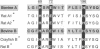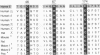Abstract
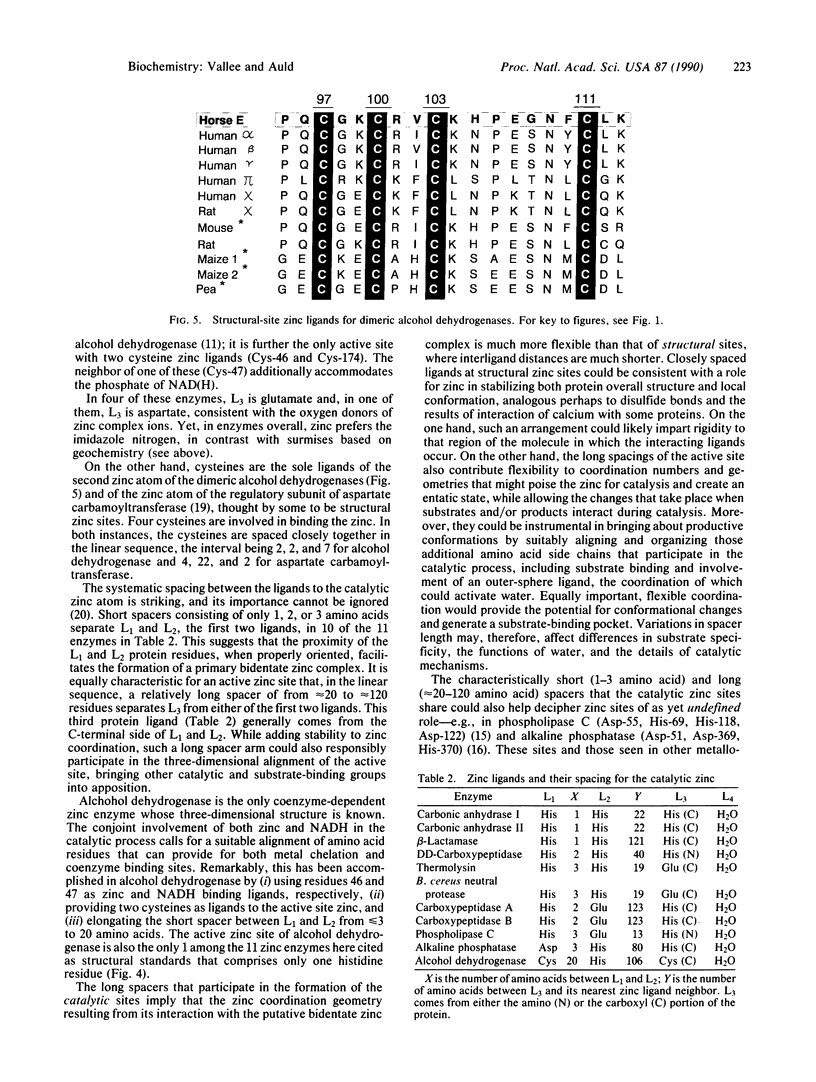
The x-ray crystallographic structures of 12 zinc enzymes have been chosen as standards of reference to identify the ligands to the catalytic and structural zinc atoms of other members of their respective enzyme families. Universally, H2O is a ligand and critical component of the catalytically active zinc sites. In addition, three protein side chains bind to the catalytic zinc atom, whereas four protein ligands bind to the structural zinc atom. The geometry and coordination number of zinc can vary greatly to accommodate particular ligands. Zinc forms complexes with nitrogen and oxygen just as readily as with sulfur, and this is reflected in catalytic zinc sites having a binding frequency of His much greater than Glu greater than Asp = Cys, three of which bind to the metal atom. The systematic spacing between the ligands is striking. For all catalytic zinc sites except the coenzyme-dependent alcohol dehydrogenase, the first two ligands are separated by a "short-spacer" consisting of 1 to 3 amino acids. These ligands are separated from the third ligand by a "long spacer" of approximately 20 to approximately 120 amino acids. The spacer enables formation of a primary bidentate zinc complex, whereas the long spacer contributes flexibility to the coordination sphere, which can poise the zinc for catalysis as well as bring other catalytic and substrate binding groups into apposition with the active site. The H2O is activated by ionization, polarization, or poised for displacement. Collectively, the data imply that the preferred mechanistic pathway for activating the water--e.g., zinc hydroxide or Lewis acid catalysis--will be determined by the identity of the other three ligands and their spacing.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dideberg O., Charlier P., Dive G., Joris B., Frère J. M., Ghuysen J. M. Structure of a Zn2+-containing D-alanyl-D-alanine-cleaving carboxypeptidase at 2.5 A resolution. Nature. 1982 Sep 30;299(5882):469–470. doi: 10.1038/299469a0. [DOI] [PubMed] [Google Scholar]
- Holmquist B., Vallee B. L. Metal substitutions and inhibition of thermolysin: spectra of the cobalt enzyme. J Biol Chem. 1974 Jul 25;249(14):4601–4607. [PubMed] [Google Scholar]
- Honzatko R. B., Crawford J. L., Monaco H. L., Ladner J. E., Ewards B. F., Evans D. R., Warren S. G., Wiley D. C., Ladner R. C., Lipscomb W. N. Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1982 Sep 15;160(2):219–263. doi: 10.1016/0022-2836(82)90175-9. [DOI] [PubMed] [Google Scholar]
- Hough E., Hansen L. K., Birknes B., Jynge K., Hansen S., Hordvik A., Little C., Dodson E., Derewenda Z. High-resolution (1.5 A) crystal structure of phospholipase C from Bacillus cereus. Nature. 1989 Mar 23;338(6213):357–360. doi: 10.1038/338357a0. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., Hög J. O., von Bahr-Lindström H., Vallee B. L. Mammalian alcohol dehydrogenases of separate classes: intermediates between different enzymes and intraclass isozymes. Proc Natl Acad Sci U S A. 1987 May;84(9):2580–2584. doi: 10.1073/pnas.84.9.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K. K., Notstrand B., Fridborg K., Lövgren S., Ohlsson A., Petef M. Crystal structure of human erythrocyte carbonic anhydrase B. Three-dimensional structure at a nominal 2.2-A resolution. Proc Natl Acad Sci U S A. 1975 Jan;72(1):51–55. doi: 10.1073/pnas.72.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljas A., Kannan K. K., Bergstén P. C., Waara I., Fridborg K., Strandberg B., Carlbom U., Järup L., Lövgren S., Petef M. Crystal structure of human carbonic anhydrase C. Nat New Biol. 1972 Feb 2;235(57):131–137. doi: 10.1038/newbio235131a0. [DOI] [PubMed] [Google Scholar]
- Messerschmidt A., Rossi A., Ladenstein R., Huber R., Bolognesi M., Gatti G., Marchesini A., Petruzzelli R., Finazzi-Agró A. X-ray crystal structure of the blue oxidase ascorbate oxidase from zucchini. Analysis of the polypeptide fold and a model of the copper sites and ligands. J Mol Biol. 1989 Apr 5;206(3):513–529. doi: 10.1016/0022-2836(89)90498-1. [DOI] [PubMed] [Google Scholar]
- Pauptit R. A., Karlsson R., Picot D., Jenkins J. A., Niklaus-Reimer A. S., Jansonius J. N. Crystal structure of neutral protease from Bacillus cereus refined at 3.0 A resolution and comparison with the homologous but more thermostable enzyme thermolysin. J Mol Biol. 1988 Feb 5;199(3):525–537. doi: 10.1016/0022-2836(88)90623-7. [DOI] [PubMed] [Google Scholar]
- Quiocho F. A., Lipscomb W. N. Carboxypeptidase A: a protein and an enzyme. Adv Protein Chem. 1971;25:1–78. doi: 10.1016/s0065-3233(08)60278-8. [DOI] [PubMed] [Google Scholar]
- Robbins A. H., Stout C. D. The structure of aconitase. Proteins. 1989;5(4):289–312. doi: 10.1002/prot.340050406. [DOI] [PubMed] [Google Scholar]
- Schmid M. F., Herriott J. R. Structure of carboxypeptidase B at 2-8 A resolution. J Mol Biol. 1976 May 5;103(1):175–190. doi: 10.1016/0022-2836(76)90058-9. [DOI] [PubMed] [Google Scholar]
- Sutton B. J., Artymiuk P. J., Cordero-Borboa A. E., Little C., Phillips D. C., Waley S. G. An X-ray-crystallographic study of beta-lactamase II from Bacillus cereus at 0.35 nm resolution. Biochem J. 1987 Nov 15;248(1):181–188. doi: 10.1042/bj2480181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee B. L., Auld D. S. Short and long spacer sequences and other structural features of zinc binding sites in zinc enzymes. FEBS Lett. 1989 Oct 23;257(1):138–140. doi: 10.1016/0014-5793(89)81805-8. [DOI] [PubMed] [Google Scholar]
- Wyckoff H. W., Handschumacher M., Murthy H. M., Sowadski J. M. The three dimensional structure of alkaline phosphatase from E. coli. Adv Enzymol Relat Areas Mol Biol. 1983;55:453–480. doi: 10.1002/9780470123010.ch6. [DOI] [PubMed] [Google Scholar]