Summary
We identified interactions between the conserved bacterial replication initiator and transcription factor DnaA and the nucleoid-associated protein Rok of Bacillus subtilis. DnaA binds directly to clusters of DnaA boxes at the origin of replication and elsewhere, including the promoters of several DnaA-regulated genes. Rok, an analog of H-NS from gamma-proteobacteria that affects chromosome architecture and Lsr2 from Mycobacteria, binds A+T-rich sequences throughout the genome and represses expression of many genes. Using crosslinking and immunoprecipitation followed by deep sequencing (ChIP-seq), we found that DnaA was associated with eight previously identified regions containing clusters of DnaA boxes, plus 36 additional regions that were also bound by Rok. Association of DnaA with these additional regions appeared to be indirect as it was dependent on Rok and independent of the DNA binding domain of DnaA. Gene expression and mutant analyses support a model in which DnaA and Rok cooperate to repress transcription of yxaJ, the yybNM operon, and the sunA-bdbB operon. Our results indicate that DnaA modulates the activity of Rok. We postulate that this interaction might affect nucleoid architecture. Furthermore, DnaA might interact similarly with Rok analogues in other organisms.
Keywords: DnaA, Rok, nucleoid-associated protein, DNA replication, transcription, Bacillus subtilis, ChIP-seq
Abbreviated summary
The bacterial replication initiator and transcription factor DnaA associates with specific chromosomal regions by binding directly to recognition sequences in the DNA. We found that in Bacillus subtilis DnaA is also associated with many chromosomal regions by associating with the nucleoide associated protein Rok (an H-NS analogue). DnaA stimulates the ability of Rok to bind DNA and together, these two proteins appear to modulate expression of several genes.
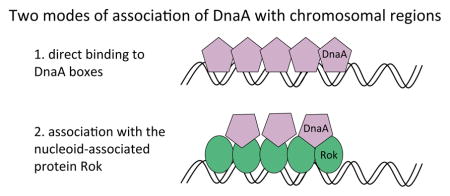
Introduction
DnaA is the conserved replication initiation protein and a transcription factor in bacteria (Messer & Weigel, 1997; Messer, 2002; Mott & Berger, 2007; Katayama et al., 2010; Leonard & Grimwade, 2010; Leonard & Grimwade, 2011). DnaA is a AAA+ ATPase that binds ATP or ADP, and the ATP-bound form is active for replication initiation (e.g., Fuller et al., 1984; Funnell et al., 1987; Sekimizu et al., 1987; Crooke et al., 1993; Kurokawa et al., 1999; Speck et al., 1999; Nishida et al., 2002; McGarry et al., 2004; Clarey et al., 2006; Erzberger et al., 2006; Duderstadt et al., 2010; Duderstadt et al., 2011). The N-terminal and AAA+ domains of DnaA contribute to oligomerization (Messer, 2002; Erzberger et al., 2006; Kawakami & Katayama, 2010; Leonard & Grimwade, 2010) and the C-terminal domain is necessary and sufficient for DNA binding (Roth & Messer, 1995; Krause et al., 1997).
DnaA binds to a 9-bp motif, the DnaA box, which occurs multiple times in the origin of replication, and elsewhere around the chromosome. In Bacillus subtilis, DnaA binds in the promoter regions of several genes and appears to directly regulate transcription, activating some genes and repressing others (Burkholder et al., 2001; Goranov et al., 2005; Ishikawa et al., 2007; Cho et al., 2008; Breier & Grossman, 2009; Veening et al., 2009; Hoover et al., 2010; Smits et al., 2011). Binding of DnaA to clusters of DnaA boxes throughout the genome might also regulate the cellular activity of DnaA and influence replication initiation (Okumura et al., 2012). DnaA is the target of multiple regulatory systems that modulate replication initiation during normal growth and in response to various cellular conditions (Mott & Berger, 2007; Katayama et al., 2010; Leonard & Grimwade, 2011; Skarstad & Katayama, 2013). Whereas DnaA is nearly ubiquitous among bacteria, the regulatory systems that control it are diverse (Katayama et al., 2010; Jonas, 2014).
We recently identified four promoters (between the genes: ywiB-sboA, yuzB-yutJ, yjcM-yjcN, and icsS-braB) that bind DnaA in vivo, but do not bind purified DnaA in vitro (Smith & Grossman, 2015). This finding indicated that additional proteins are likely to mediate DnaA binding to these regions, and potentially other regions, in vivo. The nucleoid-associated protein Rok is required for DnaA binding to these regions in vivo (Smith & Grossman, 2015).
Rok was originally identified as a repressor of comK and a negative regulator of competence development in B. subtilis (Hoa et al., 2002). Rok also regulates expression of other genes (Albano et al., 2005; Kovacs & Kuipers, 2011; Marciniak et al., 2012) and binds directly to several promoter regions in vivo and in vitro (Albano et al., 2005; Smits & Grossman, 2010). In contrast to DnaA, Rok is not known to have a well-defined binding site (Albano et al., 2005; Smits & Grossman, 2010). Rok is a nucleoid-associated protein that binds A+T-rich regions throughout the chromosome and has a role in silencing some regions of horizontally acquired DNA (Smits & Grossman, 2010). In terms of its A+T-binding preference and its role in silencing horizontally acquired DNA, Rok is analogous, but not homologous, to the nucleoid-associated proteins H-NS from E. coli and other gamma-proteobacteria that affect chromosome architecture and gene expression (Navarre et al., 2006; Dorman, 2007; Dorman, 2010) and Lsr2 from Mycobacteria (Gordon et al., 2008; Gordon et al., 2010).
In this work, we used chromatin immunoprecipitation and deep sequencing (ChIP-seq) to define the genomic regions associated with DnaA in vivo, and to determine the extent to which association of DnaA with various chromosomal regions might be mediated by other proteins, including Rok. We found that, in addition to associating with previously identified chromosomal regions containing clusters of DnaA boxes, DnaA also associated with many chromosomal regions known to be bound by the nucleoid-associated protein Rok (Albano et al., 2005; Smits & Grossman, 2010). Unlike the direct binding of DnaA to the 9-bp DnaA box binding sites, we found that DnaA binds indirectly to regions of the chromosome that bind Rok. This indirect binding appears to be due to a direct interaction between DnaA and Rok and does not require the DNA binding domain of DnaA. We also identified several genes that are regulated by both DnaA and Rok. Mutant analyses support a model in which DnaA represses these genes through its interaction with Rok. Our results indicate that the association of DnaA with chromosomal regions extends beyond those regions bound directly by DnaA, and that the regulation and functions of both DnaA and Rok may be more extensive than previously thought.
Results
Overview of ChIP-seq analysis of DnaA and Rok
We examined the chromosome-wide association of DnaA (Figs. 1, 2) and Rok (Fig. 2) at high resolution using ChIP-seq of B. subtilis cells growing exponentially in defined minimal glucose medium. We used the peak-calling algorithm SISSRs (Jothi et al., 2008; Narlikar & Jothi, 2012) and a five-fold enrichment cutoff (Experimental Procedures) to define chromosomal regions that were associated with DnaA or Rok. Antibody specificity in the immunoprecipitations was verified by performing analogous ChIP-seq experiments with anti-DnaA and anti-Rok antibodies in dnaA and rok null mutants (Experimental Procedures). The anti-DnaA and anti-Rok antibodies were highly specific. That is, there was little or no specific DNA in the immunoprecipitates from the relevant null mutants.
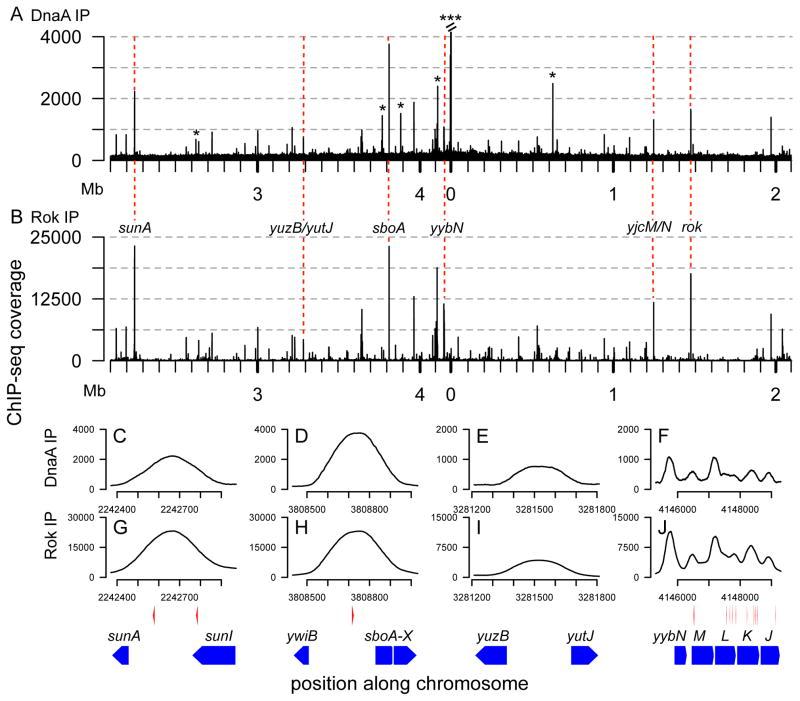
Figure 1. Comparison of in vivo and in vitro binding of DnaA to the chromosome.
A. Identification of in vivo DnaA binding sites. Wild type cells (strain AG174) were grown to mid-exponential phase and DnaA was immunoprecipitated after cross-linking with formaldehyde. ChIP-seq coverage indicates the number of sequenced fragments mapping to each nucleotide (assuming a fragment length of 250 bp; see Experimental Procedures) and is plotted on the y-axis versus the chromosomal position on the x-axis, indicated in megabases. The genomic position of 0 is set upstream of dnaA (Smith et al., 2014). Asterisks and gene names indicate previously identified regions that contain clusters of DnaA boxes. The three distinct binding regions in the oriC region are not resolved on this plot but have been well documented and are shown at greater resolution in Fig. 4 and Fig. S1. The peak at 0 Mb is truncated, as indicated by parallel slash marks at the top of the peak.
B. In vivo enrichment values for DnaA (from SISSRs; see Experimental Procedures) are compared to previously described in vitro binding data for 55 nM his-tagged DnaA in the presence of ATP (Smith & Grossman, 2015). The in vitro binding data are proportional to the number of sequenced fragments mapping to each nucleotide (assuming a fragment length of 250 bp), and were scaled to a maximum amplitude of 1. All of the 44 binding regions in Table 1 are plotted. The eight DnaA box cluster regions, plus one additional locus (region 13) that was coincident with an in vitro region, are plotted as boxed asterisks, and labeled with their region numbers, as indicated in Table 1. Data for the other 35 loci are plotted with gray diamonds.
C-H. Each panel shows a graph of the amount of DNA recovered by in vivo DnaA-ChIP (solid black curve) and in vitro DnaA-IDAP (dashed red curve) along an 800 bp chromosomal region centered on the position of maximum in vivo binding. The data for in vitro DnaA binding were with 1.4 μM DnaA-his with ATP and were described previously (Smith & Grossman, 2015). The dotted red vertical lines in panels C, D, and E indicate in vitro DnaA binding sites as determined previously (Smith & Grossman, 2015). The in vivo ChIP seq coverage was determined as described in panel A, but was then normalized to a global maximum value of 1, to facilitate comparison of the in vitro and in vivo data sets. The y-axis for each panel was adjusted based on the maximum amount of DNA recovered in the region plotted, and the in vitro and in vivo data were plotted using the same y-axis scale.
Below each graph is the chromosomal region. Red circles indicate potential DnaA boxes predicted using a PSSM with a p value cutoff of 0.0015, as described (Smith & Grossman, 2015). Gray rectangles indicate the position of genes in the regions and gene names are indicated above the rectangles and bracketed with arrowheads to indicate the direction of the open reading frame. Panels C–H correspond to binding regions 2, 21, 13, 28, 36, and 34, respectively, in Table 1.
Figure 2. Comparison of genome-wide binding of DnaA and Rok.
ChIP-seq data for DnaA (panels A, C–F; same data as in Fig. 1A, shown here for comparison), and Rok (panels B, G–J). ChIP-seq coverage is plotted on the y-axes, and indicates the number of sequenced fragments mapping to each nucleotide (assuming a fragment length of 250 bp), after the datasets were normalized to contain the same number of reads. The chromosomal position is plotted on the x-axis. In panel A, asterisks indicate the same previously identified regions that contain clusters of DnaA boxes that are labeled with gene names in Fig. 1A. The peak at 0 Mb is truncated, as indicated by parallel slash marks at the top of the peak. Dashed red lines (A and B) indicate selected regions previously identified as bound by Rok.
Association of DnaA (C–F) and Rok (G–J) with selected Rok-bound chromosomal regions in wild type cells. Each panel shows a magnified view of the data from panels A and B. The four selected chromosomal regions are shown below the graphs and include: sunA (C, G), sboA (D, H), yuzB-yutJ (E, I), and yybN (F, J). Gene regions are shown as blue pentagons with arrows indicating the direction of transcription. Putative DnaA boxes are indicated as red arrowheads above the genes and below the corresponding chromosomal positions. DnaA boxes were defined as described in Fig. 1.
ChIP-seq analysis of genome-wide binding by DnaA
We detected association of DnaA with 44 chromosomal regions in the ChIP-seq experiments (Table 1; Fig. 1A; Fig. S1). These regions included the eight DnaA binding regions that were previously identified by ChIP-chip (Breier & Grossman, 2009) and chromatin affinity purification and hybridization to DNA microarrays (ChAP-chip) (Ishikawa et al., 2007). These eight regions (indicated with asterisks in Table 1) all contain clusters of DnaA boxes, and are located in the intergenic regions between: rpmH-dnaA (the dnaA promoter region), dnaA-dnaN (directly upstream of the DNA unwinding element in oriC), gcp-ydiF, yqeG-sda, ywlC-ywlB, ywcI-vpr, yydA-yycS, and trmE-jag. These are the strongest binding regions identified by in vitro DNA affinity purification and sequencing (IDAP-Seq), indicating that DnaA has inherently high affinity for these sites (Smith & Grossman, 2015).
Table 1.
Chromosomal regions associated with DnaA in vivo 1.
| Region2 | Nearest gene(s) | Start3 | End3 | Enrichment1 |
|---|---|---|---|---|
| 1* | upstream of rpmH; upstream of dnaA * | 171 | 211 | 50 |
| 2* | downstream of dnaA; upstream of dnaN * | 1851 | 1891 | 68 |
| 3 | upstream of yceB; upstream of yceC | 312031 | 312071 | 5.0 |
| 4 | upstream of dtpT; upstream of yclG | 417831 | 417871 | 6.1 |
| 5 | downstream of phrI; upstream of yddM | 532171 | 532211 | 7.4 |
| 6* | downstream of gcp; downstream of ydiF * | 627931 | 627971 | 18 |
| 7 | downstream of ydjE; upstream of pspA | 654711 | 654751 | 5.6 |
| 8 | upstream of katA; upstream of ssuB | 944591 | 944631 | 10 |
| 9 | upstream of yhzC; upstream of comK | 1100531 | 1100571 | 7.4 |
| 10 | downstream of yjaZ; upstream of appD | 1194871 | 1194911 | 5.6 |
| 11 | upstream of yjcM; upstream of yjcN | 1248531 | 1248571 | 13 |
| 12 | downstream of ykuV; upstream of rok | 1477111 | 1477151 | 23 |
| 13 | upstream of ppsA; downstream of dacC | 1970871 | 1970911 | 22 |
| 14 | within yobI | 2040451 | 2040491 | 7.7 |
| 15 | within yobI | 2040991 | 2041031 | 6.0 |
| 16 | upstream of yosX; downstream of yosW | 2129931 | 2129971 | 12 |
| 17 | upstream of yonX; downstream of yonV | 2190471 | 2190511 | 14 |
| 18 | within sunT | 2241371 | 2242251 | 6.3 |
| 19 | upstream of sunA; downstream of sunI | 2242651 | 2242691 | 42 |
| 20 | downstream of yqgB; downstream of yqgA | 2560391 | 2560431 | 5.5 |
| 21* | upstream of yqeG; upstream of sda * | 2620011 | 2620051 | 6.8 |
| 22 | upstream of yqxI; downstream of cwlA | 2637231 | 2637271 | 6.5 |
| 23 | upstream of yraN; upstream of yraM | 2719271 | 2719311 | 9.6 |
| 24 | upstream of araA; downstream of abnA | 2921711 | 2921751 | 6.4 |
| 25 | within and upstream of argG; downstream of moaB | 2986831 | 2987171 | 5.2 |
| 26 | upstream of iscS; upstream of braB | 3000911 | 3000951 | 10 |
| 27 | downstream of malR; upstream of nupN | 3212351 | 3212391 | 12 |
| 28 | within and upstream of yutK and yuzB; upstream of yutJ | 3280691 | 3281511 | 6.0 |
| 29 | upstream of lytA; upstream of tagU | 3635911 | 3635951 | 7.4 |
| 30 | within ggaB | 3641231 | 3641271 | 12 |
| 31 | within and upstream of ggaA; downstream of tagH | 3644111 | 3645051 | 5.9 |
| 32 | upstream of glyA; downstream of ywlG | 3763271 | 3763311 | 6.5 |
| 33* | upstream of ywlC; downstream of ywlB * | 3766871 | 3766911 | 8.7 |
| 34 | upstream of ywiB; upstream of sboA | 3808711 | 3808751 | 31 |
| 35* | upstream of ywcI; upstream of vpr * | 3880431 | 3880471 | 12 |
| 36 | upstream of yxkD; upstream of yxkC | 3961871 | 3961911 | 17 |
| 37 | upstream of yxaJ; upstream of yxaI | 4077071 | 4077631 | 5.7 |
| 38 | downstream of gntZ; upstream of ahpC | 4091631 | 4091671 | 7.2 |
| 39 | within yydH | 4099151 | 4099191 | 5.8 |
| 40 | within yydD | 4104231 | 4104271 | 9.9 |
| 41* | upstream of yydA; within yyzF * | 4107771 | 4107811 | 14 |
| 42 | downstream of yybO; upstream of yybN | 4145691 | 4145731 | 9.4 |
| 43 | within yybM and yybL | 4147151 | 4147191 | 8.0 |
| 44* | upstream of trmE; downstream of jag * | 4185771 | 4185811 | 25 |
Chromosomal regions associated with DnaA in vivo were determined by ChIP-seq (Experimental Procedures). Regions that were enriched in the immunoprecipitates were identified by SISSRs (Jothi et al., 2008; Narlikar & Jothi, 2012) using a cutoff of five-fold enrichment relative to the non-immunoprecipitated control sample. Enrichment indicates the relative amount of DNA in the immunoprecipitates for the indicated region compared to the control (Experimental Procedures).
Regions are listed in order of chromosomal position. Asterisks (*) indicate regions with DnaA box cluster that were previously found to be bound by DnaA in vivo (Ishikawa et al., 2007; Breier & Grossman, 2009). Regions 1 and 2 are part of oriC. Regions 11, 26, 28, and 34 were previously reported to bind DnaA in a Rok-dependent manner (Smith & Grossman, 2015).
The start and end locations refer to the genomic coordinates of the binding peaks as determined by SISSRs based on the sequence of strain AG174 (Smith et al., 2014).
In addition to the eight DnaA box cluster regions, we found that DnaA was associated with 36 other chromosomal regions (Table 1; Fig. 1A), four of which (Table 1, regions 11, 26, 28, and 34) were previously reported (Smith & Grossman, 2015). In earlier studies of DnaA binding in vivo using ChIP-chip or ChAP-chip, none of these 36 regions were observed to bind DnaA (Ishikawa et al., 2007; Breier & Grossman, 2009), although one of the regions (sunA) was shown to bind using ChIP-PCR (Breier & Grossman, 2009). The detection of the additional in vivo binding regions here compared to previous analyses could be due to the greater sensitivity of ChIP-Seq compared to ChIP-chip (Ho et al., 2011), or other experimental differences, including the use of different anti-DnaA antiserum. Association of DnaA with these additional regions is unlikely to be an artifact of ChIP-seq sample amplification because these regions were not enriched in the control non-immunoprecipitated sample or in samples from a dnaA null mutant or a rok null mutant (see below).
Comparison of DnaA binding to DNA in vivo to that in vitro
DnaA binding in vitro is mediated by DnaA boxes in the target DNA, and the binding affinity is roughly correlated with the number and sequence of the DnaA boxes in the target region (Smith & Grossman, 2015). We sought to determine the extent to which the in vivo DnaA binding regions corresponded to in vitro DnaA binding regions and the presence of DnaA boxes. To do this, we compared the amount of DnaA binding in vitro to the fold enrichment in vivo at all 44 in vivo binding regions (Fig. 1B). We also annotated the 44 chromosomal regions associated with DnaA in vivo with potential DnaA binding sites and compared the binding profile in vivo to that determined by genome-wide binding analyses with purified DnaA in vitro (Fig. 1C–H, Fig. S1).
The eight regions containing DnaA box clusters are the regions that bind with the highest affinity in vitro (Smith & Grossman, 2015). The binding summits in the in vitro and in vivo binding data were largely superimposable, consistent with binding being driven by the interaction of DnaA with DnaA boxes both in vivo and in vitro (Fig. S1, panels 1, 2, 6, 21, 33, 35, 41, and 44). The fold enrichment in vivo was quite different for each of these eight loci (Fig. 1B). The fold enrichment of DnaA in vivo with the region upstream of the DNA unwinding element (DUE) in oriC was the highest (Fig. 1B, C), and the fold enrichment with the sda promoter region was one tenth that of the DUE (Fig. 1B, D). The fold enrichment at these eight loci did not correlate with the amount of binding in vitro (Fig. 1B), indicating that the apparent differences in association of DnaA with these eight regions in vivo and in vitro is unlikely to be due to actual differences in binding affinities. Although some of the difference may be due to crosslinking efficiencies at each region, it is also likely that the amount of DnaA binding at these loci is modulated by other proteins, by DNA structure, and-or by the nucleotide-bound state (ATP vs. ADP) of DnaA.
The remaining 36 regions associated with DnaA in vivo had only weak or no detectable binding in vitro (Smith & Grossman, 2015), and the summit of binding for most regions in vivo was not coincident with predicted DnaA boxes (Fig. S1). Specifically, only region 13 (Fig. S1) was coincident with an in vitro binding region (Fig. 1B, E). However, binding in vitro at this region is very weak (ranked 203 out of 269) whereas the binding in vivo was robust (22-fold enrichment). Some additional in vivo binding regions (regions 8, 17, 19, 22, 27, and 40) were near (45 –100 bp away) but not coincident with weak in vitro binding sites. The remaining in vivo binding regions did not appear to bind DnaA at all in vitro, and many (regions 3, 7, 15, 16, 25, 28, 29, 32, 36, 38; see Fig. S1), including yuzB-yutJ (Fig. 1F) and yxkD-yxkC (Fig. 1G) completely lacked predicted DnaA boxes within 200 bp of the binding summit.
Some other regions, including the sboA promoter (Fig. 1H), have one or more DnaA boxes (predicted by sequence analysis) near the binding summit. However, no binding is observed in vitro, most likely because the presence of a single or even a few DnaA boxes is not necessarily sufficient for DnaA binding in vitro (Smith & Grossman, 2015). Other factors, including the actual sequences of the boxes and their spacing and orientation are likely also important. In contrast to regions bound in vivo but not in vitro, many regions that bind DnaA quite well in vitro, including regions within rplB and codV, did not appear to bind at all in vivo. Overall there was very little correspondence between in vivo and in vitro binding for these 36 regions, indicating that efficient binding at these regions might require an additional DNA binding protein.
ChIP-seq analysis of genome-wide binding by Rok and comparison to that of DnaA
We noticed that many of the 36 additional DnaA binding regions had previously been found to be associated with Rok in ChIP-chip experiments (Smits & Grossman, 2010). In addition, we recently found that Rok is required for association of DnaA with at least four of these regions (yjcM-yjcN, iscS-braB, yuzB-yutJ, and ywiB-sboA; regions 11, 26, 28, and 34, respectively) in vivo (Smith & Grossman, 2015). Therefore, we performed ChIP-seq experiments with Rok to fully characterize the extent of this association. Rok is a nucleoid-associated protein that binds in vivo to many places on the B. subtilis chromosome, particularly A+T-rich regions (Smits & Grossman, 2010).
Using ChIP-seq with anti-Rok antibodies, we identified 264 chromosomal regions bound by Rok (Fig. 2B; Table S1). This binding profile is consistent with previous ChIP-chip results, including the strong correlation between Rok binding and A+T content (Smits & Grossman, 2010). Remarkably, the Rok-bound regions included all 36 DnaA binding regions that did not correspond to DnaA box clusters. In these chromosomal regions, the binding profile of Rok appeared to be similar and perhaps coincident with that of DnaA (Fig. 2; Fig. S2). In most cases the binding of each protein appeared as a single peak upstream of a gene (e.g., Fig. 2C–E, G–I). Many of the regions bound by Rok were in or upstream of operons that are regulated by Rok, including sboA, sunA, yybN, yydH, and yxaJ (this study and Albano et al., 2005).There was also binding within the coding regions of several genes (Table 1, regions 14, 15, 18, 30, 39, 40, and 43; Figs. S1, S2). There are four chromosomal loci with multiple DnaA/Rok binding sites, as defined by two DnaA/Rok binding sites within a distance of 3 kb (Fig. 2F, J; Fig. S3). Most of the binding that occurred within coding sequences (all but regions 39 and 40, which are 4.9 kb apart) were located in the these regions showing clustered binding sites
In addition to the 36 regions without clusters of DnaA boxes, we detected Rok at two regions that were previously known to be associated with DnaA and that contain multiple DnaA binding motifs: the trmE-jag intergenic region and the dnaA-dnaN intergenic region in oriC, just upstream of the DUE (Fig. S4). The possible significance of Rok binding to these regions (regions 44 and 2, respectively, in Table 1 and Figs. S1 and S2) is discussed below.
Sequence specificity of Rok binding
We used the Rok-associated regions to examine the sequence specificity of Rok binding. Previous studies, including transcriptional profiling and in vitro DNA binding (Hoa et al., 2002; Albano et al., 2005) and ChIP-chip analyses (Smits & Grossman, 2010), did not indicate a strong binding motif, possibly due to limited data sets, although a few A+T-rich motifs appeared overrepresented in Rok-bound regions (Smits & Grossman, 2010). We extracted sequences corresponding to the centers of Rok ChIP-seq binding peaks (Experimental Procedures) and searched for a motif using the ChIP-seq motif-finding tool DREME (Bailey, 2011). We identified the sequence A(T/G)AAAA as a potential binding motif (P-value of 2.1e-8). This motif was found in 200 of 264 input sequences, but since the sequence consists almost entirely of A’s, it may simply reflect the general preference of Rok for A+T-rich regions. This motif closely resembles the center of a previously identified 19-bp A+T-rich motif, which was derived from only four Rok-bound sequences and two negative control regions (Smits & Grossman, 2010). We have not further investigated the significance of this motif.
Correlation between the amounts of association of Rok and DnaA
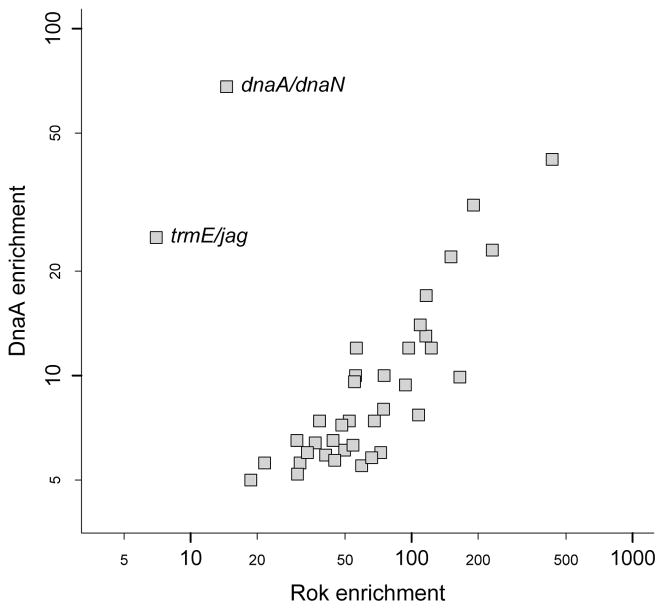
We compared the enrichment of Rok and DnaA at the 38 chromosomal regions bound by both of these proteins (Fig. 3). For the 36 regions that lack DnaA box clusters, the relative amount of DNA that was isolated in the DnaA immunoprecipitates was closely correlated with that from the Rok immunoprecipitates (correlation coefficient = 0.92; Fig. 3). The trmE-jag and dnaA-dnaN binding sites, which contained DnaA box clusters, had proportionally much weaker Rok binding than the 36 regions without clusters of DnaA boxes (Fig. 3).
Figure 3. Correlation between the relative enrichment of DnaA and Rok at regions of co-association.
In vivo fold enrichment values for Rok and DnaA are compared for the 38 regions that bind both proteins. Each data point represents one chromosomal region. Enrichment values were determined using the peak-calling algorithm SISSRs. The two regions that contain DnaA box clusters and bind DnaA independently of Rok (between dnaA-dnaN, and between trmE-jag) are indicated.
The 226 Rok-bound regions without detectable DnaA tended to be the regions with lower detectable binding by Rok (median 5.4-fold enrichment, compared to median 30.5-fold enrichment for regions with detectable DnaA). It is possible that DnaA was not bound to any of these regions. However, it is also possible that DnaA was bound to some of them, but at a level below the limit of detection, which is consistent with the fact that the weaker Rok peaks tended to be the ones that did not have detectable DnaA associated, and the fold enrichment for DnaA at each locus was significantly lower than the fold enrichment observed for Rok (Fig. 3). One region that had moderate association with Rok (peak #175 – upstream of azlB - in Table S1) had no detectable association with DnaA, indicating that other factors likely prevent DnaA from associating with this region.
rok is required for DnaA binding at regions that lack clusters of DnaA boxes
We considered three possible models that are consistent with the association of DnaA and Rok with several of the same chromosomal regions. 1) Since the ChIP-seq analyses were done on a population of cells, different subpopulations could have one or another of the proteins associated with a given region. In this way, each protein could be associated with the same region, but not necessarily in the same cells. 2) Since Rok is associated with many chromosomal regions, and there are many potential DnaA boxes throughout the genome, the association of these proteins could simply be a coincidence of independent binding to the same chromosomal regions. 3) One of these proteins could depend on the other for association.
We postulated that association of DnaA with most or all of the regions associated with both DnaA and Rok would be dependent on Rok, largely because Rok is required for DnaA to associate with at least four (iscS, yuzB, sboA, and yjcM) of these regions in vivo (Smith & Grossman, 2015). To test if association of DnaA was dependent on Rok, we used ChIP-seq to analyze genome-wide binding of DnaA in a rok null mutant (Fig. 4).
Figure 4. Genome-wide binding of DnaA in wild type and rok null mutant cells.
Wild type (AG174) and rok null mutant (WKS1038) cells were grown to mid-exponential phase and DnaA was immunoprecipitated after cross-linking with formaldehyde. Plots are as described for Figs. 1A and 2.
A. Genome-wide binding of DnaA in wild type cells (black, upper y-axis) and a rok null mutant (red, lower y-axis). Asterisks (*) indicate the eight previously identified DnaA binding sites. The three distinct binding regions (rpmH-dnaA; dnaA-dnaN; and trmE-jag) near oriC are shown at greater resolution below (Fig. 4B, C, I).
B–M. Association of DnaA with regions with DnaA box clusters (B–I) and selected regions bound by Rok (J–M) in wild type cells (black solid lines) and a rok null mutant (red dashed lines). Each panel shows a magnified view of the data from panel A. DnaA boxes (red arrowheads directly below the x-axis) and gene annotations (blue pentagons below the DnaA boxes) are shown below the corresponding chromosomal positions.
B–I. The eight DnaA box cluster regions are: rpmH-dnaA (B), dnaA-dnaN (C), gcp-ydiF (D), yqeG-sda (E), ywlC-ywlB (F), ywcI-vpr (G), yydA-yycS (H), and trmE-jag (I).
J–M. The selected Rok-bound regions are: sunA (J), sboA (K), yuzB-yutJ (L), and yybN (M), the same as those shown in Fig. 2C–J.
We found that there was little or no effect of rok at the eight regions with clusters of DnaA boxes (Fig. 4A–I). That is, association of DnaA with these regions was not dramatically altered in the rok null mutant compared to the rok+ strain. This result was expected since Rok was not detectably associated with six of these regions, and was only weakly associated with the other two (dnaA-dnaN and trmE-jag).
In contrast, Rok was required for association of DnaA with the other 36 chromosomal regions. In the rok null mutant, there was no detectable association of DnaA with the 36 regions that were associated with Rok in wild type cells (Fig. 4A, J–M). These results confirm and extend recent findings that association of DnaA with four of these chromosomal regions (sboA, yuzB-yutJ, yjcM, and iscS-braB) requires rok (Smith & Grossman, 2015). No additional DnaA-bound regions were detected in the rok null mutant, indicating that Rok was not masking potential DnaA binding regions.
The Rok-dependent binding of DnaA to regions that lack DnaA box clusters indicated that both proteins were associated with the target regions in the same cells and not in separate subpopulations of cells. The association of these proteins with the same chromosomal regions indicated that Rok and DnaA might interact directly (described below). This interaction likely accounts for the ability of DnaA to associate with regions to which it does not have inherent binding affinity.
ChIP-seq analysis of genome-wide binding of Rok in a dnaA null mutant
We wondered if the association of Rok with chromosomal regions was affected by DnaA. To evaluate this, we analyzed the genome-wide binding of Rok by ChIP-seq in a dnaA null mutant. dnaA is normally essential because of its role in replication initiation. To circumvent the need for dnaA and oriC, we used a strain (AIG200, Goranov et al., 2005) that initiates replication from a heterologous origin (oriN) using its cognate initiator protein (RepN). oriN and repN are integrated into the chromosome (Hassan et al., 1997; Moriya et al., 1997) in spoIIIJ, near oriC. The oriC region, including dnaA and dnaN, is deleted in this strain, and dnaN, which encodes the processivity clamp of DNA polymerase and is essential for growth, is expressed from a xylose-inducible promoter (Pxyl-dnaN), integrated at another region (amyE) of the chromosome. We compared this dnaA null mutant to an isogenic strain (TAW5, Merrikh & Grossman, 2011) expressing dnaA from an IPTG-inducible promoter (Pspank-dnaA) integrated into the chromosome at an ectopic site (lacA).
We found that loss of DnaA had little or no effect on association of Rok with chromosomal regions (Fig. S5). Because oriC is deleted in these strains, we could not determine if Rok binding to the oriC region required DnaA. Otherwise, in both the dnaA+ (TAW5) and dnaA null (AIG200) mutant strains, Rok was still associated with the same chromosomal regions as in dnaA+ cells. No additional Rok binding regions were identified in the dnaA null mutant, indicating that DnaA was not masking possible regions to which Rok could bind.
Independent association of DnaA and Rok with sequences in and near oriC
There were two regions, trmE-jag and dnaA-dnaN, that contain clusters of DnaA boxes and were bound by both DnaA and Rok (Fig. S4). At both of these regions, DnaA binding was independent of Rok (Fig. S4, panels A and B). In addition, Rok binding was independent of DnaA at trmE-jag (Fig. S4, panel D), but we could not determine if Rok binding to the dnaA-dnaN region was DnaA-dependent as this region was deleted in the dnaA null mutant. Given the mutual independence of Rok and DnaA binding at trmE-jag, and the Rok-independent binding of DnaA at dnaA-dnaN, it is possible that these proteins are bound in different populations of cells, or that they both bind these regions at the same time, but to slightly different sequences that could not be resolved using ChIP-seq.
The DNA unwinding element from which replication initiates is between dnaA and dnaN (Moriya et al., 1988; Moriya et al., 1992). Because Rok was associated with this region, we tested whether a rok null mutation had an effect on initiation of chromosomal replication. We measured the relative amounts of the origin and terminus regions (the marker frequency) by qPCR to determine the ori:ter ratio in cells growing exponentially in minimal and rich medium. During growth in defined minimal medium with either glucose, arabinose, or succinate as a carbon source, or in LB medium, we found that loss of rok did not reproducibly affect the ori:ter ratio (Table S2), indicating that under these conditions (and with this assay), rok was not having a detectable effect on replication.
Association of DnaA with Rok-bound chromosomal regions does not require the DNA binding domain of DnaA
The DnaA and Rok binding peaks appeared coincident at the 36 regions where association of DnaA was Rok-dependent. In addition, purified Rok binds to several of these regions in vitro (Albano et al., 2005). Together, these results indicated that DnaA likely associated with these regions via Rok. We postulated that the DNA binding domain of DnaA might not be needed for association of DnaA with these regions.
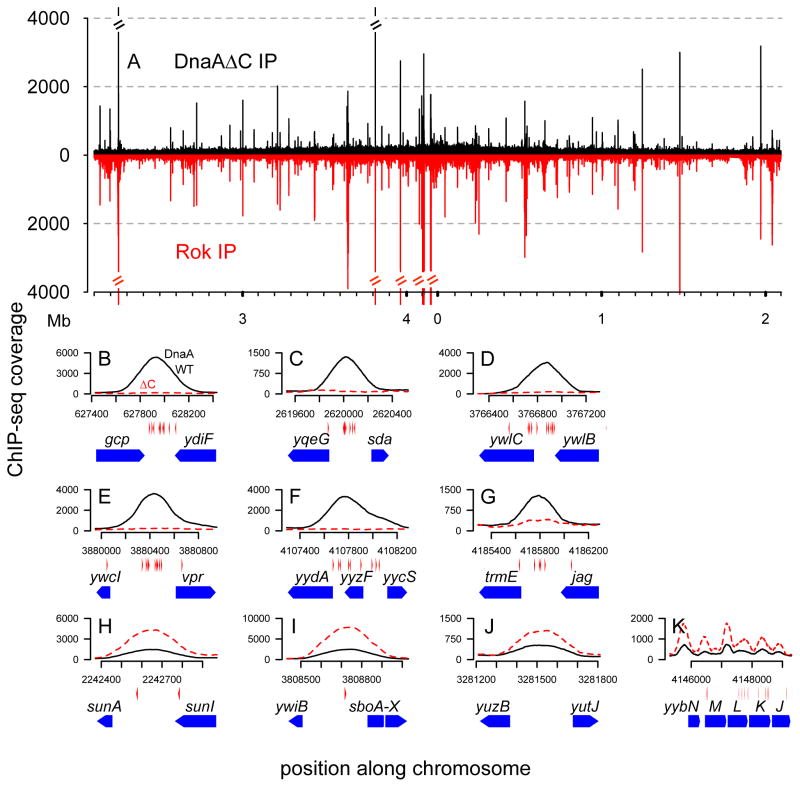
To determine if DnaA was bound directly to DNA at the Rok-associated chromosomal regions, we made a deletion in dnaA that results in loss of the C-terminal 91 amino acids of DnaA, corresponding to the conserved Domain IV region that is necessary and sufficient for DNA binding (Roth & Messer, 1995). The mutant dnaA (dnaAΔC) was expressed from an IPTG-inducible promoter (Pspank-dnaAΔC) integrated into the chromosome at an ectopic site (amyE). An isogenic control strain expressed full-length dnaA (Pspank-dnaA). We used an oriN+ strain in which the oriC region, including the native dnaA, was removed, and dnaN was constitutively expressed at its native locus from a derivative of the promoter Ppen (Experimental Procedures). We then analyzed genome-wide binding of DnaAΔC and Rok.
We found that the DNA-binding domain of DnaA was not required for association of DnaA with Rok-bound chromosomal regions (Fig. 5). DnaAΔC was associated with all 36 of the regions bound by Rok (Fig. 5A, H–K). In contrast, detectable association of DnaAΔC with the chromosomal regions containing clusters of DnaA boxes was eliminated (Fig. 5B–F), as expected for regions of direct DnaA binding. Binding to the region upstream of trmE was reduced (Fig. 5G), probably because DnaA is able to bind directly to DNA via the DnaA boxes, but is also able to bind indirectly via the Rok that is bound there. The genome-wide binding profile of Rok was similar between the dnaA+ and dnaAΔC strains (data not shown), consistent with the analysis of Rok binding in a dnaA null mutant (Fig. S5).
Figure 5. Genome-wide binding of the DNA-binding mutant DnaAΔC.
Cells expressing wild type dnaA (CAS221) or mutant dnaAΔC (CAS231) were grown to mid-exponential phase, and the indicated proteins (DnaA, DnaAΔC, Rok) were immunoprecipitated after cross-linking with formaldehyde. Plots are as described in Figs. 1A, 2, 4. The strains used contain a deletion of the dnaA-oriC region and replicate from oriN. dnaN was expressed from Ppen2028-dnaN. dnaA or dnaAΔC was ectopically expressed from Pspank with 0.1 mM IPTG. Although these strains contain engineered deletions (oriC) and insertions (dnaN and dnaA), the genome coordinates presented correspond to AG174 in order to be readily compared to other figures in this paper.
A. Genome-wide binding of DnaAΔC (black, upper y-axis) and Rok (red, lower y-axis) in cells expressing mutant dnaAΔC (CAS231).
B–K. Association of wild type DnaA (black solid lines, strain CAS221) and DnaAΔC (red dashed lines, strain CAS231) with DnaA box cluster regions (B–G) and selected regions bound by Rok (H–K). Data for dnaAΔC are from panel A. DnaA boxes (red arrowheads directly below the x-axis) and gene annotations (blue pentagons below the DnaA boxes) are shown below the corresponding chromosomal positions.
B–G. Two DnaA box cluster regions (rpmH-dnaA and dnaA-dnaN) are deleted from the strains used in these experiments and are not shown. The remaining six DnaA box cluster regions are: gcp-ydiF (B), yqeG-sda (C), ywlC-ywlB (D), ywcI-vpr (E), yydA-yycS (F), and trmE-jag (G).
H–K. The selected Rok-bound regions are: sunA (H), sboA (I), yuzB-yutJ (J), and yybN (K), the same as those shown in Fig. 2C–J and Fig. 4J–M.
The association of DnaAΔC at each of the Rok-bound regions appeared to be increased relative to that observed for full-length DnaA (Fig. 5H–K; Fig. S6). The amount of DnaAΔC in cells was approximately half (0.52 ± 0.09) that of full-length DnaA protein (as determined by quantitative Western blots), so this increased association was not due to more DnaAΔC protein. It is unclear whether the increased signal for DnaAΔC relative to full-length DnaA at Rok-bound regions reflects differences in crosslinking efficiency, or whether there is in fact increased association of DnaAΔC. If there is more DnaAΔC at these regions relative to full-length DnaA, it might be due to a global redistribution of DnaA that would otherwise have been bound to DnaA box-cluster regions, or bound nonspecifically to the chromosome.
The fact that DnaA’s DNA binding region was not required for it to associate with Rok-bound regions indicated that DnaA did not associate with these regions via canonical DNA binding. We infer that DnaA likely associates with these DNA regions via direct protein-protein interactions with Rok.
DnaAΔC requires Rok for association with DNA in vitro
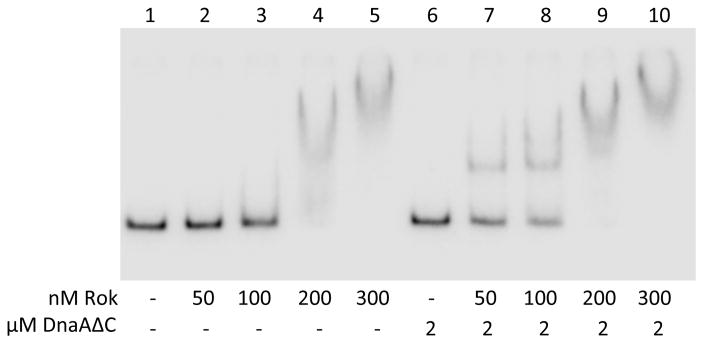
Since DnaA binding depends on Rok at many regions in vivo, we tested the model that they interact directly, using purified proteins and in vitro gel shift assays. We purified Rok and the DNA-binding mutant DnaAΔC, and measured their effects on the mobility of a 32P-labeled DNA fragment during gel electrophoresis. The DNA fragment contained the rok promoter region, which was associated with DnaA and Rok in vivo, and had previously been show to bind Rok in gel shift assays (Hoa et al., 2002; Albano et al., 2005).
When Rok was added to the DNA at increasing concentrations, the amount of the fragment that was shifted increased (Fig. 6, lanes 1–5). In addition, the decrease in mobility of the fragment became more dramatic at increasing Rok concentrations, presumably reflecting the binding of multiple Rok molecules to each fragment. (e.g., Fig. 6, lanes 4 and 5). At 300 nM Rok (Fig. 6, lanes 5), essentially all of the DNA fragment was shifted. These results are generally consistent with previous analyses of Rok binding to DNA, although somewhat higher concentrations of Rok appeared to be needed in the experiments here and the shifted species were less well-defined than in previous work (Hoa et al., 2002; Albano et al., 2005), indicating that Rok was not as stably bound during electrophoresis. The weaker binding observed here is likely due to differences in the reaction and electrophoresis conditions that were necessary to reproducibly detect effects of DnaA.
Figure 6. DnaAΔC requires Rok for association with DNA in vitro.
A 32P-labeled DNA fragment (0.1 nM) corresponding to a 342-bp region of the rok promoter was incubated in the absence of Rok (lanes 1, 6) or in the presence of increasing concentrations of Rok (50 – 300 nM) (lanes 2–5 and 7–10) in the absence (lanes 2–5) or presence of 2 μM DnaAΔC (lanes 7–10). Protein concentrations are indicated below each lane. Results are representative of at least three independent experiments.
There was no detectable binding of DnaAΔC to the DNA probe (Fig. 6, lanes 1, 6), as expected. However, addition of DnaAΔC to DNA in the presence of Rok (Fig. 6, lanes 7–10) resulted in the appearance of slower migrating bands and an increase in the amount of DNA shifted at a given Rok concentration. For example at 50 nM Rok, there was little or no detected DNA binding in the absence of DnaAΔC (Fig. 6, lane 2), but significant binding in the presence of DnaAΔC (Fig. 6, lane 7). Furthermore, the DNA mobility was decreased in the presence of DnaAΔC at a given Rok concentration (e.g., Fig. 6, lane 4 vs 9). These results indicate that DnaAΔC can act on the Rok-DNA complex to cause increased binding and a supershift, consistent with the results obtained from the ChIP analyses in vivo. The simplest interpretation of these results is that there is a direct interaction between DnaA and Rok. We did not observe any interaction between Rok and DnaA in solution using either Octet Red or gel filtration, indicating that DNA might be required for DnaA and Rok to interact.
Regulation of gene expression by DnaA and Rok
DnaA and Rok are both transcription factors. In B. subtilis, DnaA regulates expression of genes involved in DNA replication (Ogura et al., 2001), sporulation (Burkholder et al., 2001; Ishikawa et al., 2007), and the response to replication stress (Goranov et al., 2005; Ishikawa et al., 2007). Rok regulates genes involved in competence (Hoa et al., 2002), antibiotic production, and other extracellular functions (Albano et al., 2005; Kovacs & Kuipers, 2011; Marciniak et al., 2012). Preliminary analysis of gene expression in a dnaA null mutant indicated that many genes normally repressed by Rok also had increased expression in the absence of dnaA (T.A. Washington, J.L. Smith, and ADG, manuscript in preparation). Since both DnaA and Rok were associated together with several promoter regions, we wished to determine if one or both of these transcription factors affected expression of genes in these regions.
Gene expression in dnaA and rok single mutants
We compared the effects of dnaA and rok null mutations on gene expression. We used four strains, with relevant genotypes: dnaA+ rok+ (CAL2083); ΔdnaA rok+ (CAL2074); dnaA+ Δrok (CAS196); and ΔdnaA Δrok (CAS192). In each strain, oriC and dnaA were replaced with oriN and repN, and dnaN was expressed constitutively at its native locus from a derivative of the promoter Ppen (Experimental Procedures).
We used DNA microarrays to analyze global gene expression (mRNA levels) during exponential growth in defined minimal glucose medium. We focused on the 55 genes in the 36 transcription units that had Rok-dependent DnaA association. We considered these genes candidates for direct regulation by both DnaA and Rok. From these candidates, we found four transcription units adjacent to (or overlapping with) regions bound by both DnaA and Rok that had significantly altered expression in both the dnaA null mutant and in the rok null mutant (Table 2): the sun operon (sunA, sunT, bdbA, sunS, bdbB) (Table 2, lines 1–5); yxaJ (Table 2, line 6) and yxaI (Table 2, line 7), which are divergent genes with the Rok-DnaA binding region in-between; and the yybNM operon (Table 2, lines 8, 9). yybN and yybM are in an operon with three other genes (yybNMLKJ). However, there appears to be a terminator after each of the first two genes and only yybN and yybM were affected by loss of dnaA and/or rok. For the remaining DnaA-Rok binding regions, their positioning might not allow effective transcriptional regulation by one or both of these transcription factors, or the affected genes may not be expressed under the growth conditions examined here.
Table 2.
Gene expression affected by dnaA and rok.
| Gene | ΔdnaA | Δrok |
ΔdnaA Δrok |
|
|---|---|---|---|---|
| 1. | sunA | (2.0) | 5.7 | 5.1 |
| 2. | sunT | 4.7 | 21 | 14 |
| 3. | bdbA | 2.7 | 17 | 11 |
| 4. | sunS (yolJ) | 3.5 | 14 | 9.2 |
| 5. | bdbB | 3.0 | 14 | 11 |
| 6. | yxaJ | (1.8) | 2.6 | 2.8 |
| 7. | yxaI | −2.5 | −6.3 | −7.6 |
| 8. | yybN | 2.5 | 2.3 | 5.7 |
| 9. | yybM | 1.9 | 2.0 | 3.8 |
Genes are listed in order of chromosomal position. Indicated mutants (ΔdnaA, Δrok single mutants and ΔdnaA Δrok double mutant) were grown in defined minimal medium and samples taken for measurement of mRNA levels using DNA microarrays (Experimental Procedures). All strains contained a null mutation in oriC and replication was from oriN. Stains included: dnaA+ rok+ (CAL2083; genotype: rok+, dnaA+, ΔoriC, oriN+); ΔdnaA (CAL2074); Δrok (CAS196); and ΔdnaA Δrok (CAS192). Each value indicates the linear fold change in expression of the indicated gene in the indicated mutant, relative to the isogenic dnaA+ rok+ (CAL2083) cells. Negative values correspond to less gene expression in the mutant relative to CAL2083. Data are averages of 3 independent experiments, and except for the numbers in parentheses, all are statistically significant (adjusted p <0.04).
Expression of all target genes, except yxaI, was increased in the dnaA and rok mutants (Table 2), indicating that DnaA and Rok normally act, directly or indirectly, to repress expression of these genes. For example, mRNA levels of the five genes in the sun operon were increased approximately 2- to 5-fold in the dnaA null mutant and 6- to 20-fold in the rok null mutant (Table 2, lines 1–5). The sun genes are in the prophage SPβ and function in the biosynthesis and transport of the antibiotic sublancin. Expression of yxaJ and the yybNM operon was also increased in both the dnaA null mutant and the rok null mutant (Table 2, lines 6, 8–9). Negative regulation of expression of these genes by Rok (Albano et al., 2005) and DnaA (T.A. Washington, J.L. Smith, and ADG, in preparation) is generally consistent with previous findings.
yxaI and yxaJ are adjacent to and divergent from each other. Whereas yxaJ had increased expression in both the dnaA and rok mutants (Table 2, line 6), yxaI had decreased expression in both mutants (Table 2, line 7), indicating that DnaA and Rok normally function to activate expression of yxaI, either directly or indirectly. Rok is not known to directly activate transcription. Rather, Rok inhibits expression of comK, and the comK gene product is a transcription factor that activates many genes, including those needed for competence development (Hoa et al., 2002). We suspect that the effect of rok on yxaI (unlike yxaJ) is indirect. This could be similar to the indirect activation of yuaB (bslA) by Rok via an unidentified transcription factor (Kovacs & Kuipers, 2011), or through effects of Rok on expression of the transcription factor ComK (Berka et al., 2002; Hoa et al., 2002; Ogura et al., 2002; Albano et al., 2005).
Gene expression in a dnaA rok double mutant
To better characterize the potential co-regulation of genes by DnaA and Rok, we analyzed gene expression in a dnaA rok double mutant (Table 2). For most genes, rok appeared to be epistatic to dnaA; that is, the change in gene expression in the rok mutant was not further altered in the absence of dnaA (Table 2, lines 1–7). For example, expression of yxaJ and yxaI appeared to be similar in the rok dnaA double mutant and the rok single mutant (Table 2, lines 6–7).
Expression of the genes in the sun operon (sunA, sunT, sunS, bdbA, and bdbB) was similar or slightly lower in the rok dnaA double mutant than in the rok single mutant (Table 2, lines 1–5), indicating that rok is likely epistatic to dnaA. Expression in the double mutant was somewhat less de-repressed than in the rok single mutant. This could indicate that there are indirect effects of DnaA on expression of this operon.
In contrast to the examples above, expression of yybN and yybM in the rok dnaA double mutant appeared to be additive from the two single mutants (Table 2, lines 8–9), indicating that Rok and DnaA likely have independent effects.
For the genes where rok is epistatic to dnaA, a simple model is that the presence of DnaA stimulates the ability of Rok to repress gene expression (Table 2, lines 1–6). Previous studies of gene expression and Rok binding in vivo and in vitro support direct transcriptional repression of these genes by Rok (Albano et al., 2005; Smits & Grossman, 2010). We suggest that in the absence of DnaA, Rok still represses transcription, but not as well as in the presence of DnaA. Thus, there is a modest increase (de-repression) in gene expression in the absence of DnaA. However, in the absence of Rok, there is a greater increase (more de-repression) in gene expression. Loss of dnaA in the absence of rok causes no additional increase (de-repression) in gene expression.
It is also possible that the effects observed are due to much more complicated effects of dnaA and rok on gene expression. Both DnaA and Rok have indirect effects on gene expression by affecting other transcription factors. For example, DnaA activates expression of the sporulation checkpoint regulator sda (Burkholder et al., 2001; Ishikawa et al., 2007; Breier & Grossman, 2009; Veening et al., 2009; Hoover et al., 2010), and Rok represses the master regulator of competence development comK (Hoa et al., 2002) and appears to affect at least one other as yet unidentified transcription factor (Kovacs & Kuipers, 2011). We suspect that changes in gene expression in the dnaA and rok mutants described here are a reflection of both direct and indirect effects on the target genes.
At least some of the effects of dnaA and rok on expression of yybN and yybM must be indirect. There is little or no DnaA associated with yybN and yybM in the absence of rok (Fig. 4M). If there really is no DnaA without Rok and effects are direct, then Rok should be epistatic to DnaA. That is, DnaA cannot have a direct effect if it is not able to get to the chromosomal region. Since the effects of rok and dnaA appear to be additive (Table 2, lines 8–9), at least some of the effects must be indirect.
Discussion
Using ChIP-seq, we found that DnaA associates with two types of genomic regions in vivo in B. subtilis. As expected, DnaA bound directly to eight previously characterized regions (Burkholder et al., 2001; Goranov et al., 2005; Ishikawa et al., 2007; Cho et al., 2008; Breier & Grossman, 2009; Smits et al., 2011), each of which contains clusters of DnaA boxes. Binding of DnaA to these regions required the DNA-binding domain of DnaA, and did not require the nucleoid-associated protein Rok.
Additionally, we found that DnaA was associated with 36 other chromosomal regions. All of these regions were also associated with the nucleoid-associated protein Rok and association of DnaA with these 36 regions was dependent on Rok. These results are consistent with and extend the previous finding that DnaA depends on Rok for association with four chromosomal regions (Smith & Grossman, 2015). We also found that association of DnaA with the Rok-bound regions did not depend on the DNA binding domain of DnaA, indicating that the association was likely mediated by direct interaction between DnaA and Rok. In gel shift assays, purified DnaAΔC, which is incapable of direct binding to DNA, increased the apparent affinity of Rok for DNA and altered the mobility of a Rok-bound DNA fragment, indicating that DnaA associates with many chromosomal regions indirectly through an interaction with Rok. By analyzing gene expression in dnaA and rok mutants, we identified several genes that are repressed by DnaA and by Rok. Double mutant analyses were consistent with a model in which DnaA affects Rok-mediated repression at several of these genes, although more complicated indirect effects are also likely.
Together, our results are consistent with a model in which Rok binds to DNA, and DnaA then associates directly with Rok to modify Rok function. DnaA could enhance Rok-mediated repression of gene expression by causing Rok to bind more tightly to DNA. Alternatively, the presence of DnaA with Rok might enhance possible steric exclusion of RNA polymerase from the promoter. It is also possible that DnaA affects the likely function of Rok in nucleoid organization and compaction.
In B. subtilis, and likely other organisms, DnaA regulates a transcriptional response to replication stress (Goranov et al., 2005; Breier & Grossman, 2009). Several of the regions associated with both DnaA and Rok were upstream of genes previously identified as responsive to replication stress (Goranov et al., 2005). These regions include sunA, ahpC, katA, yxkC, and yjcN. Based on the Rok-dependent association of DnaA with these regions, the effects of replication stress on these genes could be due to changes in association of DnaA with Rok, or they could be due to indirect effects.
Interestingly, both Rok and DnaA are associated with two clusters of DnaA binding sites, one in the trmE-jag intergenic region near oriC, and the other just upstream of the DUE in oriC. DnaA binds to the DnaA boxes in the oriC region, promotes unwinding of the DUE, and then interacts with the newly defined DnaA-trio to stabilize the ssDNA bubble (Richardson et al., 2016). The presence of Rok in the oriC region could indicate a role in modulating replication initiation. Although we did not detect an effect of rok on replication initiation under a range of growth conditions, it is possible that the effects are either too small to measure with the assay used, or that there are other mechanisms that compensate for the loss of rok. By binding to the DUE region, perhaps bending or occluding it, or by affecting the association of DnaA with DnaA boxes or the DnaA-trio, Rok might affect the precise timing of replication initiation or the ease of origin melting. Such effects of Rok might be redundant with the functions of other DNA-binding proteins or masked by the multiple mechanisms that regulate replication initiation in B. subtilis (Noirot-Gros et al., 2002; Lee & Grossman, 2006; Rahn-Lee et al., 2009; Wagner et al., 2009; Merrikh & Grossman, 2011; Scholefield et al., 2011; Bonilla & Grossman, 2012; Scholefield et al., 2012).
Rok is an analog of the nucleoid-associated proteins H-NS of gamma-proteobacteria and Lsr2 of Mycobacteria (Smits & Grossman, 2010). H-NS has been extensively studied. It can bridge separate DNA sequences, and its activities are modulated by several other proteins (Tippner & Wagner, 1995; Dame et al., 2000; Amit et al., 2003; Dame et al., 2006; Noom et al., 2007; Dorman & Kane, 2009; Arold et al., 2010). It seems possible that Rok may be analogous to H-NS in these ways and that DnaA might affect an as-yet uncharacterized aspect of Rok function. For instance, H-NS can convert between two DNA-binding modes, bridging and stiffening (Liu et al., 2010). If Rok also has multiple DNA-binding modes, perhaps with different effects on gene regulation, the interaction with DnaA might stabilize one mode or regulate switching between modes. The effects of Rok on DNA architecture have not yet been explored, but Rok does appear to be at some of the boundaries of chromosomal domains (Marbouty et al., 2015). Based on our results, DnaA is likely to be at several of these regions too.
DnaA is highly conserved in bacteria (Ogasawara et al., 1991; Messer, 2002; Zakrzewska-Czerwinska et al., 2007), but Rok has only been identified in Bacillus species and is believed to have been acquired by horizontal gene transfer (Albano et al., 2005; Singh et al., 2012). Although Rok is not widely conserved, we speculate that the general connection between DnaA and chromosome architecture proteins may extend to other organisms. We suspect that DnaA in other organisms might interact with other nucleoid-associated proteins, including H-NS analogues. Rok and H-NS are both relatively small proteins (21 kDa and 15 kDa, respectively), predicted to be net positively charged (isoelectric points of 9.3 and 9.5, respectively). The widespread conservation of DnaA in bacteria indicates that DnaA proteins from different organisms are likely to have similar biochemical properties. For example, DnaA from E. coli and B. subtilis are 51% identical and bind the same consensus sequence, although they do not substitute for each other in vivo (Krause et al., 1997; Krause & Messer, 1999). Given the interaction between DnaA and Rok, it seems possible that DnaA could also interact with small, basic chromosome architecture proteins in other organisms. Interaction between the replication initiator and nucleoid-associated proteins analogous to Rok might then represent a broader regulatory connection between DNA replication and chromosome architecture. Beyond its role in replication initiation and gene expression, we suggest that DnaA might be involved in chromosome folding and compaction through direct interaction with nucleoid associated proteins.
Experimental Procedures
Strains and alleles
B. subtilis strains and relevant genotypes are listed in Table 3. Properties and construction of important alleles are described below.
Table 3.
B. subtilis strains used in this study.
| Strain | Relevant genotype (reference) |
|---|---|
| AG174 | trpC2 pheA1 (wild type, JH642) (Perego et al., 1988) |
| AIG200 | trp+, Δ{oriC dnaA dnaN}::spc, spoIIIJ::{oriN repN kan}, amyE::(PxylA-dnaN cat) (Goranov et al., 2005) |
| CAL2074 | Δ{oriC dnaA}::{oriN repN Ppen-2028-dnaN cat} |
| CAL2083 | Δ{oriC dnaA}::{oriN repN Ppen-2028-dnaN cat}, lacA::{Pspank-dnaA tet} |
| CAS192 | Δrok::cat::mls, Δ{oriC dnaA}::{oriN repN Ppen-2028-dnaN cat} |
| CAS196 | Δrok::cat::mls, Δ{oriC dnaA}::{oriN repN Ppen-2028-dnaN cat}, lacA::{Pspank-dnaA tet} |
| CAS221 | Δ{oriC dnaA}::{oriN repN Ppen-2028-dnaN cat}, amyE::{Pspank-dnaA spc} |
| CAS231 | Δ{oriC dnaA}::{oriN repN Ppen-2028-dnaN cat}, amyE::{Pspank-dnaAΔC spc} |
| HM57 | rok::pDG641rok (mls) (allele name rok57) (Smith & Grossman, 2015) |
| TAW5 | trp+, Δ{oriC dnaA dnaN}::spc, spoIIIJ::{oriN repN kan}, amyE::(PxylA-dnaN cat), lacA::(Pspank-dnaA tet) (Merrikh & Grossman, 2011) |
| WKS1038 | Δrok::cat (allele name rok1030) (Smits & Grossman, 2010) |
All strains are derived from AG174 and contain the trpC2 pheA1 alleles (not listed) unless otherwise indicated.
Δrok::cat
The rok open reading frame was replaced with a chloramphenicol resistance cassette (cat) to generate strain WKS1030 (Smits & Grossman, 2010) and the allele rok1030. Strain WKS1038 was the product of backcrossing genomic DNA from WKS1030 into wild type laboratory strain AG174. Strains CAS192 and CAS196 contained a derivative of this Δrok allele in which the chloramphenicol resistance gene was disrupted with an MLS (macrolide-lincosamide-streptogramin B) resistance gene (Δrok::cat::mls).
rok::pDG641rok (rok::mls) (allele rok57), contained in strain HM57, is a single crossover integration into rok of the plasmid pDG641rok. This plasmid was made using the vector pDG641 (Guerout-Fleury et al., 1995) and cloning an internal fragment of rok. The integration disrupts rok and confers a null phenotype.
Δ(oriC-dnaA-dnaN)
Strains AIG200 and TAW5 contain a deletion-insertion in which dnaA and most of dnaN are replaced with a spectinomycin resistance cassette (spc) (Goranov et al., 2005; Merrikh & Grossman, 2011). Replication is supported by insertion of the heterologous origin oriN and its initiator repN near oriC at spoIIIJ (Hassan et al., 1997; Moriya et al., 1997; Berkmen & Grossman, 2007). dnaN is expressed from the xylose-inducible promoter PxylA integrated at amyE. These strains still contain the dnaA promoter region, including the cluster of DnaA binding sites. Strains AIG200 and TAW5 contain a deviation in the ypjG-hepT region compared to that in AG174, containing tryptophan biosynthesis genes, as described previously (Berkmen & Grossman, 2007).
Strain CAL2074 and its derivatives contain a deletion-insertion in which dnaA and flanking regions are replaced with a product (generated by isothermal assembly (Gibson et al., 2009)) containing a chloramphenicol resistance cassette, oriN and repN, and a promoter driving constitutive expression of dnaN. The chloramphenicol resistance cassette (cat, including the transcription terminator) was inserted at the left end of oriC, upstream of rpmH. oriN and repN were inserted upstream of this cassette such that oriN-repN, cat, and rpmH were co-directional. A derivative of the constitutive promoter Ppen (Ppen-2028, C. Lee, unpublished data) was cloned upstream of dnaN. Ppen is derived from the B. licheniformis penicillinase gene and drives lacI expression on the amyE integration vector pDR110 (D. Rudner). Ppen-2028 carries mutations in Ppen (Ppen-2028 sequence in lowercase) between the putative −35 and −10 sequences (underlined): 5′-TTGCATTTAt ttcggtggcg tGTAATACTT TCAAA-3′ that decrease promoter activity.
dnaAΔC
Strain CAS231 contains a truncated version of dnaA that encodes a protein containing first 355 amino acids of DnaA and missing the C-terminal 91 amino acids that comprise most of the DNA binding domain, as annotated by the Conserved Domains feature of PubMed Protein (http://www.ncbi.nlm.nih.gov/protein/16077069). dnaAΔC was cloned downstream of the IPTG-inducible promoter Pspank in the NheI-linearized integration vector pDR110 using isothermal assembly. The fragments of dnaAΔC were amplified from a wild type dnaA gene that had previously been inserted into pDR110. The 5′ fragment of dnaAΔC was amplified using primers oCS105 (5′-CAATTAAGCT TAGTCGACAG CTAGCTCTAT AACAGAGAAA GACGC-3′) and oCS111 (5′-CGATTGATCC CCGGTCCTGC TA | CGTAATGA CTTTCGGTTT TGAG-3′; the vertical bar indicates the location of the deletion in dnaA). The 3′ fragment of dnaAΔC was amplified using primers oCS112 (5′-CTCAAAACCG AAAGTCATTA CG | TAGCAGGA CCGGGGATCA ATCG-3′; complementary to oCS111) and oCS106 (5′-CCACCGAATT AGCTTGCATG CGGCTAGCAG ACTGTGTATG ACTTCC-3′). The 5′ sequences in these primers correspond to regions in pDR110 and the underlined portions are sequences from within dnaA. The resulting product was transformed into AG174 cells for double-crossover integration at amyE. This intermediate oriC+ strain was then transformed with CAL2074 genomic DNA, selecting for the oriC::oriN insertion-deletion with chloramphenicol, to produce strain CAS231 (oriC- oriN+, amyE::Pspank-dnaAΔC). The same strategy was used for ectopic expression of full-length dnaA in CAS221 (oriC- oriN+, amyE::Pspank-dnaA). The full-length dnaA sequence was amplified using primers oCS105 and oCS106.
Media and growth conditions
Unless otherwise specified, all strains were grown at 30°C in S7 defined minimal medium buffered with 50 mM MOPS (Jaacks et al., 1989) and containing 1% glucose, 0.1% glutamate, trace metals, 40 μg/ml tryptophan, and 40 μg/ml phenylalanine. For growth of AIG200 and TAW5, glucose was replaced with 1% arabinose, and 0.5% xylose was used to induce expression of dnaN from the xylose-inducible promoter PxylA. To induce expression of DnaA from the LacI-repressible, IPTG-inducible promoter Pspank, 0.1 mM IPTG was added.
ChIP-seq
Immunoprecipitations were performed with anti-DnaA and anti-Rok rabbit polyclonal antiserum or with mouse monoclonal anti-myc antibodies (Invitrogen) essentially as described (Lin & Grossman, 1998; Merrikh & Grossman, 2011). Briefly, exponentially growing cells were treated with 1% formaldehyde, and the cross-linked lysates were sonicated to shear the DNA. Immunoprecipitations were performed by incubating the cross-linked lysates (after sonication) with antibodies for at least two hours at room temperature, followed by incubation with Protein A sepharose beads for at least one hour at room temperature. A control sample of non-immunoprecipitated lysate was incubated under the same conditions with Protein A sepharose beads. Immunoprecipitated (and control) material was washed and eluted from the beads, followed by reversal of cross-links by incubation at 65°C overnight. Samples were then treated with proteinase K at 37°C for at least two hours, and DNA was recovered using the Qiagen PCR purification kit. Sample preparation and single-read sequencing (40 nt) on an Illumina HiSeq were performed by the MIT BioMicro Center, essentially as described (Smith & Grossman, 2015). Seq data is available at NCBI under accession number PRJNA272948.
Antibody specificity
We used polyclonal antiserum from rabbits that had been immunized with purified Rok or DnaA (Covance). Our previous ChIP-chip experiments with DnaA used chicken anti-DnaA antibodies isolated from eggs (Breier & Grossman, 2009; Smits et al., 2011). We assessed the specificity of each antibody by performing ChIP-seq in appropriate null mutants, missing the protein of interest. There was little or no detectable precipitation of specific chromosomal regions in ChIP-seq experiments with anti-DnaA and anti-Rok antibodies from dnaA and rok null mutants, respectively. There were weak signals (three- to four-fold above background) in the anti-DnaA immunoprecipitations from a dnaA null mutant at the yoeC, yesX, dhbC, yfiZ, and yonT regions. These regions were not detectably associated with DnaA or Rok in ChIP-seq from wild type cells, and the sequence reads for these regions were not strongly symmetrical in the forward and reverse directions, indicating that they were most likely artifacts.
High-throughput sequencing analysis
We obtained approximately 7–50 million 40-nt reads for each sample. We mapped the reads to the Bacillus subtilis strain AG174 genome (Smith et al., 2014) using BWA (Burrows-Wheeler Aligner) for single-end short reads (Li & Durbin, 2009) and allowing a maximum number of alignments (n) of 2. To make comparisons across samples, we normalized the number of reads at each chromosomal position to the total number of reads for that sample. To calculate coverage at each base pair on the chromosome, we computationally extended each read by the estimated average fragment length of 250 bp (Smith & Grossman, 2015).
We used SISSRs (Jothi et al., 2008; Narlikar & Jothi, 2012) to identify enriched regions in each ChIP sample. We compared each ChIP sample to the corresponding non-immunoprecipitated (total) DNA sample. We used the following parameters: F (fragment length) = 250, e (sensitivity) = 1, m (fraction mappable) = 1, w (window) = 20, E (required number of reads) = 1 per million sample reads, L (maximum fragment length) = 400. We then optimized p (P-value) such that the enrichment fold cutoff was approximately 3. From these candidate regions, we selected those with an enrichment of at least 5-fold for DnaA and Rok and 4-fold for DnaAΔC.
To search for a Rok binding motif, we used regions that were enriched at least five-fold in the Rok immunoprecipitates as determined by SISSRs. For each region, we extracted 101 nucleotides of sequence, centered on the midpoint identified by SISSRs. These sequences were used as input for the online motif-finding tool DREME {http://meme.nbcr.net/meme/cgi-bin/meme.cgi} (Bailey, 2011), using the following parameters: comparison source = shuffled sequences, both strands used, and maximum E-value = 1.
DNA microarrays
Global mRNA levels were analyzed by hybridization to DNA microarrays as described (Goranov et al., 2009). Exponentially growing cells from at least three replicate cultures were fixed with an equal volume of −20°C methanol. RNA was purified from lysates using the Qiagen RNeasy kit. Experimental and reference RNA samples (Goranov et al., 2009) were reverse transcribed using Superscript II reverse transcriptase (Invitrogen), random hexamers, and aminoallyl-dUTP (Ambion). The cDNA was labeled by conjugation to monofunctional Cy3 or Cy5 dyes (Amersham) for reference or experimental samples, respectively. Each experimental sample was mixed with an aliquot of reference sample. Salmon testes DNA and yeast tRNA were added, and each cDNA sample was hybridized to a DNA microarray at 42°C overnight. Microarrays contained PCR products from >95% of the annotated B. subtilis ORFs spotted onto Corning GAPS slides. Microarrays were scanned with a GenePix 4000B scanner, and images were analyzed using GenePix 3.0 (Axon Instruments).
Data were analyzed using the R statistical software package Linear Models for Microarray Data (LIMMA) (Smyth et al., 2005). Spot intensities were normalized within and between arrays, and gene expression values were corrected for multiple hypothesis testing using the Benjamini-Hochberg correction option. Genes were considered differentially expressed if the adjusted P-value was <0.04.
Expression and purification of DnaAΔC and Rok
The coding sequence of dnaAΔC (i.e. the first 355 codons of dnaA) was cloned between the NcoI and NotI sites of pET-28b (Novagen), generating pCAS254, which produces DnaAΔC with a C-terminal hexa-histidine tag (DnaAΔC-his). The construct was transformed into E. coli BL21 pLysS, and expression was induced with 1 mM IPTG for 5 hr during growth in LB medium at 37°C. A pellet from 1 liter of cells was frozen at −80°C, thawed, and resuspended in lysis buffer (50 mM NaPO4 pH 7, 5 mM imidazole, 300 mM NaCl) containing AEBSF protease inhibitor (Sigma). MgCl2 (10 mM) and 3 μl Benzonase Nuclease (EMD Millipore) were added, and the lysate was stirred for 10 min. The lysate was cleared by centrifugation. The supernatant was loaded onto a 1-ml HisTALON column (Clontech). The column was washed with Talon buffer A (50 mM NaPO4 pH 7, 300 mM NaCl, 10% glycerol), followed by Talon buffer A containing 4.5 mM imidazole. The column was then eluted with a linear gradient of 4.5 to 150 mM imidazole in Talon buffer A. Fractions containing DnaAΔC were identified by SDS polyacrylamide gel electrophoresis, pooled, and combined with two volumes of buffer containing 45 mM HEPES-KOH pH 7.6, 0.75 mM EDTA, 15 mM magnesium acetate, 1.5 mM DTT, and 5% sucrose. The solution was loaded onto a 5-ml HiTrap Q FF column (GE Healthcare Life Sciences) and washed with Q buffer A (45 mM HEPES-KOH pH 7.6, 0.5 mM EDTA, 10 mM magnesium acetate, 1 mM DTT, 5% sucrose, 100 mM potassium glutamate). The column was eluted with a linear gradient to Q buffer B (Q buffer A containing 1 M potassium glutamate). The fractions containing DnaAΔC eluted in 100% Q buffer B. These fractions were pooled, and aliquots were stored at −80°C.
Rok was expressed with a C-terminal hexa-histidine tag from pED428 in E. coli M15 (Hoa et al., 2002). Expression was induced with 1 mM IPTG for 3 h during growth in LB medium at 37°C. A pellet from 500 ml of cells was resuspended in lysis buffer (50 mM NaPO4 pH 7, 4.5 mM imidazole, 300 mM NaCl) and frozen at −80°C. The cell suspension was thawed, and MgCl2 (10 mM), AEBSF protease inhibitor (Sigma), and 1.5 μl Benzonase Nuclease (EMD Millipore) were added. The suspension was stirred for 15 min, lysed with lysozyme (1 mg/ml), and stirred for an additional 15 min. The lysate was cleared by centrifugation. The supernatant was loaded onto a 1-ml HisTALON column (Clontech). The column was washed with Talon buffer A (50 mM NaPO4 pH 7, 300 mM NaCl, 10% glycerol), followed by Talon buffer A containing 25 mM imidazole. The column was then eluted with a linear gradient to Talon buffer B (Talon buffer A containing 2 M imidazole). Fractions containing Rok were identified by SDS-PAGE, pooled, and loaded onto a HiTrap Heparin HP column (GE Healthcare Life Sciences). The column was washed with Talon buffer A and eluted with a linear gradient to Heparin buffer B (Talon buffer A containing 2 M NaCl). Fractions containing Rok were identified by SDS-PAGE and pooled. The buffer for these fractions contained 750 mM NaCl, and glycerol was added to a final concentration of 10%. Protein was concentrated using a Vivaspin 6 5 kDa MWCO concentrator unit (GE Healthcare Life Sciences), and aliquots were stored at −80°C.
Using quantitative western blotting on an Odyssey infrared imager (Licor), we estimated that there were at least 30,000 molecules of Rok per cell during exponential growth in defined minimal glucose medium at 30°C. Culture samples of 15 ml were pelleted, and all but 1 ml supernatant was removed. Samples were frozen at −80°C and resuspended with the addition of 14 ml TE (10 mM Tris pH 8.0, 10 mM EDTA) with protease inhibitors (AEBSF). The optical density (proportional to the CFU/ml) of the resuspension was measured in triplicate and used for normalization. Cells were lysed by sonication rather than lysozyme treatment because lysozyme cross-reacts with the LiCor goat anti-rabbit 800 secondary antibody used for quantitation. Samples were sonicated on ice for 6 min per sample, in bursts of 0.3 sec on/off. A 450-μl aliquot was taken, and 50 μl of TE plus protease inhibitors (AEBSF) was added. Equal volumes of each sample were analyzed on a 15% SDS-PAGE gel, along with standards of purified untagged Rok. Samples were imaged and quantitated on the LiCor scanner. Protein intensities from the lysates were compared to serial dilutions of the purified protein standards. Estimates of the number of molecules per cell were calculated by normalization to optical density of the cell sample and comparison to the CFU/ml for each strain. Relative protein levels (between-strains comparisons) were made by directly comparing the western blot signal after normalization for optical density.
The amount of Rok per cell was similar to the concentrations of several other nucleoid-associated proteins. For example, there are ~20,000 molecules per cell of H-NS in Salmonella typhimurium and E. coli (Hulton et al., 1990; Ali Azam et al., 1999), ~50,000 molecules/genome of HBsu in B. subtilis (Ragkousi et al., 2000), and ~40,000 molecules/cell of HU in E. coli (Ali Azam et al., 1999). Previous estimates of approximately 1,000 – 3,000 Rok molecules/genome were based on measurement of Rok-myc compared to HBsu-myc (Smits & Grossman, 2010), which we now believe was an underestimate.
Gel electrophoresis mobility shift assay
A 342-bp region of the rok promoter was amplified from B. subtilis genomic DNA using primers HM57 (5′-CGGGATCCGC TTCTCTTTCA TTAAACAT-3′) and HM58 (5′-CGGAATTCGA TGTTTTTCCT CAATTTTAG-3′). The PCR product was purified with a PCR purification column (Qiagen), and then further purified on a 6% polyacrylamide gel. The purified probe was end-labeled with radioactive gamma-32P-ATP (Perkin Elmer) using T4 Polynucleotide Kinase (New England Biolabs). Excess label was removed using a MicroSpin G-50 column (GE Healthcare Life Sciences). The DNA probe was used at a final concentration of 0.1 nM in each reaction.
Gel shift reactions were performed in gel shift buffer (5 mM NaPO4 pH 7, 15 mM HEPES-KOH pH 7.6, 10 mM magnesium acetate, 300 mM NaCl, 100 mM potassium glutamate, 0.5 mM EDTA, 10% glycerol, 50 μg/ml bovine serum albumin, 0.5% sucrose, 0.1 mM DTT, and 0.0025% xylene cyanol) with protein concentrations as indicated in the figure legends. Reactions were incubated at room temperature for 20 min. Samples of each reaction were run on a 5% polyacrylamide gel (37:1 acrylamide/bisacrylamide) containing 0.5X Tris-borate-EDTA (TBE) and 2.5% glycerol using a Hoefer Mighty Small II electrophoresis apparatus. Gels were run in 0.5X TBE at 100 V for 3 min followed by 60 V (approximately 9 V/cm) for 3 h at 4°C. Gels were dried, exposed to a storage phosphor screen (GE Healthcare Life Sciences), and imaged on a Typhoon scanner (GE Healthcare Life Sciences). These reaction conditions were different from those used previously for analysis of Rok binding to DNA (Hoa et al., 2002; Albano et al., 2005).
Supplementary Material
Acknowledgments
We thank the MIT BioMicro Center for preparation of samples for DNA sequencing and for Illumina sequencing, C.A. Lee for construction of the oriC-oriN allele in strain CAL2074; C.A. Lee, H. Merrikh, and T. Washington for useful discussions, and C.A. Lee for comments on the manuscript.
This work was supported, in part, by the Department of Defense, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEG) Fellowship, 32 CFR 168a, to CAS and Public Health Service grant GM41934 to ADG.
References
- Albano M, Smits WK, Ho LT, Kraigher B, Mandic-Mulec I, Kuipers OP, Dubnau D. The Rok protein of Bacillus subtilis represses genes for cell surface and extracellular functions. J Bacteriol. 2005;187:2010–2019. doi: 10.1128/JB.187.6.2010-2019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit R, Oppenheim AB, Stavans J. Increased bending rigidity of single DNA molecules by H-NS, a temperature and osmolarity sensor. Biophys J. 2003;84:2467–2473. doi: 10.1016/S0006-3495(03)75051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arold ST, Leonard PG, Parkinson GN, Ladbury JE. H-NS forms a superhelical protein scaffold for DNA condensation. Proc Natl Acad Sci U S A. 2010;107:15728–15732. doi: 10.1073/pnas.1006966107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka RM, Hahn J, Albano M, Draskovic I, Persuh M, Cui X, Sloma A, Widner W, Dubnau D. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol Microbiol. 2002;43:1331–1345. doi: 10.1046/j.1365-2958.2002.02833.x. [DOI] [PubMed] [Google Scholar]
- Berkmen MB, Grossman AD. Subcellular positioning of the origin region of the Bacillus subtilis chromosome is independent of sequences within oriC, the site of replication initiation, and the replication initiator DnaA. Mol Microbiol. 2007;63:150–165. doi: 10.1111/j.1365-2958.2006.05505.x. [DOI] [PubMed] [Google Scholar]
- Bonilla CY, Grossman AD. The primosomal protein DnaD inhibits cooperative DNA binding by the replication initiator DnaA in Bacillus subtilis. J Bacteriol. 2012;194:5110–5117. doi: 10.1128/JB.00958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier AM, Grossman AD. Dynamic association of the replication initiator and transcription factor DnaA with the Bacillus subtilis chromosome during replication stress. J Bacteriol. 2009;191:486–493. doi: 10.1128/JB.01294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104:269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Cho E, Ogasawara N, Ishikawa S. The functional analysis of YabA, which interacts with DnaA and regulates initiation of chromosome replication in Bacillus subtils. Genes Genet Syst. 2008;83:111–125. doi: 10.1266/ggs.83.111. [DOI] [PubMed] [Google Scholar]
- Clarey MG, Erzberger JP, Grob P, Leschziner AE, Berger JM, Nogales E, Botchan M. Nucleotide-dependent conformational changes in the DnaA-like core of the origin recognition complex. Nat Struct Mol Biol. 2006;13:684–690. doi: 10.1038/nsmb1121. [DOI] [PubMed] [Google Scholar]
- Crooke E, Thresher R, Hwang DS, Griffith J, Kornberg A. Replicatively active complexes of DnaA protein and the Escherichia coli chromosomal origin observed in the electron microscope. J Mol Biol. 1993;233:16–24. doi: 10.1006/jmbi.1993.1481. [DOI] [PubMed] [Google Scholar]
- Dame RT, Noom MC, Wuite GJ. Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature. 2006;444:387–390. doi: 10.1038/nature05283. [DOI] [PubMed] [Google Scholar]
- Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman CJ. H-NS, the genome sentinel. Nat Rev Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- Dorman CJ. Horizontally acquired homologues of the nucleoid-associated protein H-NS: implications for gene regulation. Mol Microbiol. 2010;75:264–267. doi: 10.1111/j.1365-2958.2009.06996.x. [DOI] [PubMed] [Google Scholar]
- Dorman CJ, Kane KA. DNA bridging and antibridging: a role for bacterial nucleoid-associated proteins in regulating the expression of laterally acquired genes. FEMS Microbiol Rev. 2009;33:587–592. doi: 10.1111/j.1574-6976.2008.00155.x. [DOI] [PubMed] [Google Scholar]
- Duderstadt KE, Chuang K, Berger JM. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478:209–213. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duderstadt KE, Mott ML, Crisona NJ, Chuang K, Yang H, Berger JM. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J Biol Chem. 2010;285:28229–28239. doi: 10.1074/jbc.M110.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- Fuller RS, Funnell BE, Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984;38:889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Funnell BE, Baker TA, Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem. 1987;262:10327–10334. [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Goranov AI, Breier AM, Merrikh H, Grossman AD. YabA of Bacillus subtilis controls DnaA-mediated replication initiation but not the transcriptional response to replication stress. Mol Microbiol. 2009;74:454–466. doi: 10.1111/j.1365-2958.2009.06876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goranov AI, Katz L, Breier AM, Burge CB, Grossman AD. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc Natl Acad Sci U S A. 2005;102:12932–12937. doi: 10.1073/pnas.0506174102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon BR, Imperial R, Wang L, Navarre WW, Liu J. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J Bacteriol. 2008;190:7052–7059. doi: 10.1128/JB.00733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon BR, Li Y, Wang L, Sintsova A, van Bakel H, Tian S, Navarre WW, Xia B, Liu J. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2010;107:5154–5159. doi: 10.1073/pnas.0913551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- Hassan AK, Moriya S, Ogura M, Tanaka T, Kawamura F, Ogasawara N. Suppression of initiation defects of chromosome replication in Bacillus subtilis dnaA and oriC-deleted mutants by integration of a plasmid replicon into the chromosomes. J Bacteriol. 1997;179:2494–2502. doi: 10.1128/jb.179.8.2494-2502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JW, Bishop E, Karchenko PV, Negre N, White KP, Park PJ. ChIP-chip versus ChIP-seq: lessons for experimental design and data analysis. BMC Genomics. 2011;12:134. doi: 10.1186/1471-2164-12-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa TT, Tortosa P, Albano M, Dubnau D. Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol Microbiol. 2002;43:15–26. doi: 10.1046/j.1365-2958.2002.02727.x. [DOI] [PubMed] [Google Scholar]
- Hoover SE, Xu W, Xiao W, Burkholder WF. Changes in DnaA-dependent gene expression contribute to the transcriptional and developmental response of Bacillus subtilis to manganese limitation in Luria-Bertani medium. J Bacteriol. 2010;192:3915–3924. doi: 10.1128/JB.00210-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulton CS, Seirafi A, Hinton JC, Sidebotham JM, Waddell L, Pavitt GD, Owen-Hughes T, Spassky A, Buc H, Higgins CF. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell. 1990;63:631–642. doi: 10.1016/0092-8674(90)90458-q. [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Ogura Y, Yoshimura M, Okumura H, Cho E, Kawai Y, Kurokawa K, Oshima T, Ogasawara N. Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method. DNA Res. 2007;14:155–168. doi: 10.1093/dnares/dsm017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaacks KJ, Healy J, Losick R, Grossman AD. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K. To divide or not to divide: control of the bacterial cell cycle by environmental cues. Curr Opin Microbiol. 2014;18:54–60. doi: 10.1016/j.mib.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- Kawakami H, Katayama T. DnaA, ORC, and Cdc6: similarity beyond the domains of life and diversity. Biochem Cell Biol. 2010;88:49–62. doi: 10.1139/o09-154. [DOI] [PubMed] [Google Scholar]
- Kovacs AT, Kuipers OP. Rok regulates yuaB expression during architecturally complex colony development of Bacillus subtilis 168. J Bacteriol. 2011;193:998–1002. doi: 10.1128/JB.01170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Messer W. DnaA proteins of Escherichia coli and Bacillus subtilis: coordinate actions with single-stranded DNA-binding protein and interspecies inhibition during open complex formation at the replication origins. Gene. 1999;228:123–132. doi: 10.1016/s0378-1119(99)00009-8. [DOI] [PubMed] [Google Scholar]
- Krause M, Ruckert B, Lurz R, Messer W. Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J Mol Biol. 1997;274:365–380. doi: 10.1006/jmbi.1997.1404. [DOI] [PubMed] [Google Scholar]
- Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 1999;18:6642–6652. doi: 10.1093/emboj/18.23.6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Grossman AD. The chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) contribute to accurate chromosome partitioning, separation of replicated sister origins, and regulation of replication initiation in Bacillus subtilis. Mol Microbiol. 2006;60:853–869. doi: 10.1111/j.1365-2958.2006.05140.x. [DOI] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE. Regulating DnaA complex assembly: it is time to fill the gaps. Curr Opin Microbiol. 2010;13:766–772. doi: 10.1016/j.mib.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE. Regulation of DnaA assembly and activity: taking directions from the genome. Annu Rev Microbiol. 2011;65:19–35. doi: 10.1146/annurev-micro-090110-102934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbouty M, Le Gall A, Cattoni DI, Cournac A, Koh A, Fiche JB, Mozziconacci J, Murray H, Koszul R, Nollmann M. Condensin- and Replication-Mediated Bacterial Chromosome Folding and Origin Condensation Revealed by Hi-C and Super-resolution Imaging. Mol Cell. 2015;59:588–602. doi: 10.1016/j.molcel.2015.07.020. [DOI] [PubMed] [Google Scholar]
- Marciniak BC, Trip H, Fusetti F, Kuipers OP. Regulation of ykrL (htpX) by Rok and YkrK, a novel type of regulator in Bacillus subtilis. J Bacteriol. 2012;194:2837–2845. doi: 10.1128/JB.00324-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry KC, V, Ryan T, Grimwade JE, Leonard AC. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc Natl Acad Sci U S A. 2004;101:2811–2816. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh H, Grossman AD. Control of the replication initiator DnaA by an anti-cooperativity factor. Mol Microbiol. 2011;82:434–446. doi: 10.1111/j.1365-2958.2011.07821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol Rev. 2002;26:355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- Messer W, Weigel C. DnaA initiator--also a transcription factor. Mol Microbiol. 1997;24:1–6. doi: 10.1046/j.1365-2958.1997.3171678.x. [DOI] [PubMed] [Google Scholar]
- Moriya S, Atlung T, Hansen FG, Yoshikawa H, Ogasawara N. Cloning of an autonomously replicating sequence (ars) from the Bacillus subtilis chromosome. Mol Microbiol. 1992;6:309–315. doi: 10.1111/j.1365-2958.1992.tb01473.x. [DOI] [PubMed] [Google Scholar]
- Moriya S, Fukuoka T, Ogasawara N, Yoshikawa H. Regulation of initiation of the chromosomal replication by DnaA-boxes in the origin region of the Bacillus subtilis chromosome. EMBO J. 1988;7:2911–2917. doi: 10.1002/j.1460-2075.1988.tb03149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya S, Hassan AK, Kadoya R, Ogasawara N. Mechanism of anucleate cell production in the oriC-deleted mutants of Bacillus subtilis. DNA Res. 1997;4:115–126. doi: 10.1093/dnares/4.2.115. [DOI] [PubMed] [Google Scholar]
- Mott ML, Berger JM. DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- Narlikar L, Jothi R. ChIP-Seq data analysis: identification of protein-DNA binding sites with SISSRs peak-finder. Methods Mol Biol. 2012;802:305–322. doi: 10.1007/978-1-61779-400-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Nishida S, Fujimitsu K, Sekimizu K, Ohmura T, Ueda T, Katayama T. A nucleotide switch in the Escherichia coli DnaA protein initiates chromosomal replication: evidnece from a mutant DnaA protein defective in regulatory ATP hydrolysis in vitro and in vivo. J Biol Chem. 2002;277:14986–14995. doi: 10.1074/jbc.M108303200. [DOI] [PubMed] [Google Scholar]
- Noirot-Gros MF, Dervyn E, Wu LJ, Mervelet P, Errington J, Ehrlich SD, Noirot P. An expanded view of bacterial DNA replication. Proc Natl Acad Sci U S A. 2002;99:8342–8347. doi: 10.1073/pnas.122040799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noom MC, Navarre WW, Oshima T, Wuite GJ, Dame RT. H-NS promotes looped domain formation in the bacterial chromosome. Curr Biol. 2007;17:R913–914. doi: 10.1016/j.cub.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Ogasawara N, Moriya S, Yoshikawa H. Initiation of chromosome replication: structure and function of oriC and DnaA protein in eubacteria. Res Microbiol. 1991;142:851–859. doi: 10.1016/0923-2508(91)90065-i. [DOI] [PubMed] [Google Scholar]
- Ogura M, Yamaguchi H, Kobayashi K, Ogasawara N, Fujita Y, Tanaka T. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J Bacteriol. 2002;184:2344–2351. doi: 10.1128/JB.184.9.2344-2351.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Imai Y, Ogasawara N, Moriya S. Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J Bacteriol. 2001;183:3833–3841. doi: 10.1128/JB.183.13.3833-3841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura H, Yoshimura M, Ueki M, Oshima T, Ogasawara N, Ishikawa S. Regulation of chromosomal replication initiation by oriC-proximal DnaA-box clusters in Bacillus subtilis. Nucleic Acids Res. 2012;40:220–234. doi: 10.1093/nar/gkr716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M, Spiegelman GB, Hoch JA. Structure of the gene for the transition state regulator, AbrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- Ragkousi K, Cowan AE, Ross MA, Setlow P. Analysis of nucleoid morphology during germination and outgrowth of spores of Bacillus species. J Bacteriol. 2000;182:5556–5562. doi: 10.1128/jb.182.19.5556-5562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn-Lee L, Gorbatyuk B, Skovgaard O, Losick R. The conserved sporulation protein YneE inhibits DNA replication in Bacillus subtilis. J Bacteriol. 2009;191:3736–3739. doi: 10.1128/JB.00216-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson TT, Harran O, Murray H. The bacterial DnaA-trio replication origin element specifies single-stranded DNA initiator binding. Nature. 2016;534:412–416. doi: 10.1038/nature17962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Messer W. The DNA binding domain of the initiator protein DnaA. EMBO J. 1995;14:2106–2111. doi: 10.1002/j.1460-2075.1995.tb07202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield G, Errington J, Murray H. Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 2012;31:1542–1555. doi: 10.1038/emboj.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield G, Whiting R, Errington J, Murray H. Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol Microbiol. 2011;79:1089–1100. doi: 10.1111/j.1365-2958.2010.07507.x. [DOI] [PubMed] [Google Scholar]
- Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50:259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- Singh PK, Ramachandran G, Duran-Alcalde L, Alonso C, Wu LJ, Meijer WJ. Inhibition of Bacillus subtilis natural competence by a native, conjugative plasmid-encoded comK repressor protein. Environ Microbiol. 2012;14:2812–2825. doi: 10.1111/j.1462-2920.2012.02819.x. [DOI] [PubMed] [Google Scholar]
- Skarstad K, Katayama T. Regulating DNA replication in bacteria. Cold Spring Harb Perspect Biol. 2013;5:a012922. doi: 10.1101/cshperspect.a012922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Goldberg JM, Grossman AD. Complete genome sequences of Bacillus subtilis subsp. subtilis laboratory strains JH642 (AG174) and AG1839. Genome Announc. 2014;2:e00663–00614. doi: 10.1128/genomeA.00663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Grossman AD. In Vitro Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA. PLoS Genet. 2015;11:e1005258. doi: 10.1371/journal.pgen.1005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits WK, Grossman AD. The transcriptional regulator Rok binds A+T-rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis. PLoS Genet. 2010;6:e1001207. doi: 10.1371/journal.pgen.1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits WK, Merrikh H, Bonilla CY, Grossman AD. Primosomal proteins DnaD and DnaB are recruited to chromosomal regions bound by DnaA in Bacillus subtilis. J Bacteriol. 2011;193:640–648. doi: 10.1128/JB.01253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- Speck C, Weigel C, Messer W. ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 1999;18:6169–6176. doi: 10.1093/emboj/18.21.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippner D, Wagner R. Fluorescence analysis of the Escherichia coli transcription regulator H-NS reveals two distinguishable complexes dependent on binding to specific or nonspecific DNA sites. J Biol Chem. 1995;270:22243–22247. doi: 10.1074/jbc.270.38.22243. [DOI] [PubMed] [Google Scholar]
- Veening JW, Murray H, Errington J. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 2009;23:1959–1970. doi: 10.1101/gad.528209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JK, Marquis KA, Rudner DZ. SirA enforces diploidy by inhibiting the replication initiator DnaA during spore formation in Bacillus subtilis. Mol Microbiol. 2009;73:963–974. doi: 10.1111/j.1365-2958.2009.06825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska-Czerwinska J, Jakimowicz D, Zawilak-Pawlik A, Messer W. Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol Rev. 2007;31:378–387. doi: 10.1111/j.1574-6976.2007.00070.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.