Abstract
Given the capacity of self-renewal and multilineage differentiation, stem cells are promising sources for use in regenerative medicines as well as in the clinical treatment of certain hematological malignancies and degenerative diseases. Complex networks of cellular signaling pathways largely determine stem cell fate and function. Small molecules that modulate these pathways can provide important biological and pharmacological insights. However, it is still challenging to identify the specific protein targets of these compounds, to explore the changes in stem cell phenotypes induced by compound treatment and to ascertain compound mechanisms of action. To facilitate stem cell related small molecule study and provide a better understanding of the associated signaling pathways, we have constructed a comprehensive domain-specific chemogenomics resource, called StemCellCKB (http://www.cbligand.org/StemCellCKB/). This new cloud-computing platform describes the chemical molecules, genes, proteins, and signaling pathways implicated in stem cell regulation. StemCellCKB is also implemented with web applications designed specifically to aid in the identification of stem cell relevant protein targets, including TargetHunter, a machine-learning algorithm for predicting small molecule targets based on molecular fingerprints, and HTDocking, a high-throughput docking module for target prediction and systems-pharmacology analyses. We have systematically tested StemCellCKB to verify data integrity. Target-prediction accuracy has also been validated against the reported known target/compound associations. This proof-of-concept example demonstrates that StemCellCKB can (1) accurately predict the macromolecular targets of existing stem cell modulators and (2) identify novel small molecules capable of probing stem cell signaling mechanisms, for use in systems-pharmacology studies. StemCellCKB facilitates the exploration and exchange of stem cell chemogenomics data among members of the broader research community.
Graphical abstract
INTRODUCTION
Stem cells are capable of self-renewal and multilineage differentiation, thereby offering an indispensable cellular source for stem cell therapies and regenerative medicine.1 Given stem cells’ endogenous characteristics and biological activity in vivo,2 they offer a promising regenerative medicine strategy to replace, engineer, as well as regenerate human cells, tissues, or organs in order to restore or establish normal function.3,4 However, the greater potential of stem cell-based therapies is not fully realized due to the scarcity of these cells as well as the complicated stem cell signaling mechanisms and poor understanding of their expansion and differentiation behaviors in response to proliferative stimuli. Among all stem cell types, for example, hematopoietic stem cell (HSC) is most widely used in clinical treatment.5 The first clinical use of HSC in transplantation occurred over 50 years ago,6 and HSC transplantation continues to be an indispensable therapy for hematological malignancies.7,8 However, it is difficult to separate stem cells from solid tissues and organs because the ratio of stem cells to mature cells in adults is extremely low and specific stem cell markers have not yet been definitively identified.
Furthermore, stem cell fate choices include self-renewal, differentiation, quiescence, apoptosis, or exhaustion. Complex signaling pathway networks precisely modulate these choices. For example, the Wnt,9 Notch,10 Hedgehog,11 and transforming growth factor (TGF)12 pathways have been implicated in the homeostasis, self-renewal, and multilineage differentiation of normal stem cells. Disrupting the complicated signaling pathway networks that govern homeostasis can cause malignant transformation, producing cancer stem cells;13 consequently, understanding these networks is vitally important. Recent advances in stem cell biology are increasingly dependent on this understanding. For example, new technologies can reprogram somatic cells to behave like embryonic stem cells (ESC). The resulting pluripotent cells can then differentiate into specified cell types14 and so have great potential for use in regenerative therapies. In addition, stem cell manipulation has historically depended on genetic techniques like gene overexpression or silencing via viral infection. These experimental procedures are slow, complicated, tedious, and quite often tumorigenic and, so, are not amenable to the large-scale, quality-controlled cell production that clinical therapies require.15
Given the endogenous characteristics and biological activity of stem cells in vivo,2 small molecules that modulate stem cell self-renewal and committed differentiation may achieve specificity and efficacy that are not possible with conventional methods. Thus, a chemical approach provides a greater potential for stem cell regulation and has emerged as a promising alternative because it is far simpler to control compound exposure to cells, tissues, and organs than it is to manipulate genes.16 Careful adjustment of the administered concentrations and use of combinations of stem cell regulating small molecules may allow for rapid, spatial, and reversible effects. Small molecule modulation has initiated a new trend in stem cell biology.17,18 However, the large-scale clinical use of treated cells may present a safety risk because some of these compounds have unknown targets and mechanisms of action. The number of published studies focusing on stem cell-related signaling pathways and small molecule modulators is steadily increasing, while generating the experimental data required to find, associate, and validate reported active compounds can be slow and challenging.2,19
The points given above briefly describe the potential and the challenges of the emerging multidimensional data and information from the fields of stem cell biology/biochemistry, chemical biology, and regenerative medicine research from the past decades. It is anticipated that the stem cell research data and information will continue growing rapidly and will involve scientists in various fields. Thus, there is a critical need to establish an integrated “big-data” type platform to facilitate research endeavors in the broader stem cell scientific community.
In fact, computational predictions and systems biology modeling can guide, supplement, and accelerate experimental studies. We, and many others, have previously reported several computational approaches for target prediction and modeling. Examples of these include, Connecting Map, which uses gene-expression signatures to connect small molecules, genes, and diseases,20 Similarity Ensemble Approach (SEA), which relates protein pharmacology with ligand chemistry,21 STITCH, a resource to explore known and predicted interactions of chemicals and proteins,22 and AlzPlatform, an Alzheimer’s disease (AD) specific chemogenomics database for analyzing the polypharmacological networks of anti-AD drugs,23 just to name a few. These platform technologies have been used successfully in drug-abuse systems-pharmacology research,24 as well as in studies describing the network pharmacology and molecular mechanism of an herbal medicine25 and were built using several of our established algorithms. These algorithms enable virtual screening, protein-target identification, and network-polypharmacology studies19 and include a GPU-accelerated machine-learning algorithm for ligand-specificity and functionality prediction,26–29 a molecular-fingerprint-based drug-target identification program,30 a cannabinoid-receptor homology-modeling protocol, and a virtual-screening work-flow.31,32
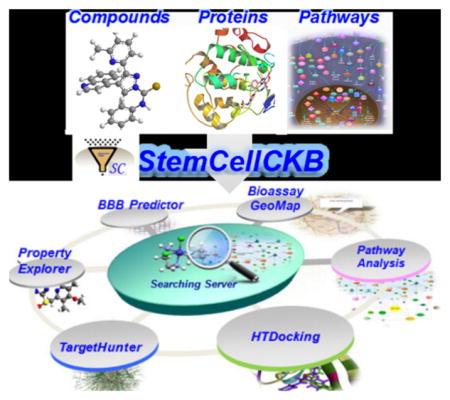
Here we describe an integrated cloud-computing server, which we have named StemCellCKB (http://www.cbligand.org/StemCellCKB),33 that was developed in response to the needs described above. The platform assembles a large repertoire of stem cell related chemogenomics data, including databases of genes, protein targets, and small molecule compounds with associated bioactivity, bioassay, and reference information. StemCellCKB also enables cloud-computing and sourcing services, exploiting powerful online computing tools for target identification (e.g., our TargetHunter and HTDocking programs24,30), drug repurposing, and polypharmacological analyses of small molecule stem cell modulators (Figure 1). StemCellCKB is a valuable platform for investigating and sharing information about stem cell-related proteins/genes and small molecule compounds on a chemogenomics scale, with the ultimate goal of facilitating identification of systems-poly-pharmacology mechanisms to enable stem cell chemical biology.
Figure 1.
StemCellCKB overview, including integrated computing and data-mining functions. The COMPOUND module includes two chemical records that describe small molecules specific to stem cell targets and small molecules involved in stem cell pathways. The TARGET module contains a list of proteins relevant to stem cell regulation and the HTDocking tool. The PATHWAY module describes signaling pathways relevant to stem cell regulation. The TOOLS module enables prediction of chemical properties, toxicity, and targets for small molecule stem cell modulators.
MATERIALS AND METHODS
Constructing the Stem Cell Database
StemCellCKB was constructed using the same technologies we developed for our AlzPlatform23 and drug-abuse chemogenomics databases.24 The backend consists of a MySQL database version 5.1,34 an Apache web server version 2.2.14,35 and our in-house chemoinformatics tools. We populated the MySQL database with information about stem cell-related genes, proteins, pathways, compounds, and bioassays by mining public databases and the literature. The current version of StemCellCKB provides the following information and data records:
Genes/Proteins/Pathways
We compiled a list of proteins relevant to key pathways involved in the regulation of stem cell function, as reported in the literature, patents, and some databases (e.g., DrugBank version 4.3,36,37 ClinicalTrials.gov,38 MetaCore version 6.24,39 PubChem,40 and PubMed41). These target genes and proteins were then mapped to their respective UniprotKB IDs. To allow users to further explore other potential pathways involved in stem cell regulation, all the pathways associated with these proteins/genes have been hyper-linked to the public KEGG database release 77.1.42–44
Chemicals
To identify compounds that target these stem cell related proteins/genes, we thoroughly searched both the literature and the ChEMBL database (version 20).45,46 Drugs in clinical trials were also considered in hopes that they can be repurposed for stem cell research. We also collected any bioassay data that confirms interactions between these molecules and the respective target proteins.
User-Friendly Web Interface
An easy-to-use web interface was created using the PHP version 5.3.1.47 This interface provides an effective and efficient search engine for data retrieval. The user can either draw molecular structures in the browser using JSME v2013-10-1348 or upload and submit a file describing small-molecule structures in the SMILES, sdf, mol, or cdx formats. On the backend, OpenBabel version 2.3.249 serves as the small molecule structure search engine.
Chemoinformatics Tools
Two chemoinformatics tools have been incorporated into the online platform to facilitate stem cell-related drug design and target identification:
TargetHunter
StemCellCKB includes our online target-identification program, TargetHunter30,50 for predicting the targets and potential off-targets of submitted compounds. TargetHunter uses a powerful data-mining algorithm (TAMO-SIC) for target prediction that exploits an important principle of medicinal chemistry: compounds with structural similarities often have similar physicochemical properties and biological profiles. Bioassay GeoMap, integrated into TargetHunter, provides a search function to locate potential collaborators nearby who have established bioassays to experimentally validate target predictions.
HTDocking
StemCellCKB also includes an online high-throughput molecular docking program HTDocking,51 for identifying possible interactions between protein targets and small molecules. Briefly, all available PDB structure files of stem cell-related target proteins were downloaded from the Protein Data Bank. HTDocking first docks query compounds into each of these potential targets using AutoDock Vina version 1.1.252 and then ranks them by the docking scores, which are roughly proportional to binding affinities. As we described previously,23 the docking score is calculated as pKi, where pKi = −log(predicted Ki) and the predicted Ki = exp(1000ΔG/ (1.987 191 7 × 298.15). ΔG (kJ mol−1) is the best binding affinity value predicted by AutoDock Vina. A good docking score (greater than 6.0, or predicted Ki < 1000 nM) indicates that the protein is a candidate target of the queried small molecule.
In addition, StemCellCKB provides toxicity predictions53 with the Toxtree package version 2.5.0,54 PAINS55 remover56 to filter out protein-reactive compounds, and a property calculator57 for the calculation of molecular properties and Lipinski’s rule of five.58
RESULTS
Database Construction and Analysis
Stem cells play critical roles in regenerative medicine and the clinical treatment of various diseases (e.g., hematological malignancy, degenerative diseases, cancer, etc.).5 Small molecules have emerged as novel and potent agents for stem cell expansion and differentiation, as well as somatic cell reprogramming.17 We have constructed a new online platform called StemCellCKB that focuses on the three most commonly studied stem cell types: hematopoietic stem cell (HSC), embryonic stem cell (ESC), and induced pluripotent stem cell (iPS). Figure 1 shows an overview of the StemCellCKB user interface, including the available functional modules and computational tools/programs.
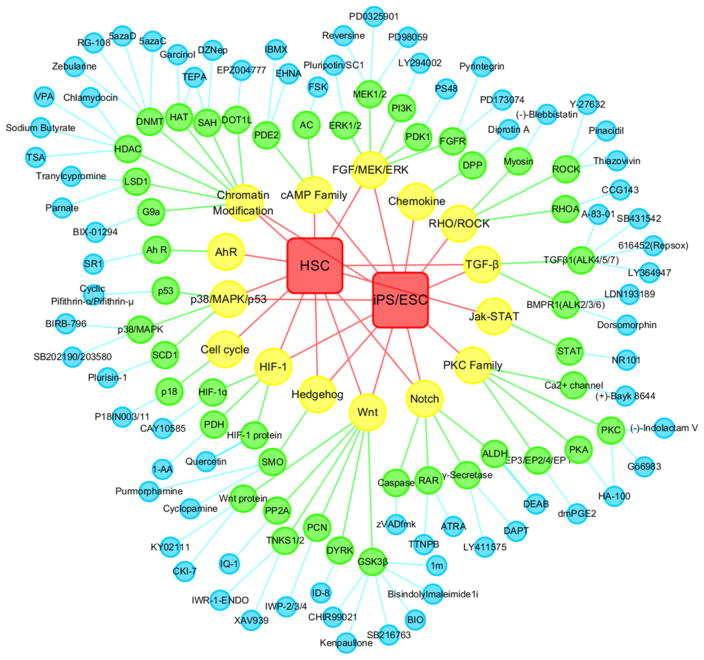
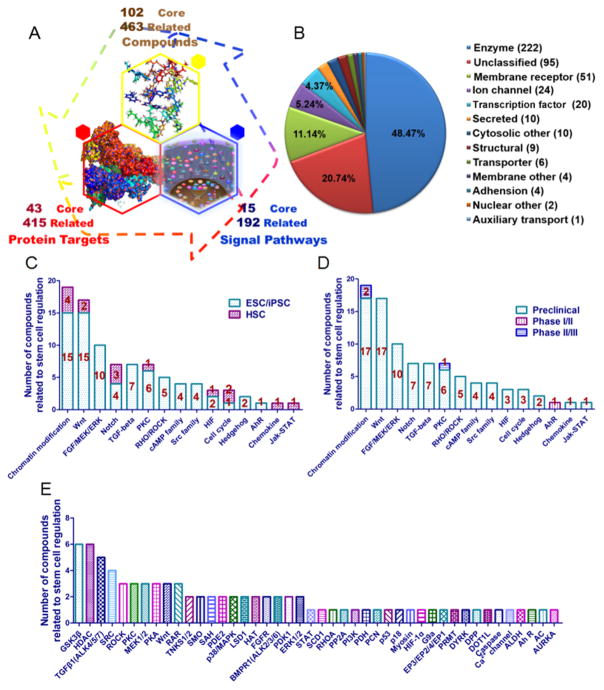
To date, the StemCellCKB describes 458 genes and proteins, as well as 207 relevant pathways and bioassays. The catalogued proteins include: (i) 222 enzymes, such as glycogen synthase kinase-3 beta (GSK3β), cyclin-dependent kinase family (CDK), mitogen-activated protein kinase family (MAPK), tyrosine-protein kinase JAK (JAK), and signal transducer and activator of transcription (STAT); (ii) 51 membrane receptors, such as C-X-C chemokine receptor (CXCR); (iii) 24 ion channels, such as the Bcl protein; (iv) 20 transcription factors, such as hypoxia-inducible factor 1 (HIF1); (v) 10 secreted proteins, such as vascular endothelial growth factor (VEGF), interleukin (IL), and fibroblast growth factor (FGF); (vi) 10 cytosolic proteins, such as cyclin-dependent kinase inhibitor (CKI); (vii) 9 structural proteins, such as collagen; and (viii) 112 miscellaneous proteins. StemCellCKB also includes information about 102 stem cell related small molecules (Figure 2, blue nodes) with known biological effects on HSC, ESC, or iPS (Figure 2, red rectangles).
Figure 2.
Interaction network of stem cells, pathways, targets, and compounds. Red rectangles represent stem cell types. Yellow nodes represent core signaling pathways involved in stem cell regulation. Green nodes represent core protein targets associated with stem cell modulation. Blue nodes represent known core stem cell-related compounds.
Using this database, we performed an in-depth analysis of the protein targets and corresponding pathways modulated by these compounds (Figures 2 and 3). Using the StemCellCKB compound/target mapping function, we mapped out 44 “core proteins” (Figure 2, green nodes) involved in 15 key signaling pathways (Figure 2, yellow nodes). Those proteins and signaling pathways that are associated with at least one compound tested for the bioactivities on stem cell regulation were defined as core proteins and key pathways, respectively. Among these 15 signaling pathways, the following have biological effects on both HSC and iPS/ESC: Wnt, Notch, transforming growth factor (TGF), p38/MAPK/p53, HIF, Hedgehog, FGF/mitogen-activated protein kinase kinase (MEK) /extracellular signal regulated kinase (ERK), chromatin modification, cell cycle, and the cyclic adenosine mono-phosphate (cAMP) family. In contrast, aryl hydrocarbon receptor (AhR) and JAK/STAT affected only HSC; RHO/ RHO kinase (ROCK), protein kinase C (PKC), and chemo-kine affected only iPS/ESC.
Figure 3.
Chemogenomics data in StemCellCKB. (A) Association among compounds, protein targets, and signal pathways involved in stem cell regulation. (B) StemCellCKB target classification. (C) Compounds known to be involved in stem cell regulation were plotted according to stem cell type and the corresponding target pathways. (D) Compounds known to be involved in stem cell regulation were plotted according to their development phases and associated pathways. (E) Compounds known to be involved in stem cell regulation were plotted according to their protein targets.
To enable future stem cell drug repositioning and target discovery, we also included some signaling pathways in the database that are thought to play roles in HSC or iPS/ESC regulation but are not currently associated with known small molecule modulators (e.g., the Ras, mTOR, and chemokine signaling pathways). In addition to the 102 small molecules known to be capable of directing stem cell manipulation, StemCellCKB also describes 463 additional small molecules that have not been tested to alter stem cell phenotypes. These additional compounds are included because their target proteins are involved in stem cell-related signaling pathways; consequently, they may have future utility in stem cell research. These compounds and their targets are summarized in Figure 3A and B, respectively.
A survey of these compounds is illustrated in Figure 3C, organized according to interacting targets, signaling pathways, and stem cell types. For example, Wnt signaling may be the most important pathway, as 17 of the 102 associated compounds (16%) are known to regulate stem cell fate or to induce somatic cell reprogramming through this pathway. Among all Wnt signaling protein targets, GSK3β may be most critical for stem cell regulation because it is associated with six compounds: SB216763, kenpaullone,59 CHIR99021,60,61 bisin-dolylmaleimide1i,62 BIO,63 and 1m64 (Figure 3E). Other related targets in this signaling pathway include the Wnt protein, protein phosphatase 2A (PP2A), tankyrase 1/2 (TNKS1/2), porcupine (PCN), and dual specificity tyrosine phosphorylation regulated kinase (DYRK). Chromatin modification proteins are also prominent, with 19 associated compounds. Histone deacetylase (HDAC) and DNA methyl-transferase (DNMT) are already useful targets for the clinical treatment of many diseases, as well as for stem cell regulation. For example, HDAC inhibitors (e.g., valproic acid (VPA),65 sodium butyrate,66 trichostatin A (TSA),67 etc.) and DNMT modulators (e.g., zebularine,68 RG-108,69 5azaD,67 and 5azaC70) have been used in stem cell research. Transforming growth factor beta receptor family (TGFβR, also known as anaplastic lymphoma kinase, ALK) also contains key targets, especially in manipulating iPS/ESC, with four associated compounds: SB431542,71 LY364947,72 A-83-01,66 and 616452/repsox.60 Cell cycle regulator (CKI) also plays a vital role in stem cell manipulation; two inhibitors of CKI p18INK4C, P18IN003 and P18IN011, are known to induce HSC expansion.19 Other molecules that induce HSC expansion are advancing through clinical trials, including TEPA (copper chelate),73 SR1 (AhR antagonist),74 and Nicord (SIRT1 inhibitor).75 These compounds have completed phase I/II and are now in phase II/III multicenter clinical trials76 (Figure 3D).
StemCellCKB also features an integrated cloud-computing service with intrinsic scalability and convenient features like search functions and useful tools for further expansion. It provides a fast and effective method to identify proteins involved in stem cell regulation, to identify chemicals with known activities, to find new chemicals for stem cell manipulation, and to determine which chemical cocktails might best act in concert. In summary, StemCellCKB facilitates drug research and development aimed at stem cell regulation.
StemCellCKB Database Validation
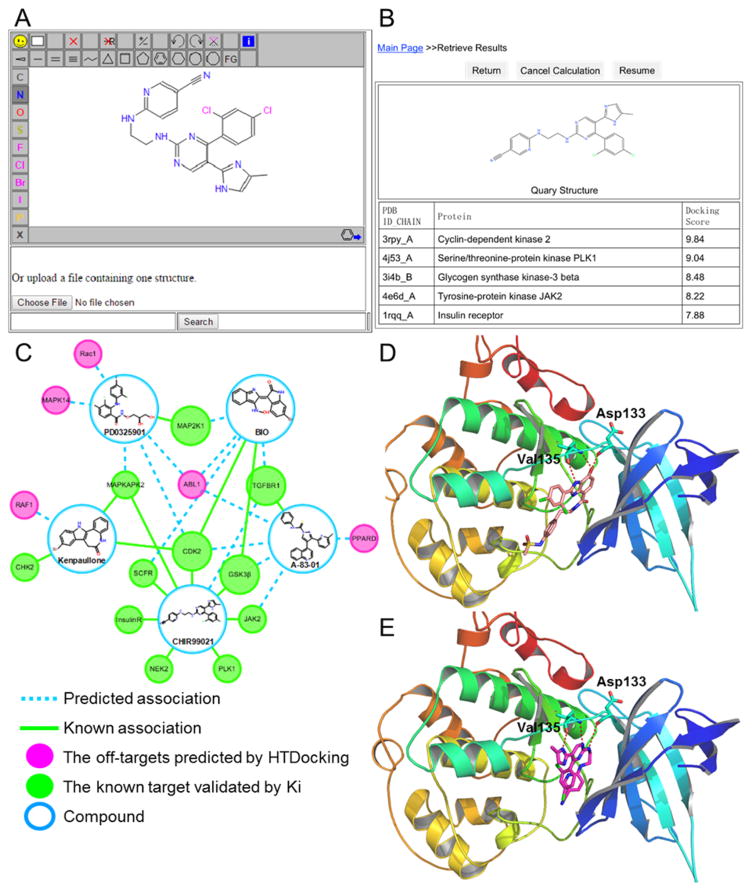
Target identification sheds critical light on small molecule mechanisms of action, and systems pharmacology suggests that small molecules often have multiple targets. StemCellCKB facilitates stem cell relevant target identification by providing two integrated online programs: TargetHunter30 and HTDocking.24 To illustrate how the HTDocking component can be used to identify potential targets and explore mechanisms of action, we first validated our predictions using a set of known drug-target associations. Using the online StemCellCKB interface, we submitted SDF files describing a number of known small molecules (Figure 4A). The HTDocking program automatically calculated the docking scores associated with each compound positioned within the binding pockets of the respective targets. The targets were ranked according to docking scores and displayed on the results webpage (Figure 4B). These rankings were consistent with experimental data, as shown in Figure 4C, where green nodes indicate predicted protein targets which were already biologically validated and pink nodes indicate predicted protein targets with no experimental validation. The predicted and experimental Ki values, as well as pKi values of the known stem cell modulators (per journal articles and databases such as ChEMBL), are summarized in Table 1, with the primarily corresponding protein targets marked with “*”. Indeed, some of the predicted associations have already been reported in the literature,59–61,63,66 further demonstrating HTDocking reliability.
Figure 4.
StemCellCKB Database validation by target identification. (A) Chemical-structure input window of the HTDocking server, with CHIR99021 displayed. (B) HTDocking results. (C) Polypharmacology analysis of known stem cell drugs. Green nodes represent the known or biologically validated protein targets of the compounds, and pink nodes represent new targets predicted using HTDocking. (D) Interaction between Z48 (salmon stick) and GSK3β (rainbow cartoon) in the active site (PDB ID: 3I4B). (E) CHIR99021 (magenta stick) docked into the active site of GSK3β (rainbow cartoon; PDB ID: 3I4B).
Table 1.
Comparison of the Experimental Ki Values and Docking Scores of Known Stem Cell-Related Compounds
| compound ID | protein ID | Ki (nM) | −log(Ki × 10−9) | docking score |
|---|---|---|---|---|
| CHIR99021 | CDK2 | 1584.89 | 5.80 | 9.84 |
| PLK1 | 7943.28 | 5.10 | 9.04 | |
| GSK3β* | 3.98 | 8.40 | 8.48 | |
| JAK2 | 1584.89 | 5.80 | 8.22 | |
| InsulinR | 5011.87 | 5.30 | 7.88 | |
| MAPAPK2 | 7943.28 | 5.10 | 7.74 | |
| Nek2 | 10000 | 5.0 | 7.04 | |
| BIO | GSK3β* | 5 | 8.30 | 5.24 |
| CDK2 | inhibition | N/A | 6.34 | |
| PD0325901 | MAP2K1* | 0.6 | 9.22 | 7.04 |
| A-83-01 | TGFβR1* | 12 | 7.92 | 7.81 |
| Kenpaulone | CDK2* | 100 | 7.00 | 6.84 |
| CHK2 | 630.96 | 6.20 | 6.36 | |
| MAPAPK2 | 7943.28 | 5.10 | 6.07 |
The reported original protein target of each compound is given with
The Ki values are all from ChEMBL database results.
The predicted targets merit further experimental validation (Figure 4C). As a specific example, consider CHIR99021, the most potent GSK3β inhibitor (Ki = 3.98 nM, ChEMBL 1201862). CHIR99021 is known to increase both human and murine HSC expansion ex vivo and to induce somatic cell reprogramming when combined with other compounds.61 The associated HTDocking score was 8.48, supporting the theory that CHIR99021 interacts with GSK3β (Table 1). The predicted CHIR99021/GSK3β interaction is shown in Figure 4D. Similar to the crystallographic binding mode of Z48, a known GSK3β inhibitor (Figure 4D, salmon sticks; PDB ID: 3I4B), CHIR99021 (Figure 4E, magenta sticks) may form hydrogen bonds with the GSK3β residues Asp133 and Val135 (Figure 4E, rainbow cartoon). The ChEMBL database describes in vitro binding assays that validate this prediction, as is the case for most StemCellCKB-predicted targets with experimentally validated ligands (Table 1). CHIR99021 may additionally interact with other targets, including CDK, serine/ threonine-protein kinase (PLK1), JAK2, insulin receptor (InsulinR), mitogen activated protein kinase activated protein kinase 2 (MAPAPK2), and serine/threonine-protein kinase (NEK2), which all had docking scores above 7.0 (Figure 4C, green nodes, and Table 1). Other known compound-target associations (e.g., PD0325901 with MEK, A-83-01 with TGFβR1, Kenpaullone with CDK2, and BIO with GSK3β) were also correctly identified using our HTDocking approach (Figure 4C, green nodes).
Understanding compound–target interactions enables poly-pharmacology, wherein novel chemical combinations yield improved pharmacological effects with reduced side effects.23 For example, the binding pocket of GSK3β is very similar to that of CDK2.77 An ideal small molecule would potentiate cell cycle activation by inhibiting GSK3β without modulating CDK activity, which is essential for cell cycle progression. Further studies could focus on finding useful combinations of GSK3β inhibitors and cell cycle activators (e.g., CKI inhibitors).
DISCUSSION
Human stem cells typically face several fate choices such as self-renewal, differentiation, quiescence, homing, aging, apoptosis, and eventually cell death. A better understanding of the mechanisms that govern stem cell regulation would give rise to stem cell therapies,78 as such, a comprehensive stem cell-specific chemical-genomics knowledgebase for systems pharmacology study of small molecules and their targets would certainly benefit chemical biology research of stem cell.
StemCellCKB offers an integrated platform for both chemical biology and mechanism exploration. To demonstrate its utility, we used the platform to study GSK3β, an important constituent of the Wnt/β-catenin signaling pathway. Wnt signaling, required for the proper development of most tissues and organisms, is highly conserved in mammals79 and β-catenin is known to be the main signal transducer in the canonical Wnt signaling pathway. A complex comprised of GSK3β, adenomatous polyposis coli protein (APC), Axin, and casein kinase I (CK1) regulates the β-catenin pool via phosphorylation.80 In embryonic hematopoiesis, β-catenin activation is required for arterial specification and HSC generation,81,82 and in adult hematopoiesis, β-catenin improves HSC generation in a dose-dependent manner.83 Small molecule GSK3β inhibitors could activate β-catenin and therefore increase HSC expansion.63,84 The convenient data visualization features built into Stem-CellCKB confirmed that GSK3β is the known target of numerous stem cell regulating compounds.
The current study also demonstrates that the target-based HTDocking program can successfully identify previously uncharacterized small-molecule/protein-target interactions. We validated HTDocking by comparing the predicted and experimental Ki values of known stem cell related small molecules. As a complementary method, the ligand-based TargetHunter tool predicts potential targets and off-targets using our established chemogenomics database.30 Target-Hunter is especially useful when there is no available high-quality protein structure of the target of interest. Both of our established programs facilitate drug repositioning and sideefiect prediction.
Due to the complexity of the signaling network, manipulating stem cell function requires simultaneous targeting of multiple stem cell signaling pathways. For example, by screening a chemical library, Hou et al. recently identified a cocktail of four small molecules, each targeting a different pathway, that together transformed mouse somatic cells into pluripotent stem cells.60 However, chemical library screening is time-consuming and costly. StemCellCKB accelerates stem cell-relevant polypharmacology analyses, target identification, and drug-mechanism exploration by bridging the knowledge gap between biology and chemistry. By assembling information from multiple sources, StemCellCKB provides domain-specific data and tools to help users explore combinations of signaling pathways, protein targets, and relevant small molecules, potentially enabling the development of future multicompound cocktails that together are more effective than a single agent at stem cell regulation.
We should point out that due to the limited understanding of stem cell regulation, the StemCellCKB may miss some important protein targets and pathways that also play key roles in stem cell regulation, which may further affect the performance of the computational tools. To address this issue, we will continue to update and refine the database records. We also would like to mention that TargetHunter and HTDocking can be complementary to each other. TargetHunter is based on the similarity between small molecules and HTDocking is based on the fitness of a small molecule into protein binding pockets. As such, it is very easy to find the target(s) of a small molecule if the bioactivities of its analogs have been reported. On the other hand, HTDocking can be used to identify previously undiscovered targets (or novel targets). It should be noted that while the StemCellCKB may provide the target ranking based on either similarity threshold or docking score, the selection of target prediction for validation is subjective and may depend on the user’s expertise, as well as the availability of bioassays.
CONCLUSION
In summary, StemCellCKB (http://www.cbligand.org/StemCellCKB)33 is an integrative, publically accessible platform that includes stem cell relevant data and tools for target identification and system pharmacology research. It is a one-stop resource for searching, organizing, and validating stem cell-related information that is otherwise only available from scattered sources (e.g., PubChem, ChEMBL, and many other databases). The server will integrate computational, chemical, biological, and clinical knowledge and so will be useful for those studying stem cell system biology, polypharmacology, chemo-genomics, and computer-aided drug design.
Acknowledgments
Research reported in this publication was supported by the NIH grants R21HL109654 and P30DA035778, (X.-Q.X.), the National Basic Research Program of China (nos. 2016YFA0100602 and 2012CB966600), the National Natural Science Foundation of China (NSFC 81421002, 81570100, 81500086, 81430004, and 81600085), the key technologies R & D program of Tianjin (13RCGFSY19500), and the China National Scholarship.
Footnotes
The authors declare no competing financial interest.
References
- 1.Watt FM, Hogan B. Out of Eden. stem cells and their niches. Science. 2000;287:1427–30. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 2.Li W, Li K, Wei W, Ding S. Chemical approaches to stem cell biology and therapeutics. Cell stem cell. 2013;13:270–283. doi: 10.1016/j.stem.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimmeler S, Ding S, Rando TA, Trounson A. Translational strategies and challenges in regenerative medicine. Nat Med. 2014;20:814–821. doi: 10.1038/nm.3627. [DOI] [PubMed] [Google Scholar]
- 4.Cheng T. Toward ‘SMART’ stem cells. Gene Ther. 2008;15:67–73. doi: 10.1038/sj.gt.3303066. [DOI] [PubMed] [Google Scholar]
- 5.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell stem cell. 2012;10:120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Thomas ED, Lochte HL, Jr, Lu WC, Ferrebee JW. Intravenous infusion of bone marrow in patients receiving radiation and chemotherapy. N Engl J Med. 1957;257:491–496. doi: 10.1056/NEJM195709122571102. [DOI] [PubMed] [Google Scholar]
- 7.Bari S, Seah KKH, Poon Z, Cheung AMS, Fan X, Ong S-Y, Li S, Koh LP, Hwang WYK. Expansion and Homing of Umbilical Cord Blood Hematopoietic Stem and Progenitor Cells for Clinical Transplantation. Biol Blood Marrow Transplant. 2015;21:1008–1019. doi: 10.1016/j.bbmt.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 8.Pineault N, Abu-Khader A. Advances in umbilical cord blood stem cell expansion and clinical translation. Exp Hematol. 2015;43:498–513. doi: 10.1016/j.exphem.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, Reya T. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 10.Katoh M, Katoh M. Notch ligand, JAG1, is evolutionarily conserved target of canonical WNT signaling pathway in progenitor cells. Int J Mol Med. 2006;17:681–685. [PubMed] [Google Scholar]
- 11.van den Brink GR, Bleuming SA, Hardwick JC, Schepman BL, Offerhaus GJ, Keller JJ, Nielsen C, Gaffield W, van Deventer SJ, Roberts DJ, Peppelenbosch M. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat Genet. 2004;36:277–282. doi: 10.1038/ng1304. [DOI] [PubMed] [Google Scholar]
- 12.Cheng T, Shen H, Rodrigues N, Stier S, Scadden DT. Transforming growth factor β1 mediates cell-cycle arrest of primitive hematopoietic cells independent of p21Cip1/Waf1or p27Kip1. Blood. 2001;98:3643–3649. doi: 10.1182/blood.v98.13.3643. [DOI] [PubMed] [Google Scholar]
- 13.Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042–4045. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 15.Tenzen T, Zembowicz F, Cowan CA. Genome modification in human embryonic stem cells. J Cell Physiol. 2010;222:278–281. doi: 10.1002/jcp.21948. [DOI] [PubMed] [Google Scholar]
- 16.Xu T, Zhang M, Laurent T, Xie M, Ding S. Concise review: chemical approaches for modulating lineage-specific stem cells and progenitors. Stem Cells Transl Med. 2013;2:355. doi: 10.5966/sctm.2012-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walasek MA, van Os R, de Haan G. Hematopoietic stem cell expansion: challenges and opportunities. Ann N Y Acad Sci. 2012;1266:138–150. doi: 10.1111/j.1749-6632.2012.06549.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Li W, Laurent T, Ding S. Small molecules, big roles–the chemical manipulation of stem cell fate and somatic cell reprogramming. J Cell Sci. 2012;125:5609–5620. doi: 10.1242/jcs.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao Y, Yang P, Shen H, Yu H, Song X, Zhang L, Zhang P, Cheng H, Xie Z, Hao S, et al. Small-molecule inhibitors targeting INK4 protein p18INK4C enhance ex vivo expansion of haematopoietic stem cells. Nat Commun. 2015;6:6328. doi: 10.1038/ncomms7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet J-P, Subramanian A, Ross KN. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 21.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 22.Kuhn M, Szklarczyk D, Franceschini A, von Mering C, Jensen LJ, Bork P. STITCH 3: zooming in on protein–chemical interactions. Nucleic Acids Res. 2012;40:D876–D880. doi: 10.1093/nar/gkr1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Wang L, Lv M, Pei R, Li P, Pei Z, Wang Y, Su W, Xie X-Q. AlzPlatform: an alzheimer’s disease domain-specific chemogenomics knowledgebase for polypharmacology and target identification research. J Chem Inf Model. 2014;54:1050–1060. doi: 10.1021/ci500004h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie X-Q, Wang L, Liu H, Ouyang Q, Fang C, Su W. Chemogenomics knowledgebased polypharmacology analyses of drug abuse related G-protein coupled receptors and their ligands. Front Pharmacol. 2014;5:3. doi: 10.3389/fphar.2014.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheng S, Wang J, Wang L, Liu H, Li P, Liu M, Long C, Xie C, Xie X, Su W. Network pharmacology analyses of the antithrombotic pharmacological mechanism of Fufang Xueshuantong Capsule with experimental support using disseminated intravascular coagulation rats. J Ethnopharmacol. 2014;154:735–744. doi: 10.1016/j.jep.2014.04.048. [DOI] [PubMed] [Google Scholar]
- 26.Sandeep G, Nagasree KP, Hanisha M, Kumar MMK. AUDocker LE: A GUI for virtual screening with AUTODOCK Vina. BMC Res Notes. 2011;4:445. doi: 10.1186/1756-0500-4-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma C, Lazo JS, Xie X-Q. Compound acquisition and prioritization algorithm for constructing structurally diverse compound libraries. ACS Comb Sci. 2011;13:223–231. doi: 10.1021/co100033m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma C, Wang L, Xie X-Q. Ligand classifier of adaptively boosting ensemble decision stumps (LiCABEDS) and its application on modeling ligand functionality for 5HT-subtype GPCR families. J Chem Inf Model. 2011;51:521–531. doi: 10.1021/ci100399j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma C, Wang L, Yang P, Myint KZ, Xie X-Q. LiCABEDS II Modeling of ligand selectivity for G-protein-coupled cannabinoid receptors. J Chem Inf Model. 2013;53:11–26. doi: 10.1021/ci3003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Ma C, Wipf P, Liu H, Su W, Xie X-Q. TargetHunter: an in silico target identification tool for predicting therapeutic potential of small organic molecules based on chemo-genomic database. AAPS J. 2013;15:395–406. doi: 10.1208/s12248-012-9449-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng Z, Alqarni MH, Yang P, Tong Q, Chowdhury A, Wang L, Xie X-Q. Modeling, molecular dynamics simulation, and mutation validation for structure ofcannabinoid receptor 2based on known crystal structures of GPCRs. J Chem Inf Model. 2014;54:2483–2499. doi: 10.1021/ci5002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie XQ, Chen JZ, Billings EM. 3D structural model of the G-protein-coupled cannabinoid CB2 receptor. Proteins: Struct, Funct, Genet. 2003;53:307–319. doi: 10.1002/prot.10511. [DOI] [PubMed] [Google Scholar]
- 33.StemCellCKB. [accessed 2016/01/07]; http://www.cbligand.org/StemCellCKB.
- 34. [accessed 2015/10/08];MySQL software. http://www.mysql.com.
- 35. [accessed 2015/ 10/07];Apache web server. http://www.apache.org/
- 36.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2007;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. [accessed 2015/ 10/07];DrugBank database. http://www.drugbank.ca/
- 38. [accessed 2016/01/07];clinicaltrialsgov database. http://clinicaltrials.gov/
- 39. [accessed 2016/ 01/08];MetaCore database. http://portal.genego.com/
- 40. [accessed 2015/10/08];PubChem database. http://pubchem.ncbi.nlm.nih.gov/
- 41. [accessed 2015/10/08];PubMed database. http://www.ncbi.nlm.nih.gov/pubmed.
- 42.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. [accessed 2015/ 11/07];KEGG database. http://www.genome.jp/kegg.
- 45. [accessed 2015/11/06];ChEMBL database. http://www.ebi.ac.uk/chembl/
- 46.Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, Kruger FA, Light Y, Mak L, McGlinchey S, Nowotka M, Papadatos G, Santos R, Overington JP. The ChEMBL bioactivity database: an update. Nucleic Acids Res. 2014;42:D1083–D1090. doi: 10.1093/nar/gkt1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. [accessed 2015/11/07];PHP language. http://www.php.net/
- 48.Ertl P. Molecular structure input on the web. J Cheminf. 2010;2:1–9. doi: 10.1186/1758-2946-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. [accessed 2016/01/09];TargetHunter. http://www.cbligand.org/TargetHunter.
- 51. [accessed 2016/01/08];HTDocking. http://www.cbligand.org/StemCelldb/docking_search.php.
- 52.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2009;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. [accessed 2016/01/07];Online toxicity predictor. http://cbligand.org/Tox.
- 54.Patlewicz G, Jeliazkova N, Safford R, Worth A, Aleksiev B. An evaluation of the implementation of the Cramer classification scheme in the Toxtree software. SAR QSAR Environ Res. 2008;19:495–524. doi: 10.1080/10629360802083871. [DOI] [PubMed] [Google Scholar]
- 55.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J Med Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 56. [accessed 2015/11/07];PAINS compound remover. http://cbligand.org/PAINS.
- 57. [accessed 2015/11/07];Chemical property calculator. http://www.cbligand.org/cbid/Property_Explorer.php.
- 58.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Delivery Rev. 2012;64:4–17. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 59.Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, Bouchez LC, et al. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc Natl Acad Sci U S A. 2009;106:8912–8917. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 61.Huang J, Nguyen-McCarty M, Hexner EO, Danet-Desnoyers G, Klein PS. Maintenance of hematopoietic stem cells through regulation of Wnt and mTOR pathways. Nat Med. 2012;18:1778–1785. doi: 10.1038/nm.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bone HK, Damiano T, Bartlett S, Perry A, Letchford J, Ripoll YS, Nelson AS, Welham MJ. Involvement of GSK-3 in regulation of murine embryonic stem cell self-renewal revealed by a series of bisindolylmaleimides. Chem Biol. 2009;16:15–27. doi: 10.1016/j.chembiol.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 63.Ko KH, Holmes T, Palladinetti P, Song E, Nordon R, O’Brien TA, Dolnikov A. GSK-3β inhibition promotes engraftment of ex vivo-expanded hematopoietic stem cells and modulates gene expression. Stem Cells. 2011;29:108–118. doi: 10.1002/stem.551. [DOI] [PubMed] [Google Scholar]
- 64.Bone HK, Nelson AS, Goldring CE, Tosh D, Welham MJ. A novel chemically directed route for the generation of definitive endoderm from human embryonic stem cells based on inhibition of GSK-3. J Cell Sci. 2011;124:1992–2000. doi: 10.1242/jcs.081679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Felice L, Tatarelli C, Mascolo MG, Gregorj C, Agostini F, Fiorini R, Gelmetti V, Pascale S, Padula F, Petrucci MT. Histone deacetylase inhibitor valproic acid enhances the cytokine-induced expansion of human hematopoietic stem cells. Cancer Res. 2005;65:1505–1513. doi: 10.1158/0008-5472.CAN-04-3063. [DOI] [PubMed] [Google Scholar]
- 66.Zhou J, Su P, Li D, Tsang S, Duan E, Wang F. High-Efficiency Induction of Neural Conversion in Human ESCs and Human Induced Pluripotent Stem Cells with a Single Chemical Inhibitor of Transforming Growth Factor Beta Superfamily Receptors. Stem Cells. 2010;28:1741–1750. doi: 10.1002/stem.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saraf S, Araki H, Petro B, Park Y, Taioli S, Yoshinaga KG, Koca E, Rondelli D, Mahmud N. Ex vivo expansion of human mobilized peripheral blood stem cells using epigenetic modifiers. Transfusion. 2015;55:864–874. doi: 10.1111/trf.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hayakawa K, Hirosawa M, Tabei Y, Arai D, Tanaka S, Murakami N, Yagi S, Shiota K. Epigenetic switching by the metabolism-sensing factors in the generation of orexin neurons from mouse embryonic stem cells. J Biol Chem. 2013;288:17099–17110. doi: 10.1074/jbc.M113.455899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi Y, Desponts C, Do JT, Hahm HS, Schöler HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell stem cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 70.Li J, Zhu K, Wang Y, Zheng J, Guo C, Lai H, Wang C. Combination of IGF-1 gene manipulation and 5-AZA treatment promotes differentiation of mesenchymal stem cells into cardiomyocyte-like cells. Mol Med Rep. 2014;11:815–820. doi: 10.3892/mmr.2014.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Osakada F, Jin Z-B, Hirami Y, Ikeda H, Danjyo T, Watanabe K, Sasai Y, Takahashi M. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J Cell Sci. 2009;122:3169–3179. doi: 10.1242/jcs.050393. [DOI] [PubMed] [Google Scholar]
- 72.Jaremko KL, Marikawa Y. Regulation of developmental competence and commitment towards the definitive endoderm lineage in human embryonic stem cells. Stem Cell Res. 2013;10:489–502. doi: 10.1016/j.scr.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 73.Peled T, Glukhman E, Hasson N, Adi S, Assor H, Yudin D, Landor C, Mandel J, Landau E, Prus E, et al. Chelatable cellular copper modulates differentiation and self-renewal of cord blood–derived hematopoietic progenitor cells. Exp Hematol. 2005;33:1092–1100. doi: 10.1016/j.exphem.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 74.Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, Walker JR, Flaveny CA, Perdew GH, Denison MS, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peled T, Shoham H, Aschengrau D, Yackoubov D, Frei G, Rosenheimer N, Lerrer B, Cohen HY, Nagler A, Fibach E, Peled A. Nicotinamide, a SIRT1 inhibitor, inhibits differentiation and facilitates expansion of hematopoietic progenitor cells with enhanced bone marrow homing and engraftment. Exp Hematol. 2012;40:342–355. doi: 10.1016/j.exphem.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 76.Horwitz ME, Frassoni F. Improving the outcome of umbilical cord blood transplantation through ex vivo expansion or graft manipulation. Cytotherapy. 2015;17:730–738. doi: 10.1016/j.jcyt.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 77.Berg S, Bergh M, Hellberg S, Högdin K, Lo-Alfredsson Y, Söderman P, von Berg S, Weigelt T, Ormö M, Xue Y, et al. Discovery of novel potent and highly selective glycogen synthase kinase-3β (GSK3β) inhibitors for Alzheimer’s disease: design, synthesis, and characterization of pyrazines. J Med Chem. 2012;55:9107–9119. doi: 10.1021/jm201724m. [DOI] [PubMed] [Google Scholar]
- 78.Daley GQ. The promise and perils of stem cell therapeutics. Cell stem cell. 2012;10:740–749. doi: 10.1016/j.stem.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10:468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 80.Behrens J, Jerchow B-A, Würtele M, Grimm J, Asbrand C, Wirtz R, Kühl M, Wedlich D, Birchmeier W. Functional interaction of an axin homolog, conductin, with β-catenin, APC, and GSK3β. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 81.Corrigan PM, Dobbin E, Freeburn RW, Wheadon H. Patterns of Wnt/Fzd/LRP gene expression during embryonic hematopoiesis. Stem Cells Dev. 2009;18:759–772. doi: 10.1089/scd.2008.0270. [DOI] [PubMed] [Google Scholar]
- 82.Cattelino A, Liebner S, Gallini R, Zanetti A, Balconi G, Corsi A, Bianco P, Wolburg H, Moore R, Oreda B, et al. The conditional inactivation of the β-catenin gene in endothelial cells causes a defective vascular pattern and increased vascular fragility. J Cell Biol. 2003;162:1111–1122. doi: 10.1083/jcb.200212157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luis TC, Naber BA, Roozen PP, Brugman MH, de Haas EF, Ghazvini M, Fibbe WE, van Dongen JJ, Fodde R, Staal FJ. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell stem cell. 2011;9:345–356. doi: 10.1016/j.stem.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 84.MacDonald BT, Tamai K, He X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]