Abstract
The hypothalamic arcuate-median eminence complex (Arc-ME) controls energy balance, fertility, and growth through molecularly distinct cell types, many of which remain unknown. To catalog cell types in an unbiased way, we profiled gene expression in 20,921 individual cells in and around the adult mouse Arc-ME using Drop-seq. We identify 50 transcriptionally distinct Arc-ME cell populations, including a rare tanycyte population at the Arc-ME diffusion barrier, a novel leptin-sensing neuronal population, multiple AgRP and POMC subtypes, and an orexigenic somatostatin neuronal population. We extended Drop-seq to detect dynamic expression changes across relevant physiological perturbations, revealing cell type-specific responses to energy status, including distinctly responsive subtypes of AgRP and POMC neurons. Finally, integrating our data with human GWAS data implicates two previously unknown neuronal subtypes in the genetic control of obesity. This resource will accelerate biological discovery by providing insights into molecular and cell type diversity from which function can be inferred.
The arcuate hypothalamus (Arc) is an evolutionarily conserved brain region with diverse roles in mammalian physiology, including energy homeostasis, reproduction, and neuroendocrine control of growth hormone and prolactin release. Aside from its functional diversity, Arc is known for its unique anatomical relationship with the blood brain barrier (BBB) 1, which protects the cell bodies and dendrites of arcuate neuroendocrine neurons while allowing their axons to enter BBB-free areas of the adjoining median eminence (ME); these axons release signals into fenestrated capillaries that carry blood to the pituitary. Bloodborne signals can also diffuse from ME to Arc, giving Arc privileged access to peripheral hormones, nutrients, and other metabolic signals 1. This access is dynamically regulated by tanycytes 2, a specialized type of ependymal cell lining the third ventricle that extends processes throughout the Arc and ME. Together the Arc and ME form an anatomically unique and functionally important complex, the Arc-ME.
The varied functions of Arc-ME are supported by molecularly specialized neuron subtypes. For instance, orexigenic AgRP neurons and anorexigenic POMC neurons control feeding 3, 4, while KISS1 neurons regulate serum luteinizing hormone levels 5. Despite decades of research on Arc-ME, a complete census of its cell types is not available, with several functional Arc neuron populations having no known markers 6, 7. Studies relying on immunohistochemistry or in situ hybridization have been limited by the number of proteins or transcripts that can be simultaneously visualized in individual cells and by a strong bias towards known markers. With recent advances in transcriptomic technology, thousands of cells can be profiled individually, enabling discovery of cell types with reduced bias (e.g., 8–10).
Using Drop-seq we systematically catalogued cell types from in and around mouse Arc-ME, identifying 34 distinct neuronal (24 from Arc-ME) and 36 non-neuronal populations (26 from Arc-ME) from 20,921 individual cell profiles. We determine specific markers that can be used both to identify cell types and to infer their function. Among our findings, we uncover several distinct subsets of AgRP and POMC neurons, a novel group of leptin-responsive neurons, and an undescribed orexigenic role for Arc somatostatin neurons. We further used Drop-seq to assess cell-type specific responses to fasting and high-fat diet, revealing energy status-sensitive populations and reinforcing the functional heterogeneity of AgRP and POMC subtypes. Lastly, we show how the increased detail provided by such profiles improves the ability to connect GWAS genes to relevant cell types. Together our results demonstrate how such a molecular census can be used to transform our understanding of a complex tissue and the biological processes it regulates.
RESULTS & DISCUSSION
Unbiased transcriptomics identifies 50 distinct Arc-ME cell types
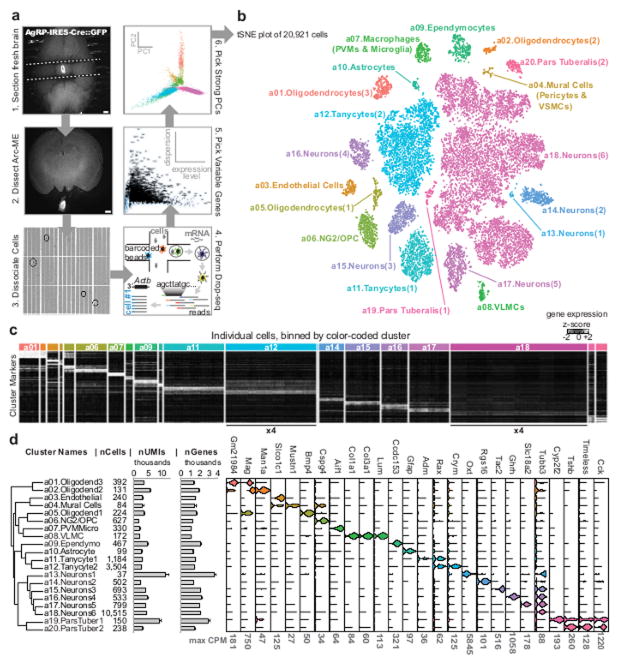
Using Drop-seq 8 we profiled 20,921 transcriptomes from acutely dissociated Arc-ME cells of adult mice under various feeding conditions: ad libitum access to standard mouse chow, low-fat diet, or high-fat diet; overnight fasting, with or without subsequent refeeding (Figures 1A, S1A). After correcting for batch effects, we performed principal component (PC) analysis, dimensionality reduction with spectral t-distributed stochastic neighbor embedding (tSNE), and density-based clustering (Figure 1A; see Supplemental Experimental Methods). Our initial analysis identified 20 distinct clusters (Figures 1B, 1C). Each cluster contained cells from each feeding condition and sample batch, indicating the transcriptional identities of these cell clusters are stable across those experimental conditions (Figure S1A, Table S1). Using expression patterns of cell-type specific marker genes, we assigned a single identity to each cluster: neurons (Tubb3+); ependymocytes (Ccdc153+); and tanycytes (Rax+); oligodendrocyte lineage cells (Mag+); oligodendrocyte precursor cells/NG2 cells (Cspg4+); macrophages (Aif1+); endothelial cells (Slco1c1+); mural cells (Mustn1+); and astrocytes (Gfap+; Figure 1D, Table S2).
Figure 1. Overview of all cell types.
(A) Schematic of Arc-ME single-cell transcriptomics. (B) Spectral tSNE plot of 20,921 cells, colored per density clustering and annotated according to known cell types. (C) Heatmap of top marker genes for each cluster. The two largest clusters, a12 and a18, were reduced to ¼ size to better visualize the smaller clusters (D) Dendrogram showing relatedness of cell clusters, followed by (from left to right): cluster identification numbers; cells per cluster; mean ± S.E.M. unique molecular identifiers (UMIs) per cluster; mean ± S.E.M. genes detected per cluster; and violin plots showing expression of cell type marker genes.
We also identified clusters of non-neural cells from tissues adjoining the Arc-ME – i.e., vascular and leptomeningeal cells (VLMCs; Lum+ 9) and pituitary cells from pars tuberalis (Tshb+). Exemplifying our detection of novel markers, we find that pars tuberalis cells highly express the gene encoding cholecystokinin (Cck; Figures 1D, S1B, C). Of note, CCK has been shown to control pituitary release of prolactin 11, a well-known function of the pars tuberalis 12.
In many cases, a previously-described cell type was represented by multiple clusters. We detected oligodendrocyte markers (e.g., Olig1, Cd9, Pllp) in 4 clusters, each corresponding to distinct stages of oligodendrocyte differentiation 9 (Figure S1D). Neurons and tanycytes also formed multiple clusters, consistent with their known heterogeneity (see below). We subclustered each original non-neuronal cluster (Figure S1E, Table S3) yielding a total of 36 subclusters, revealing additional heterogeneity – e.g., the “mural cell” cluster (Figure 1B) comprises pericytes and vascular smooth muscle cells (Figure S1F) and a “macrophage” cluster (Figure 1B) consists of transcriptionally-related microglia and perivascular macrophages (Figure S1G).
Our subclustering revealed 8 clusters of ependymal cells (Vim+, Sox2+; Figure S2A–B). Ependymal cells line the ventricular surface of the hypothalamus and are broadly categorized as either ependymocytes, multi-ciliated cells that secrete and move cerebrospinal fluid (CSF), or tanycytes, mono-ciliated cells with basal processes extending throughout the mediobasal hypothalamus and median eminence 1. Tanycytes have a variety of functions, from glucose sensing and neurogenesis to controlling the chemical exchange between brain parenchyma, CSF, and bloodstream 1, 13. Studies in rodents have described 4 tanycyte subtypes (α1, α2, β1, and β2) occupying distinct regions along the third ventricle 13 (Figure 2A). While markers for these subtypes have been suggested 13, it is unclear whether these markers truly distinguish between molecularly distinct cellular subtypes.
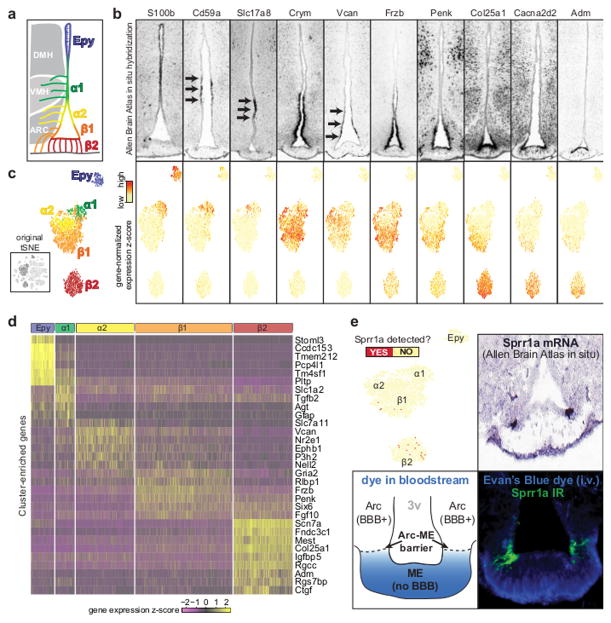
Figure 2. Ependymal cell types.
(A) Illustration of known subtypes of hypothalamic ependymal cells, their approximate anatomical locations, and the orientation of their processes - ependymocyte have cilia in the ventricle and tanycytes have basal processes in the brain parenchyma and median eminence (B) Marker gene expression shown by ependymal cell feature plot (top) and in situ hybridization of coronal brain sections (Allen Mouse Brain Atlas; bottom). Ependymal cell feature plot derived from tSNE plot shown in Figure 1B. Genes were selected from those differentially expressed among ependymal cell clusters. (C) Annotation of ependymal cell clusters based on anatomical localization of marker genes. Figure was derived from tSNE plot shown in Figure 1B (thumbnail). (D) Heatmap of single-cell expression of cluster-enriched transcripts. (E) Top left, ependymal cell feature plot re-colored to indicate cells with any amount of Sprr1a transcript. Top right, Sprr1a in situ hybridization in a coronal brain section (Allen Mouse Brain Atlas). Bottom left, schematic of an experiment to define the diffusion barrier between Arc and ME. Bottom right, confocal micrograph comparing SPRR1A immunoreactivity to the location of the Arc/ME diffusion barrier, visualized by extravasation of intravascular Evan’s Blue; micrograph is representative of 2 mice.
To trace the anatomical origin of our ependymal cell clusters, we cross-referenced each’s markers with in situ hybridization data from the Allen Mouse Brain Atlas (ABA;http://mouse.brain-map.org) 14. We found many are expressed in well-defined regions along the third ventricle (examples in Figure 2B), allowing us to assign each cluster to the ependymal cell subtype occupying those regions (Figure 2C). Our results confirm and extend functional categorization of ependymal cell subtypes by anatomical position: we validate several ependymal subtype markers (Figure S2B) and discover novel markers for each subtype (Figures 2D, S2C–D). Our data nearly double the number of ependymal cell subtypes thought to exist (Figure S2C–D) and provide insight into each’s possible functions.
While many genes were expressed in gradients along the third ventricle, some showed very restricted patterns of expression. For instance, Sprr1a is found only at the border between Arc and ME (Figure 2E), where tanycytes are thought to form a diffusion barrier 1, 2. Using an intravenous injection of Evan’s blue to mark this diffusion barrier, we found that SPRR1A immunoreactive tanycytes are located precisely at the Arc-ME barrier (Figure 2E). Small proline-rich (SPRR) proteins, including SPRR1A, are crucial constituents of the cornified envelope, the diffusion barrier in the skin 15. By identifying Sprr1a as a specific marker for these tanycytes, our results provide a genetic “handle” to develop tools for investigating the role of Sprr1a and Sprr1a+ tanycytes in the Arc-ME diffusion barrier.
Neuron-specific clustering reveals novel types and subtypes of known Arc-ME populations
At least 6 distinct types of neurons have been identified in the Arc-ME. Among these are two neuroendocrine populations – growth hormone releasing hormone (GHRH) and tuberoinfundibular/tuberohypophyseal dopaminergic (TIDA/THDA) neurons – that control pituitary release of growth hormone and prolactin, respectively. In addition, there are at least four types of centrally-projecting neurons, including agouti-related peptide (AgRP) neurons and pro-opiomelanocortin (POMC) neurons, which play vital but opposing roles in energy balance, and Kisspeptin/Neurokinin B/Dynorphin (“KNDy”) neurons, which regulate fertility. Somatostatin (SST) neurons have also been described as distinct 16, but their function is not well understood. Other functional types of neurons exist in the Arc-ME as well, including a thermogenic population known for its expression of the rat insulin II promoter (RIP)-Cre transgene as well as a leptin-sensing GABAergic population distinct from AgRP neurons 6, 7, 17, 18.
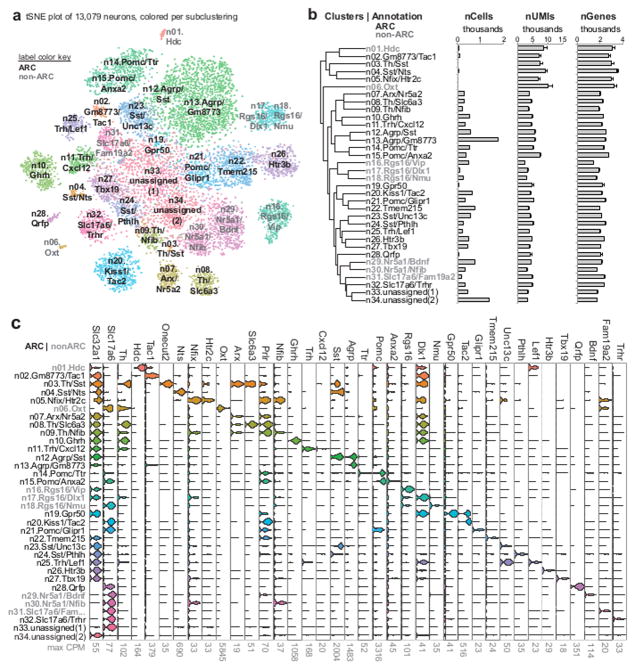
To further characterize neuronal diversity in the Arc-ME, we performed additional rounds of clustering on the 13,079 neurons in our dataset, identifying 34 clusters (Figures S3A, B and 3A, B). Many of these clusters are defined by unique candidate markers (Figure 3C; Table S4), though some lacked a unique marker and are annotated instead with a combination of markers. Two clusters, containing a combined 14% of the neurons in our dataset, appeared to be heterogenous, lacking any strong and specific markers, and so are annotated as “unassigned.” The reason(s) for lack of assignment are not clear but could include technical issues with sample quality or purity (suggested by the lower UMI/gene counts in this population), or could reflect true biological ambiguity. However, due to a lack of specific markers or specific combination of markers for these neurons, we could not investigate them further.
Figure 3. Neuronal cell types.
(A) Spectral tSNE plot of 13,079 neurons, colored according to the results of iterative subclustering, and labeled according to expression of either a specific marker gene or a specific combination of marker genes. Clusters with gray labels most likely originated from regions surrounding the Arc-ME (see Supplemental Figure 5A–D). (B) Dendrogram showing relatedness of neuronal clusters, followed by (from left to right): cluster identification numbers; histograms of neurons per cluster; mean ± S.E.M. unique molecular identifiers (UMIs) per cluster; mean ± S.E.M. unique genes detected per cluster. (C) Violin plots of known and novel markers of neuron subtypes in and around the Arc-ME, with maximum counts per million (CPM) below.
Importantly, we detected markers of all 6 previously described types of Arc-ME neurons (Agrp, Pomc, Sst, Kiss1/Tac2, Ghrh, Th). In total, 14 clusters expressed those markers (2 Agrp+, 3 Pomc+, 5 Sst+, and 6 Th+ clusters), revealing molecular heterogeneity even within well-studied arcuate populations. We observe Th+/Sst+ and Agrp+/Sst+ co-expressing clusters that represent “overlaps” between Arc-ME neuronal populations thought to be distinct. Also, while six neuronal clusters are potentially dopaminergic (Th+, little to no Dbh), only two of these expressed prolactin receptor (Prlr) and dopamine transporter (Slc6a3), representing true TIDA/THDA populations (Figure 3C; clusters n03.Th/Sst, n08.Th/Slc6a3) 19, 20.
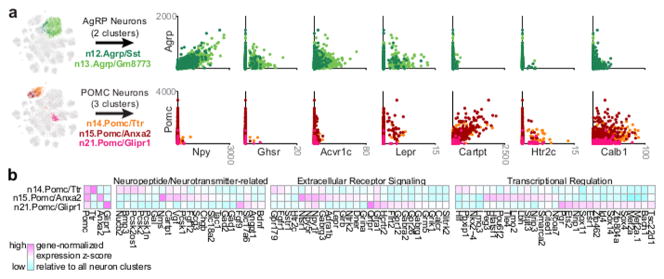
To validate our clustering, we checked expression of genes previously shown as enriched in AgRP neurons and POMC neurons, two well-studied Arc-ME neuronal populations. While we detect leptin receptor transcript (Lepr) in both AgRP neurons and POMC neurons, Npy, Ghsr, and Acvr1c are highly enriched in AgRP neurons, as are Cartpt, Htr2c, and Calb1 in POMC neurons (Figure 4A). Markers from our unbiased clustering therefore concur with previous studies of these neuronal subpopulations (e.g., 21).
Figure 4. AgRP Neurons and POMC Neurons.
(A) Left, selection of AgRP neurons and POMC neurons for analysis of gene co-expression. Right, co-expression of Agrp or Pomc with genes known to be enriched in AgRP neurons and/or POMC neurons. Values are in counts per million (CPM). (B) Differentially expressed genes related to neuropeptide/transmitter signaling and transcriptional regulation in three subtypes of POMC neurons.
POMC neurons have been shown to have functional heterogeneity 22, but it is unclear whether these subtypes are transcriptionally distinct. Our analysis found three distinct subtypes of POMC neurons, each defined by a set of enriched transcripts (Figure S4). Some of these transcripts encoded secreted peptides, receptors, and transcriptional regulators that may underlie their transcriptional identity (Figure 4B). Notably, among POMC subtypes, Lepr transcript is largely restricted to n15.Pomc/Anxa2 subtype, whereas Htr2c is predominantly expressed by the other POMC subtypes (Figure 4B). Thus, these transcriptionally distinct subtypes of POMC neurons likely correspond to the leptin-sensing and serotonin-sensing/Htr2c+ subtypes defined by previous studies 22.
Roughly half the neuronal clusters did not express markers of known Arc-ME neurons. These “unknown” clusters could represent neurons from outside Arc-ME, inadvertently included during dissection (Figure S5A–B). Indeed, we found transcripts enriched in 8 neuronal clusters, or 17% of all neurons, that ABA shows are expressed in regions neighboring Arc-ME, suggesting that these 8 neuronal clusters originated from neighboring regions: ventromedial hypothalamus, VMH (“SF-1”/Nr5a1+, Fezf1+; Figure S5C; 3 clusters); suprachiasmatic nucleus, SCN (Rgs16+, Figure S5C; 3 clusters); tuberomammillary nucleus, TMN (Hdc+, Figure 3C; 1 cluster); and retrochiasmatic area (Oxt+, Figure 3C; 1 cluster). Conversely, another 24 neuronal clusters (70% of neurons) expressed transcripts enriched in Arc-ME relative to neighboring regions, indicating these clusters likely came from Arc-ME (Figure S4D). The two “unassigned” clusters had heterogeneous expression of regional marker genes and could not be confidently attributed to Arc-ME or surrounding regions.
While inclusion of non-Arc neurons represents dissection artifact, it is noteworthy for two reasons. First, the presence of cells from beyond the rostral, caudal, and lateral boundaries of the Arc suggests that our samples included the full extent of the Arc. Second, the fact that non-Arc neurons clustered apart from Arc neurons indicates regional identity genes strongly influence clustering and can be used post hoc to determine origin.
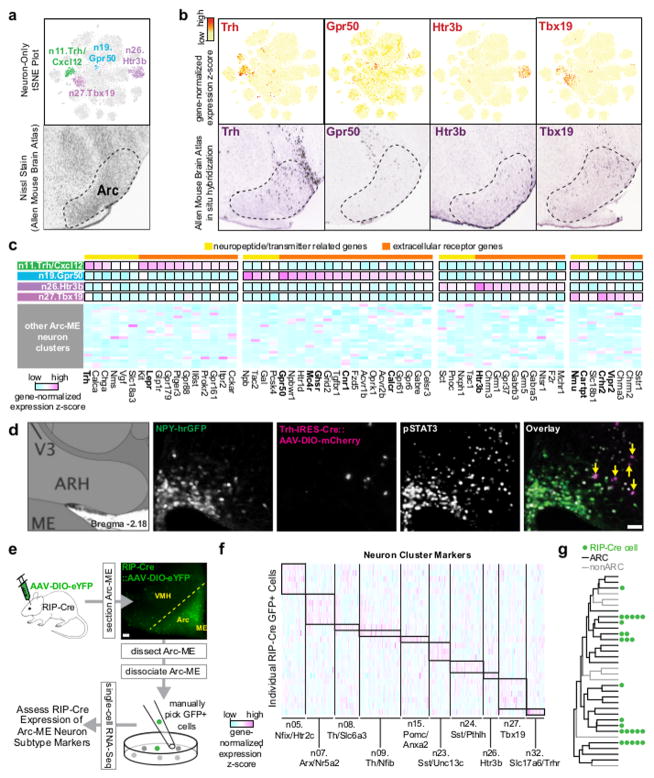
After excluding non-Arc neuronal clusters and those expressing markers of previously described Arc-ME neuron populations, 12 neuronal clusters remained, representing novel Arc-ME neurons. Some of the markers identified for these novel types were previously detected in rodent Arc-ME but not known to mark distinct types of neurons (Qrfp, Nts, Trh, Tbx19; Figure 3C) 23–26. We highlight four of the novel neuron types here: n11.Trh/Cxcl12, n19.Gpr50, n26.Htr3b and n27.Tbx19 (Figure 5A). ABA data confirms their marker genes are transcribed in Arc (Figure 5B). Focusing on neuropeptides and receptor expression profiles, we find that one novel type, n19.Gpr50 neurons, expressed several receptor genes linked to energy balance, including ghrelin receptor (Ghsr), melanocortin 4 receptor (Mc4r), cannabinoid receptor (Cnr1), and calcitonin receptor (Calcr) (Figure 5C). In addition, n27.Tbx19 neurons are enriched for corticotrophin-releasing hormone 2 receptor (Crhr2) (Figure 5C), potentially linking these neurons with known cardiovascular effects of CRH2 receptor agonism in Arc-ME 27, 28.
Figure 5. Novel subtypes of Arc-ME neurons.
(A) Top, neuron-only tSNE plot re-colored to indicate four novel neuron subtypes selected for further analysis: n11.Trh/Cxcl12 neurons, n19.Gpr50 neurons, n26.Htr3b neurons, and n27.Tbx19 neurons; bottom, Nissl stain of sagittal Arc-ME (Allen Mouse Brain Atlas). (B) Expression of marker genes shown by re-coloring of neuron-only tSNE plot (top) and by in situ hybridization of sagittal brain sections (Allen Mouse Brain Atlas; bottom). (C) Heatmap of neuropeptide and receptor genes enriched in four novel neuron subtypes. Other Arc-ME neuron subtypes included for comparison are not labeled but are in numerical order (by n#). (D) Leptin-induced pSTAT3 immunofluorescence in the caudal Arc-ME of a fasted Trh-IRES-Cre mouse in which cells were labeled by Cre-dependent AAV-mCherry (micrograph representative of 4 mice). Yellow arrows indicate pSTAT3+/mCherry+ cells. Scale bar, 50μm. (E) Single-cell RNA-Seq of eYFP labeled RIP-Cre+ neurons that were acutely dissociated and manually isolated from Arc-ME of 2 adult male mice. Micrograph representative of 2 mice. Scale bar, 50μm. (F) RIP-Cre neuron expression of Arc-ME neuronal subtype markers. (G) Dendrogram of Arc-ME neuron subtypes with green dots indicating the subtypes most similar to RIP-Cre neurons based on marker expression.
The n11.Trh/Cxcl12 cluster had the highest expression of leptin receptor (LepR; Lepr) among all Arc-ME clusters (Figure 5C). To confirm functional LepR expression in vivo, we administered leptin to fasted mice and compared fluorescent TRH reporter expression (Trh-IRES-Cre::AAV-DIO-mCherry) with pSTAT3 immunofluorescence, which marks leptin-sensing neurons 29. Most mCherry+ neurons in the Arc-ME were also pSTAT3 immunofluorescent after leptin treatment, confirming their sensitivity to leptin (Figure 5D; 62 of 106 mCherry+ neurons (59%) were pSTAT3+; n=4 mice). Our results thus identify n11.Trh/Cxcl12 neurons as a novel GABAergic (Slc32a1+; see above) leptin-sensing Arc-ME population. Of note, we previously found that leptin controls body weight largely through its action on an uncharacterized population of non-AgRP GABAergic neurons that control the activity of POMC neurons 6 and AgRP neurons 18. Our data strongly suggest that n11.Trh/Cxcl12 neurons may be this missing leptin-sensing population, exemplifying how this transcriptomic resource can fill in known gaps in Arc-ME knowledge. Future studies will address whether these LepR+ TRH neurons provide synaptic input to POMC or AgRP neurons, as this circuit could be key to leptin-mediated control of body weight.
Arc-ME contains neurons marked by the RIP-Cre transgene that are distinct from AgRP neurons and POMC neurons 17. These so-called “RIP-Cre neurons” sense leptin and drive energy expenditure but do not affect feeding 7. However, other than their lack of Agrp and Pomc expression, these RIP-Cre neurons are transcriptionally uncharacterized. We performed single-cell RNA-seq on RIP-Cre neurons manually isolated from acutely dissociated Arc-ME (Figure 5E). As expected 7, nearly all of the 25 RIP-Cre neurons we profiled appeared to be GABAergic, containing GABAergic marker transcript Slc32a1 (21/25 neurons), and roughly half had Lepr transcript (12/25 neurons; Figure S5E).
To determine which Arc-ME neuronal cluster(s) each RIP-Cre neuron was most similar to, we examined RIP-Cre neuron expression of Arc-ME neuronal cluster markers and matched each RIP-Cre neuron to a Drop-seq neuronal cluster based on its expression of cluster markers. RIP-Cre neurons are quite heterogeneous, with individual neurons localized to 10 different clusters, though 23/25 RIP-Cre neurons mapped to 3 distinct transcriptional clades (Figures 5F–G). The most common clusters represented were n07.Arx/Nr5a2, n27.Tbx19, and n05.Nfix/Ht2rc. Of these, the only GABAergic cluster that expressed Lepr at a significant level was the n27.Tbx19 neurons (Figure 5C), making it the likeliest candidate to mediate the previously characterized role for RIP-Cre neurons in energy expenditure. Consistent with this hypothesis, n27.Tbx19 neurons show enriched expression of Cartpt and Nmu, which encode neuropeptides that induce thermogenesis when administered centrally, as well as Vipr2, knockouts of which are lean with increased metabolic rate 30–32. These results demonstrate how this transcriptional database can be used to classify and characterize individual Arc-ME neurons of interest.
Transcriptional relationship predicts anatomical and functional similarities between AgRP neurons and SST neurons
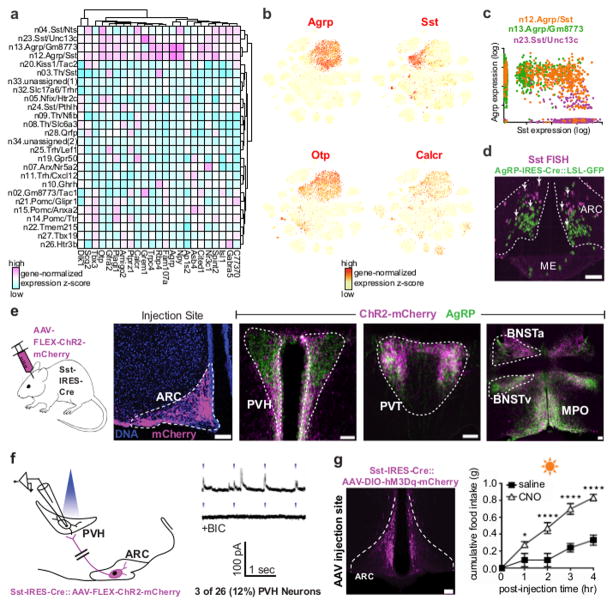
We observed that GABAergic (Slc32a1+) neuron clusters n13.Agrp/Gm8773 and n23.Sst/Unc13c were co-enriched with several transcripts (Figure 6A), including those encoding a transcription factor (Otp) and the calcitonin receptor (Calcr) (Figures 6A, B). In addition, we found a cluster of neurons (n12.Agrp/Sst) highly expressing both Agrp and Sst (Figure 6C) with a transcriptional profile that includes both n23.Sst/Unc13c and n13.Agrp/Gm8773 cluster markers (Figure S6A). Overall, 9% of Agrp+ neurons also expressed Sst, and 19% of all Sst+ neurons also expressed Agrp, above gene-specific expression thresholds (calculated as in ref. 10). Interestingly, transcripts distinguishing n12.Agrp/Sst neurons from other AgRP neurons (n13.Agrp/Gm8773) were largely enriched in other Sst+ neuron clusters as well, suggesting a common somatostatinergic gene program (Figure S6B). To confirm that Agrp/Sst neurons exist in vivo, we compared fluorescent in situ hybridization (FISH) for Sst mRNA to Agrp-IRES-Cre::loxSTOPlox-GFP immunofluorescence, which marks AgRP neurons 33. The results show that 5% of AgRP neurons are Sst+ (38 of 779 neurons), and 9% of Sst+ neurons are AgRP neurons (38 of 442 neurons; representative section in Figure 6D), confirming that Agrp/Sst neurons exist in vivo. While Agrp/Sst neurons represent a small minority of all Arc-ME AgRP neurons, our findings provide the first evidence for transcriptional subtypes of AgRP neurons. Of note, a recent study found that deleting Crhr1 from AgRP neurons alters hepatic glucose production and thermogenesis but not feeding or body weight 34. Our data shows Crhr1 is predominantly expressed by the n12.Agrp/Sst subtype (Figure S6B), predicting this subtype mediates AgRP neuron control of glucoregulation and metabolism specifically.
Figure 6. Similarities between AgRP neurons and SST neurons.
(A) Heatmap of genes co-enriched in AgRP neurons and SST neurons. (B) Expression of two co-enriched genes, Otp and Calcr, as well as Agrp and Sst shown by re-coloring of neuron-only tSNE plots. (C) Co-expression of Agrp and Sst by individual cells in three neuron clusters: n12.Agrp/Sst, n13.Agrp/Gm8773, and n23.Sst/Unc13c. (D) Representative micrograph comparing Agrp-IRES-Cre::loxSTOPlox-GFP immunofluorescence to Sst mRNA in situ hybridization. White arrows indicate co-labeled cells. (E) Axon projection patterns of AgRP neurons and Arc-ME SST neurons. From left to right, micrographs of mCherry immunofluorescence where Cre-dependent AAV-Channelrhodopsin2(ChR2)-mCherry was injected in the Arc-ME of a Sst-IRES-Cre mouse, and comparison of mCherry and AgRP immunofluorescence in paraventricular hypothalamus (PVH); paraventricular thalamus (PVT); bed nucleus of the stria terminalis, anterior (aBNST) and ventral (vBNST) parts; and medial preoptic nucleus (MPO). Micrographs are representative of 3 mice. Scale bar, 100μm. (F) Left, schematic of channelrhodopsin-assisted circuit mapping (CRACM) from ChR2+ Arc-ME SST (ARCSST) to unidentified PVH neurons (n=2 mice). Right, representative patch-clamp recordings of PVH neurons during photostimulation of ARCSST neuron axons in PVH, in the absence (top) or presence (bottom) of the bath-applied GABA A receptor antagonist bicuculline (BIC); blue dash indicates light pulse. (G) Effect of chemogenetic stimulation of Arc-ME ARCSST neurons on daytime food intake. Left, micrograph of mCherry immunofluorescence where Cre-dependent AAV-hM3Dq-mCherry was injected bilaterally in the Arc-ME of a Sst-IRES-Cre mouse; representative of 5 mice; scale bar, 100μm. Right, cumulative food intake in 4hr period after i.p. injection of either the hM3Dq ligand, clozapine N-oxide (CNO), or vehicle (saline). N=5 mice; data shown as mean ± SEM. *p < 0.05, ****p < 0.0001; two-way ANOVA followed by Sidak’s multiple comparisons test
The transcriptional similarities between AgRP neurons and SST neurons led us to consider other potential similarities, such as in their synaptic circuitry and function. Previous studies in rat showed that Arc SST (ARCSST) neurons and AgRP neurons both innervate the paraventricular hypothalamus (PVH) 35. To confirm this in mice and further compare AgRP neuron and ARCSST neuron circuitry, we injected Arc-ME of Sst-IRES-Cre mice with Cre-dependent AAV-channelrhodopsin2(ChR2)-mCherry and performed AgRP immunofluorescence to visualize ARCSST axons and AgRP axons respectively. We found that ARCSST neurons and AgRP neurons have similar projection patterns, both innervating regions where AgRP neurons control feeding, such as the paraventricular hypothalamus (PVH), paraventricular thalamus (PVT), and bed nucleus of the stria terminalis (BNST; Figures 6E, S6D) 36.
The PVH is a major synaptic target of AgRP neurons in their control of feeding behavior 37, 38. Since we observed ARCSST axons in PVH, we tested whether they form functional synapses on PVH neurons. Specifically, we performed channelrhodopsin-assisted circuit mapping, CRACM 39, with mice in which ChR2 was selectively expressed in ARCSST neurons (see above; n=2 mice). Photo-stimulating ARCSST axons in the PVH evoked time-locked post-synaptic currents in ~12% PVH neurons (3 of 26), indicating that ARCSST neurons release fast-acting neurotransmitters onto a subset of PVH neurons (Figure 6F). Bath application of a GABA receptor antagonist, bicuculline abolished the light-evoked post-synaptic currents, revealing the GABAergic nature of these synapses (Figure 6F). Thus, ARCSST neurons, like AgRP neurons, directly inhibit a subset of PVH neurons.
We next determined whether chemogenetic activation of ARCSST neurons could affect feeding, as has been robustly shown for AgRP neurons 4. After injecting Arc-ME of Sst-IRES-Cre mice with a Cre-dependent AAV expressing hM3Dq, a mutant Gq-coupled muscarinic receptor 40, we activated ARCSST neurons using the hM3Dq ligand, clozapine-N-oxide, (CNO; i.p.; n=5 mice). Administering CNO to these mice acutely and significantly increased feeding (Figure 6G), thus establishing ARCSST neurons as a novel orexigenic population whose activation is sufficient to drive feeding behavior. Interestingly, previous studies have shown that adult mice can recover 3 weeks after total ablation of AgRP neurons 41. Whether this recovery results from an increased role of ARCSST neurons to compensate for AgRP neuron loss is a question for further investigation. Additional experiments are also needed to determine which of the 5 Sst+ Arc-ME neuron subtypes is/are responsible for the feeding phenotype we observed. Our gene expression data provide subtype-specific markers from which to design genetic tools to specifically manipulate each Sst+ subtype (Figures 2, S6E, F).
In vivo metabolic stress triggers generalized as well as cell type- and subtype-specific changes in gene expression
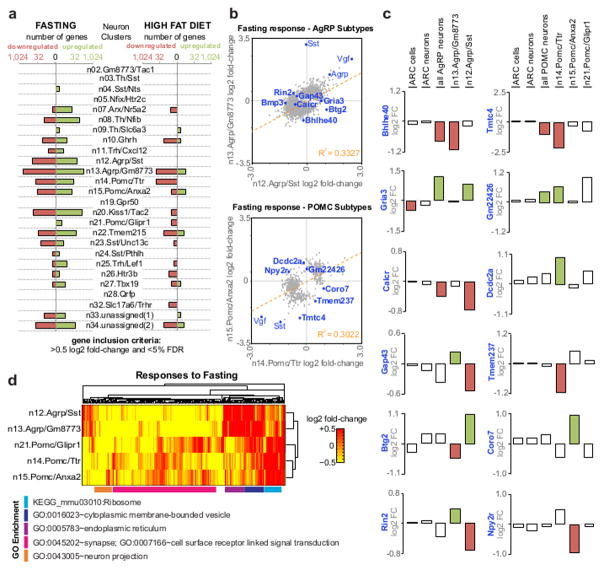
Studies of how in vivo perturbations affect gene expression are typically performed at the tissue level, or at best, on samples of cells pooled on the basis of transgenic reporter expression. Such studies may be limited by their dependence on the availability/fidelity of transgenic reporter animals and, more importantly, their presumption of homogeneity within a target cell population. Unbiased single-cell approaches such as Drop-seq have the potential to overcome these limitations but so far have been used mostly to parse cell types from complex tissues, where transcriptional differences are large and static. To find out whether dynamic transcriptional changes can be assessed, we performed Drop-seq profiling on Arc-ME from mice across different feeding conditions/energy states: ad libitum fed or fasted overnight, or one week of low-fat diet (10% calories from fat; LFD) or high-fat diet (60% calories from fat; HFD). We identified thousands of genes significantly up- or down-regulated in response to fasting and HFD across individual non-neuronal (Figure S7A; Table S5, S6) and neuronal populations (Figure 7A; Table S7, S8). Generally, fasting induced a stronger transcriptional response than HFD (i.e., greater number and amplitude of changes; Figures 7A, S7B), and the neuronal types most responsive to fasting also tended to be most responsive to HFD (Figure 7A), suggesting energy state–responsive neuronal types.
Figure 7. Transcriptional responses to energy imbalance.
(A) Histograms showing the number of genes significantly up- or down-regulated in response to fasting and high fat diet in each Arc-ME neuron subtype. (B) Comparison of fasting responses of AgRP neuron subtypes (top) and POMC neuron subtypes (bottom). Genes plotted were significantly affected by fasting (false-discovery rate (FDR), <0.25) in at least one AgRP neuron subtype (top) or POMC neuron subtype (bottom). While subtypes generally show significant correlation (R2 = 0.33 for AgRP neuron subtypes and R2 = 0.30 for POMC neuron subtypes), there are many individual genes that are differentially affected by fasting (e.g., within top-left and bottom-right quadrants). (C) Examples of genes affected by fasting only in one subtype of AgRP neurons and POMC neurons, or affected oppositely between subtypes. For comparison, average fold-change values are also shown for all Arc-ME cells, all Arc-ME neurons, and all AgRP or POMC neurons. Bars are shaded to indicate the gene was differentially expressed at FDR<0.25 (D) Heatmap of gene expression fold-change values for genes significantly affected in at least one AgRP or POMC subtype. Genes are clustered by gene expression similarities across AgRP and POMC subtypes. GO terms with highest significance for each cluster are shown.
To globally validate gene expression changes, we compared them to a recently published RNA-seq study on pooled neurons marked by AgRP or POMC transgenic reporter expression 21. We found strong correlations between AgRP neuron subtypes and pooled AgRP neurons, and between POMC neuron subtypes and pooled POMC neurons (Figure S7C, D), demonstrating that Drop-seq detects gene expression changes found by more conventional methods.
To further test whether the AgRP and POMC neuronal subtypes we found are functionally distinct, we compared their transcriptional responses to fasting. Interestingly, while most genes changed similarly across the AgRP and POMC neuronal populations (e.g. Agrp, Pomc, Vgf), many genes were differentially regulated between their subtypes (Figure 7B, C). Some of these were fasting-sensitive genes identified in pooled AgRP and POMC neuron samples 21, but which we found to be fasting-sensitive only in one subtype (in AgRP neurons: Bhlhe40, Gria3, Calcr; in POMC neurons: Tmtc4, Gm22426; Figure 7C). We also identified gene-regulatory events that were discordant in different subtypes and had been missed in a bulk-tissue analysis, a real benefit of the single-cell resolution of Drop-seq.. For example, fasting did not significantly alter the activity-dependent synaptogenic marker gene Gap43 in our pooled AgRP neuron data (Figure 7C) nor in published pooled data 21, but the Drop-seq analysis revealed that Gap43 was significantly upregulated by fasting in the n13.Agrp/Gm8773 neurons and significantly down-regulated in n12.Agrp/Sst neurons (Figure 7C; see also Btg2, Rin2). Overall, 647 genes changed significantly (FDR<0.05) in expression in one AgRP neuron subtype but not in the AgRP neuron population as a whole. We found similar examples of subtype-specific transcriptional responses in POMC neurons, including the Neuropeptide Y receptor gene Npy2r (Figure 7C; see also Dcdc2a, Tmem237, Coro7). In total, fasting significantly (FDR<0.05) altered expression of 42 genes in one or two POMC neuron subtypes that were not significantly altered in the whole POMC population. The observed differences in how individual AgRP and POMC subtypes respond to fasting underscores the necessity of a single-cell approach to identifying such heterogeneity and shows that our transcriptionally-defined cell subtypes make distinct functional responses to the same stimulus.
While fasting affects a subset of genes in the two AgRP subtypes differently, the transcriptional responses of these subtypes are broadly similar to each other and different from responses of the POMC subtypes (Figure 7D). The POMC subtypes show greater functional heterogeneity in their fasting responses with the Pomc/Ttr and Pomc/Anxa2 showing high correlation that is distinct from Pomc/Glipr1 neurons (Figure 7D). Extending these comparisons to all Arc-ME neurons (Figure S7E), we find that AgRP neurons and Th/Slc6a3 neurons have similar transcriptional responses to fasting that are distinct from other Arc-ME neurons. Specifically, AgRP neurons and Th/Slc6a3 (TIDA/THDA) neurons strongly upregulate endoplasmic reticulum genes but not the ribosomal and protein translation genes upregulated by other Arc-ME neuron types. Interestingly, fasting is known to activate AgRP neurons and TIDA/THDA neurons 42, 43, which may explain at least some of the similarity in their transcriptional response.
Also notable, many of the fasting induced changes in AgRP neurons were inversely regulated by HFD (Figure S7F). A specific account of all responsive genes and gene sets is beyond the scope of this manuscript; although we focus on responses of a few neuronal subtypes to fasting and feeding, we provide differential expression data for all the Arc-ME cell types, many of which are known to be important in regulating responses to fasting and HFD (Tables S5–S8). These data should provide a rich resource for identification of genes whose dynamic expression in specific cell types may be important for that cell type’s functional response. Overall, these results show that single cell profiling by Drop-seq can robustly and comprehensively detect changes in gene expression across perturbations, and with unprecedented sensitivity, specificity, and scale. Diet-sensitive cell types and genes yield an array of new translational targets for obesity treatment.
Cell-type specific expression profiles connect candidate GWAS genes to novel cell types
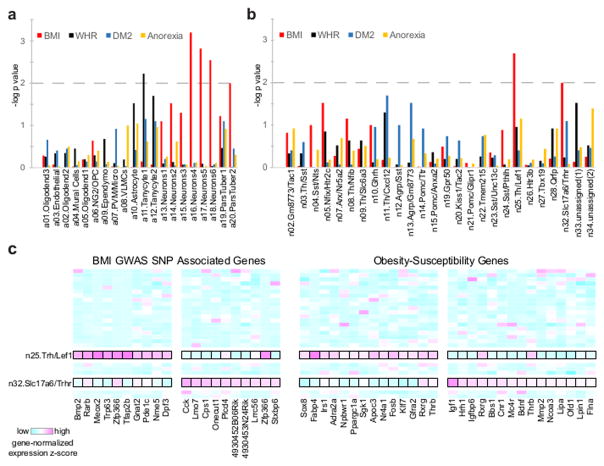
Genome-wide association studies (GWAS) have uncovered thousands of SNPs conferring disease risk, but determining the gene(s) each SNP regulates and the cell type(s) in which that gene acts remains a significant challenge, and is a major barrier to deriving biological insight from such data. DEPICT (Data-driven Expression Prioritized Integration for Complex Traits) is an analytical tool designed to systematically prioritize tissue type(s) based on enriched expression of GWAS-associated genes 44. Previously, body mass index (BMI) GWAS-linked loci genes were shown to have enriched expression in central nervous system (CNS) tissues 45. However, this analysis was limited by the tissue-level resolution of available expression data, and it remained unclear what specific CNS cell type(s) were involved. We therefore used DEPICT with our Drop-seq data to determine which, if any, of our identified cell types mediate GWAS signals for body weight (BMI).
Normalizing to the expression profiles of the other Arc-ME cell populations, we found significant enrichment of BMI-linked gene expression in neuronal, but not non-neuronal, populations (Figure 8A). This neuronal enrichment is specific, as it was not observed for anorexia, DM2, or waist-hip ratio GWAS loci, the last of which was previously shown to be enriched in adipose tissue and adipocytes 46. To hone in on the signal, we focused DEPICT on the Arc-ME neuron types and found two types, n25.Trh/Lef1 and n32.Slc17a6/Trhr, with significant BMI GWAS enrichment (Figure 8B). In addition to the BMI-linked genes, we also observe that these neuron types are enriched for expression of many previously described obesity-susceptibility genes (Figure 8C, Table S9) 47. These analyses thus identify two types of Arc-ME neurons, newly described in this study, as clear candidates for future study in body weight regulation, and prioritize a set of obesity-associated genes whose regulation in these neurons may affect phenotype.
Figure 8. DEPICT predicts specific neuronal subtypes affecting BMI.
(A) DEPICT predicts transcripts from BMI-associated loci (p < 3×10−3), but not WHR, DM2, or anorexia-linked loci, are enriched in neurons. The dotted line shows statistically significant enrichment. (B) DEPICT predicts transcripts from BMI-associated loci (P < 1×10−2) are enriched in n25.Trh/Lef1 and n32.Slc17a6/Trhr neuron clusters. (C) Heatmap of n25.Trh/Lef1 and n32.Slc17a6/Trhr neuron cluster expression of genes near BMI-linked loci or related to obesity susceptibility.
A parallel study from our lab also implicated the n32.Slc17a6/Trhr subpopulation in energy balance. Specifically, Fenselau and colleagues identified a population of Slc17a6+ and Oxtr+ Arc-ME neurons that are both necessary and sufficient for satiety 48. Chemogenetic activation of Arc-ME neurons expressing Slc17a6-IRES-Cre and Oxtr-Cre rapidly suppressed feeding – a timescale not seen with similar activation of POMC neurons 48. Our Drop-seq data show that, among non-POMC neurons in the Arc-ME, Slc17a6 and Oxtr are predominantly co-expressed by a single population, the n32.Slc17a6/Trhr neurons. These results thus implicate n32.Slc17a6/Trhr neurons in body weight control as predicted by our DEPICT analysis.
We also analyzed GWAS data for other phenotypes potentially related to Arc-ME function: growth (height), fertility (menarche, menopause), and glucose regulation (DM2). We found enriched expression of menarche-associated genes in neurons, specifically the n04.Sst/Nts neuron subtype (Figure S8A–B). Notably, Nts encodes neurotensin, a neuropeptide that can directly stimulate GnRH neurons, contributing to luteinizing hormone (LH) surge 49. We observe enrichment for menopause-associated genes in pars tuberalis, an endocrine population that includes cells expressing the reproductive hormones LH and follicle-stimulating hormone (FSH) (Figure S8A). Lastly, we observe highly significant enrichment for height-associated gene expression in multiple cell types including endothelial, mural and VLMC populations, as well as in pars tuberalis cells and many neuron types (AgRP subtypes; n14.Pomc/Ttr; n20.Kiss1/Tac2; n22.Tmem215) (Figures S8A–B). These results agree with and refine previous DEPICT height analyses showing enrichment in cardiovascular, endocrine, skeletal, respiratory, and urogenital tissues 50.
In summary, we have extended unbiased single cell profiling to an extremely complex brain region with myriad important physiological functions. This approach reveals many Arc-ME cell types and subtypes not previously described, despite intensive study of this region. It is important to consider that our cell-type census may not be complete. Our study may have missed rare cell types, cells that do not survive dissociation, or particularly large cells that were filtered out of the cell suspension (see Online Methods). Nonetheless, we establish a resource that provides molecular profiles of many Arc-ME cell types in exceptional detail.
These data will enable rational design and discovery of tools to manipulate Arc-ME cell types with unprecedented specificity. We show for the first time that Drop-seq can assess dynamic transcriptional responses comprehensively across cell types in vivo, and that our ab initio-inferred cell types respond differently to in vivo perturbations, affirming the close relationship between a cell’s transcriptional and functional identities. Finally, we show how detailed cell type transcriptional profiles can unlock the full power of GWAS studies. Overall these studies form a cornucopia of testable hypotheses and provide the means to address them. We look forward to seeing how this resource is used to guide future studies.
ONLINE METHODS
All animal care and experimental procedures were approved in advance by the National Institute of Health and Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee.
Mice
The Drop-seq experiments used a total of 53 adult (4–12 week old) virgin male and female mice randomly assigned to experimental groups and processed in 5 sample batches (see Figure S1A). Pubertal stage and day of estrous cycle were not determined. Mice were generated by crossing Agrp-IRES-Cre 33 mice or Pomc-Cre mice 51 with transgenic loxSTOPlox-GFP mice, all of which had been back-crossed to C57BL6/J background for at least 9 generations. The CCK and ARCSST immunofluorescence experiments used Cck-IRES-Cre::loxSTOPlox-GFP, Sst-IRES-Cre::loxSTOPlox-GFP, Sst-IRES-Cre heterozygous adult (6–12 week old) male mice derived from crossing Cck-IRES-Cre 52 and Sst-IRES-Cre 52 homozygote mice (Jackson Laboratories, JAX; strains 012706 and 013044, respectively) with loxSTOPlox-GFP homozygote mice or wildtype mice (C57BL6/J and mixed background, respectively). The leptin/pSTAT3 immunofluorescence experiments used adult male Trh-IRES-Cre 53 and Trh-IRES-Cre::Npy-hrGFP 54 mice (mixed background). The in situ hybridization experiment used an Agrp-IRES-Cre::loxSTOPlox-GFP adult male mouse bred from Agrp-IRES-Cre and loxSTOPlox-GFP mice (both of C57BL/6J background). The electrophysiological studies used adult male and female Sst-IRES-Cre mice bred from Sst-IRES-Cre homozygote mice (JAX) and wildtype mice (mixed background). The food intake studies used Sst-IRES-Cre adult males bred from Sst-IRES-Cre homozygote mice (JAX strain 013044) and wildtype mice (mixed background). The Arc-ME RIP-Cre studies used adult male RIP-Cre heterozygote mice 55 (JAX strain 003573).
Housing and diets
Mice were housed at 22–24°C with a 12 hour light:12 hour dark cycle and ad libitum access to standard mouse chow (Teklad F6 Rodent Diet 8664; 4.05 kcal per g, 3.3 kcal per g metabolizable energy, 12.5% kcal from fat; Harlan Teklad) and water. For high-fat diet (HFD) experiments, mice were switched from standard chow to high-fat chow (Research Diets, Inc. D12492, 60 kcal% fat) or to a control, low-fat chow (Research Diets, Inc. D12450B, 10 kcal% fat) for 1 week prior to sampling. All diets were provided as pellets.
Generation of Arc-ME single cell suspensions
Brains were rapidly extracted, cooled in ice-cold DMEM/F12 media for 5 minutes, then placed, ventral surface up, into a chilled stainless steel brain matrix (catalog # SA-2165, Roboz Surgical Instrument Co., Gaithersburg, MD). Using GFP fluorescence to demarcate the arcuate’s rostral and caudal boundaries, brains were blocked to obtain a single coronal section containing the entire GFP+ arcuate, ~2mm thick. The Arc-ME was micro-dissected by knife cuts at its visually approximated dorsolateral borders and pooled by experimental condition. For example, in batch #5, we pooled tissue from all fasted males into a single sample prior to digestion and dissociation.
Pooled tissue samples were digested in papain solution with DNAse for 1hr at 37°C with gentle agitation, according to a published protocol 56 except that an entire vial of papain was used in a 1.6mL final reaction volume. Digestion was stopped by dilution with “EBSS #2” 56, with 10% fetal bovine serum used in place of the ovomucoid protease inhibitor. Tissue was centrifuged at 300×g for 5 min, re-suspended in the modified EBSS #2, then gently triturated using a series of 5 Pasteur pipettes with tips fire-polished to incrementally smaller openings. The resulting cell suspension was divided between two microfuge tubes, diluted with Drop-seq buffer (Macosko et al., 2015) to a volume of 1.5mL per tube. Cells were then washed twice by centrifugation at 300×g for 5 min, followed by re-suspension in Drop-seq buffer. Drop-seq buffer consisted of: 5% weight/volume trehalose; 1x Hank’s Buffered Salt Solution (magnesium- and calcium-free); 2.13mM MgCl2; 2mM MgSO4; 1.26mM CaCl2; 1mM glucose; 0.01% bovine serum albumin (molecular biology grade). Cells were filtered through a 20um mesh, diluted to 220 cells/uL based on cell concentration estimates from a hemacytometer, and then kept on ice until use.
Drop-seq generation of single cell sequencing libraries
Drop-seq was performed as in reference 8 but with the following modifications: first, flow rates of 2.1 mL/hr were used for each aqueous suspension, and 12 mL/hr for the oil (Macosko et al., 2015). Second, libraries were sequenced on the Illumina NextSeq500, using 1.7 pM in a volume of 1.2 mL HT1 and 3 mL of 0.3 μM Read1CustSeqB (GCCTGTCCGCGGAAGCAGTGGTATCAACGCAGAGTAC). Read 1 was 20 bp (bases 1–12 cell barcode, bases 13–20 UMI), read 2 (paired end) was 60 bp, and index primer was 8bp (on multiplexed samples).
Drop-seq read alignment and generation of digital expression data
Raw sequence data was first filtered to remove all read pairs with a barcode base quality of less than 10. The second read (60 bp) was then trimmed at the 5′ end to remove any TSO adapter sequence, and at the 3′ end to remove polyA tails of length 6 or greater, then aligned to either the mouse (mm10) genome using STAR v2.4.0 a with default setting. Uniquely mapped reads were grouped by cell barcode. To digitally count gene transcripts, a list of UMIs in each gene, within each cell, was assembled, and UMIs within ED = 1 were merged together. The total number of unique UMI sequences was counted, and this number was reported as the number of transcripts of that gene for a given cell. All cell barcodes in which 800 or more genes were detected were used in downstream analysis, while the remaining cell barcodes were discarded, resulting in a 21,241-cell barcode data set.
Unsupervised dimensionality reduction and clustering
Mouse Arc-ME suspensions were processed through Drop-Seq in 11 separate groups over six separate batches/days, and each sequenced separately. Raw digital expression matrices were generated for the seven sequencing runs. The full 21,241 cells were merged together in a single matrix. Gene expression was normalized to library size and genes showing expression in >50 cells were retained for clustering. Prior to clustering, batch effect correction was performed using the removeBatchEffect function of edgeR 57. All calculations and data were then performed in log space (i.e., ln(CPM +1)).
We used Seurat software to perform clustering as per reference 58. We identified genes that were most variable across the entire data set, controlling for the known relationship between mean expression and variance. We calculated the mean and dispersion (variance/mean) for each gene across all cells, and placed genes into 300 bins based on their average expression. Within each bin, we then identified outlier genes whose dispersion was greater than the median dispersion value plus the difference between the median and minimum dispersion. This process yielded a total of 2251 significantly variable genes.
Principal Components Analysis (PCA) was performed using the prcomp function in R, after scaling and centering the data along each variable gene. To distinguish PCs for further analysis, we plotted the cumulative variance accounted for by each successive PC. Such data display a “knee” at a PC number after which successive PCs explain diminishing degrees of variance. Empirically we found that downstream clustering analyses were optimized when using this PC cutoff. 25 PCs were chosen for further All-cell clustering analyses and used as input for t-Distributed Stochastic Neighbor Embedding (tSNE) 59, implemented by the Seurat software package with “perplexity” parameter set to the default, 30, as per reference 8. The tSNE procedure returns a two-dimensional embedding of single cells, with cells that have similar expression signatures of genes within our variable set localizing near each other in the embedding. To identify cell types, a density clustering we used a density clustering approach implemented in the DBSCAN R package 60, setting the reachability distance parameter (eps) to 3.62. Clusters with fewer than 10 cells and those containing expression markers for more than one canonical cell type (e.g., neuron, oligodendrocyte, tanycyte), representing cell doublets (2 cells in a single droplet) were removed. As a result of these steps, we were able to assign 20,921 cells (98.5% of our data) into 20 cell type clusters.
Average gene expression for each cluster was determined and Euclidean distances between all pairs was calculated, using this data as input for complete-linkage hierarchical clustering and dendrogram assembly. To identify marker genes, we compared each of the clusters using pairwise differential expression analysis using Bioconductor package EdgeR with settings recommended for data with batch effect as described in 4.2 section of the manual. For heatmaps, cluster markers were considered if average FC expression compared to other included clusters was > 2.
For subclustering, each cluster was iteratively clustered as above until post-hoc testing of subclusters showed <10 genes showing average expression difference greater than 1 natural log value between clusters with a Bonferroni corrected p<0.01), thresholds based on those of reference 8. For neuronal subclustering, all cells from the 6 “neuron” groups from initial clustering (clusters a13-a18) were first combined and then iteratively subclustered as above. Perplexity was set to the default (30) for all subclustering.
Statistical Analysis
For differential expression between cell type clusters as well as for Fasting vs. Fed comparison, we performed pairwise differential expression analysis using Bioconductor package EdgeR with settings recommended for data with batch effect as described in 4.2 section of the manual. For 10% Diet/HFD comparison, EdgeR was run with standard settings as these data were from a single batch. For heatmaps, clusters showing at least one significant change (FDR<0.25) in both directions were included. As EdgeR does not account for gene dropout rate, we utilized the SCDE package, which adopts a Bayesian approach fitting individual error models to account for stochastic detection of low-expressing genes 61. We perfomed SCDE on a subset of our data to compare detection of differentially expressed genes marking clusters (pair-wise comparisons of AgRP (n12 and n13) and POMC (n14, n15, and n21) subclusters for which there are well-validated cluster markers). To reduce complexity to a manageable computation time, we used setting to model error using the nearest 100 cells (knn.error.models()) and modified batch correction methodology from Fisher’s exact test to a Chi-square test to account for large number of cells.
Our results showed that fold changes are highly correlated across the statistical methods and that there is large overlap in significantly differentially expressed genes. When averaging across the 10 comparisons, EdgeR calls 69 ± 6% (mean ± S.D.) of the DEgenes that SCDE calls, and SCDE calls 49 ± 10% of the DE genes that EdgeR does (mean ± S.D) (FDR<25% for SCDE and FDR<0.1% for EdgeR). For our benchmark set of marker genes, SCDE showed 92% sensitivity while EdgeR performed at 100% sensitivity. We conclude from these results that EdgeR performs adequately for Drop-seq differential expression analysis and recommend using a threshold of FDR <0.1% from the EdgeR results we provide in Tables S2–S8 for determination of robust differential expression that are likely to meet statistical significance using the more stringent SCDE.
Food intake was analyzed with two-way ANOVA followed by Sidak’s multiple comparisons test.
No statistical methods were used to pre-determine sample sizes, but sample size in the feeding study is comparable to previous studies 4. Data distribution was assumed to be normal but this was not formally tested. Other than the random assignment of mice to experimental groups, no randomization was performed. Data collection and analysis were not performed blind to the conditions of the experiments.
Sex-of-origin prediction for cells from mixed-sex samples
For each of the batch 5 experimental conditions, cells were pooled prior to Drop-seq. In order to predict whether each cell came from female or male, we used the MLSeq package with random forest algorithm to determine a model for predicting sex-of-origin. Using 310 genes showing differential expression (EdgeR, with FDR<0.25, FC>.5) between sexes in both fasted and fed conditions in batch 6 (where sex is known), we created a model on 80% of batch 6 cells (randomly selected) using standard recommended options. The resulting model on batches 1, 2, 3, 4, and the remaining 20% of the batch 6 showed 90% accuracy, with equal rates of misassignment for male and female cells.
Gene ontology analyses
Gene set enrichment analysis was carried out using Bioconductor package RDAVIDWebService (https://www.bioconductor.org/packages/devel/bioc/html/RDAVIDWebService.html). Hierarchical clustering of fold changes across clusters was performed, and all nodes including >10% of genes and <50% of genes were tested for enrichment in DAVID. Non-overlapping nodes showing the highest significance for a gene set were selected. The background set was defined as genes having at least one UMI across all samples 62. Top scoring gene sets were chosen to include in figures.
Single-cell RNA-seq
Mice were anaesthetized with isoflurane and then decapitated. Brains were quickly extracted and immediately chilled in ice-cold, carbogen-saturated (95% O2, 5% CO2) high sucrose solution (238mM sucrose, 26mM NaHCO3, 2.5mM KCl, 1.0mM NaH2PO4, 5.0mM MgCl2, 10.0mM CaCl2, 11mM glucose). Chilled brains were sectioned coronally at 300μm intervals on a Leica VT1000S Vibratome (Leica, Wetzlar, Germany) and incubated in oxygenated artificial cerebrospinal fluid (aCSF; 126mM NaCl, 21.4mM NaHCO3, 2.5mM KCl, 1.2mM NaH2PO4, 1.2mM MgCl2, 2.4mM CaCl2, 10mM glucose) at 34°C for 30min. The Arc-ME was visualized by fluorescence stereoscope, micro-dissected, then enzymatically dissociated as described above, except that cells were re-suspended in “Medium solution” 56 instead of Drop-seq buffer. The resulting cell suspension was plated in a 35mm plastic culture dish, and GFP-positive cells were individually collected and washed by glass micropipette according to a published protocol 63 except that “Medium solution” was used in place of artificial cerebrospinal fluid. Each cell was transferred to a separate qPCR tube cap containing a 2uL drop of PBS with 10% recombinant RNase inhibitor (Clontech). Droplets were confirmed by microscope to contain a single cell and excess buffer was aspirated away. To control for mRNA contamination during cell collection, an equivalent volume of cell-picking buffer was collected and processed along with the single-cell samples.
Cells and the buffer control sample were frozen at −80°C until processed for cDNA synthesis and amplification 64. After 20 cycles of amplification by polymerase chain reaction (PCR), cDNA was purified with AMPure XP (1:1 ratio; Beckman-Coulter, Brea, CA), eluted in 30uL PCR-grade water, and then analyzed by quantitative PCR (qPCR) for expression of a housekeeping gene, Actb or Rplp0 (catalog # Mm.PT.58.28904620.g and Mm.PT.58.43894205, respectively; Integrated DNA Technologies, IDT, Coralville, IA, U.S.A.). As quality control we excluded from sequencing any sample with a Ct value greater than 1 standard deviation from the mean for their batch, and we excluded any sample batches showing contamination (defined as qPCR detection of housekeeping gene expression in the cell-picking buffer control sample). Samples passing quality control were diluted to 150pg/uL and input to Nextera XT library preparation kit (96 indices, Illumina), following the manufacturer’s instructions except at ¼ reaction volume. Library concentration was analyzed by fluorometry (Qubit 2.0; high-sensitivity dsDNA kit; ThermoFisher) and then pooled for equimolar representation. The pool concentration was measured by KAPA library quant qPCR according to the manufacturer’s instructions (KAPA Biosciences, Wilmington, MA). The qPCR product and the 20nM standard were analyzed for size (base pairs) by ethidium bromide-agarose gel electrophoresis. The size of the 20nM standard was divided by that of the library pool, and this value was multiplied by the library pool concentration to get a final, size-corrected concentration. The library pool was then diluted to 2nM for sequencing according to manufacturer’s instructions.
Viral injections
Stereotaxic injections were performed as previously described 7. Briefly, mice were anesthetized with xylazine (5mg per kg) and ketamine (75mg per kg) diluted in saline and placed into a stereotaxic apparatus (model 963, David Kopf Instruments, Tujunga, CA). For postoperative care, mice were injected intraperitoneally with meloxicam (0.5mg per kg). After exposing the skull via a small incision, a small hole was drilled for injection. A pulled-glass pipette was inserted into the brain and virus was injected by an air pressure system. A micromanipulator (model S48 stimulator, Grass Technologies, Rockland, MA) was used deliver the injection at 5nl per min and the pipette was withdrawn 5 min after injection. For electrophysiology and tracing, AAV1-CBA-Flex-ChR2(H134R)-mCherry (University of Pennsylvania School of Medicine, Philadephia, PA) was injected unilaterally into the arcuate (2–5nL; from Bregma, AP: −1.35mm, DV: −6.00mm, ML: ±0.2mm). For in vivo chemogenetic experiments, AAV8—hSyn-DIO-hM3Dq-mCherry (University of North Carolina Vector Core, Chapel Hill, NC) was bilaterally injected into the arcuate (2–5nL, coordinates as above). For profiling of Arc-ME RIP-Cre neurons, AAV8-EF1a-DIO-eYFP (University of North Carolina Vector Core, Chapel Hill, NC) was injected into arcuate (100nL, coordinates as above).
Evan’s blue dye injection
After anesthetizing the mouse with 7% chloral hydrate (350mg/kg), 100μL of Evan’s Blue dye solution (1% w/v in sterile saline) was injected into the tail vein 20 minutes prior to sacrifice, as per reference 65. Brains were quickly extracted, blocked into a 4mm coronal section containing the entire arcuate hypothalamus (0mm to −4mm Bregma), and submerged in 10% neutral buffered formalin (PFA) for 1 week at room temperature, protected from light. Brain blocks were transferred to 20% sucrose overnight at room temperature and then cut by freezing microtome into 35μm thick coronal sections for immunohistochemistry.
In situ hybridization
To generate riboprobe templates, cDNA from mouse hypothalamus was used to PCR amplify the mouse somatostatin (Sst) fragment corresponding to bases 89–488 (Genbank accession NM_009215.1) with the T7 RNA polymerase recognition site incorporated into the product for subsequent transcription. Standard in vitro transcription methods using T7 polymerase (Promega, Madison, WI) and digoxigenin-UTP RNA labeling mix (Roche, Basel, Switzerland) were used to produce both sense and antisense riboprobes for Sst mRNA. Prior to hybridization riboprobes were diluted in hybridization cocktail (Amresco, Solon, OH) with tRNA.
Brain tissue was cryo-sectioned coronally at 12 μm, thaw-mounted onto electrostatically clean slides, and stored at −80°C. Prior to hybridization, sections were post-fixed in 4% paraformaldehyde, rinsed in 0.1M PBS (pH 7.4), equilibrated in 0.1M triethanolamine (pH 8.0), acetylated in triethanolamine containing 0.25% acetic anhydride, and hybridized overnight at 56°C with Sst riboprobe. The next day slides were treated with RNase A, and stringently washed in 0.5x SSC at 60°C for 30min. Following washing, slides were incubated with an antibody against digoxigenin conjugated to horseradish peroxidase (Roche) overnight at 4°C. Riboprobe signal was further enhanced using the TSA-plus biotin system (Perkin Elmer) and streptavidin Alexa Fluor 647 conjugate (Invitrogen, Carlsbad, CA) was used for fluorescent visualization of Sst expression. Fluorescent image capture and analysis was done using an VS120 Slide Scanner (Olympus, Tokyo, Japan) and OlyVIA image analysis software.
Leptin treatment
Leptin was administered to induce STAT3 phosphorylation as previously described 6. Briefly, mice were fasted overnight, injected the following morning with 4mg/kg recombinant leptin (intraperitoneally, i.p.; Dr. A. F. Parlow, National Hormone and Peptide Program, NHPP, Harbor-UCLA Medical Center, Torrance, CA), and then sacrificed for immunohistochemistry 1 hour later.
Immunohistochemistry
Mice were terminally anesthetized with 7% chloral hydrate (350mg/kg) diluted in isotonic saline and transcardially perfused with phosphate-buffered saline (PBS) followed by 10% neutral buffered formalin (PFA). Brains were removed, stored in the same fixative for 2hr, transferred into 20% sucrose at 4°C overnight, and cut into 40μm coronal sections on a freezing microtome. Brain sections were washed twice in 0.1M phosphate-buffered saline (PBS) with Tween-20, pH 7.4 (PBST), blocked in 3% normal donkey serum/0.25% Triton X-100 in PBS for 1hr at room temperature, and incubated overnight at room temperature in blocking solution containing primary antisera: rabbit anti-TSHb, 1:1000 (catalog # AFP-1274789, NHPP); goat anti-AgRP, 1:1000 (catalog # GT15023, Neuromics, Minneapolis, MN); rabbit anti-dsRed (mCherry), 1:1000, (catalog # 632496, Clontech, Mountain View, CA); chicken anti-GFP, 1:1000 (catalog # A10262, Invitrogen, Carlsbad, CA); rabbit anti-SPRR1A 66, 1:500, a kind gift of Dr. Stephen Strittmatter (Yale University, New Haven, CT); 1:100 rabbit anti-phospho-Stat3 (Tyr705) (D3A7) XP (catalog # 9145S, Cell Signaling, Danvers, MA). Validation data for each antibody are available from its manufacturer. The morning after incubation in primary antibody, sections were washed three times in PBS and then incubated in the Alexa fluorophore-conjugated donkey secondary antisera, 1:1000 (Molecular Probes, Eugene, OR) for 2hr in the dark at room temperature. An exception was the Alexa 647 donkey anti-rabbit secondary antibody used for pSTAT3 immunofluorescence, which required a higher concentration (1:100). Sections were washed 3 times in PBS, mounted onto gelatin-coated slides (Southern Biotech; Birmingham, AL), coverslipped with Vectashield Anti-fade Mounting Medium with DAPI (Vector Labs, Burlingame, CA) and sealed with nail polish. Fluorescent images were captured with Olympus VS120 slide scanner microscope and with a confocal microscope (Zeiss LSM510 Upright Confocal System).
Electrophysiology
Animals were deeply anesthetized with isoflurane, decapitated and brains were quickly removed into ice-cold cutting solution consisting of (in mM): 93 NMDG, 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 5 sodium ascorbate, 2 thiourea, 3 sodium pyruvate, 10 MgSO4, 0.5 CaCl2 (pH 7.3 adjusted with HCl), oxygenated with 95% O2, 5% CO2, measured osmolarity 310–320mOsm per L. Then, 300μm-thick coronal sections were cut with a Leica VT1000S vibratome and incubated in oxygenated cutting solution at 34°C for 10min. Next, slices were transferred to oxygenated aCSF consisting of (in mM): 126 NaCl, 21.4 NaHCO3, 2.5 KCl, 1.2 NaH2PO4, 1.2 MgCl2, 2.4 CaCl2, 10 glucose and recovered for 30min at 34°C. Slices were stored in oxygenated aCSF at room temperature (20–24°C) for at least 60min prior to recording. A single slice was placed in the recording chamber where it was continuously superfused at a rate of 3–4mL per min with oxygenated aCSF. Neurons were visualized with a SliceScope microscope (Scientifica, Uckfield, U.K.) equipped with infrared-differential interference contrast and fluorescence optics.
Recordings were obtained using borosilicate glass microelectrodes (5–7MΩ) filled with a Cs+-based low Cl− internal solution containing (in mM) 135 CsMeSO3, 10 HEPES, 1 EGTA, 4 MgCl2, 4 Na2-ATP, 0.4 Na2-GTP, 10 Na2-phosphocreatine (pH 7.3 adjusted with CsOH; 295mOsm per kg; ECl = −70 mV) from unidentified neurons in the PVH. Photostimulation-evoked IPSCs were recorded in whole-cell voltage-clamp mode, with membrane potential clamped and 0mV in presence of CNQX (10μM) and D-AP5 (50μM). To confirm whether post-synaptic currents were mediated by GABA A receptors, bicuculline was applied to the bath (10μM) during recordings.
Recordings were made using a Multiclamp 700B amplifier, and data were filtered at 2kHz and digitized at 10kHz. To photostimulate channelrhodopsin2-positive fibers, a LED light source (473nm; CoolLED, Andover, U.K.) was used, as previously described 7. The light output was controlled by a programmable pulse stimulator, Master-8 (A.M.P.I., Jerusalem, Israel) and pClamp 10.2 software (Axon Instruments, Foster City, CA). The photostimulation-evoked IPSCs detection protocol consisted of four blue light pulses administered 1 sec apart during the first 4s of an 8s sweep, repeated for a total of 30 sweeps. Evoked IPSCs with short latency (≤ 6ms) upon light stimulation were considered as light-driven.
Food intake studies
All mice were singly housed for at least 2.5wk following surgery and handled for 10 consecutive days before the assay to reduce stress response. Feeding studies were performed in home cages with ad libitum food access. The day before the experiment, mice were provided with fresh cages to avoid leftover of food spilling in the bedding.
Mice were intraperitoneally administered clozapine-N-oxide (CNO; diluted in isotonic saline) at 1mg per kg of body weight, or an equivalent volume of isotonic saline. For light cycle measurements, animals were injected with either CNO or saline at 9:00am and food was weighed 1hr, 2hr, 3hr and 4hr later. A full trial consisted of assessing food intake from the study subjects after they received injections of saline on day 1 and CNO on day 2. For dark cycle measurements, animals were injected with either saline or CNO at 6:00pm and food was weighed 1hr, 2hr, 3hr and 4hr later. A full trial consisted of assessing food intake from the study subjects after they received injections of saline on day 1 and CNO on day 2. Following feeding studies, mice were sacrificed for immunohistochemical assessment of AAV-Syn1-DIO-hM3Dq-mCherry expression in the Arc. Mice in which mCherry was expressed in cell bodies outside of the Arc were excluded from analysis of behavioral data.
Classification of oligodendrocyte clusters and RIP-Cre neurons
To compare oligodendrocyte clusters to maturational stages, we generated a heatmap showing single-cell expression of oligodendrocyte stage-specific marker genes identified by reference 9. We then assigned each oligodendrocyte cluster to a maturational stage based on how well it visually matched the stage-specific marker profile. We used a similar process to assign RIP-Cre neurons to neuronal clusters. Specifically, we generated a heatmap showing each RIP-Cre neuron’s expression of neuronal cluster marker genes and then visually matched each RIP-Cre neuron to a cluster.
DEPICT analysis
DEPICT version 1, release 194 (https://github.com/perslab/depict), was used to prioritize Arc-ME cell clusters based on GWAS summary statistics for body mass index 45, height 50, waist hip ratio adjusted for body mass index 46, type 2 diabetes 67, menopause 68, menarche 69 and anorexia 70 (Supplemental Table 8). For body mass index, waist hip ratio and height, we applied the same association p value cutoffs as used in refs. 45, 46, 50. For the traits with no published DEPICT analyses (i.e., menarche, menopause and anorexia), we used GWAS association p value cutoff <10−5 (STXXX). Default DEPICT settings were used to construct loci: (a) input SNPs were clumped using PLINK version 1.90 71 enforcing a clumping distance of 500 kb and/or linkage disequilibrium r2 threshold of <0.1, (b) loci were formed around the index SNP of each clump using r2 >0.5 as the locus definition, (c) genes were mapped to loci, and (d) loci containing the same gene(s) were merged into single loci – for details, see reference 61.1000 Genomes Project phase 1 genotype data 72 were used to perform clumping and compute locus boundaries based on human genome build 19. Association signals on sex chromosomes were omitted from the analyses. Normalized Arc-ME single cell expression data were averaged across Seurat clusters, mapped from mouse gene symbols to mouse Ensembl 73 gene identifiers (using Ensembl version 83) and to the human Ensembl gene identifiers (using Ensembl version 82). Mouse gene identifiers mapping to several mouse Ensembl identifiers were discarded and the human gene with the highest degree of mouse homology was retained in instances where a mouse gene mapped to several human genes (code at https://github.com/perslab/campbell-2016/). Expression levels of mouse genes mapped to the same human gene were averaged. The resulting cell cluster Arc-ME gene expression levels were transformed to standard normal distributions prior to the DEPICT analyses. DEPICT was run using default settings.
Supplementary Material
Acknowledgments
Funding was provided by: US National Institutes of Health grants to B.L. (R01 DK096010, R01 DK089044, R01 DK071051, R01 DK075632, R37 DK053477, BNORC Transgenic Core P30 DK046200, BADERC Transgenic Core P30 DK057521), E.R. (R01 DK102170, R01 DK085171, R01 DK102173), E.R. and L.T. (BNORC Functional Genomics Core P30 DK046200), L.T. (BADERC Pilot and Feasibility grant NIH 2P30DK057521-16), and J.R. (F32 DK103387); a Department of Defense grant to L.T. (Discovery Award W81XWH-15-1-0251); an American Heart Association Postdoctoral Fellowship to J.C. (14POST20100011); the Lundbeck Foundation and the Benzon Foundation (T.H.P.); the Stanley Center for Psychiatric Research (S.M.); and the Stanley-MGH Fellowship in Psychiatric Neuroscience (E.M.). qPCR and confocal imaging were done at BIDMC’s Molecular Medicine Core and Confocal Imaging Core, respectively. We gratefully acknowledge Zongfang Yang, Joseph Madara, and Chen Wu for technical assistance, Alastair Garfield for editorial advice, and Peter Kharchenko for advice on SCDE.
Footnotes
ACCESSION CODES
Raw and fully processed single-cell RNA-Seq data on RIP-Cre neurons are available at GEO accession number GSE90806. Raw Drop-seq data are available at GEO accession number XXXXXXX.
AUTHOR CONTRIBUTIONS
J.C., L.T., E.M., E.R. & B.L. designed the study. J.C., A.V., and L.T. prepared samples for Drop-seq. L.T., D.T., J.C., E.M., and M.G. did Drop-seq. D.T., M.G., and L.T. made Drop-seq libraries. J.C. did single-cell RNA-seq. L.T., J.C., A.L., and E.M. analyzed transcriptomic data. J.R. did in situ hybridization. J.C. and H.F. did histology and imaging, with advice from B.L. H.F. did electrophysiology. H.F. and A.V. did stereotaxic injections and feeding studies. J.C., L.T., and A.L. prepared figures. J.C., L.T., E.R., and B.L. wrote the manuscript with input from all authors.
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare no competing financial interests.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon request.
References
- 1.Rodriguez EM, Blazquez JL, Guerra M. The design of barriers in the hypothalamus allows the median eminence and the arcuate nucleus to enjoy private milieus: the former opens to the portal blood and the latter to the cerebrospinal fluid. Peptides. 2010;31:757–776. doi: 10.1016/j.peptides.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Langlet F, et al. Tanycytic VEGF-A boosts blood-hypothalamus barrier plasticity and access of metabolic signals to the arcuate nucleus in response to fasting. Cell Metab. 2013;17:607–617. doi: 10.1016/j.cmet.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aponte Y, Atasoy D, Sternson SM. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nat Neurosci. 2011;14:351–355. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krashes MJ, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 2011;121:1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han SY, McLennan T, Czieselsky K, Herbison AE. Selective optogenetic activation of arcuate kisspeptin neurons generates pulsatile luteinizing hormone secretion. Proc Natl Acad Sci U S A. 2015;112:13109–13114. doi: 10.1073/pnas.1512243112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vong L, et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong D, et al. GABAergic RIP-Cre neurons in the arcuate nucleus selectively regulate energy expenditure. Cell. 2012;151:645–657. doi: 10.1016/j.cell.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marques S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Usoskin D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 11.Vijayan E, Samson WK, McCann SM. In vivo and in vitro effects of cholecystokinin on gonadotropin, prolactin, growth hormone and thyrotropin release in the rat. Brain Res. 1979;172:295–302. doi: 10.1016/0006-8993(79)90539-0. [DOI] [PubMed] [Google Scholar]
- 12.Morgan PJ, Williams LM. The pars tuberalis of the pituitary: a gateway for neuroendocrine output. Rev Reprod. 1996;1:153–161. doi: 10.1530/ror.0.0010153. [DOI] [PubMed] [Google Scholar]
- 13.Goodman T, Hajihosseini MK. Hypothalamic tanycytes-masters and servants of metabolic, neuroendocrine, and neurogenic functions. Front Neurosci. 2015;9:387. doi: 10.3389/fnins.2015.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 15.Tesfaigzi J, Carlson DM. Expression, regulation, and function of the SPR family of proteins. A review. Cell Biochem Biophys. 1999;30:243–265. doi: 10.1007/BF02738069. [DOI] [PubMed] [Google Scholar]
- 16.Everitt BJ, et al. The hypothalamic arcuate nucleus-median eminence complex: immunohistochemistry of transmitters, peptides and DARPP-32 with special reference to coexistence in dopamine neurons. Brain Res. 1986;396:97–155. doi: 10.1016/s0006-8993(86)80192-5. [DOI] [PubMed] [Google Scholar]
- 17.Choudhury AI, et al. The role of insulin receptor substrate 2 in hypothalamic and beta cell function. J Clin Invest. 2005;115:940–950. doi: 10.1172/JCI24445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garfield AS, et al. Dynamic GABAergic afferent modulation of AgRP neurons. Nat Neurosci. 2016 doi: 10.1038/nn.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyons DJ, Hellysaz A, Broberger C. Prolactin regulates tuberoinfundibular dopamine neuron discharge pattern: novel feedback control mechanisms in the lactotrophic axis. J Neurosci. 2012;32:8074–8083. doi: 10.1523/JNEUROSCI.0129-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demaria JE, et al. Dopamine transporters participate in the physiological regulation of prolactin. Endocrinology. 2000;141:366–374. doi: 10.1210/endo.141.1.7281. [DOI] [PubMed] [Google Scholar]
- 21.Henry FE, Sugino K, Tozer A, Branco T, Sternson SM. Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. Elife. 2015;4 doi: 10.7554/eLife.09800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sohn JW, et al. Serotonin 2C receptor activates a distinct population of arcuate pro-opiomelanocortin neurons via TRPC channels. Neuron. 2011;71:488–497. doi: 10.1016/j.neuron.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahn D, Abrams GM, Zimmerman EA, Carraway R, Leeman SE. Neurotensin neurons in the rat hypothalamus: an immunocytochemical study. Endocrinology. 1980;107:47–54. doi: 10.1210/endo-107-1-47. [DOI] [PubMed] [Google Scholar]
- 24.Nishiyama T, et al. Hypothalamic thyrotropin-releasing hormone (TRH)-containing neurons involved in the hypothalamic-hypophysial-thyroid axis. Light microscopic immunohistochemistry. Brain Res. 1985;345:205–218. doi: 10.1016/0006-8993(85)90996-5. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, et al. Tbx19, a tissue-selective regulator of POMC gene expression. Proc Natl Acad Sci U S A. 2001;98:8674–8679. doi: 10.1073/pnas.141234898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takayasu S, et al. A neuropeptide ligand of the G protein-coupled receptor GPR103 regulates feeding, behavioral arousal, and blood pressure in mice. Proc Natl Acad Sci U S A. 2006;103:7438–7443. doi: 10.1073/pnas.0602371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chitravanshi VC, Kawabe K, Sapru HN. Mechanisms of cardiovascular actions of urocortins in the hypothalamic arcuate nucleus of the rat. Am J Physiol Heart Circ Physiol. 2013;305:H182–191. doi: 10.1152/ajpheart.00138.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reyes TM, et al. Urocortin II: a member of the corticotropin-releasing factor (CRF) neuropeptide family that is selectively bound by type 2 CRF receptors. Proc Natl Acad Sci U S A. 2001;98:2843–2848. doi: 10.1073/pnas.051626398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaisse C, et al. Leptin activation of Stat3 in the hypothalamus of wild-type and ob/ob mice but not db/db mice. Nat Genet. 1996;14:95–97. doi: 10.1038/ng0996-95. [DOI] [PubMed] [Google Scholar]
- 30.Kong W, et al. A role for arcuate cocaine and amphetamine-regulated transcript in hyperphagia, thermogenesis, and cold adaptation. FASEB J. 2003;17:1688–1690. doi: 10.1096/fj.02-0805fje. [DOI] [PubMed] [Google Scholar]
- 31.Asnicar MA, et al. Vasoactive intestinal polypeptide/pituitary adenylate cyclase-activating peptide receptor 2 deficiency in mice results in growth retardation and increased basal metabolic rate. Endocrinology. 2002;143:3994–4006. doi: 10.1210/en.2002-220354. [DOI] [PubMed] [Google Scholar]
- 32.Novak CM, Zhang M, Levine JA. Neuromedin U in the paraventricular and arcuate hypothalamic nuclei increases non-exercise activity thermogenesis. J Neuroendocrinol. 2006;18:594–601. doi: 10.1111/j.1365-2826.2006.01454.x. [DOI] [PubMed] [Google Scholar]
- 33.Tong Q, Ye CP, Jones JE, Elmquist JK, Lowell BB. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nat Neurosci. 2008;11:998–1000. doi: 10.1038/nn.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuperman Y, et al. CRFR1 in AgRP Neurons Modulates Sympathetic Nervous System Activity to Adapt to Cold Stress and Fasting. Cell Metab. 2016;23:1185–1199. doi: 10.1016/j.cmet.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moga MM, Saper CB. Neuropeptide-immunoreactive neurons projecting to the paraventricular hypothalamic nucleus in the rat. J Comp Neurol. 1994;346:137–150. doi: 10.1002/cne.903460110. [DOI] [PubMed] [Google Scholar]
- 36.Betley JN, Cao ZF, Ritola KD, Sternson SM. Parallel, redundant circuit organization for homeostatic control of feeding behavior. Cell. 2013;155:1337–1350. doi: 10.1016/j.cell.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atasoy D, Betley JN, Su HH, Sternson SM. Deconstruction of a neural circuit for hunger. Nature. 2012;488:172–177. doi: 10.1038/nature11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garfield AS, et al. A neural basis for melanocortin-4 receptor-regulated appetite. Nat Neurosci. 2015;18:863–871. doi: 10.1038/nn.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 40.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci U S A. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Q, Boyle MP, Palmiter RD. Loss of GABAergic signaling by AgRP neurons to the parabrachial nucleus leads to starvation. Cell. 2009;137:1225–1234. doi: 10.1016/j.cell.2009.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn TM, Breininger JF, Baskin DG, Schwartz MW. Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nat Neurosci. 1998;1:271–272. doi: 10.1038/1082. [DOI] [PubMed] [Google Scholar]
- 43.Hsueh YC, Cheng SM, Pan JT. Fasting stimulates tuberoinfundibular dopaminergic neuronal activity and inhibits prolactin secretion in oestrogen-primed ovariectomized rats: involvement of orexin A and neuropeptide Y. J Neuroendocrinol. 2002;14:745–752. doi: 10.1046/j.1365-2826.2002.00835.x. [DOI] [PubMed] [Google Scholar]
- 44.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Locke AE, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vimaleswaran KS, et al. Candidate genes for obesity-susceptibility show enriched association within a large genome-wide association study for BMI. Hum Mol Genet. 2012;21:4537–4542. doi: 10.1093/hmg/dds283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fenselau H, et al. A rapidly acting glutamatergic ARC-->PVH satiety circuit postsynaptically regulated by alpha-MSH. Nat Neurosci. 2016 doi: 10.1038/nn.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boules M, Li Z, Smith K, Fredrickson P, Richelson E. Diverse roles of neurotensin agonists in the central nervous system. Front Endocrinol (Lausanne) 2013;4:36. doi: 10.3389/fendo.2013.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balthasar N, et al. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42:983–991. doi: 10.1016/j.neuron.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krashes MJ, et al. An excitatory paraventricular nucleus to AgRP neuron circuit that drives hunger. Nature. 2014;507:238–242. doi: 10.1038/nature12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van den Pol AN, et al. Neuromedin B and gastrin-releasing peptide excite arcuate nucleus neuropeptide Y neurons in a novel transgenic mouse expressing strong Renilla green fluorescent protein in NPY neurons. J Neurosci. 2009;29:4622–4639. doi: 10.1523/JNEUROSCI.3249-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Postic C, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 56.Saxena A, et al. Trehalose-enhanced isolation of neuronal sub-types from adult mouse brain. Biotechniques. 2012;52:381–385. doi: 10.2144/0000113878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Maaten L, Hinton G. Visualizing Data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- 60.Esther M, Kriegel H-P, Sander J, Xu X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. 2nd International Conference on Knowledge Discovery and Data Mining (KDD-96); Institute for Computer Science, University of Munich; 1996. [Google Scholar]
- 61.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fresno C, Fernandez EA. RDAVIDWebService: a versatile R interface to DAVID. Bioinformatics. 2013;29:2810–2811. doi: 10.1093/bioinformatics/btt487. [DOI] [PubMed] [Google Scholar]
- 63.Hempel CM, Sugino K, Nelson SB. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nat Protoc. 2007;2:2924–2929. doi: 10.1038/nprot.2007.416. [DOI] [PubMed] [Google Scholar]
- 64.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 65.Mullier A, Bouret SG, Prevot V, Dehouck B. Differential distribution of tight junction proteins suggests a role for tanycytes in blood-hypothalamus barrier regulation in the adult mouse brain. J Comp Neurol. 2010;518:943–962. doi: 10.1002/cne.22273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bonilla IE, Tanabe K, Strittmatter SM. Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J Neurosci. 2002;22:1303–1315. doi: 10.1523/JNEUROSCI.22-04-01303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Day FR, et al. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. 2015;47:1294–1303. doi: 10.1038/ng.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perry JR, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boraska V, et al. A genome-wide association study of anorexia nervosa. Mol Psychiatry. 2014;19:1085–1094. doi: 10.1038/mp.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Genomes Project C et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yates A, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.