–1139. doi: 10.1093/aje/kwp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buuren S, Groothuis-Oudshoorn K. Mice: Multivariate imputation by chained equations in r. Journal of Statistical Software. 2011;45(3):1–67. [Google Scholar]
- Wang H, Li R, Tsai C. Tuning parameter selectors for the smoothly clipped absolute deviation method. Biometrika. 2007;94(3):553–568. doi: 10.1093/biomet/asm053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Lagakos S. Inference after variable selection using restricted permutation methods. Canadian Journal of Statistics. 2009;37(4):625–644. doi: 10.1002/cjs.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Song P, Zhu J. Doubly regularized reml for estimation and selection of fixed and random effects in linear mixed-effects models. The University of Michigan Department of Biostatistics Working Paper Series. 2010;89:1–10. [Google Scholar]
- White IR, Royston P, Wood AM. Multiple imputation using chained equations: issues and guidance for practice. Statistics in medicine. 2011;30(4):377–399. doi: 10.1002/sim.4067. [DOI] [PubMed] [Google Scholar]
- Wood A, White I, Royston P. How should variable selection be performed with multiply imputed data? Statistics in medicine. 2008;27(17):3227–3246. doi: 10.1002/sim.3177. [DOI] [PubMed] [Google Scholar]
- Yang X, Belin T, Boscardin W. Imputation and variable selection in linear regression models with missing covariates. Biometrics. 2005;61(2):498–506. doi: 10.1111/j.1541-0420.2005.00317.x. [DOI] [PubMed] [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2006;68(1):49–67. [Google Scholar]
- Yucel R. Multiple imputation inference for multivariate multilevel continuous data with ignorable non-response. Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences. 2008;366(1874):2389–2403. doi: 10.1098/rsta.2008.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Lin X. Variance component testing in generalized linear mixed models for longitudinal/clustered data and other related topics. Random effect and latent variable model selection. 2008;192:19–36. [Google Scholar]
- Zhu H, Zhang H. Generalized score test of homogeneity for mixed effects models. The Annals of Statistics. 2006;34(3):1545–1569. [Google Scholar]