Abstract
Objective
The objective of this study is to investigate the role and underlying mechanism of Olfactomedin 2 (Olfm2) in SMC phenotypic modulation and vascular remodeling.
Approach and Results
Platelet-derived growth factor-BB (PDGF-BB) induces Olfm2 expression in primary SMCs while modulating SMC phenotype as shown by the down-regulation of SMC marker proteins. Knockdown of Olfm2 blocks PDGF-BB-induced SMC phenotypic modulation, proliferation and migration. Conversely, Olfm2 overexpression inhibits SMC marker expression. Mechanistically, Olfm2 promotes the interaction of serum response factor with the runt-related transcription factor 2 that is induced by PDGF-BB, leading to a decreased interaction between serum response factor and myocardin, causing a repression of SMC marker gene transcription and consequently SMC phenotypic modulation. Animal studies show that Olfm2 is up-regulated in balloon-injured rat carotid arteries. Knockdown of Olfm2 effectively inhibits balloon injury-induced neointima formation. Importantly, knockout of Olfm2 in mice profoundly suppresses wire injury-induced neointimal hyperplasia while restoring SMC contractile protein expression, suggesting that Olfm2 plays a critical role in SMC phenotypic modulation in vivo.
Conclusions
Olfm2 is a novel factor mediating SMC phenotypic modulation. Thus, Olfm2 may be a potential target for treating injury-induced proliferative vascular diseases.
Keywords: Olfactomedin 2, smooth muscle, phenotypic modulation, vascular remodeling
Vascular smooth muscle cells (SMCs) do not terminally differentiate and can undergo transition between a quiescent contractile phenotype and a proliferative synthetic phenotype in response to pathological stimuli such as vascular injury.1 This phenotypic plasticity of SMCs is essential for vascular development and remodeling. Fully differentiated SMCs express high levels of SM contractile proteins such as SM myosin heavy chain (SMMHC, MYH11), SM22α (TAGLN), and α-SM actin (α-SMA, ACTA2) that are required to fulfill their principal contractile function. Proliferating SMCs, in contrast, express reduced levels of contractile proteins. Consequently, changes of SMC contractile gene expression are often used to mark SMC phenotype alteration.1
Platelet-derived growth factor-BB (PDGF-BB) is one of the key modulators for SMC phenotypes.1 In cultured SMCs, PDGF-BB represses the expression of SMC markers while increasing SMC proliferation and migration rates.2–4 In vivo, the expression of PDGF-BB and PDGF-β receptor is increased at the site of vascular lesions, and inhibition of their signaling has been reported to reduce neointimal thickening.5–9 The PDGF-BB-induced repression of SMC genes at the transcriptional level is serum response factor (SRF)-dependent manner.10–13
SRF, a widely expressed MADS (MCM1, Agamous, Deficiens, SRF) box transcription factor, binds as a homodimer to a DNA consensus sequence known as a CArG box [CC(A/T)6 GG], which is found in the 5’ promoter and intronic regions of SMC-specific genes.14 SRF binding to CArG box is a critical step for activation of SMC-specific genes in vascular SMC differentiation. High SRF expression facilitates SRF-CArG binding. Transforming growth factor-β (TGF-β), a potent stimulator of SMC differentiation, has been shown to induce SRF expression via undefined mechanisms.15, 16 Moreover, SRF can interact with other transcription factors or cofactors to promote SMC differentiation. Among them, Myocardin (Myocd) is the most powerful SRF cofactor that is expressed in smooth and cardiac muscle lineages throughout embryonic development and adulthood.17 Myocd stimulates SRF-dependent transcription by interacting with the MADS domain of SRF.18, 19 Myocd is up-regulated at the transcriptional level in the late stage of TGF-β-induced SMC differentiation from mesenchymal stem cells.20, 21 In addition to their expression level being regulated in SMC differentiation, both SRF and Myocd may undergo post-translational modification such as phosphorylation, which affects SRF/Myocd activity in SMC differentiation.22–24 Recent studies have revealed that runt-related transcription factor 2 (Runx2), a key osteogenic transcription factor, serves as a corepressor independent of its DNA binding by interacting with SRF and disrupting the formation of SRF/Myocd complex, which contributes to the phenotypic modulation of SMCs.25, 26 The precise molecular mechanisms whereby Runx2 interferes with SRF/Myocd complex formation, however, remain largely unknown.
Olfactomedin 2 (Olfm2) belongs to the family of olfactomedin domain-containing proteins consisting of at least 13 members in mammals.27–29 It is well established that Olfm2 is a developmentally regulated gene and involved in early eye development and function.30–32 Our previous study indicates that Olfm2 regulates TGF-β-induced SMC differentiation from mesenchymal stem cells.33 However, it remains unknown if Olfm2 plays a role in mature SMCs in artery, especially if it is involved in SMC phenotypic modulation, a process of SMC dedifferentiation during pathological vascular remodeling. In an analysis of PDGF-BB-induced phenotypic modulation of primary SMCs, we unexpectedly found that Olfm2 expression was markedly up-regulated by PDGF-BB, which prompted us to hypothesize that Olfm2 is involved in PDGF-BB-induced SMC phenotypic modulation. Indeed, our studies showed that knockdown of Olfm2 suppressed PDGF-BB-induced SMC phenotypic modulation. Olfm2 appeared to mediate SRF-Runx2 interaction, which prevented Myocd from binding to SRF, leading to a repressed transcription of SMC marker genes and consequently SMC phenotypic modulation. Consistently, a significant reduction in intimal hyperplasia and an increased level of SMC contractile protein were observed in wire-injured arteries of Olfm2 knockout (Olfm2−/−) mice, indicating that Olfm2 is a novel regulator of SMC phenotypic modulation and vascular remodeling.
Materials and Methods
Materials and Methods are available in the online-only Data Supplement.
Results
Olfm2 was upregulated during SMC phenotypic modulation both in vitro and in vivo
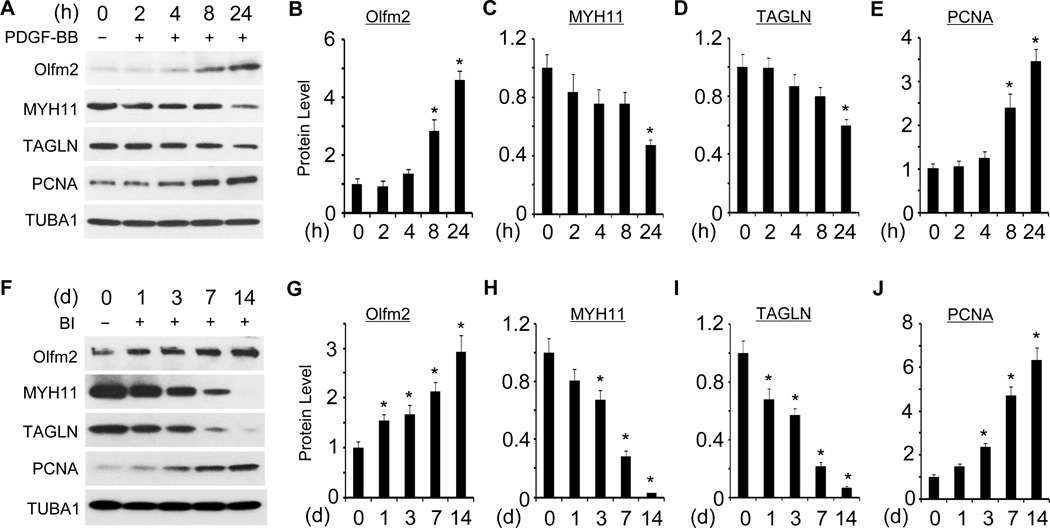
Since Olfm2 is involved in TGF-β-induced SMC differentiation from mesenchymal progenitors in vitro,33 we examined the expression of Olfm2 in primary rat SMCs (RASMCs) treated with PDGF-BB, a SMC phenotype modulator.1 As shown in Figure 1, Olfm2 was markedly up-regulated 8 h after PDGF-BB treatment, a time earlier than the reduction of SMC marker genes that occurred 24 h after the treatment (Figure 1A–1D). Olfm2 expression also correlated with the induction of proliferating cell nuclear antigen (PCNA), a proliferation marker (Figure 1A and 1E). Importantly, Olfm2 was expressed at a low level in normal arteries, but its expression was time-dependently up-regulated in balloon-injured (BI) rat carotid arteries (Figure 1F) along with the reduction of SMC marker proteins and induction of PCNA in the arteries. Since GTEx database suggests that Olfm2 mRNA is not expressed in several human arteries (http://www.gtexportal.org/home/gene/OLFM2), we verified the specificity of Olfm2 antibody used in our studies. As shown in Figure IA in the online-only Data Supplement, the Olfm2 antibody detected a single band in SMC isolated from mouse carotid arteries and rat aorta but not in Olfm2 knockout mouse carotid arteries, indicating that the Olfm2 antibody is specific and that Olfm2 was indeed expressed in aorta or carotid artery. Importantly, Olfm2 was mainly expressed in the neointimal and medial SMC in the balloon-injured artery (Figure IB in the online-only Data Supplement). In fact, Olfm2 was up-regulated as early as one day after the balloon injury preceding the alteration of MYH11, TAGLN and PCNA expression (Figure 1F–1J), indicating that Olfm2 may be involved in SMC phenotypic modulation in vivo, which eventually leads to vascular remodeling.
Figure 1. Olfm2 was upregulated in SMC phenotypic modulation both in vitro and in vivo.
A, PDGF-BB induced Olfm2 expression in primary RASMCs. Serum-starved RASMCs were treated with vehicle (−) or PDGF-BB (+, 20 ng/ml) for the time indicated. Western blotting was performed to examine the expression of Olfm2, SMC markers, and proliferation marker PCNA. B–E, Quantification of Olfm2, SMC markers and PCNA expression shown in A by normalizing to the TUBA1 level. *, P < 0.05 vs. vehicle-treated group (0 h) (n=3). F, Olfm2 expression was increased in rat carotid arteries after balloon injury (BI). Tissue lysates from uninjured (−) or balloon-injured (BI, +) carotid arteries with different days (d) were prepared for Western blotting analysis of Olfm2, SMC marker, and PCNA expression. G–J, Quantification of the protein expression shown in F by normalizing to the TUBA1 level. *, P < 0.05 vs. uninjured group (d 0) (n=3).
Olfm2 was required for PDGF-BB-induced SMC phenotypic modulation in vitro
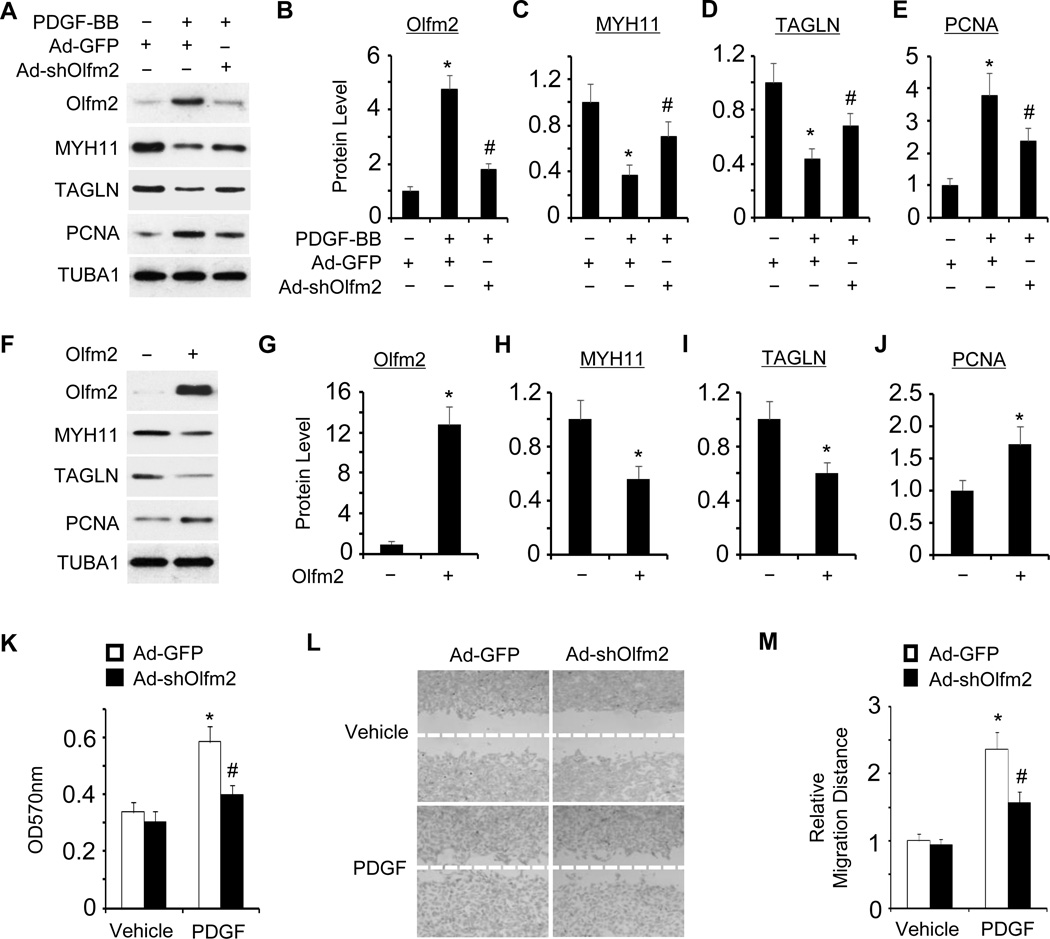
In order to determine whether or not Olfm2 plays a role in the modulation of SMC phenotype, we knocked down Olfm2 expression using an adenoviral vector expressing Olfm2 shRNA (Ad-shOlfm2) and examined the expression of SMC markers as well as PCNA in PDGF-BB-treated RASMCs. As shown in Figure 2A–2E, Knockdown of Olfm2 reversed the expression of PDGF-BB-suppressed SMC markers and inhibited the expression of PDGF-BB-induced PCNA, suggesting that Olfm2 was essential for PDGF-BB-induced SMC phenotypic modulation. On the other hand, the forced expression of Olfm2 suppressed the SMC marker expression and increased PCNA expression (Figure 2F–2J), suggesting that Olfm2 was able to mediate a synthetic SMC phenotype. Since phenotypically modulated SMCs exhibit higher growth rates and migratory activity compared to contractile SMCs,1 we examined whether Olfm2 has effects on SMC proliferation and migration. As shown in Figure 2K–2M, Olfm2 knockdown significantly inhibited PDGF-BB-induced SMC proliferation (Figure 2K) and migration (Figure 2L–2M). These results demonstrate that Olfm2 is a novel factor for SMC phenotypic modulation in vitro.
Figure 2. Olfm2 was essential for PDGF-BB-induced SMC phenotypic modulation.
A, Olfm2 knockdown reversed the effect of PDGF-BB on SMC marker and PCNA expression. RASMCs were transduced with Ad-GFP or Ad-shOlfm2 followed by vehicle (−) or PDGF-BB (+, 20 ng/ml) treatment for 24 h. Western blotting was performed to detect the expression of proteins as indicated. B–E, Quantification of the protein expression shown in A by normalizing to the TUBA1 level. *, P < 0.01 vs. Ad-GFP group with vehicle treatment (n=3). #, P < 0.01 vs. Ad-GFP group with PDGF-BB treatment (n=3). F, Forced Olfm2 expression promoted SMC phenotypic modulation. RASMCs were transfected with control or Olfm2 expression plasmid. Cell lysates were collected for Western blotting of the proteins indicated. G–J, Quantification of the protein expression shown in F by normalizing to the TUBA1 level. *, P < 0.01 vs. control group (n=3). K, Olfm2 knockdown diminished PDGF-BB-induced SMC proliferation. Ad-GFP- or Ad-shOlfm2-transduced RASMCs were seeded on 96-well plates and incubated with vehicle or PDGF-BB for 24 h. MTT assay was performed. *, P < 0.01 vs. Ad-GFP group with vehicle treatment. #, P < 0.01 vs. Ad-GFP group with PDGF-BB treatment (n=6). OD refers to the optical density normalized to the blank (medium only). L, Olfm2 knockdown diminished PDGF-BB-induced SMC migration. Ad-GFP- or Ad-shOlfm2-transduced RASMCs were seeded on 24-well plates containing a wound healing insert followed by incubation with vehicle or PDGF-BB for 24 h. Wound-healing assay was performed. M, Quantification of the relative migration distance shown in L. *, P < 0.01 vs. Ad-GFP group with vehicle treatment. #, P < 0.01 vs. Ad-GFP group with PDGF-BB treatment (n=3).
Olfm2 was essential for injury-induced SMC phenotypic modulation and vascular remodeling
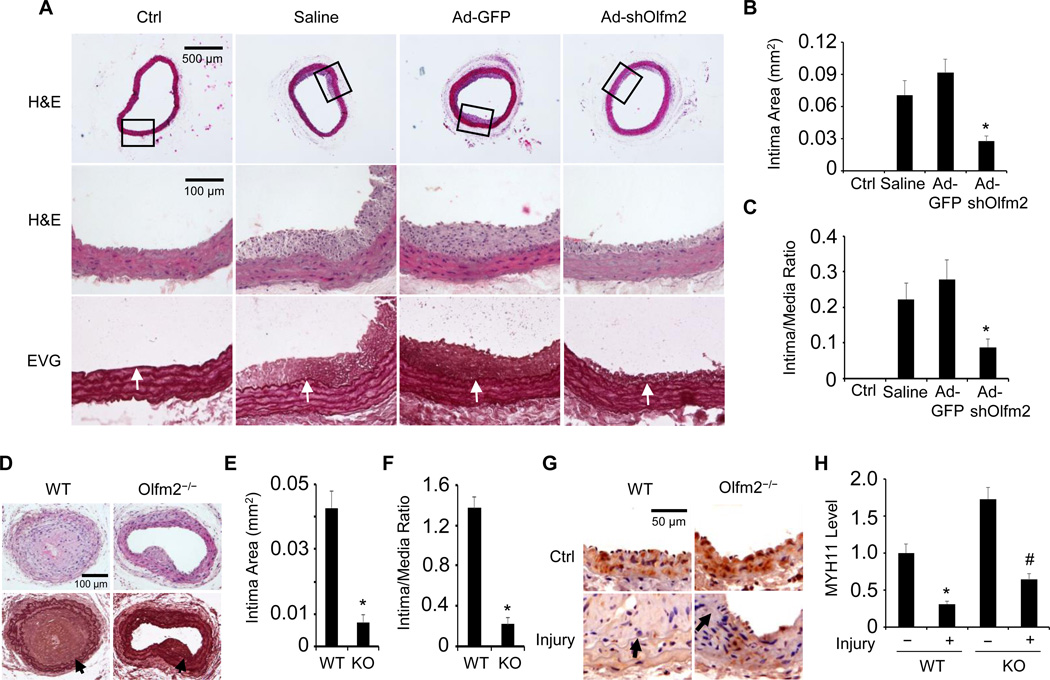
To investigate if Olfm2 is functionally important in vascular remodeling in vivo, we knocked down Olfm2 expression in arterial SMCs by incubating the endothelium-denuded rat carotid arteries with adenovirus expressing Olfm2 shRNA (Ad-shOlfm2) for 20 min immediately after the balloon injury. Olfm2 expression was dramatically induced in neointimal SMCs, However, its expression was markedly inhibited in Olfm2 shRNA-expressing vessels 14 days after the injury (Figure IC in the online-only Data Supplement). Importantly, exogenously introduced Ad-shOlfm2 significantly reduced neointima formation with a decrease in intima area by 68% and a reduction of intima/media area ratio by 64% (Figure 3A–3C). These results suggest an essential role of Olfm2 in balloon injury-induced vascular remodeling.
Figure 3. Olfm2 deficiency inhibited injury-induced neointima formation and SMC phenotype modulation.
A, Knockdown of Olfm2 inhibited injury-induced neointima formation. The balloon-injured rat carotid arteries were incubated with sterile saline solution, Ad-GFP and Ad-shOlfm2 as indicated. 14 days later, the artery samples were collected and stained with hematoxylin and eosin (H&E) or Elastica van Gieson solution (EVG). Arrows point to internal elastic lamina. B–C, Morphometric quantification of the intima area (C) and intima/media area ratio (D). *, P < 0.01 vs. saline- or Ad-GFP-treated arteries (n=5). D, Olfm2 knockout blocked wire injury-induced neointima formation. Carotid artery sections with 14 day wire injury were stained with H&E (upper panel) and Elastica van Gieson solution (lower panel). Arrows point to internal elastic lamina. E–F, Morphometric quantification of intima area and intima/media area ratio in the injured arteries of WT and Olfm2−/− (KO) mice. *, P < 0.01 vs. injured WT mouse arteries (n=5). G, Olfm2 deficiency preserved MYH11 expression in wire-injured arteries. Artery sections were immunostained for MYH11. MYH11 expression was visualized using DAB. Arrows point to internal elastic lamina. H, Quantification of MYH11 expression shown in G by normalizing to the staining intensity in the WT control (Ctrl) arteries. *, P < 0.01 vs. the Ctrl WT (−) arteries; #, P < 0.01 vs. the injured WT arteries (n=5).
Since adenoviral shRNA transduction may only suppress Olfm2 expression in medial SMCs that directly contact with the virus, we performed carotid artery wire injury in Olfm2 knockout (Olfm2−/−) mice to further confirm the role of Olfm2 in vascular remodeling. As shown in Figure IIA and IIB in the online-only Data Supplement, Olfm2 expression was completely blocked in Olfm2−/− mouse carotid artery. Consistent with the results obtained by using Olfm2 shRNA (Figure 3A–3C), Olfm2−/− significantly suppressed the injury-induced neointima formation (Figure 3D). Thus, the intima area and intima/media area ratio in Olfm2−/− arteries were reduced by 83% and 85%, respectively, compared with the WT control (Figure 3E and 3F). These data further demonstrate that Olfm2 is critically important for the injury-induced vascular remodeling.
In order to determine if Olfm2 regulates SMC phenotypic modulation in vivo following vascular injury, we examined the expression of SMC-specific contractile protein MYH11 in both WT and Olfm2−/− mouse carotid arteries with and without wire injury. As shown in Figure 3G and 3H, injury attenuated the MYH11 expression in both WT and Olfm2−/− mouse arteries. However, Olfm2−/− significantly preserved the MYH11 level in arterial SMCs as compared with the injured WT arteries (Figure 3G and 3H). Consistently, Olfm2−/− led to a marked decrease in the PCNA-positive cells in injured mouse arteries (Figure IIC and IID in the online-only Data Supplement), suggesting that Olfm2 is important for SMC proliferation in vivo. Together, these data indicate that Olfm2 plays an important role in injury-induced SMC phenotypic modulation in vivo.
Olfm2 inhibited Myocd binding to SRF
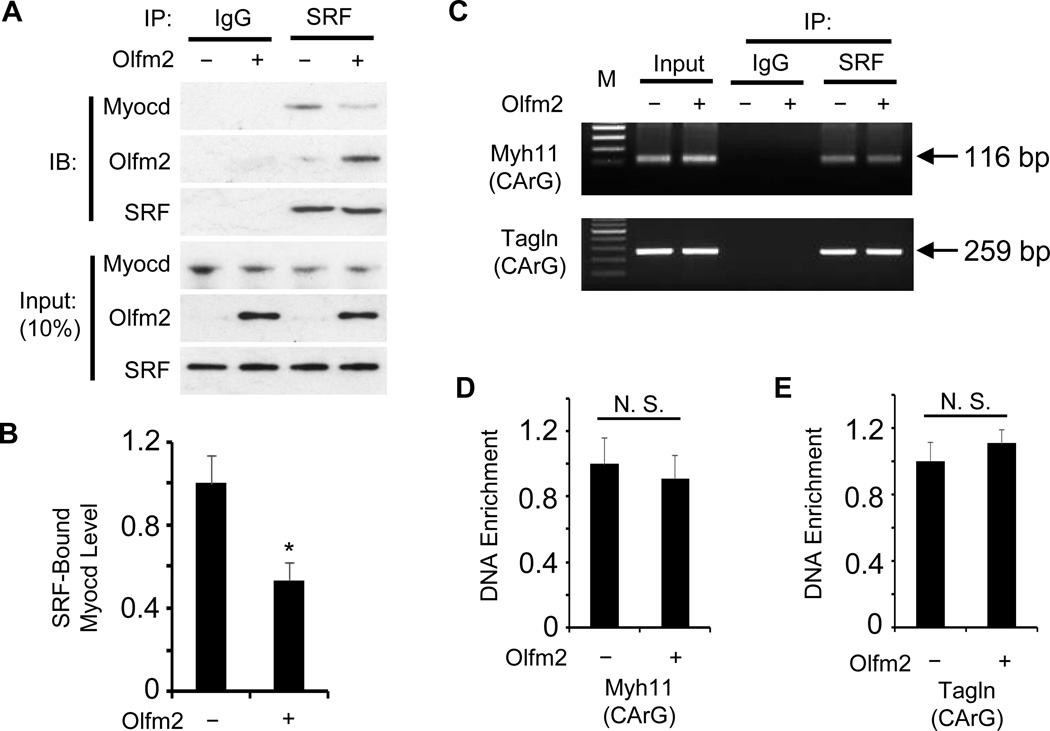
Myocd is a critical SRF coactivator regulating SMC contractile protein expression.34, 35 SMC phenotypic modulation is regulated by multiple mechanisms including suppression of Myocd and SRF expression, disruption of Myocd-SRF interaction, and interference with SRF binding to promoters of SMC-specific markers such as Myh11 and Tagln.1, 36 Since our previous study indicates that Olfm2 physically interacts with SRF during SMC differentiation,33 we tested if Olfm2 causes a disruption of SRF-Myocd interaction during SMC phenotypic modulation. As shown in Figure 4A and 4B, forced expression of Olfm2 inhibited the physical binding of Myocd to SRF by 54% in cultured SMCs. Olfm2 appeared not to affect SRF or Myocd expression (Figure 4A), suggesting that Olfm2 disrupted SRF-Myocd interaction not by altering SRF or Myocd expression. Moreover, there was no change in the binding affinity of SRF to Myh11 and Tagln promoters in Olfm2-expressing SMCs (Figure 4C–4E). These results indicate that the dissociation between SRF and Myocd is a major mechanism underlying Olfm2 function in the SMC phenotypic modulation.
Figure 4. Olfm2 inhibited SRF-Myocd interaction.
A, Forced Olfm2 expression diminished Myocd binding to SRF. Primary RASMCs were transfected with control (−) or Olfm2 expression plasmid (+) followed by Co-IP with IgG (blank control) or SRF antibody. The immunoprecipitated complex was immunoblotted (IB) with the antibodies as indicated. B, Quantification of SRF-bound Myocd by normalizing to the input Myocd level shown in A. *, P < 0.01 vs. control plasmid-transfected (−) group (n=3). C, Olfm2 had no effect on SRF binding to the promoters of Myh11 and Tagln. RASMCs were transfected with control (−) or Olfm2 expression plasmid (+) followed by ChIP assay with IgG (blank control) or SRF antibody, and semi-quantitative PCR was performed to amplify the immunoprecipitated chromatin fragment with primers flanking the binding site of SRF (CArG box) in Myh11 and Tagln promoters. D–E, Real-time quantitative PCR was performed to detect the binding of SRF to CArG box in Myh11 (D) and tagln (E) promoters by normalizing to the input DNA levels. N.S.: non-significant (n=3).
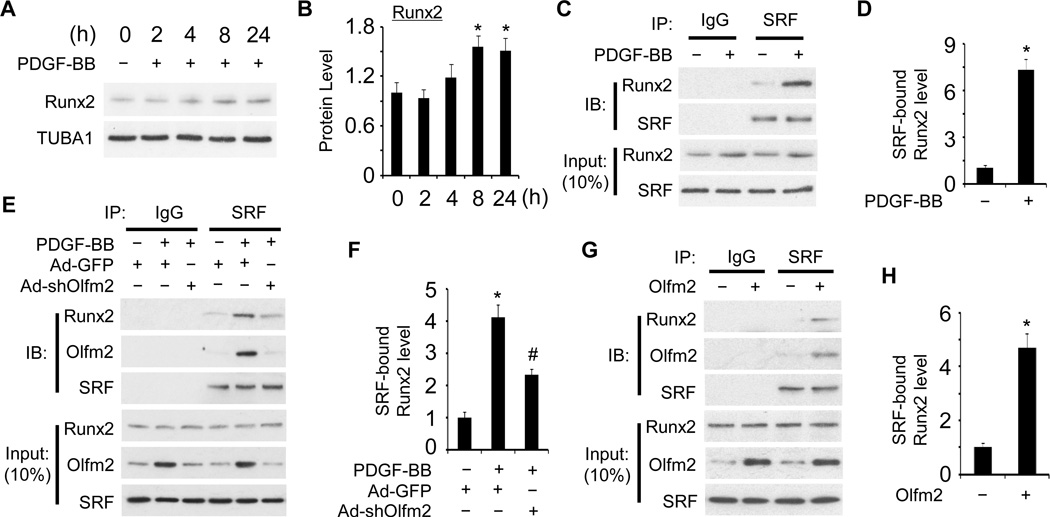
Olfm2 mediated Runx2 binding to SRF
It has been reported that Runx2 binds to SRF and competitively inhibits Myocd binding to SRF, and thus regulates SMC phenotypic modulation.25, 26 We therefore tested if Olfm2 is involved in the Runx2 activity in the SRF-Myocd interaction. Interestingly, PDGF-BB only induced a modest increase in Runx2 expression (1.5 fold) 24 h after the induction (Figure 5A and 5B). Similarly, a marginal, but not significant increase in Runx2 expression was observed in balloon-injured rat carotid arteries (Figure III in the online-only Data Supplement). These findings suggest that regulation of Runx2 expression may not be a major mechanism controlling SMC phenotypic modulation both in vitro and in vivo. In contrast, PDGF-BB stimulation resulted in a 7.2-fold increase in SRF-Runx2 binding without affecting SRF expression (Figure 5C and 5D), suggesting that other factors are involved in enhancing the SRF-Runx2 binding. Olfm2 appeared to be one of these factors. Knockdown of Olfm2 attenuated the PDGF-BB-induced SRF-Runx2 interaction (Figure 5E and 5F), indicating that Olfm2 was required for SRF-Runx2 interaction during SMC phenotypic modulation. Consistently, forced expression of Olfm2 promoted SRF-Runx2 interaction (Figure 5G and 5H). These results demonstrate that Olfm2 regulates SMC phenotypic modulation by facilitating SRF-Runx2 binding.
Figure 5. Olfm2 regulated SRF-Runx2 interaction.
A, PDGF-BB induced Runx2 expression. Serum-starved RASMCs were treated with vehicle (−) or PDGF-BB (+, 20 ng/ml) for the times indicated. Western blotting was performed to examine Runx2 expression. B, Quantification of Runx2 shown in A by normalizing to the TUBA1 level. *, P < 0.05 vs. vehicle-treated group (0 h) (n=3). C, PDGF-BB dramatically promoted SRF-Runx2 interaction. Serum-starved RASMCs were treated with vehicle (−) or PDGF-BB (+, 20 ng/ml) for 24 h followed by Co-IP with IgG or SRF antibody. The immunoprecipitated lysate was immunoblotted (IB) with the antibodies indicated. D, Quantification of SRF-bound Runx2 by normalizing to the input SRF level shown in C. *, P < 0.01 vs. vehicle-treated group (−) (n=3). E, Olfm2 was essential for PDGF-BB-induced SRF-Runx2 binding. RASMCs were transduced with Ad-GFP or Ad-shOlfm2 followed by vehicle (−) or PDGF-BB (+, 20 ng/ml) treatment. Co-IP and IB were performed with antibodies as indicated. F, Quantification of SRF-bound Runx2 by normalizing to the input SRF level shown in E. *, P < 0.01 vs. vehicle-treated group (−); #, P < 0.01 vs. Ad-GFP group with PDGF-BB treatment (n=3). G, Olfm2 mediated SRF-Runx2 binding. RASMCs were transfected with control (−) or Olfm2 expression plasmid (+) followed by Co-IP and IB with antibodies as indicated. H, Quantification of SRF-bound Runx2 by normalizing to the input SRF level shown in G. *, P < 0.01 vs. control plasmid-transfected (−) group (n=3).
In addition to Runx2, Kruppel-like factor 4 (KLF4) also interacts with SRF to mediate PDGF-BB-induced SMC phenotypic modulation.10, 37 Therefore, we tested if Olfm2 has effects on KLF4. As shown in Figure IVA–IVC in the online-only Data Supplement, KLF4 was expressed at a low level in SMCs and Olfm2 did not regulate KLF4 expression. Moreover, Olfm2 did not alter KLF4 binding to SRF in KLF4-expressing SMCs (Figure IVD and IVE in the online-only Data Supplement). Therefore, Olfm2 and KLF4 may regulate SMC phenotypic modulation independently. Together, these results indicate that Olfm2 couples Runx2, but not KLF4, to block SRF-Myocd interaction and thus modulate SMC phenotype.
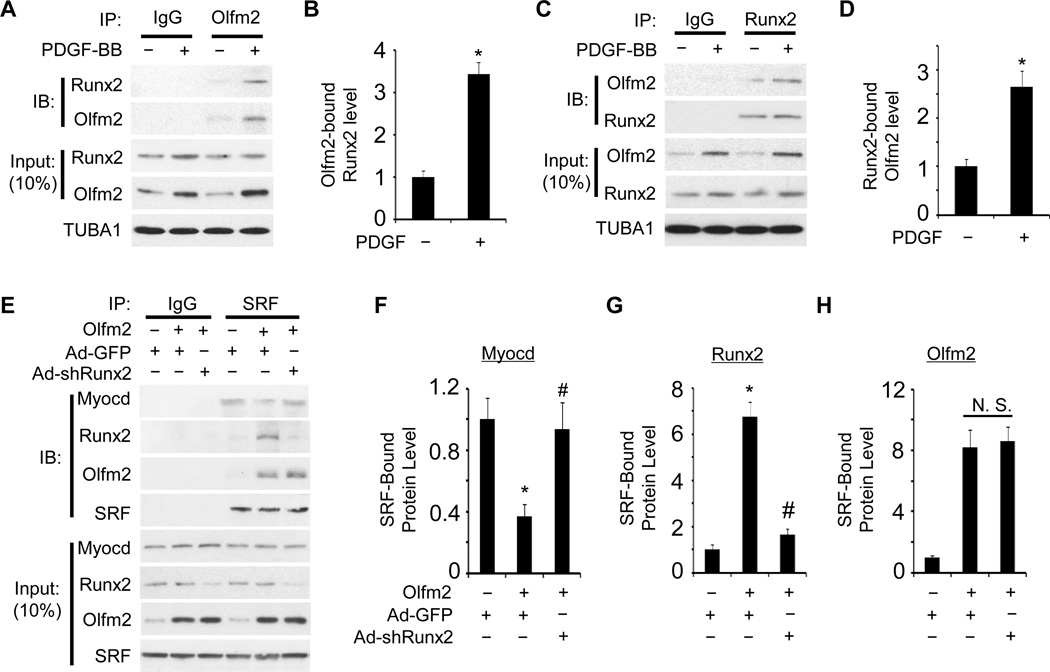
Runx2 physically interacted with Olfm2 and mediated its function in inhibiting SRF-Myocd binding
As Olfm2 mediated SRF-Runx2 interaction, we test if Olfm2 interacts with Runx2. Co-IP assays showed that Olfm2 physically interacted with Runx2 in SMCs (Figure 6A and 6C) and PDGF-BB significantly enhanced their interaction (Figure 6A–6D), which was likely due to the increased expression of Olfm2 by PDGF-BB. These results suggest that Olfm2 may mediate Runx2-SRF binding through its binding with both Runx2 and SRF.
Figure 6. Olfm2 physically interacted with Runx2 to inhibit Myocd-SRF binding.
A and C, Olfm2 bound to Runx2. RASMCs were treated with vehicle (−) or PDGF-BB (+, 20 ng/ml) for 24 h followed by Co-IP with Olfm2 antibody (A) and by the reverse Co-IP with Runx2 antibody (C). The immunoprecipitated lysates were immunoblotted (IB) with the antibodies as indicated. B and D, Quantification of Olfm2-bound Runx2 (B) and Runx2-bound Olfm2 (D) by normalizing to the corresponding TUBA1 level shown in A and C, respectively *, P < 0.01 vs. corresponding vehicle-treated group (−) (n=3). E, Knockdown of Runx2 restored Olfm2-inhibited SRF-Myocd binding. Ad-GFP- or Ad-shRunx2-transduced RASMCs were transfected with Olfm2 expression plasmid followed by Co-IP with IgG or SRF antibody. The immunoprecipitated lysates were immunoblotted (IB) with the antibodies as indicated. F–H, Quantification of SRF-bound proteins as indicated in E by normalizing to the input SRF level. *, P < 0.01 vs. Ad-GFP group without Olfm2; #, P < 0.01 vs. Ad-GFP group with Olfm2 transfection (n=3).
Since both Olfm2 and Runx2 can disrupt SRF-Myocd interaction, we sought to determine if Olfm2-mediated dissociation of Myocd from SRF was dependent on Runx2. As shown in Figure 6E–6H, knockdown of Runx2 by its specific shRNA restored the Myocd-SRF interaction that was inhibited by Olfm2, indicating that Runx2 played an essential role in mediating the Olfm2 disruption of SRF-Myocd binding during SMC phenotypic modulation.
To verify if the effect of Olfm2 on Myocd-SRF interaction via Runx2 actually impacts SMC marker gene transcription, we tested if endogenous Olfm2 has effects on Runx2 and Myocd association with the CArG box in endogenous SMMHC promoter in PDGF-BB-treated RASMCs. ChIP analyses showed that shRNA knockdown of Olfm2 inhibited the Runx2 while promoted the Myocd association with the CArG box (Figure V in the online-only Data Supplement). These data further support that Olfm2 facilitates Runx2 binding to SRF that directly interacts with CArG box, which competitively inhibits Myocd interaction with SRF.
Discussion
SMC phenotypic modulation contributes to the pathogenesis of a variety of cardiovascular disorders including restenosis following angioplasty, atherosclerosis and hypertension.1 However, the precise molecular mechanisms underlying this process are not fully understood. In this study, we have identified Olfm2 as a novel regulator for SMC phenotypic modulation both in vitro and in vivo. Olfm2 expression is significantly increased both in PDGF-BB-treated SMCs and in injured arteries (Figure 1). Importantly, Olfm2 deficiency restores the suppression of SMC marker proteins induced by PDGF-BB or vascular injury (Figure 2 and Figure 3), indicating that Olfm2 promotes SMC phenotypic modulation both in vitro and in vivo. Interestingly, the uninjured carotid arteries in Olfm2−/− mice exhibit higher expression of SMC-specific marker proteins than those in WT mice (Figure 3), suggesting that a low level of Olfm2 is required for preventing an excessive expression of contractile proteins in quiescent SMCs although Olfm2 is needed for SMC differentiation from the progenitors. The suppressive role of Olfm2 in SMC marker gene expression is further supported by the facts that ectopic expression of Olfm2 inhibits MYH11 and TAGLN expression in cultured SMCs (Figure 2). Consistent with these findings, Olfm2 deficiency also suppresses PDGF-BB-induced SMC proliferation and migration in vitro as well as injury-induced neointima formation/vascular remodeling in vivo (Figure 2 and Figure 3).
Olfm2 appears to regulate SMC phenotypic modulation by promoting Runx2 binding to SRF. Runx2 has been shown to occupy the Myocd docking site on SRF, and thus competitively inhibiting the association of Myocd with SRF, which subsequently leads to the repression of SMC marker gene transcription.25 Olfm2 physically interacts with both Runx2 and SRF (Figure 5 and Figure 6), which renders Olfm2 capable of carrying Runx2 toward SRF, mediating Runx2/SRF interaction, and consequently disrupting SRF/Myocd association. Indeed, Knockdown of Olfm2 blocks PDGF-BB-induced Runx2/SRF interaction (Figure 5). It seems that Runx2 binds to SRF with a relatively low affinity, and Olfm2 serves as a “glue” to secure Runx2 adhesion to SRF in order to effectively compete with Myocd for the docking site on SRF. Equally, Runx2 is also essential for mediating Olfm2 function in SMC phenotypic modulation because knockdown of Runx2, without affecting Olfm2 expression, rescues the SRF-Myocd binding impaired by Olfm2 (Figure 6). It appears that Olfm2 and Runx2 work coordinately in PDGF-BB-induced SMC phenotypic modulation, i.e., Olfm2 provides physical support for Runx2 association with SRF, whereas Runx2 diminishes the availability of docking sites for Myocd and therefore prevents Myocd from binding to SRF.
Facilitating Runx2 binding to SRF and consequently disrupting Myocd-SRF binding appears to be the major mechanism underlying Olfm2 function in SMC phenotypic modulation because Olfm2 neither modifies Myocd and SRF expression nor interferes with SRF/CArG box association (Figure 4). Since it is known that the dissociation of myocardin from SRF normally results in a reduction in SRF-CArG binding, the absence of this effect by Olfm2 must be due to the binding of Runx2 to the SRF. Indeed, Runx2 has been shown to increase SRF binding to CArG boxes in both Myh11 and Tagln promoters.25 In addition, although KLF4 plays a critical role in SMC phenotypic modulation, Olfm2 appears to work in a KLF4-independent manner (Online Figure IV). Unlike the function of Olfm2 in disrupting SRF/Myocd interaction, KLF4 has been shown to reduce Myocd expression, SRF binding to CArG box as well as SRF/Myocd binding,10, 38, 39 indicating that Olfm2 and KLF4 regulate SMC phenotypic modulation via distinct mechanisms.
Our previous study revealed that Olfm2 regulated TGF-β-induced SMC differentiation from mesenchymal stem cells.33 It is paradoxical that Olfm2 plays dual roles in both SMC differentiation and phenotypic modulation. The possible explanation is that the roles of Olfm2 depend on the cellular context: In SMC precursors like mesenchymal stem cells, the expression of Runx2 is undetectable, and Runx2 is not induced by TGF-β (data not shown). As a result, Olfm2 does not impair the SRF/Myocd interaction. Instead, it promotes the dissociation of SRF from HES-related repressor protein 1 (HERP1), a transcriptional repressor for SMC gene, to enhance SRF-CArG box interaction.33 However, Runx2 is substantially present in mature SMCs (Figure 5 and Online Figure III). The binding of Olfm2 with Runx2 directs Olfm2 function toward modulating SMC phenotype by enhancing Runx2-SRF interaction, thus disrupting Myocd-SRF binding. Interestingly, although TGF-β induces strong expression of Olfm2 and SMC markers in mesenchymal progenitor cells, it fails to significantly induce Olfm2 or MYH11 expression in mature rat aortic SMC (Figure VI in the online-only Data Supplement). These paradoxical results suggest that cellular environments could influence the protein function dramatically. Olfm2 may promote or suppress SMC marker gene expression depending on its cooperation with either HERP1 during normal SMC differentiation or Runx2 in the pathological vascular remodeling process.
Taken together, our present study demonstrates that Olfm2 is a novel regulator for SMC phenotypic modulation both in vitro and in vivo. Therefore, targeting Olfm2 may hinder injury-induced vascular remodeling and thus improve vascular repair.
Supplementary Material
Highlights.
Olfm2 is up-regulated in phenotypically modulated smooth muscle.
Olfm2 knockdown inhibits PDGF-BB-induced smooth muscle phenotypic modulation.
Olfm2 knockdown suppresses neointima formation in balloon-injured rat carotid arteries.
Olfm2 knockout blocks vascular remodeling in wire-injured mouse carotid arteries.
Olfm2 mediates Runx2-SRF interaction, replacing the myocardin binding to SRF.
Acknowledgments
We are grateful to Dr. Afia Sultana at the National Eye Institute, NIH, for providing Olfm2−/− mice.
Sources of funding
This work was supported by grants from National Institutes of Health (HL123302, HL119053 and HL135854).
Non-standard Abbreviations and Acronyms
- Ad
adenoviral vector
- GFP
green florescent protein
- HERP1
HES-related repressor protein 1
- KLF4
Kruppel-like factor 4
- Myocd
myocardin
- Olfm2
Olfactomedin 2
- PCNA
proliferating cell nuclear antigen
- PDGF-BB
platelet-derived growth factor-BB
- Runx2
runt-related transcription factor 2
- SMC
smooth muscle cell
- RASMC
Rat smooth muscle cell
- SMMHC
smooth muscle myosin heavy chain
- SM22α
smooth muscle 22α
- SRF
serum response factor
- TGF-β
transforming growth factor-β
Footnotes
Disclosures: None
References
- 1.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 2.Dandre F, Owens GK. Platelet-derived growth factor-bb and ets-1 transcription factor negatively regulate transcription of multiple smooth muscle cell differentiation marker genes. Am J Physiol Heart Circ Physiol. 2004;286:H2042–H2051. doi: 10.1152/ajpheart.00625.2003. [DOI] [PubMed] [Google Scholar]
- 3.Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- 4.Raines EW. Pdgf and cardiovascular disease. Cytokine Growth Factor Rev. 2004;15:237–254. doi: 10.1016/j.cytogfr.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Banai S, Wolf Y, Golomb G, Pearle A, Waltenberger J, Fishbein I, Schneider A, Gazit A, Perez L, Huber R, Lazarovichi G, Rabinovich L, Levitzki A, Gertz SD. Pdgf-receptor tyrosine kinase blocker ag1295 selectively attenuates smooth muscle cell growth in vitro and reduces neointimal formation after balloon angioplasty in swine. Circulation. 1998;97:1960–1969. doi: 10.1161/01.cir.97.19.1960. [DOI] [PubMed] [Google Scholar]
- 6.Ferns GA, Raines EW, Sprugel KH, Motani AS, Reidy MA, Ross R. Inhibition of neointimal smooth muscle accumulation after angioplasty by an antibody to pdgf. Science. 1991;253:1129–1132. doi: 10.1126/science.1653454. [DOI] [PubMed] [Google Scholar]
- 7.Libby P, Warner SJ, Salomon RN, Birinyi LK. Production of platelet-derived growth factor-like mitogen by smooth-muscle cells from human atheroma. N Engl J Med. 1988;318:1493–1498. doi: 10.1056/NEJM198806093182303. [DOI] [PubMed] [Google Scholar]
- 8.Uchida K, Sasahara M, Morigami N, Hazama F, Kinoshita M. Expression of platelet-derived growth factor b-chain in neointimal smooth muscle cells of balloon injured rabbit femoral arteries. Atherosclerosis. 1996;124:9–23. doi: 10.1016/0021-9150(95)05742-0. [DOI] [PubMed] [Google Scholar]
- 9.Leppanen O, Janjic N, Carlsson MA, Pietras K, Levin M, Vargeese C, Green LS, Bergqvist D, Ostman A, Heldin CH. Intimal hyperplasia recurs after removal of pdgf-ab and -bb inhibition in the rat carotid artery injury model. Arteriosclerosis, thrombosis, and vascular biology. 2000;20:E89–E95. doi: 10.1161/01.atv.20.11.e89. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. The Journal of biological chemistry. 2005;280:9719–9727. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida T, Gan Q, Shang Y, Owens GK. Platelet-derived growth factor-bb represses smooth muscle cell marker genes via changes in binding of mkl factors and histone deacetylases to their promoters. American journal of physiology. Cell physiology. 2007;292:C886–C895. doi: 10.1152/ajpcell.00449.2006. [DOI] [PubMed] [Google Scholar]
- 12.Mack CP. Signaling mechanisms that regulate smooth muscle cell differentiation. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:1495–1505. doi: 10.1161/ATVBAHA.110.221135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawai-Kowase K, Owens GK. Multiple repressor pathways contribute to phenotypic switching of vascular smooth muscle cells. American journal of physiology. Cell physiology. 2007;292:C59–C69. doi: 10.1152/ajpcell.00394.2006. [DOI] [PubMed] [Google Scholar]
- 14.Miano JM. Serum response factor: Toggling between disparate programs of gene expression. J Mol Cell Cardiol. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 15.Hautmann MB, Madsen CS, Owens GK. A transforming growth factor beta (tgfbeta) control element drives tgfbeta-induced stimulation of smooth muscle alpha-actin gene expression in concert with two carg elements. The Journal of biological chemistry. 1997;272:10948–10956. doi: 10.1074/jbc.272.16.10948. [DOI] [PubMed] [Google Scholar]
- 16.Hirschi KK, Lai L, Belaguli NS, Dean DA, Schwartz RJ, Zimmer WE. Transforming growth factor-beta induction of smooth muscle cell phenotpye requires transcriptional and post-transcriptional control of serum response factor. The Journal of biological chemistry. 2002;277:6287–6295. doi: 10.1074/jbc.M106649200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 18.Cao D, Wang Z, Zhang CL, Oh J, Xing W, Li S, Richardson JA, Wang DZ, Olson EN. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Molecular and cellular biology. 2005;25:364–376. doi: 10.1128/MCB.25.1.364-376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: Versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 20.Xie WB, Li Z, Miano JM, Long X, Chen SY. Smad3-mediated myocardin silencing: A novel mechanism governing the initiation of smooth muscle differentiation. The Journal of biological chemistry. 2011;286:15050–15057. doi: 10.1074/jbc.M110.202747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerrero F, Herencia C, Almaden Y, Martinez-Moreno JM, Montes de Oca A, Rodriguez-Ortiz ME, Diaz-Tocados JM, Canalejo A, Florio M, Lopez I, Richards WG, Rodriguez M, Aguilera-Tejero E, Munoz-Castaneda JR. Tgf-beta prevents phosphate-induced osteogenesis through inhibition of bmp and wnt/beta-catenin pathways. PloS one. 2014;9:e89179. doi: 10.1371/journal.pone.0089179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer D, Chang D, Marx J, Wei L, Olson EN, Parmacek MS, Balasubramanyam A, Schwartz RJ. Serum response factor mads box serine-162 phosphorylation switches proliferation and myogenic gene programs. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4516–4521. doi: 10.1073/pnas.0505338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blaker AL, Taylor JM, Mack CP. Pka-dependent phosphorylation of serum response factor inhibits smooth muscle-specific gene expression. Arteriosclerosis, thrombosis, and vascular biology. 2009;29:2153–2160. doi: 10.1161/ATVBAHA.109.197285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taurin S, Sandbo N, Yau DM, Sethakorn N, Kach J, Dulin NO. Phosphorylation of myocardin by extracellular signal-regulated kinase. The Journal of biological chemistry. 2009;284:33789–33794. doi: 10.1074/jbc.M109.048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka T, Sato H, Doi H, Yoshida CA, Shimizu T, Matsui H, Yamazaki M, Akiyama H, Kawai-Kowase K, Iso T, Komori T, Arai M, Kurabayashi M. Runx2 represses myocardin-mediated differentiation and facilitates osteogenic conversion of vascular smooth muscle cells. Molecular and cellular biology. 2008;28:1147–1160. doi: 10.1128/MCB.01771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohyama Y, Tanaka T, Shimizu T, Matsui H, Sato H, Koitabashi N, Doi H, Iso T, Arai M, Kurabayashi M. Runx2/smad3 complex negatively regulates tgf-beta-induced connective tissue growth factor gene expression in vascular smooth muscle cells. J Atheroscler Thromb. 2012;19:23–35. doi: 10.5551/jat.9753. [DOI] [PubMed] [Google Scholar]
- 27.Zeng LC, Han ZG, Ma WJ. Elucidation of subfamily segregation and intramolecular coevolution of the olfactomedin-like proteins by comprehensive phylogenetic analysis and gene expression pattern assessment. FEBS letters. 2005;579:5443–5453. doi: 10.1016/j.febslet.2005.08.064. [DOI] [PubMed] [Google Scholar]
- 28.Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: Possible mechanisms of action and functions in normal development and pathology. Molecular neurobiology. 2009;40:122–138. doi: 10.1007/s12035-009-8076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anholt RR. Olfactomedin proteins: Central players in development and disease. Frontiers in cell and developmental biology. 2014;2:6. doi: 10.3389/fcell.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JA, Anholt RR, Cole GJ. Olfactomedin-2 mediates development of the anterior central nervous system and head structures in zebrafish. Mech Dev. 2008;125:167–181. doi: 10.1016/j.mod.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sultana A, Nakaya N, Senatorov VV, Tomarev SI. Olfactomedin 2: Expression in the eye and interaction with other olfactomedin domain-containing proteins. Investigative ophthalmology & visual science. 2011;52:2584–2592. doi: 10.1167/iovs.10-6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sultana A, Nakaya N, Dong L, Abu-Asab M, Qian H, Tomarev SI. Deletion of olfactomedin 2 induces changes in the ampa receptor complex and impairs visual, olfactory, and motor functions in mice. Experimental neurology. 2014;261:802–811. doi: 10.1016/j.expneurol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi N, Guo X, Chen SY. Olfactomedin 2, a novel regulator for transforming growth factor-beta-induced smooth muscle differentiation of human embryonic stem cell-derived mesenchymal cells. Mol Biol Cell. 2014;25:4106–4114. doi: 10.1091/mbc.E14-08-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: A component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 35.Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, Lu MM, Owens GK, Parmacek MS. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Molecular and cellular biology. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang J, Parmacek MS. Modulation of smooth muscle cell phenotype: The other side of the story. Circulation research. 2012;111:659–661. doi: 10.1161/CIRCRESAHA.112.277368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo X, Shi N, Cui XB, Wang JN, Fukui Y, Chen SY. Dedicator of cytokinesis 2, a novel regulator for smooth muscle phenotypic modulation and vascular remodeling. Circulation research. 2015;116:e71–e80. doi: 10.1161/CIRCRESAHA.116.305863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of srf binding to carg box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng B, Han M, Wen JK. Role of kruppel-like factor 4 in phenotypic switching and proliferation of vascular smooth muscle cells. IUBMB Life. 2010;62:132–139. doi: 10.1002/iub.298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.