Abstract
Clear Cell Renal Cell Carcinoma (CC-RCC) is the most lethal of all genitourinary cancers. The functional loss of the von Hippel-Lindau (VHL) gene occurs in 90% of CC-RCC, driving cancer progression. The objective of this study was to identify chemical compounds that are synthetically lethal with VHL deficiency in CC-RCC. An annotated chemical library, the Library of Pharmacologically Active Compounds (LOPAC), was screened in parallel on VHL-deficient RCC4 cells and RCC4VHL cells with re-introduced VHL cDNA. The ROCK inhibitor, Y-27632, was identified and validated for selective targeting of VHL-deficient CC-RCC in multiple genetic backgrounds by clonogenic assays. Downregulation of ROCK1 by siRNA selectively reduced the colony forming ability of VHL-deficient CC-RCC, thus mimicking the effect of Y-27632 treatment, whereas downregulation of ROCK2 had no effect. In addition, two other ROCK inhibitors, RKI 1447 and GSK 429286, selectively targeted VHL-deficient CC-RCC. CC-RCC treatment with ROCK inhibitors is cytotoxic and cytostatic based on BrdU assay, Propidium Iodide (PI) staining, and growth curves; and blocks cell migration based on transwell assay. Importantly, knockdown of Hypoxia Inducible Factor (HIF) β in the VHL-deficient CC-RCC had a protective effect against Y-27632 treatment, mimicking VHL reintroduction. On the other hand, CC-RCCVHL cells were sensitized to Y-27632 treatment in hypoxia (2% O2). These results suggest that synthetic lethality between ROCK inhibition and VHL deficiency is dependent on HIF activation. Moreover, HIF1α or HIF2α overexpression in CC-RCCVHL cells is sufficient to sensitize them to ROCK inhibition. Finally, Y-27632 treatment inhibited growth of subcutaneous 786-OT1 CC-RCC tumors in mice. Thus, ROCK inhibitors represent potential therapeutics for VHL-deficient CC-RCC.
Keywords: ROCK1, Clear Cell Renal Cell Carcinoma, Synthetic Lethality, VHL, HIF
Introduction
Renal cancer is the most deadly of all genitourinary cancers with 62,700 new cases and 14,240 deaths projected to occur in 20161. While surgical resection is often curative at early stages, metastatic renal cancer remains a devastating disease with a 5-year survival rate of less than 20%1,2. The poor survival rate is due to renal cancer’s resistance to radiotherapy3, chemotherapy2, and immunotherapy2, which has been linked to multidrug resistance mechanisms4 and the lack of common solid tumor mutations5.
Clear Cell Renal Cell Carcinomas (CC-RCCs) account for 90% of all renal cancer cases, and the tumor-suppressor VHL is functionally lost in up to 90% of CC-RCC tumors6. VHL loss occurs early in the disease and drives its pathogenesis6. VHL is an E3 ubiquitin ligase that targets multiple proteins for proteasomal degradation, including the Hypoxia Inducible Factor (HIF) α subunits and the Epidermal Growth Factor Receptor (EGFR)7. Thus, upon VHL loss, CC-RCCs upregulate expression of EGFR and other Receptor Tyrosine Kinases (RTKs), as well as HIFs, in turn upregulating proangiogenic genes, like Vascular Endothelial Growth Factor (VEGF). As a consequence, CC-RCCs are highly vascularized and aggressive. Accordingly, the majority of approved CC-RCC therapies inhibit angiogenesis. The RTK inhibitors (RTKi) sunitinib8, sorafenib9, and axitinib10, which block VEGFR and Platelet Derived Growth Factor Receptor (PDGFR), prolong progression-free survival for a median of 5 months when compared to placebo9,11 or standard of care treatments like interferon α12. Another class of CC-RCC therapeutics is represented by mammalian target of rapamycin inhibitors (mTORi) everolimus13 and temsirolimus14, which prolong progression-free survival for a median of 3 months when used as single agents compared to standard of care. While these treatments offer significant clinical benefit, resistance to both RTKi and mTORi therapeutics develops quickly creating the need for new and improved therapeutics15–17.
In this study we relied on a “synthetic lethality” approach to identify new therapeutics for VHL-deficient CC-RCC. A large body of evidence supports the use of synthetic lethality screens for identifying specific chemical compounds or small hairpin RNAs (shRNAs) that cause cell death and/or inhibit cell proliferation in combination with a particular cancer mutation18,19. The principle underlying such screens is that cancer cells with a specific mutation will be more sensitive to targeted inhibition of a certain pathway than normal cells that are do not the same mutation. Thus, the resulting synthetic lethality compounds represent excellent candidates for therapies that target mutation-bearing cancer cells, but spare normal tissues. Several synthetic lethality screens have been conducted in CC-RCC to date20–25. Each of these screens utilized the loss of the VHL tumor suppressor to identify compounds that are selectively targeting VHL-deficient CC-RCCs. The synthetic lethality screens relied on “matched” cell lines, which were created by introducing either a vector control or the wild-type VHL cDNA to VHL-deficient CC-RCC19. These matched cell lines were then used in chemical or shRNA library screens to identify chemical compounds or shRNAs that selectively target VHL-deficient CC-RCCs, while sparing their VHL-reconstituted “matched” counterparts. While both unannotated chemical library20–24 and shRNA25 screens have been conducted, to date no screens have been conducted using an annotated chemical library.
In this study we screened the annotated Library of Pharmacologically Active Compounds (LOPAC). This approach simultaneously revealed the exact molecular pathways responsible for selective targeting of VHL-deficient cells, and chemical compounds that inhibit them. Herein we report a chemical hit identified in a LOPAC screen, Y-27632 an inhibitor of the Rho-associated coiled-coil-containing protein kinase (ROCK) that selectively targets VHL-deficient CC-RCC. The ROCK proteins are regulated by the small GTP-binding protein Rho and are best known for their role in regulating cell morphology and motility by controlling actin-myosin contractile force26. This role is mediated through phosphorylation of their downstream substrates, including Myosin Light Chain (MLC), Myosin Light Chain 2 (MLC2), MYosin Phosphatase Target 1 (MYPT1), and LIM Kinases (LIMK)26. ROCK signaling is commonly upregulated in bladder27, testicular28, breast29, prostate30, and renal cancer31, and has been shown to contribute to tumor metastasis in bladder27, breast32, and prostate cancer30. In addition, certain ROCK substrates induce cell proliferation30, apoptosis33, and inhibit autophagy34. The two ROCK isoforms, ROCK1 and ROCK2, are differentially expressed throughout the body with ROCK1 being expressed ubiquitously and ROCK2 being expressed predominantly in the brain, muscle, heart, and lungs35. Although the two isoforms are highly homologous and have redundant functions, they also have unique functions and substrates26.
In the present study we show that ROCK inhibitors selectively target VHL-deficient CC-RCC to reduce cell proliferation, induce cell death, and block migration, which is mediated through inhibition of ROCK1 and not ROCK2. Our studies also reveal that HIF over-activation caused by VHL loss is both necessary and sufficient to cause synthetic lethality with ROCK inhibitors. Importantly, treatment with ROCK inhibitors blocks tumor growth in vivo, validating ROCK inhibitors as potential therapeutics for VHL-deficient CC-RCC.
Results
Identification of chemical hit Y-27632 targeting VHL-deficient CC-RCC
To identify novel chemical compounds that selectively target VHL-deficient CC-RCC, we screened the LOPAC composed of 1,280 compounds annotated with their protein targets (unpublished data), which allowed us to identify not only the chemicals, but also the molecular pathways necessary for survival/proliferation of VHL-deficient CC-RCC. The screen utilized the RCC4±VHL matched cell lines. RCC4 cells lack both alleles of VHL and as a consequence HIF1α and HIF2α expression and activity are dramatically elevated compared to cell lines expressing VHL tumor suppressor6,36,37. RCC4VHL cells were generated by stably transfecting full-length wild type VHL cDNA to RCC438. Both RCC4 and RCC4VHL cells were labeled with Enhanced Yellow Fluorescent Protein (EYFP) and the matched cell lines were treated in parallel with the LOPAC compounds at concentrations ranging from 0.3µM to 20µM in 384-well plates. Fluorescence intensity, a surrogate measure of cell numbers per well, was measured 96 hours following the treatment. The ROCK inhibitor Y-27632 (structure shown in Supplemental Figure 1a) was identified in this screen and selectively targeted VHL-deficient RCC4 while sparing RCC4VHL. The structures of the other two ROCK inhibitors, used later in this study, are shown in Supplemental Figure 1b,c. We validated Y-27632 as a “hit” by fluorescence-based viability assay (Figure 1a).
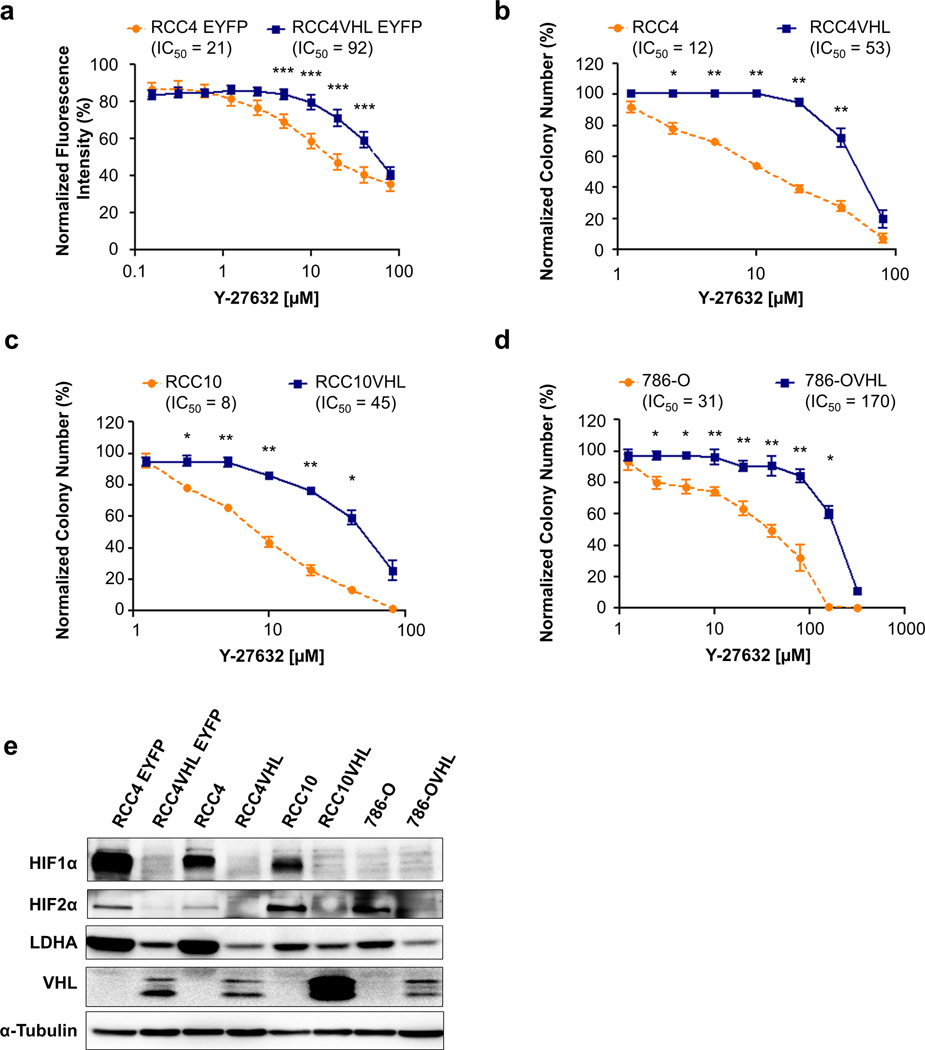
Figure 1. The ROCK inhibitor Y-27632 causes synthetic lethality with VHL loss in multiple CC-RCC cell lines.
(a) The LOPAC hit Y-27632 was validated in the RCC4-EYFP and RCC4VHL-EYFP matched cell lines, showing selective toxicity towards VHL-deficient cells. Each dose of Y-27632 within each experiment was tested in quadruplicate, and the experiment was repeated three times. Fluorescence intensity of EYFP-labeled cells was used as a surrogate for cell number. (b–d) Clonogenic assays in (b) RCC4±VHL, (c) RCC10±VHL, and (d) 786-O±VHL matched cell lines confirming that Y-27632 causes synthetic lethality with VHL loss in multiple CC-RCC genetic backgrounds. Each dose of Y-27632 within each experiment was tested in duplicate, and the experiment was repeated three times. IC50s are indicated. Statistical analysis in (a–d) was performed using a paired t-test between the matched cell lines at each dose (* p < 0.05, ** p < 0.01, *** p < 0.001), SEMs are shown. (e) Western blot showing the effect of VHL re-expression in CC-RCC cell lines on HIF1α and HIF2α expression, and the expression of their downstream target LDHA. α-tubulin serves as a loading control.
To further validate Y-27632 as a chemical hit we conducted clonogenic assays on RCC4 and RCC4VHL cell lines (Figure 1b and Supplemental Figure 2a). Importantly, VHL-deficient RCC4 cells were 4–5 times more sensitive to Y-27632 treatment than RCC4VHL in both assays (Figure 1a–b).
Treatment with ROCK inhibitor Y-27632 selectively targets VHL-deficient CC-RCCs of multiple genetic backgrounds
Next, we tested if the synthetic lethality effect could be reproduced in multiple genetic backgrounds. We repeated the clonogenic assays in two more VHL matched CC-RCC cell lines based on RCC10 expressing both HIF1α and HIF2α and 786-O expressing only HIF2α (Figure 1c–d and Supplemental Figure 2b–c). Similar to the results obtained in RCC4, Y-27632 treatment specifically targeted the VHL-deficient RCC10 and 786-O cell lines, while sparing the CC-RCCVHL. Y-27632 treatment not only reduced colony numbers selectively in VHL-deficient CC-RCC (Supplemental Figure 2a–c), but also caused reduced colony staining intensity, due to a reduction in cell numbers per colony (Supplemental Figure 2d–f). For each CC-RCC/CC-RCCVHL cell line pair the IC50 for VHL-deficient CC-RCC was approximately five times lower than for CC-RCCVHL (Figure 1b–d). VHL expression in each CC-RCCVHL cell line was confirmed by Western blot analysis, and it caused a reduction in HIF1α and HIF2α expression compared to the respective CC-RCC cell line (Figure 1e).
Y-27632’s ability to inhibit ROCK activity was assayed via Western blot analysis of MYPT1 Thr696 phosphorylation (ROCK substrate39). Y-27632 treatment for 2 hours was effective at inhibiting MYPT1 phosphorylation (Supplemental Figure 3). Interestingly, VHL-deficient CC-RCC have decreased basal MYPT1 phosphorylation in comparison to CC-RCCVHL. Together these results indicate that Y-27632 inhibits ROCK in CC-RCC and selectively targets VHL-deficient CC-RCC while sparing VHL-reconstituted CC-RCC in multiple genetic backgrounds.
Synthetic lethality occurs through inhibition of ROCK1
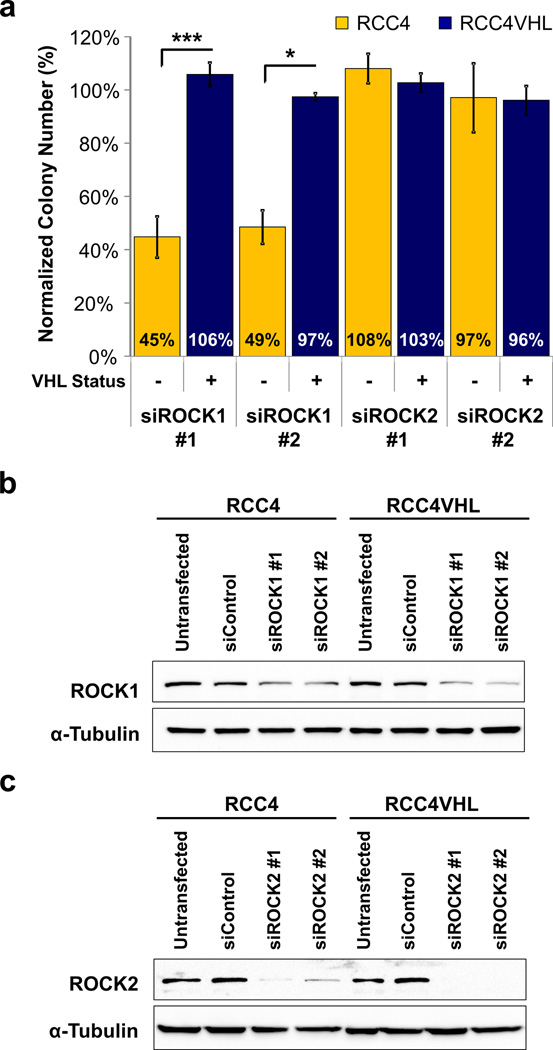
We aimed to confirm that the synthetic lethal effect of Y-27632 is “on-target” through blocking ROCKs as annotated in LOPAC. Since Y-27632 inhibits both ROCK family members: ROCK1 and ROCK2, we sought to determine which ROCK was responsible for the synthetic lethal effect. To do this we knocked down ROCK1 or ROCK2 with selective siRNAs. To control for the off-target effects of the siRNAs we used two siRNAs to knockdown ROCK1 (siROCK1#1, siROCK1#2) and two to knockdown ROCK2 (siROCK2#1, siROCK2#2). Our data showed that knockdown of ROCK1, but not ROCK2, reduced the colony forming ability and colony size of VHL-deficient RCC4 cells, sparing RCC4VHL cells, thus mimicking the effect of Y-27632 treatment (Figure 2a, Supplemental Figure 4). The ROCK1 and ROCK2 knockdowns were confirmed by Western blot analysis. Importantly, the knockdowns were equal or greater in the RCC4VHL cells for each siRNA used (Figure 2b–c). These data were reproduced in 786-O and 786-OVHL matched cell lines (Supplemental Figure 5). In summary, siRNA knockdown of ROCK1, but not ROCK2, selectively inhibits colony formation of VHL-deficient CC-RCC.
Figure 2. Synthetic lethality of Y-27632 with VHL loss is mimicked by siRNA downregulation of ROCK1, not ROCK2.
RCC4±VHL matched cell lines were transfected with siRNAs targeting ROCK1, ROCK2, or non-targeting siRNA control (siControl). Twenty-four hours after transfection cells were plated for a clonogenic assay. Each transfection was done in triplicate, followed by clonogenic assays conducted in triplicate, and the experiments were repeated at least two times. (a) Transfection with siROCK1, but not siROCK2, resulted in significant reduction in RCC4 colony numbers in comparison to RCC4VHL. Thus, ROCK1 downregulation mimics the effect of Y-27632 treatment on viability of RCC4 cells, making it a likely target for Y-27632 causing synthetic lethality effect. Statistical analysis was performed using a paired t-test comparing numbers of colonies in each siROCK group to siControl. SEMs are shown. (b–c) The degree of each target knockdown by its specific siRNA (as indicated) was assessed by Western blot.
RKI 1447 and GSK 429286 ROCK inhibitors target VHL-deficient CC-RCC
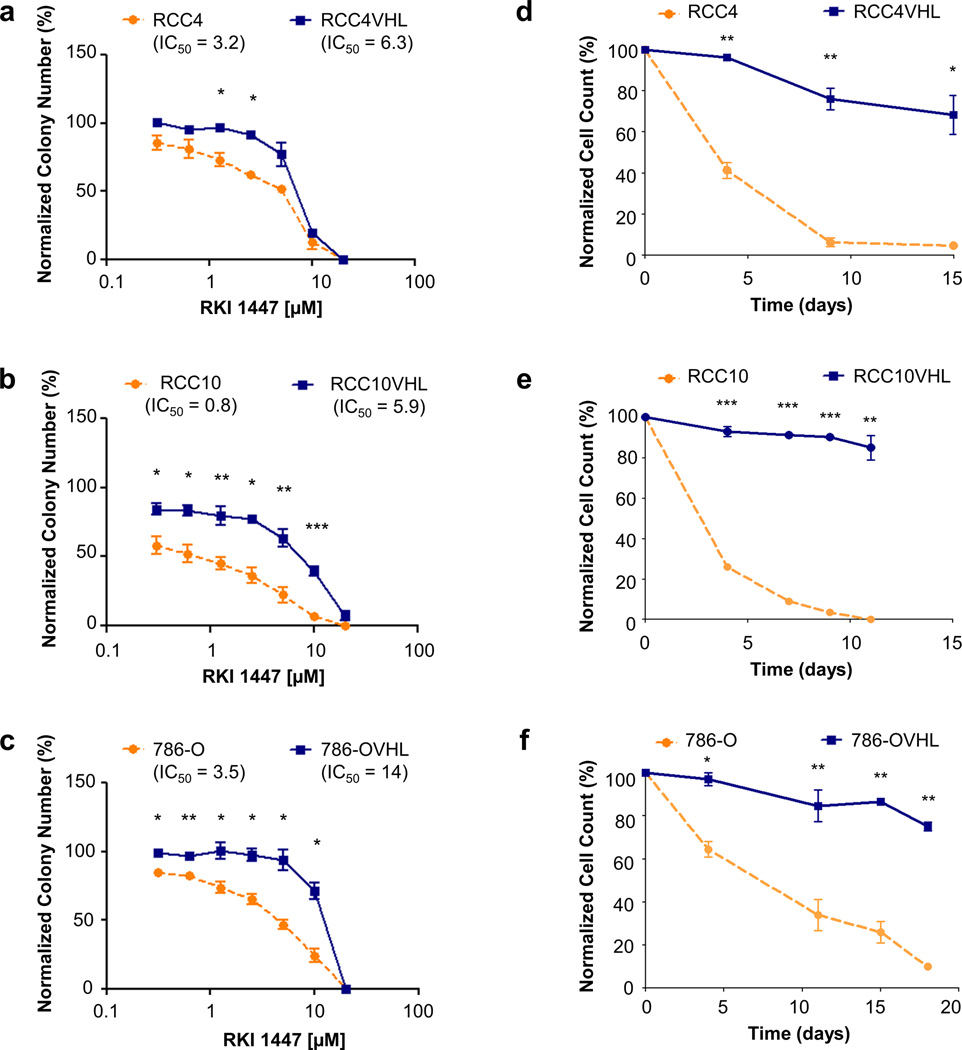
Since there are several commercially available ROCK inhibitors, and all of them differ in their potency and selectivity towards ROCK1 versus ROCK2, we tested two additional ROCK inhibitors in clonogenic assays: RKI 1447 (structure shown in Supplemental Figure 1b) and GSK 429286 (structure shown in Supplemental Figure 1c). Since RKI 1447 showed the strongest potency it was tested in all three matched cell lines. RKI 1447, similar to Y-27632 treatment and ROCK1 knockdown, selectively reduced the number of colonies and cells per colony in the VHL-deficient CC-RCC (Figure 3a–c, Supplemental Figure 6). The potencies of Y-27632, RKI 1447, and GSK 429286 were compared in RCC10±VHL in Figure 1c, Figure 3b, and Supplemental Figure 7 respectively. The overall inhibitor potencies based on IC50s are as follows: RKI 1447 (0.8µM) > GSK 429286 (6.4µM) > Y-27632 (8.2µM). Since GSK 429286 was less potent than RKI 1447 we did not test it further. We also observed that repeat, daily, treatment with 2µM RKI 1447 led to an enhanced synthetic lethal effect in each of the VHL-deficient CC-RCC, while minimally affecting matched VHL-expressing CC-RCCVHL (Figure 3d–f). For visual comparison representative images were obtained on day 14 of treatment for RCC4 and 786-O, and day 9 of treatment for RCC10 (Supplemental Figure 8). Thus, multiple ROCK inhibitors specifically target VHL-deficient CC-RCC.
Figure 3. ROCK inhibitor RKI 1447 causes synthetic lethality with VHL deficiency similar to Y-27632.
Clonogenic assays in RCC4±VHL (a), RCC10±VHL (b) and 786-O±VHL (c) matched cell lines confirmed the synthetic lethality of ROCK inhibitors with VHL loss. Each dose of RKI 1447 within each experiment was tested in duplicate, and each experiment was repeated three times. (d-f) Long term repeat administration of RKI 1447 enhances the synthetic lethality effect. Repeated daily treatment of RCC4±VHL (d), RCC10±VHL (e) and 786-O±VHL (f) with 2µM RKI 1447 causes VHL-deficient RCC cell numbers to decline, while RCCVHL cells continue to proliferate. Daily treatment with DMSO was used as a control. Cells were counted and passaged at 1:10 when the DMSO-treated VHL-expressing cells became >80% confluent. Statistical analysis was performed using a paired t-test between the matched cell lines at each dose (* p < 0.05, ** p < 0.01, *** p < 0.001), SEMs are shown.
Treatment with ROCK inhibitors reduces CC-RCC proliferation and induces cell death
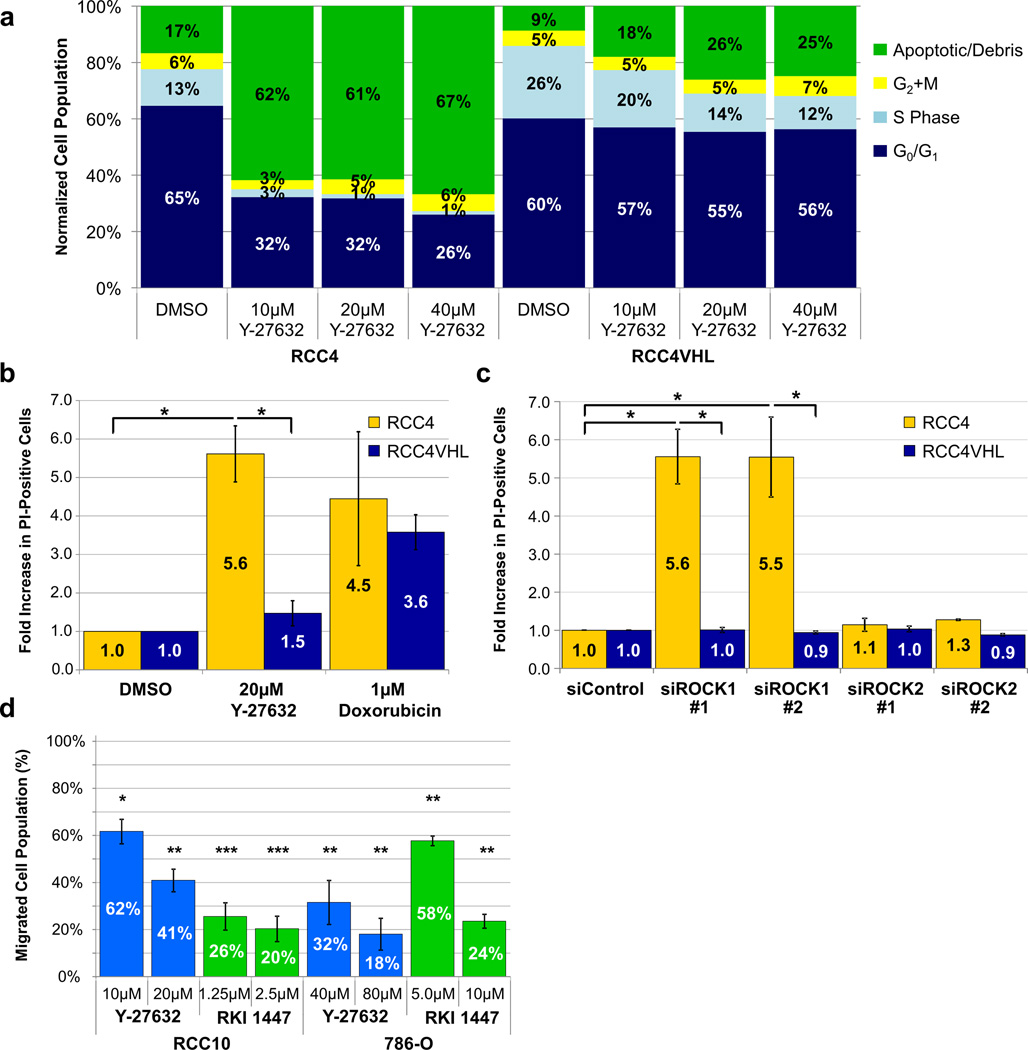
The results from the clonogenic assays pointed to both cell death (reduced colony numbers) and proliferation defect (reduced colony size) as biological outcomes of Y-27632 treatment (Figure 1b–d, Supplemental Figure 2). To confirm these biological outcomes, we assessed cell cycle progression using a FITC-Bromodeoxyuridine (BrdU) assay. Treatment of RCC4 and RCC4VHL cells with Y-27632 at 10µM, 20µM, and 40µM resulted in an increase in the apoptotic/debris population and a decrease in the S phase and G0/G1 phase populations, but the effects were more pronounced in RCC4 than in RCC4VHL (Figure 4a, Supplemental Figure 9). To determine if apoptosis was responsible for the increase in the apoptotic/debris population we assessed if Y-27632 stimulated caspase 3 cleavage in CC-RCC cells by Western blot analysis. Our results show that Y-27632 induced caspase 3 cleavage in both RCC4 and RCC4VHL, but did not induce caspase 3 cleavage in RCC10±VHL or 786-O±VHL over the basal level, thus ruling out apoptosis as a cause of selective cell death in VHL-deficient CC-RCC (Supplemental Figure 10).
Figure 4. ROCK inhibition in VHL-deficient CC-RCC cells decreases proliferation, induces cell death, and blocks cell migration.
(a) BrdU assay reveals that Y-27632 treatment is both cytotoxic and cytostatic in RCC4. RCC4 cells acquire a large fraction of apoptotic/debris cells and greatly reduce the S phase upon treatment with Y-27632 for 72 hours as opposed to RCC4VHL. The graph shows the representative experiment of two experiments performed (b) RCC4 cells treated with 20µM Y-27632 for 24 hours show a more than five-fold increase in cell death while RCC4VHL are minimally affected. Cells were stained with the vital dye PI and imaged at 4x. The number of PI-positive cells was then counted for each field. The data was normalized to DMSO treated cells. (c) Knockdown of ROCK1, but not ROCK2, induces cell death in the VHL-deficient RCC4. 48hours post siRNA transfection, RCC4 matched cells were stained with PI and imaged. Knockdown of ROCK1 resulted in over a 5 fold increase in PI-positive cells. The data was normalized to siControl treated cells. (b–c) Each experiment was performed in triplicate. (d) ROCK inhibition blocks CC-RCC migration in a transwell assay. RCC10 or 786-O cells were treated with Y-27632, RKI 1447 or DMSO vehicle (as indicated) for 6 hours. The assay was performed in duplicate and the experiment was repeated three times. Statistical analysis in (b–d) was performed using a paired t-test comparing each normalized dose to the negative control (* p < 0.05, ** p < 0.01, *** p < 0.001), SEMs are shown.
To confirm that Y-27632 treatment induces cell death, we treated RCC4 and RCC4VHL cells with 20µM Y-27632 for 24 hours and then stained the cells with propidium iodide (PI). Imaging of the RCC4 cells showed a 5.4-fold increase in the number of PI-positive dead cells, while RCC4VHL showed a 1.5-fold increase (Figure 4b, Supplemental Figure 11a). Additionally, siRNA knockdown of ROCK1, but not ROCK2, resulted in a more than 5-fold increase in PI-positive dead cells (Figure 4c, Supplemental Figure 11b). Together these results indicate that ROCK1 inhibition induces cell death and blocks proliferation in VHL-deficient CC-RCC.
ROCK inhibition blocks CC-RCC migration
Due to the known role of ROCKs in the regulation of cell adhesion, migration, and invasion32,40,41, we decided to assess the contribution of ROCKs to CC-RCC migration. First, we noticed that treatment with Y-27632 results in a change in cell morphology (cells become elongated and spindly), likely due to ROCKs role in regulating actin cytoskeleton reorganization and actomyosin contraction26 (Supplemental Figure 12). When we stopped the compound treatment at 48 hours, cells reverted to their non-elongated phenotype (Supplemental Figure 12). Second, both Y-27632 and RKI 1447 caused a dramatic reduction of RCC10 and 786-O cell migration in a transwell migration assay (Figure 4d). To rule out the cytotoxic/proliferation-inhibitory effect of Y-27632 and RKI 1447 on migrating cells, we conducted all of the migration experiments at short 6-hour time points. At 6 hours the live cell numbers were assessed by PI vital dye exclusion flow cytometry and no changes were detected (Supplemental Figure 13). Thus, ROCK inhibitors have the potential to reduce CC-RCC primary tumor growth through their cytotoxic and cytostatic effects and may inhibit metastasis through blocking cell migration.
Synthetic lethality between ROCK inhibition and VHL deficiency is dependent on HIFs
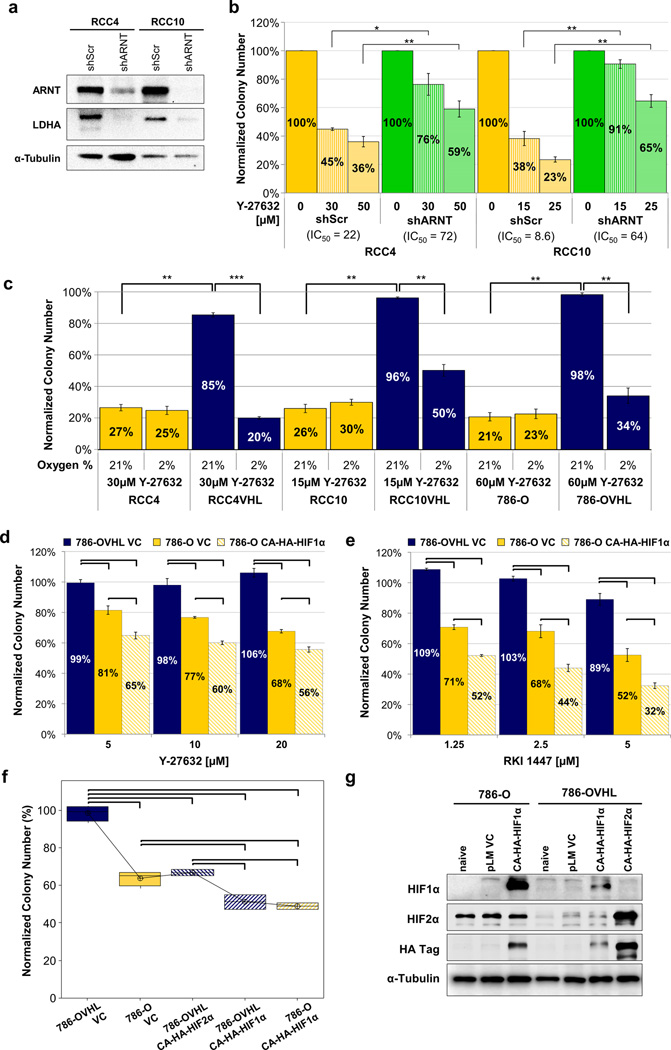
One of the best-studied consequences of VHL loss/mutation in CC-RCC is the massive stabilization and activation of HIF1α and HIF2α18–20 (Figure 1e). Thus, we hypothesized that the synthetic lethal effect between ROCK inhibition and VHL deficiency would be dependent on HIF activation. To test this hypothesis, we acquired RCC4 and RCC10 cell lines where we stably knocked down HIFβ, also known as Aryl hydrocarbon Receptor Nuclear Translocator (ARNT) with a specific shRNA. Since ARNT forms a heterodimer with either HIF1α or HIF2α, and is essential for HIF transcriptional activity, its knockdown inhibited HIF activity. This resulted in a reduction of the Lactate DeHydrogenase A (LDHA) HIF target gene expression (Figure 5a). As predicted, knockdown of ARNT in the VHL-deficient RCC4 and RCC10 cell lines had a protective effect against Y-27632 treatment (Figure 5b), mimicking VHL reintroduction. These results indicate that synthetic lethality between ROCK inhibition and VHL deficiency is dependent on HIF activation.
Figure 5. The synthetic lethal interaction between VHL loss and ROCK inhibition is HIF-dependent.
(a) Western blot showing the efficiency of ARNT knockdown by shRNA (shARNT) in VHL-deficient RCC4 and RCC10. The scrambled shRNA was used as a control (shScr). ARNT inhibition causes a decrease in HIF1α and HIF2α activity as evidenced by the decrease in expression of a HIF-target gene LDHA. (b) Clonogenic assay showing that CC-RCCs transduced with shARNT exerted resistance to ROCK inhibition in comparison to shScr transduced CC-RCCs. In that respect, CC-RCCs transduced with shARNT behaved similarly to the cell lines with reintroduced VHL. Each treatment was normalized to the DMSO control. (c) Cells from the CC-RCC±VHL were treated with Y-27632 at the indicated concentrations, plated for clonogenic assays, and replicate plates were subjected to either normoxia (21% O2) or hypoxia (2% O2). Each assay was performed in duplicate and repeated three times. Colony numbers were normalized to the vehicle control. RCC4VHL, RCC10VHL, and 786-OVHL cells were sensitized to ROCK inhibition in hypoxia. Statistical analysis was performed using a paired t-test (* p < 0.05, ** p < 0.01, *** p < 0.001), SEMs are shown. (d–e) Clonogenic assay showing that HIF-1α expression sensitizes 786-O cells to Y-27632 (d) and RKI 1447 (e). Each dose was compared statistically using a one-way ANOVA followed by Tukey’s Posthoc. Tie bars indicate significant differences with a p-value < 0.01. (f) Clonogenic assay showing that non-degradable HIF1α and HIF2α expression are sufficient to induce the synthetic lethal effect with 20µM Y-27632 treatment. Statistical analysis was conducted using a one-way ANOVA (p < 0.001) followed by Tukey’s Posthoc. There were 3 statistically significant groups: 786OVHL > 786O, 786OVHL HIF2a > 786OVHL HIF1a, 786O HIF1a.
To further confirm our findings, cells from the matched cell lines RCC4±VHL, RCC10±VHL, and 786-O±VHL were treated with Y-27632, plated for clonogenic assays, and replicate plates were subjected to either normoxia (21% O2, low HIF level and activity) or hypoxia (2% O2, high HIF level and activity). Each Y-27632 treatment was normalized to the DMSO vehicle control in both normoxia and hypoxia groups. The normalized colony numbers for Y-27632-treated VHL-deficient CC-RCC cell lines were not affected by oxygen concentration (Figure 5c). In contrast, CC-RCCVHL were sensitized to Y-27632 treatment in hypoxia having reduced colony-forming ability in hypoxia compared to normoxia (Figure 5c). Hypoxic induction of HIF1α and HIF2α was confirmed by Western blot analysis (Supplemental Figure 14). These results confirm that the synthetic lethal interaction between ROCK inhibition and VHL deficiency is HIF dependent.
Since 786-O cells were the most resistant to Y-27632 out of the three matched cell lines tested (Figure 1b–d) and they do not express HIF1α, while RCC4 and RCC10 do, we hypothesized that HIF1α re-expression in 786-O would sensitize them to Y-27632. To test this hypothesis we generated a 786-O cell line expressing a non-degradable constitutively active HA-tagged HIF1α (CA-HA-HIF1α). The 786-O CA-HA-HIF1α cells showed increased sensitivity to both Y-27632 (Figure 5d) and RKI 1447 (Figure 5e) when compared to the 786-O vector control expressing cell line. Additionally, we generated 786-OVHL cells expressing either the CA-HA-HIF1α or CA-HA-HIF2α. Expression of either HIF1α or HIF2α in 786-OVHL was sufficient to cause the synthetic lethal effect with ROCK inhibition, with 786-OVHL CA-HA-HIF1α showing a more pronounced effect than 786-OVHL CA-HA-HIF2α (Figure 5f–g). Altogether these results indicate that the synthetic lethal interaction of VHL loss with ROCK inhibition is due to the resulting constitutive activation of HIF in VHL-deficient CC-RCC.
Y-27632 inhibits tumor growth in vivo
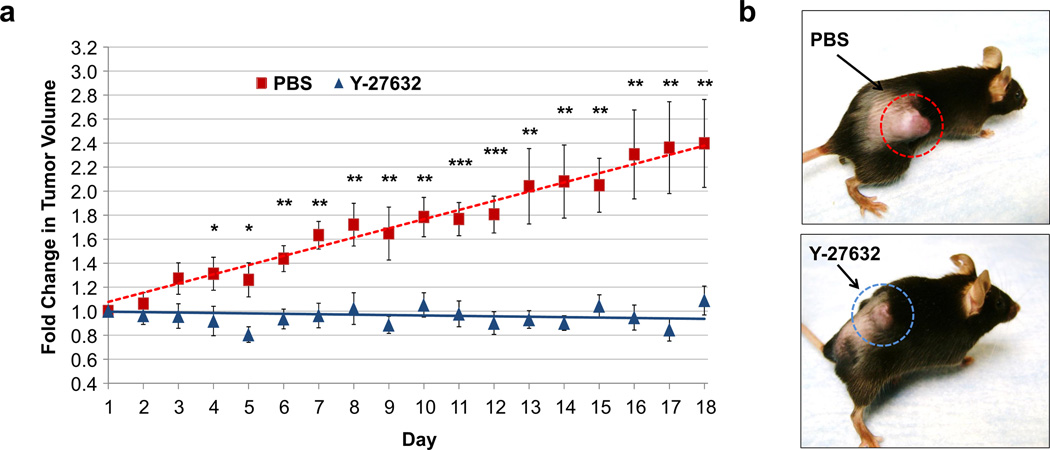
786-OT1 cells were isolated from a 786-O tumor grown in a RAG1 mouse and re-established in vitro to acquire a cell sub-line capable of fast growth in vivo. 786-OT1 were injected subcutaneously (sc) into the right flank of 18 RAG1 mice. After the tumors reached ~500 mm3, the mice were randomized and either daily treated with a vehicle control (PBS) or 10 mg/kg Y-27632 via intraperitoneal (ip) injection for 18 days. Y-27632 was selected for in vivo experiments based on abundant literature reporting its maximum tolerated dose and treatment regimens in mouse experiments41–44 in comparison to RKI 1447 used in a single study40. Tumor size was measured every day during treatment, and tumor volume constantly increased in the control group (n = 9), whereas tumor volume remained static in the Y-27632 group (n = 9) (Figure 6). The treatment was well tolerated with no weight loss in the mice (Supplemental Figure 15). The antitumor effects of Y-27632 support the concept that ROCK inhibitors can be used to selectively target VHL-deficient CC-RCC in vivo.
Figure 6. Y-27632 inhibits tumor growth in vivo.
(a) 5 × 106 786-OT1 cells were injected into the right flank of 18 NOG mice. After one month, mice were randomized into two groups. Mice were treated daily with PBS vehicle (n = 9) or 10 mg/kg Y-27632 (n = 9) by ip injection. The fold change in tumor volume was analyzed statistically using a One-way ANOVA with treatment as the factor (*p < 0.05, **p < 0.01, ***p < 0.005), SEMs are shown. The solid line represents the linear trend fit of the data for each treatment group. (b) Representative images of a control mouse (top) and a Y-27632-treated mouse (bottom) on day 14 of treatment are shown.
Discussion
In this study we identified a synthetic lethal interaction between the ROCK inhibitor, Y-27632, and the loss of VHL in CC-RCC. We have focused on validating ROCK inhibitors (Y-27632 and RKI 1447), which exhibited cytotoxic and cytostatic effects on VHL-deficient CC-RCC, making them candidate novel therapeutics for CC-RCC. First, the vast majority of CC-RCCs have lost VHL expression/function6 making over 90% of CC-RCC potentially sensitive to ROCK inhibition. Second, we have shown that the ROCK inhibitor Y-27632 suppresses CC-RCC tumor growth in vivo. Third, ROCK inhibitors reduce CC-RCC cell migration, indicating that they may have the potential to inhibit CC-RCC metastasis. Finally, ROCK1 and ROCK2 knockout mice are viable, indicating that both are dispensable under physiological conditions26, predicting no normal tissue toxicity. Since we have shown that synthetic lethality of ROCK inhibition with VHL deficiency occurs primarily through ROCK1, one future direction would be to acquire ROCK inhibitors specifically targeting ROCK1 and not ROCK2.
Previous synthetic lethality studies of VHL deficiency have identified “hits” being HIF-dependent20,22 and HIF-independent21,25. Our data in Figure 5 indicates that the synthetic lethal interaction of ROCK inhibitors with VHL loss is HIF-dependent. Exposure of VHL-reconstituted CC-RCC to hypoxia conferred sensitivity to ROCK inhibitors, while the knockdown of ARNT in VHL-deficient CC-RCC conferred resistance to ROCK inhibitors. Re-expression of non-degradable HIFs also sensitized 786-OVHL cells to ROCK inhibition. Importantly, VHL-deficient CC-RCC patient’s tumors differ in their repertoire of HIF subunits: 69% of patients express both HIF1α and HIF2α, while 31% express only HIF2α45. ROCK inhibition is synthetically lethal in both tumor types; although the cell lines RCC4 and RCC10 expressing both HIF1α and HIF2α are more sensitive to ROCK inhibition than 786-O expressing HIF2α only. In support of the role of HIF1α in the sensitization to ROCK inhibition the same increase in sensitivity to ROCK inhibitors was observed in the cell lines expressing HIF1α (786-OVHL HIF1α and 786-OHIF1α) over those only expressing HIF2α (786-O and 786-OVHL HIF2α).
The dependence of the synthetic lethal effect of ROCK inhibition on HIF overexpression is important since ROCK inhibitors may serve as potential therapeutics for other types of solid tumors besides CC-RCC where both HIF and ROCK are overactive. In addition to CC-RCC31, ROCK overexpression occurs commonly in multiple cancer types46 including lung35, breast29,32, osteosarcoma47, and prostate cancer30. On the other hand, a large fraction of solid tumors possesses hypoxic regions (where HIFs are stabilized48) or stabilize HIF1α and HIF2α by VHL independent mechanisms, including Phosphatase and TENsin homolog (PTEN)-loss or Harvey Rat Sarcoma Viral Oncogene Homolog (H-Ras) activation48,49. Thus, we predict that ROCK inhibitors will be effective against several more tumor types besides CC-RCC.
The crosstalk between HIF and ROCK has been investigated previously50–53. On the one hand, two studies show that RhoA and ROCK1 are HIF target genes in breast cancer50 and trophoblast cells51. In addition, Turcotte et al. showed that RhoA expression and activity are hypoxia inducible in renal cancer, although it does not depend on HIF activity52. If this regulation is maintained in renal cancer, the loss of VHL would be predicted to induce ROCK1 upregulation. We do not see increased ROCK1 expression (Figure 2b–c, Supplemental Figure 5) and actually observe decreased phosphorylation of the ROCK substrate MYPT1 in VHL-deficient cells (Supplemental Figure 3), thus not supporting this type of regulation. On the other hand, multiple studies show that the Rho/ROCK pathway stimulates HIF activity by multiple mechanisms, which are likely to be cell type specific52–54. This data for CC-RCC is missing and the crosstalk needs to be investigated.
In summary ROCK1 inhibition is synthetically lethal with VHL loss in CC-RCC, and ROCK inhibitors could serve as novel therapeutics for the disease. ROCK inhibitors would complement currently approved angiogenesis inhibitors since ROCK inhibitors selectively induce tumor cell death, reduce proliferation and migration, ultimately leading to inhibition of tumor growth and potentially metastasis.
Materials and Methods
Cell culture and chemical treatments
The CMV-EYFP labeled RCC4±VHL matched cell lines were previously described22. Cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM; Caisson Labs #25–500, North Logan, UT) + 10% Fetal Bovine Serum (FBS; Omega Scientific #FB-12, Tarzana, CA) + 1% Penicillin/Streptomycin (Caisson Labs #25–512). in 5%CO2, 21%O2 at +37°C. Y-27632 and GSK 429286 were obtained from Tocris (Minneapolis, MN). RKI 1447 was obtained from Selleck Chemicals (Houston, TX). All compounds were diluted in DiMethyl SulfOxide (DMSO) and serially diluted for each experiment.
Cell viability assay based on measurements of fluorescence intensity
RCC4 EYFP and RCC4VHL EYFP were plated at 5,000 cells per well in black 96-well tissue culture plates in FBS-free DMEM media. The following day, DMSO vehicle or varying compound concentrations were prepared in 20% FBS DMEM by serial dilution and an equal volume was added to the cells. Cells were incubated for 72 hours. Wells were washed with Phosphate Buffered Saline (PBS). Then, 100µL of PBS was added to each well and fluorescence intensity was measured on a BioTek Synergy HT Microplate Reader (Winooski, VT) at 488nm. Each experiment was performed three times in quadruplicate per treatment.
Clonogenic cell survival assay
Clonogenic assays were performed plating 300 cells/plate as previously described36.
Long term repeat treatment experiments
RCC4 EYFP, RCC4VHL EYFP, 786-O and 786-OVHL were plated at 5×104 cells per well into a 6-well plate and treated with vehicle (DMSO) or 2µM RKI 1447 daily. Each day the media was aspirated and fresh media with DMSO vehicle or 2µM RKI 1447 was added to each well. When the vehicle control plate was at 80% confluency, the cells were passaged 1:10 into new plates. Due to the different growth kinetics each cell line was passaged and counted at different time points: RCC4 on day 4, 9, and 15; RCC10 on 4, 7, 9, 11; and 786-O on 4, 11, 15, and 18.
Gene knockdowns by siRNAs
RCC4±VHL were plated at 200,000 cells per well of a 6-well plate in FBS-free DMEM. The following day the cells were transfected with 6 µL DharmaFECT1 (Dharmacon, Lafayette, CO) and up to 2 nM siRNA accordingly to manufacturer’s protocol. The siRNAs for ROCK1 (#1: SASI_Hs01_00065573 and #2: SASI_Hs01_00065570), ROCK2 (#1: SASI_Hs01_00204253 and #2: SASI_Hs01_00204251), and MISSION(R) Universal Negative Control #1 siRNA were obtained from Sigma (St. Louis, MO). The following day, transfected cells were plated for the clonogenic cell survival assay. Replicate plates were lysed after 72 hours and ROCK1 and ROCK2 expression analyzed by Western blot.
Western blot analysis
After treatments, cells were lysed and Western blot was conducted as previously described55. Proteins were visualized using primary antibodies recognizing HIF1α (BD Biosciences, #610959, San Jose, CA), HIF2α (Novus Biological, #NB100–122, Littleton, CO), α-tubulin (Fitzgerald, #10R-842, North Acton, MA), MYPT1-P Thr696 (EMD Millipore, #ABS45, Temecula, CA), MYPT1, (Abcam, #ab32393, Cambridge, MA), Cleaved Caspase 3, VHL (Cell Signaling, #9661, #2738, Danvers, MA), ROCK1, ROCK2 (Thermo Scientific, #PA5–22262, #PA5–21131, Grand Island, NY); and Horseradish peroxidase conjugated Goat anti-Rabbit IgG and Goat anti-Mouse IgG secondary antibodies (Thermo Scientific, #31460, #31430). Blots were imaged using a Bio Rad ChemiDoc XRS+ (BioRad, Hercules, CA).
PI-immunofluorescence staining
RCC4±VHL cells were cultured in the presence of DMSO, 20µM Y-27632, or 1µM Doxorubicin. After 24 hours, 1 µg/ml PI was added to each well and the cells were imaged on a Nikon TI-E at 4x and the PI-positive cells were counted per field. For the siRNA experiments, siRNAs were transfected at 5 nM following Dharmafect’s protocol and imaged at 10x after 48 hours. Each transfection was conducted in triplicate.
Transwell migration assay
8.0µm Polyethylene terephthalate Transwells (Corning, Corning, NY) were coated with Fibronectin as previously described55. 70,000 RCC10 or 35,000 786-O cells were used per transwell.
Cell cycle analysis
105 cells were seeded per well of a 6-well plate and treated the following day with vehicle (DMSO) or Y-27632 for 72 hours. BrdU analysis was performed using the FITC BrdU Flow Kit (BD Biosciences, Catalog #559619) following the manufacturer’s protocol.
shRNA expression constructs, lentivirus packaging, and infection of target cells
HEK 293T cells were transfected with lentiviral plasmids (pLKO.1shARNT: 5‘AAATAAACCATCTGACTTCTC3’ (OpenBiosystems, Huntsville, AL)) or pLKO.1shScrambled (Addgene, Cambidge, MA, #1864) along with packaging plasmids, pVSVG and ΔR8.2, as previously described56.
Tumor growth analysis
Briefly, 18 RAG1 (B6.129S7-Rag1tm1Mom/J, Jackson Labs) mice (11–20 weeks old) were injected sc into the right flank with 5×106 786-OT1 cells. Before each injection, cells were resuspended in 50 µl PBS/matrigel (BD Bioscience # 354248) mixture at 50/50 ratio. One month post-injections, when the tumors had reached the size of ~500mm3, littermates were randomized into two groups. 10 mg/kg Y-27632 or PBS diluent was administered ip daily for 18 days. Tumor size was measured daily with a digital caliper. On day 18 the mice were sacrificed. Tumor volume was calculated using the formula: V=(a)(b2/2), where “a” is the shorter measurement of length/width. Every measurement for each mouse was normalized to the day 1 measurement to show the fold change over time. Statistical analysis was performed using a one-way ANOVA between the two groups per day.
Growth curves and statistical analysis
Dose response and cell growth curves were generated using GraphPad Prism. IC50 values were calculated by transforming the X axis using X=Log(X), normalizing the transformed data to the vehicle control with 0 as 0%, and then fitting the normalized transformed data with a nonlinear trend line either using a normalized response (“log(inhibitor) vs. normalized response”) or a variable slope (“log(inhibitor) vs. normalized response – Variable slope”). The correct nonlinear trendline was selected using GraphPad’s comparison of fits, which directly compares both fit lines statistically using an extra sum-of-squares F test. The fit line is not shown in the figures. The IC50 values for each experiment were then calculated from the Best-fit values. Statistical analysis was conducted in Minitab 16 using a paired t-test or ANOVA between cell lines with a p-value of less than 0.05 considered statistically significant. All error bars represent the SEMs.
Supplementary Material
Acknowledgments
This work was funded by ACS-IRG-98-279-10 and NCI R03 CA202563-01 to OVR, NIH T32 (2T32CA009054-36A1) to JMT, NIH T32 NS82174-3 to IN, and NIH ICTS (UL1 TR001414) to LJN. The authors thank Dr. David Fruman and Dr. Hung Fan for critical reading of the manuscript and anonymous reviewers for constructive critique.
Abbreviations
- ARNT
Aryl hydrocarbon Receptor Nuclear Translocator
- BrdU
Bromodeoxyuridine
- CC-RCC
Clear Cell Renal Cell Carcinoma
- DMEM
Dulbecco’s Modified Eagle’s Medium
- DMSO
DiMethyl SulfOxide
- EGFR
Epidermal Growth Factor Receptor
- EYFP
Enhanced Yellow Fluorescent Protein
- FBS
Fetal Bovine Serum
- HIF
Hypoxia Inducible Factor
- ip
intraperitoneal
- LDHA
Lactate DeHydrogenase A
- LOPAC
Library of Pharmacologically Active Compounds
- mTORi
Mammalian Target of Rapamycin Inhibitors
- MYPT1
MYosin Phosphatase Target 1
- PBS
Phosphate Buffered Saline
- PI
Propidium Iodide
- QRT-PCR
Quantitative Real Time Polymerase Chain Reaction
- ROCK
Rho-Associated, coiled-coil-containing protein kinase
- RTK
Receptor Tyrosine Kinase
- RTKi
RTK Inhibitors
- sc
subcutaneous
- shRNA
small hairpin RNA
- siRNA
small interfering RNA
- VEGF
Vascular Endothelial Growth Factor
- VHL
Von Hippel Lindau
Footnotes
Conflict of Interest: The authors disclose no potential conflicts of interest.
Author contributions: OVR and JMT designed the study. JMT, QHN, MS, MWP, IN, LJN, ACL, OVR conducted experiments and analyzed the data. JMT and OVR wrote the paper.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2016. Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Motzer RJ, Russo P. Systemic therapy for renal cell carcinoma. J Urol. 2000;163:408–417. [PubMed] [Google Scholar]
- 3.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 4.Hodorova I, Rybarova S. Multidrug resistance proteins in renal cell carcinoma. Folia Biol. 2008;192:187–192. doi: 10.14712/fb2008054060187. [DOI] [PubMed] [Google Scholar]
- 5.Jonasch E, Futreal Pa, Davis IJ, Bailey ST, Kim WY, Brugarolas J, et al. State of the Science: An Update on Renal Cell Carcinoma. Mol Cancer Res. 2012;10:859–880. doi: 10.1158/1541-7786.MCR-12-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowey CL, Rathmell WK. VHL gene mutations in renal cell carcinoma: Role as a biomarker of disease outcome and drug efficacy. Curr Oncol Rep. 2009;11:94–101. doi: 10.1007/s11912-009-0015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Yang H. The Roles of VHL-Dependent Ubiquitination in Signaling and Cancer. Front Oncol. 2012;2:1–7. doi: 10.3389/fonc.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motzer RJ, Hutson TE, Tomczak P. Sunitinib versus Interferon Alfa in Metastatic Renal-Cell Carcinoma. N Engl JMed. 2007;356:115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 9.Escudier B, Eisen T, Stadler W, Szczylik C, Oudard S, Siebels M, et al. Sorafenib in Advanced Clear-Cell Renal-Cell Carcinoma. N Engl JMed. 2007;456:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 10.Escudier B, Gore M. Axitinib for the management of metastatic renal cell carcinoma. Drugs R D. 2011;11:113–126. doi: 10.2165/11591240-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sternberg CN, Davis ID, Mardiak J, Szczylik C, Lee E, Wagstaff J, et al. Pazopanib in locally advanced or metastatic renal cell carcinoma: Results of a randomized phase III trial. J Clin Oncol. 2010;28:1061–1068. doi: 10.1200/JCO.2009.23.9764. [DOI] [PubMed] [Google Scholar]
- 12.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Oudard S, et al. Overall survival and updated results for sunitinib compared with interferon alfa in patients with metastatic renal cell carcinoma. J Clin Oncol. 2009;27:3584–3590. doi: 10.1200/JCO.2008.20.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, et al. Phase 3 trial of everolimus for metastatic renal cell carcinoma: Final results and analysis of prognostic factors. Cancer. 2010;116:4256–4265. doi: 10.1002/cncr.25219. [DOI] [PubMed] [Google Scholar]
- 14.Hudes Carducci M, Tomczak P, Dutcher J, Figlin R, Kapoor A, et al. The Global ARCC G. Temsirolimus, interferon alpha or both for advanced renal cell carcinoma. N Engl J Med. 2007;356:2271–2281. doi: 10.1056/NEJMoa066838. [DOI] [PubMed] [Google Scholar]
- 15.Pinto A. Adjuvant Therapy for Renal Cell Carcinoma. Clin Genitourin Cancer. 2014:1–5. doi: 10.1016/j.clgc.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009;10:992–1000. doi: 10.1016/S1470-2045(09)70240-2. [DOI] [PubMed] [Google Scholar]
- 17.Buczek M, Escudier B, Bartnik E, Szczylik C, Czarnecka A. Resistance to tyrosine kinase inhibitors in clear cell renal cell carcinoma: from the patient’s bed to molecular mechanisms. Biochim Biophys Acta. 2014;1845:31–41. doi: 10.1016/j.bbcan.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Chan D, Giaccia AJ. Harnessing synthetic lethal interactions in anticancer drug discovery. Nat Rev Drug Discov. 2011;10:351–364. doi: 10.1038/nrd3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson JM, Nguyen QH, Singh M, Razorenova O. Approaches to Identifying Synthetic Lethal Interactions in Cancer. Yale J Biol Med. 2015;88:1–11. [PMC free article] [PubMed] [Google Scholar]
- 20.Chan Da, Sutphin PD, Nguyen P, Turcotte S, Lai EW, Banh A, et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med. 2011 doi: 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turcotte S, Chan D, Sutphin PD, Hay MP, Denny Wa, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14:90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutphin PD, Chan D, Li JM, Turcotte S, Krieg AJ, Giaccia AJ. Targeting the loss of the von Hippel-Lindau tumor suppressor gene in renal cell carcinoma cells. Cancer Res. 2007;67:5896–5905. doi: 10.1158/0008-5472.CAN-07-0604. [DOI] [PubMed] [Google Scholar]
- 23.Woldemichael GM, Turbyville TJ, Vasselli JR, Linehan WM, McMahon JB. Lack of a functional VHL gene product sensitizes renal cell carcinoma cells to the apoptotic effects of the protein synthesis inhibitor verrucarin A. Neoplasia. 2012;14:771–777. doi: 10.1593/neo.12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolff NC, Pavía-Jiménez A, Tcheuyap VT, Alexander S, Vishwanath M, Christie A, et al. High-throughput simultaneous screen and counterscreen identifies homoharringtonine as synthetic lethal with von Hippel-Lindau loss in renal cell carcinoma. Oncotarget. 2015;6:16951–16962. doi: 10.18632/oncotarget.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bommi-Reddy A, Almeciga I, Sawyer J, Geisen C, Li W, Harlow E, et al. Kinase requirements in human cells: III Altered kinase requirements in VHL−/− cancer cells detected in a pilot synthetic lethal screen. Proc Natl Acad Sci USA. 2008;105:16484–16489. doi: 10.1073/pnas.0806574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi J, Wu X, Surma M, Vemula S, Zhang L, Yang Y, et al. Distinct roles for ROCK1 and ROCK2 in the regulation of cell detachment. Cell Death Dis. 2013;4:e483. doi: 10.1038/cddis.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamai T, Tsujii T, Arai K. Significant Association of Rho / ROCK Pathway with Invasion and Metastasis of Bladder Cancer Significant Association of Rho / ROCK Pathway with Invasion and Metastasis of Bladder Cancer 1. 2003;9:2632–2641. [PubMed] [Google Scholar]
- 28.Kamai T, Yamanishi T, Shirataki H, Takagi K, Asami H, Ito Y, et al. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin Cancer Res. 2004;10:4799–4805. doi: 10.1158/1078-0432.CCR-0436-03. [DOI] [PubMed] [Google Scholar]
- 29.Lane J, Martin T, Watkins G, Mansel R, Jiang W. The expression and prognostic value of ROCK I and ROCK II and their role in human breast cancer. Int J Oncol. 2008;33:585–593. [PubMed] [Google Scholar]
- 30.Zhang C, Zhang S, Zhang Z, He J, Xu Y, Liu S. ROCK has a crucial role in regulating prostate tumor growth through interaction with c-Myc. Oncogene. 2013:1–10. doi: 10.1038/onc.2013.505. [DOI] [PubMed] [Google Scholar]
- 31.Abe H, Kamai T, Tsujii T, Nakamura F, Mashidori T, Mizuno T, et al. Possible role of the RhoC/ROCK pathway in progression of clear cell renal cell carcinoma. Biomed Res. 2008;29:155–161. doi: 10.2220/biomedres.29.155. [DOI] [PubMed] [Google Scholar]
- 32.Liu S, Goldstein RH, Scepansky EM, Rosenblatt M. Inhibition of rho-associated kinase signaling prevents breast cancer metastasis to human bone. Cancer Res. 2009;69:8742–8751. doi: 10.1158/0008-5472.CAN-09-1541. [DOI] [PubMed] [Google Scholar]
- 33.Liao JK, Seto M, Noma K. Rho Kinase (ROCK) Inhibitors. J Cardiovasc Pharmacol. 2007;50:17–24. doi: 10.1097/FJC.0b013e318070d1bd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mleczak A, Millar S, Tooze Sa, Olson MF, Chan EYW. Regulation of autophagosome formation by Rho kinase. Cell Signal. 2013;25:1–11. doi: 10.1016/j.cellsig.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Nakagawa O, Fujisawa K, Ishizaki T, Saito Y, Nakao K, Narumiya S. ROCK-I and ROCK-II, two isoforms of Rho-associated coiled-coil forming protein serine/threonine kinase in mice. FEBS Lett. 1996;392:189–193. doi: 10.1016/0014-5793(96)00811-3. [DOI] [PubMed] [Google Scholar]
- 36.Razorenova O, Castellini L, Colavitti R, Edgington LE, Nicolau M, Huang X, et al. The apoptosis repressor with a CARD domain (ARC) gene is a direct hypoxia-inducible factor 1 target gene and promotes survival and proliferation of VHL-deficient renal cancer cells. Mol Cell Biol. 2014;34:739–751. doi: 10.1128/MCB.00644-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 38.Chan D, Sutphin PD, Denko NC, Giaccia AJ. Role of prolyl hydroxylation in oncogenically stabilized hypoxia-inducible factor-1alpha. J Biol Chem. 2002;277:40112–40117. doi: 10.1074/jbc.M206922200. [DOI] [PubMed] [Google Scholar]
- 39.Amin E, Dubey BN, Zhang SC, Gremer L, Dvorsky R, Moll JM, et al. Rho-kinase: Regulation, (dys)function, and inhibition. Biol Chem. 2013;394:1399–1410. doi: 10.1515/hsz-2013-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel R, Forinash KD, Pireddu R, Sun Y, Sun N, Martin MP, et al. RKI-1447 is a potent inhibitor of the Rho-associated ROCK kinases with anti-invasive and antitumor activities in breast cancer. Cancer Res. 2012;72:5025–5034. doi: 10.1158/0008-5472.CAN-12-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isler D, Ozaslan M, Karagoz ID, Kilic IH, Karakok M, Taysi S, et al. Antitumoral effect of a selective Rho-kinase inhibitor Y-27632 against Ehrlich ascites carcinoma in mice. Pharmacol Rep. 2014;66:114–120. doi: 10.1016/j.pharep.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Rikitake Y, Kim H-H, Huang Z, Seto M, Yano K, Asano T, et al. Inhibition of Rho kinase (ROCK) leads to increased cerebral blood flow and stroke protection. Stroke. 2005;36:2251–2257. doi: 10.1161/01.STR.0000181077.84981.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inan S, Büyükafşar K. Antiepileptic effects of two Rho-kinase inhibitors, Y-27632 and fasudil, in mice. Br J Pharmacol. 2008;155:44–51. doi: 10.1038/bjp.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagatoya K, Moriyama T, Kawada N, Takeji M, Oseto S, Murozono T, et al. Y-27632 prevents tubulointerstitial fibrosis in mouse kidneys with unilateral ureteral obstruction. 2002;61:1684–1695. doi: 10.1046/j.1523-1755.2002.00328.x. [DOI] [PubMed] [Google Scholar]
- 45.Gordan J, Lal P, Dondeti V, Letrero R. HIF-α Effects on c-Myc Distinguish Two Subtypes of Sporadic VHL-Deficient Clear Cell Renal Carcinoma. Cancer Cell. 2008;14:435–446. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lochhead P, Wickman G, Mezna M, Olson MF. Activating ROCK1 somatic mutations in human cancer. Oncogene. 2010;29:2591–2598. doi: 10.1038/onc.2010.3. [DOI] [PubMed] [Google Scholar]
- 47.Liu X, Choy E, Hornicek FJ, Yang S, Yang C, Harmon D, et al. ROCK1 as a potential therapeutic target in osteosarcoma. J Orthop Res. 2011;29:1259–1266. doi: 10.1002/jor.21403. [DOI] [PubMed] [Google Scholar]
- 48.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 49.Kapitsinou PP, Haase VH. The VHL tumor suppressor and HIF: insights from genetic studies in mice. Cell Death Differ. 2008;15:650–659. doi: 10.1038/sj.cdd.4402313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilkes DM, Xiang L, Lee SJ, Chaturvedi P, Hubbi ME, Wirtz D, et al. Hypoxia-inducible factors mediate coordinated RhoA-ROCK1 expression and signaling in breast cancer cells. Proc Natl Acad Sci U S A. 2013:1–10. doi: 10.1073/pnas.1321510111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Greijer AE, van der Groep P, Kemming D, Shvarts A, Semenza GL, Meijer GA, et al. Up-regualtion of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor I (HIF-I) J Pathol. 2005;206:291–304. doi: 10.1002/path.1778. [DOI] [PubMed] [Google Scholar]
- 52.Turcotte S. HIF-1alpha mRNA and protein upregulation involves Rho GTPase expression during hypoxia in renal cell carcinoma. J Cell Sci. 2003;116:2247–2260. doi: 10.1242/jcs.00427. [DOI] [PubMed] [Google Scholar]
- 53.Takata K, Morishige K-I, Takahashi T, Hashimoto K, Tsutsumi S, Yin L, et al. Fasudil-induced hypoxia-inducible factor-1alpha degradation disrupts a hypoxia-driven vascular endothelial growth factor autocrine mechanism in endothelial cells. Mol Cancer Ther. 2008;7:1551–1561. doi: 10.1158/1535-7163.MCT-07-0428. [DOI] [PubMed] [Google Scholar]
- 54.Mizukami Y, Fujiki K, Duerr E-M, Gala M, Jo W-S, Zhang X, et al. Hypoxic regulation of vascular endothelial growth factor through the induction of phosphatidylinositol 3-kinase/Rho/ROCK and c-Myc. J Biol Chem. 2006;281:13957–13963. doi: 10.1074/jbc.M511763200. [DOI] [PubMed] [Google Scholar]
- 55.Razorenova O, Finger E, Colavitti R, Chernikova S, Boiko A, Chan C, et al. VHL loss in renal cell carcinoma leads to up-regulation of CUB domain-containing protein 1 to stimulate PKC{delta}-driven migration. Proc Natl Acad Sci U S A. 2011;108:1931–1936. doi: 10.1073/pnas.1011777108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Razorenova O, Ivanov V, Budanov V, Chumakov P. Virus-based reporter systems for monitoring transcriptional activity of hypoxia-inducible factor 1. Gene. 2005;350:89–98. doi: 10.1016/j.gene.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.