Abstract
Objective
Triggering receptor expressed on myeloid cells (TREM)-1 has recently been recognized as one of the potent amplifiers of acute and chronic inflammation. However, exact role of TREM-1 in regard to insulin insensitivity is unknown.
Methods
We examined the mRNA transcripts and protein expression of TREM-1, TREM-2 and TREM-1/TREM-2 ratio in the tissue biopsies (liver, omentum and subcutaneous fat) and blood samples (neutrophils and monocytes) of subjects with obesity and diabetes (SO+D+; n=15), subjects with obesity but not diabetes (SO+D−; n=7), and compared with the subjects without obesity (BMI < 30) and diabetes (SO−D−; n=5).
Results
The immunofluorescence and RT-PCR revealed significant increase in TREM-1, decrease in TREM-2, and increase in the TREM1/TREM2 ratio in SO+D+ group compared to other groups. Overall, increased liver TREM-1 expression and soluble-TREM-1 were found in SO+D+ group compared to SO+D− group (100% vs 57.14%, r=0.582; P=0.023). TREM-1 was significantly increased in all subjects with obesity, those had HOMA-IR index of >2.
Conclusions
TREM-1 was found to be significantly higher in tissues biopsies and blood of subjects with obesity. Greater expression and activity of TREM-1 suggests a possible role in the underlying pathophysiology of obesity and associated co-morbidities.
Keywords: Diabetes, Inflammation, Insulin resistance, Metabolic-syndrome, Obesity, TREM-1
Introduction
Obesity has long been considered a chronic low grade inflammatory condition (1). Obesity activates the innate immune system and results in macrophage and T cell infiltration in expanded adipose tissue (2, 3). This induces the secretion of inflammatory cytokines, including TNF-α, IL-6 and monocyte chemoattractant protein-1 (MCP-1) from adipocytes, associated with the pathogenesis of obesity-induced insulin resistance (4).
Triggering receptor expressed on myeloid cells (TREM)-1 belongs to the immunoglobulin superfamily and is expressed on monocytes/macrophages and neutrophils (5, 6). These receptors are activated by various pro-inflammatory cytokines and are mainly involved in the acute phase of inflammation (6). Activation of TREM-1 results in secretion of various proinflammatory cytokines, which leads to the activation of cell surface molecules involved in extravasation, cell activation and co-stimulation (5). Recently, several authors have investigated the role of TREM-1 in chronic inflammatory conditions such as upper respiratory airway disease (7, 8).
Similar to TREM-1, TREM-2 is also expressed on monocytes/macrophages (9, 10) but contrary to TREM-1, in most cases increased expression and activity of TREM-2 are anti-inflammatory (11, 12). Since obesity is a chronic low-grade inflammatory condition, TREM-1 and TREM-2 can serve as potential marker for insulin resistance. The aim of the study was to examine the correlation between insulin resistance, sTREM-1 and the ratio of TREM-1 and TREM-2 in the subjects with obesity (with or without diabetes).
Methods
Patient selection
This prospective study was approved by the Institutional Review Board (IRB) of Creighton University. After IRB-approved informed consent was obtained, 27 patients in the surgery clinic and scheduled for surgery at Creighton University Medical Center and Immanuel Medical Center were enrolled in the study. Study participation was voluntary. Of the 27 patients in 3 groups, 15 subjects with obesity and diabetes (SO+D+) ; 7 subjects with obesity but not diabetes (SO+D−) and 5 subjects without obesity and diabetes (SO−D−; controls) those who were undergoing elective non-weight loss surgery (ex-hernia, fundoplication) and elective gall bladder surgery. Power analysis was carried out for sample size among groups based on BMI with a power (1-β) equal to 0.80 and (α) equal to 5%. A total of 22 subjects with obesity were enrolled as per inclusion and exclusion criteria (Supporting information in Table 1).
Tissue and blood collection
Tissue biopsies for liver, omentum and subcutaneous fat were obtained during procedure and immediately transported to the lab in the University of Wisconsin solution and maintained at 4°C. Blood was collected pre-operatively into vacutainer tubes that contained ethylene diamine tetra acetic acid for biochemical, flow cytometry and ELISA studies.
Clinical data and biochemical analysis
The clinical and biochemical data was collected from the chart review. The variables collected include patient demographics and co-morbid conditions are given in Table 1 and biochemical parameters are given in Table S1.
Table 1.
Demographics and co-morbid conditions of study population
| Clinical data | SO−D− (5) | SO+D− (7) | SO+D+ (15) | P value |
|---|---|---|---|---|
| Demographics | ||||
| Gender (Male/ Female) |
1/4 | 1/6 | 6/9 | |
| Age (year) | 44.4 ± 15.02 | 36.71 ± 6.79 | 45.28 ± 10.17 | |
| BMI | 26.46 ± 1.71 | 46.95 ± 7.82 | 47.60 ± 9.31 | NS |
| Height (feet' inches") |
5.64 ± 0.20 | 5.34 ± 0.26 | 5.65 ± 0.28 | |
| Weight (pounds) | 150.70 ± 8.92 | 284.57 ± 55.38 | 312.69 ± 81.60 | |
| Co-morbid conditions | ||||
| Hypertension | 1 (20%) | 4 (57.1) | 13 (86.7%) | |
| Hyperlipidemia | -- | 1 (14.3%) | 6 (40%) | |
| Sleep apnea | -- | 2 (28.6%) | 9 (60%) | NS |
| Smokers | Former- 3 (60%) No - 2 (40%) |
Former- 4 (57.1%) No - 3 (42.8%) |
Former- 7 (46.6) No - 8 (53.3%) |
|
Demographics and co-morbidities were compared between SO+D− and SO+D+ subjects using student t-test for continuous variables and Fisher’s exact test or Pearson’s χ2 for categorical variables. A total of 22 subjects with obesity were enrolled as per inclusion and exclusion criteria. Inclusion criteria: All subjects with morbid obesity (BMI >40 and BMI <65) (age >21 years and <65 years) who were undergoing bariatric surgery. Exclusion criteria: Patients with prior systemic inflammatory conditions (SLE, rheumatoid arthritis and systemic sclerosis), malignant conditions and patients on immunosuppressive medications were not included in the study. Patients on non-steroidal anti-inflammatory agents were advised to discontinue at least 1-week before the surgery or switched to another pain medication. The subjects required to quit smoking at least three months or even in some cases more than a year ago before the surgery were included in the study. Blood nicotine levels were checked to ensure the subjects were quit before they were allowed to continue in the program. These levels were also checked every month after to ensure that these subjects did not relapse. Not significant (NS), data for age, body mass index (BMI), height and weight are presented as mean values ± SD; the sample size is shown as number (percentage). Data in co-morbid condition show number of subjects (%) in that group. Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Fatty liver grading
The Hematoxylin and Eosin stained biopsy samples of liver were reviewed blindly by a board certified pathologist for fatty liver grading and inflammation (Table S2).
ELISA
The levels of FFA (μM/ml, ab65341), Insulin (μIU/ml, ab100578), TNF-α (ab100572), IL-1β (ab100654), IL-6 (ab100562), sTREM-1 (pg/ml, ab100659) and sTREM-2 (MBS928570) were measured by ELISA kits as per manufacture's protocol.
Flow cytometry (FACS)
The expression of TREM-1 and TREM-2 on the monocytes (CD45+CD14+HLA-DR+) and neutrophils (CD45+CD16+) were characterized in the flow cytometry studies. The fluorescent conjugated antibody panel used for the determination of monocytes and neutrophils are given in Table S3. The purified cell populations were examined in FACS using the standard protocol in our laboratory. Isotypes for each fluorochrome were used for the negative control (Figure S1). OneComp eBeads (eBioscience 01-1111-42) with fluorescent conjugated antibody were used for the positive control. Cell populations were analyzed using Flow-Jo (v10) software. The information regarding gating of the cells is shown in Figure 1.
Figure 1.
TREM-1 and TREM-2 expression in monocytes and neutrophils of the study subjects. The protein expression of TREM-1 and TREM-2 in monocytes and neutrophils were compared between SO−D−, SO+D− and SO+D+ groups using One-way ANOVA for continuous variables. Cell phenotype was performed using anti-human antibodies for monocytes (CD45+, CD14+, HLA-DR+) represented in figure A and neutrophils (CD45+, CD16+) in figure B. A) The representative figures for monocytes gating; live granulocytes were gated using FSC/SSC. Then, CD45+ were gated and analyzed for CD14 and HLA-DR expression. The double positive populations (CD14+ HLA-DR+) were further gated for expression of TREM-1 and TREM-2 Aa) SO−D−, Ab) SO+D− and Ac) SO+D+. B) The representative figures for neutrophils gating; live monocytes were gated using FSC/SSC. Then, CD45+ were gated and analyzed for CD16 expression. CD16+ populations were further gated for expression of TREM-1 and TREM-2 Ba) SO−D−, Bb) SO+D− and Bc) SO+D+. C) The expression of TREM-1 and TREM-2 levels in monocytes (%). Data are shown as mean ± SD (N= 5 SO−D−; 7 SO+D−; 15 SO+D+); D) The expression of TREM-1 and TREM-2 levels in neutrophils (%). Data are shown as mean ± SD (N= 5 SO−D−; 7 SO+D−; 15 SO+D+); *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
RNA isolation, cDNA synthesis, and Real-Time PCR
Total RNA isolation was done using TRI reagent (Trizol reagent, Sigma, St Louis, MO, USA). Further, the cDNA was synthesized and Real-time PCR (CFX96, BioRad Laboratories, and Hercules, CA, USA) was performed in triplicate using SYBR Green Master Mix. The primers for TREM-1, TREM-2, TNF-α, IL-1β, IL-6, CRP, IL-5, CD64 and GAPDH were obtained from Integrated DNA Technologies and the sequences are given in Table S4 (Coralville, IA, USA). The relative expressions of target genes were stated as fold ratio using 2−ΔΔCT Method. Supporting information is provided in Figure 2-3.
Figure 2.
Fold change in mRNA transcripts and protein levels for TREM-1 and TREM-2 relative to GAPDH in the study subjects. The expression of TREM-1, TREM-2 and TREM-1/TREM-2 ratio levels in biopsy samples and serum were compared between SO−D−, SO+D− and SO+D+ groups using One-way ANOVA for continuous variables. The TREM-1 and TREM-2 expression fold changes in SO+D− and SO+D+ were calculated, when compared to SO−D− fold ratio were stated as 1. Increased in folds of TREM-1 or TREM-2 expressions were regarded as up regulation, likewise decrease in folds as down regulation. To describe the balance between pro and anti-inflammatory state, we analysed TREM-1/TREM-2 ratio in all the study subjects. A) Gene Expression study in Liver biopsy sample (Aa) TREM-1; (Ab) TREM-2; (Ac) TREM-1/TREM-2 ratio. B) Gene Expression study in Omentum biopsy sample (Ba) TREM-1; (Bb) TREM-2; (Bc) TREM-1/TREM-2 ratio. C) Gene Expression study in Subcutaneous biopsy sample (Ca) TREM-1; (Cb) TREM-2; (Cc) TREM-1/TREM-2 ratio. The cDNA prepared from mRNA was subjected to Q-PCR with gene specific primers for TREM-1 and TREM-2 and the fold change relative to GAPDH as housekeeping gene are shown. Results were expressed as fold change in SO+D− and SO+D+ compared to SO−D− group. D) Quantification of sTREM-1 and sTREM-2 (in pg/ml) using ELISA in the study populations. Da) sTREM-1; Db) sTREM-2; Dc) serum TREM-1/TREM-2 ratio. E) Protein Expression study in Liver biopsy sample (Ea) TREM-1; (Eb) TREM-2; (Ec) TREM-1/TREM-2 ratio. F) Protein Expression study in Omentum biopsy sample (Fa) TREM-1; (Fb) TREM-2; (Fc) TREM-1/TREM-2 ratio. G) Protein Expression study in Subcutaneous biopsy sample (Ga) TREM-1; (Gb) TREM-2; (Gc) TREM-1/TREM-2 ratio. The protein samples were isolated from the biopsy tissues and quantified using Bicinchoninic acid assay. The protein samples (50μg/well) were electrophoresed by using SDS–PAGE and transferred to nitrocellulose membrane Immunoblot analysis was carried out using rabbit anti-human TREM-1 or TREM-2 and mouse anti-GAPDH as primary antibody and HRP conjugated goat anti-rabbit IgG and HRP conjugated rabbit anti-mouse IgG as secondary antibodies, respectively. The protein expression was detected by incubating the membrane to chemiluminescence solution. The exposure time was adjusted to keep the integrated optical densities within a linear and non-saturated range. The band intensity was measured by densitometric analysis using Image J software. GAPDH was used to normalize the target and the relative expressions were stated as fold ratio. Data were shown as mean ± SD (N= 5 SO− D−; 7 SO+D−; 15 SO+D+); *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Figure 3.
Expression of mRNA transcripts for classical inflammatory genes TNF-α, IL-1β, IL-6, IL-5, CRP and CD64 in the study subjects. The expression of TNF-α, IL-1β, IL-6, IL-5, CRP and CD64 levels in biopsy samples were compared between SO−D−, SO+D− and SO+D+ groups using One-way ANOVA for continuous variables. The expression fold changes in SO+D− and SO+D+ were calculated, when compared to SO−D− fold ratio were stated as 1. Increased in folds of TNF-α, IL-1β, IL-6, IL-5, CRP and CD64 expressions were regarded as up regulation, likewise decrease in folds as down regulation. A) Gene expression study in omentum biopsy sample (Aa) TNF-α; (Ba) IL-1 β; (Ca) IL-6; (Da) IL-5; (Ea) CRP; (Fa) CD64. (B) Gene expression study in subcutaneous biopsy sample (Ab) TNF-α; (Bb) IL-1 β; (Cb) IL-6; (Db) IL-5; (Eb) CRP; (Fb) CD64. C) Gene expression study in liver biopsy sample (Ac) TNF-α; (Bc) IL-1 β; (Cc) IL-6; (Dc) IL-5; (Ec) CRP; (Fc) CD64. The cDNA prepared from mRNA was subjected to Q-PCR with gene specific primers and the fold change relative to GAPDH as housekeeping gene is shown. Results were expressed as fold change in SO+D− and SO+D+ compared to SO−D− group. Data were shown as mean ± SD (N= 5 SO−D−; 7 SO+D−; 15 SO+D+); *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Western blotting
Protein was isolated and quantified from the biopsy samples. Immunoblot analysis was carried as per standard protocol using rabbit anti-TREM-1 (1:1000; ab93717), rabbit anti-TREM-2 (1:1000; ab175262) mouse anti-GAPDH (NB300221) as primary antibody and HRP conjugated goat anti-rabbit IgG (1:4000; NB7160) and HRP-conjugated rabbit anti-mouse IgG (1:4000; NB7544) as secondary antibodies. Supporting information is provided in Figure 2.
Immunofluorescence (IF) study
Immunofluorescence staining was done as per standard protocol using goat anti-TREM-1 and rabbit anti-TREM-2 (Santa Cruz Biotech) at 1:50 dilution as primary antibody and Alexa Fluor 594 (red) and Alexa Fluor 488 (green) conjugated secondary antibodies (Invitrogen, Grand Island, NY, USA) at 1:200 dilution. The details on the image analysis are in Figures 4-5.
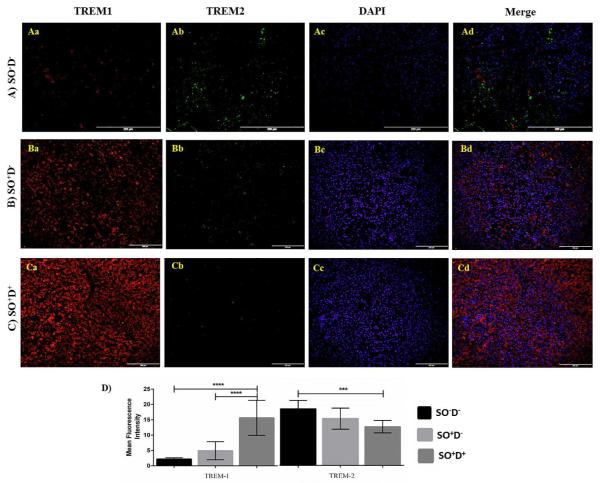
Figure 4.
Immunofluorescence staining of TREM-1 and TREM-2 in liver biopsy sections of study populations. The protein expression levels of TREM-1 and TREM-2 in liver biopsy sections were compared between SO−D−, SO+D− and SO+D+ groups using One-way ANOVA for continuous variables. Dual fluorescence staining was done using anti-human TREM-1 (Alexa 594-Red) and TREM-2 (Alexa 488-green) antibodies to co-localize TREM-1 and TREM-2, counterstained with DAPI. Immunofluorescence microscopy at 20X was done with an Olympus inverted fluorescent microscope (Olympus BX51). Negative controls were run by using isotypes for each fluorochrome. Fluorescence intensity for TREM-1 and TREM-2 was measured in the stained slides using Image-J software and mean fluorescence intensity (MFI) was calculated. (A) Immunofluorescence staining in SO−D− group Aa) TREM-1; Ab) TREM-2; Ac) DAPI; Ad) Merge. (B) Immunofluorescence staining in SO+D− group Ba) TREM-1; Bb) TREM-2; Bc) DAPI; Bd) Merge. (C) Immunofluorescence staining in SO+D+ group Ca) TREM-1; Cb) TREM-2; Cd) DAPI; Ce) co-localization. D) Fluorescence density of TREM-1 and TREM-2 in the tissue sections of study populations. Data are shown as mean ± SD (N= 5 SO−D−; 7 SO+D−; 15 SO+D+); *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
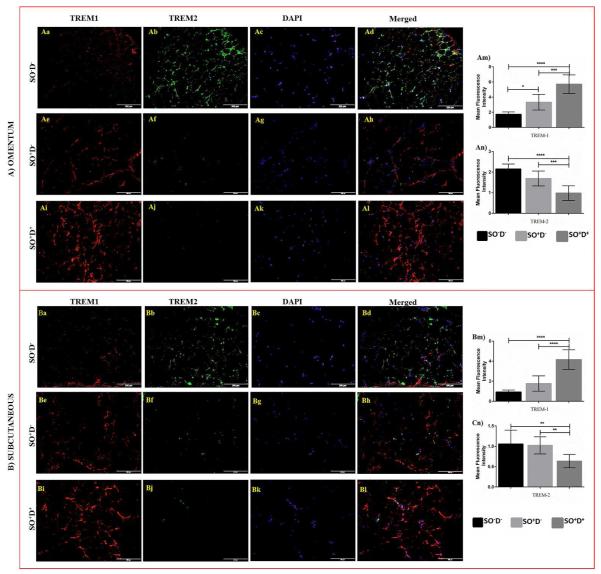
Figure 5.
Immunofluorescence staining of TREM-1 and TREM-2 in omentum and subcutaneous biopsy sections of study populations. The protein expression levels of TREM-1 and TREM-2 in omentum and subcutaneous biopsy sections were compared between SO−D−, SO+D− and SO+D+ groups using One-way ANOVA for continuous variables. Dual fluorescence staining was done by using anti-human TREM-1 (Alexa 594-Red) and TREM-2 (Alexa 488-green) antibodies to co-localize TREM-1 and TREM-2, counterstained with DAPI. Immunofluorescence microscopy at 20X was done with an Olympus inverted fluorescent microscope (Olympus BX51). Negative controls were run by using isotypes for each fluorochrome. Fluorescence intensity for TREM-1 and TREM-2 was measured in the stained slides using Image-J software and mean fluorescence intensity (MFI) was calculated. A) Immunofluorescence staining in omentum biopsy samples for TREM-1 (panels Aa, Ae, Ai), TREM-2 (panels Ab, Af, Aj), DAPI (panels Ac, Ag, Ak) and co-localization of TREM-1 and TREM-2 (panels Ad, Ah, Al). Fluorescence density for TREM-1 (panel Am) and TREM-2 (panels An) in the omentum tissue sections of study populations. (B) Immunofluorescence staining in subcutaneous biopsy samples for TREM-1 (panels Ba, Be, Bi), TREM-2 (Bb, Bf, Bj), DAPI (panels Bc, Bg, Bk) and co-localization of TREM-1 and TREM-2 (Bd, Bh, Bl). Fluorescence density for TREM-1 (panel Bm) and TREM-2 (panels Bn) in the omentum tissue sections of study populations. Data were shown as mean ± SD (N= 5 SO−D−; 7 SO+D−; 15 SO+D+); *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Statistical analysis
The mRNA and protein expression of TREM-1, TREM-2 and TREM-1/TREM-2 ratio in biopsy tissues and blood were compared between SO−D−, SO+D− and SO+D+ groups using One-way ANOVA for continuous variables. Demographics, co-morbidities, biochemical profile and fatty liver grading comparison were done between SO+D− and SO+D+ group using student t-test for continuous variables and Fisher’s exact test or Pearson’s χ2 for categorical variables. Among the categorical variables of the subjects, the increased expression of TREM-1, down regulation of TREM-2 and TREM-1/TREM-2 ratio were analyzed between SO+D− and SO+D+ groups. Subject’s categorical variables with strong correlation between TREM-1/TREM-2 ratio (in biopsy samples) and co-morbid conditions or fatty liver grading were also been analyzed among the SO+D− and SO+D+ groups. Fisher’s exact test or Pearson’s χ2 test were again used to analyze the association between sTREM-1/sTREM-2 ratio and biochemical profiles among SO+D− and SO+D+ groups.. Data are presented as mean ± SD and number (percentage) of patients. All the data were analyzed by SPSS v21 and Graph-Pad Prism. *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
Results
Demographics, co-morbid conditions and biochemical profile
The details of demographics and co-morbid conditions are shown in Table 1. The analysis of biochemical profiles is shown in Table S1.
Grading of fatty liver
Histopathological analysis of biopsy samples (liver, omental fat, and subcutaneous fat) are shown in Figure S2. Hepatosteatosis was present in all SO+D+ and in 85.7% of SO+D− (Table S5). It was noteworthy that 40% of SO+D+ had grade III (66-100%) hepatosteatosis. However, liver function test (AST and ALT), was normal in all the study populations.
Increased expression of TREM-1 on monocytes and neutrophils in SO+D+ group
In monocytes (Figure 1a), TREM-1 expression was significantly greater in SO+D+ group compared to SO+D− (78.13% ± 10.32 vs 57.60% ± 7.86, p<0.01) and SO−D− groups (78.13% ± 10.32 vs 44.52% ± 13.05, p<0.0001). However, TREM-2 expression were significantly lower in SO+D+ compared to SO+D− (27.26% ± 15.98 vs 49.39% ± 6.05, p<0.01) and SO−D− (27.26% ± 15.98 vs 58.47% ± 9.67, p<0.01) (Figure 1c).
In neutrophils (Figure 1b), TREM-1 expression was significantly greater in SO+D+ (89.61% ± 8.24 vs 66.65% ± 7.23, p<0.05) and SO+D− (86.90% ± 10.16 vs 66.65% ± 7.23, p<0.05) compared to SO−D− group. TREM-2 expression in neutrophils was also significantly lower in SO+D+ (23.26 ± 14.16% vs 63.21% ± 12.16, p<0.001) and SO+D− (29.03% ± 9.65 vs 63.21% ± 12.16, p<0.01) compared to SO−D− (Figure 1d). Further, the monocytes and neutrophils single and double positive for TREM-1 and TREM-2 are shown in Figure S3.
Increased expression of TREM-1 mRNA transcripts in the tissues of SO+D+ group
Overall, TREM-1 increased expression and TREM-2 down regulation were observed in subjects with obesity compared to subjects without obesity (Figure 2). TREM-1 mRNA expression folds were significantly higher in SO+D+ liver (4.96 ± 2.33 vs 2.47 ± 1.39, p<0.05; 4.96 ± 2.33 vs 1.36 ± 0.52, p<0.01) (Figure 2Aa), omentum (5.62 ± 1.94 vs 3.56 ± 1.46, p<0.05; 5.62 ± 1.94 vs 1.06 ± 0.19, p<0.0001) (Figure 2Ba), and subcutaneous (4.25 ± 1.86 vs 2.17 ± 1.14, p<0.05; 4.25 ± 1.86 vs 1.17 ± 0.10, p<0.01) (Figure 2Ca) tissue biopsies compared to SO+D− and SO−D− group, respectively. The mRNA expression of TREM-1 was significantly increased only in the omentum (not significant in liver and subcutaneous) of SO+D− compared to SO−D− group (3.56 ± 1.46 vs 1.06 ± 0.19, p<.05) (Figure 2Ba).
TREM-2 mRNA expression was significantly downregulated in liver (0.74 ± 0.24 vs 1.22 ± 0.29, p<0.01) (Figure 2Ab), omentum (0.63 ± 0.14 vs 1.06 ± 0.15, p<0.001) (Figure 2Bb), and subcutaneous (0.74 ± 0.22 vs 1.06 ± 0.13, p<0.05) (Figure 2Cb) tissues of SO+D+ compared to SO−D−. The significant downregulation of TREM-2 was also observed in liver (0.74 ± 0.24 vs 1.06 ± 0.32, p<0.05) between SO+D+ and SO+D− groups (Figure 2Ab).
The ratio of TREM-1/TREM-2 transcripts followed the trend of TREM-1 expression in the liver, omentum and subcutaneous tissues of SO+D+. The TREM-1/TREM-2 ratio was significantly higher in liver (6.88 ± 3.38 vs 1.09 ± 0.27, p<0.01) (Figure 2Ac), omentum (9.19 ± 3.60 vs 1.03 ± 0.33, p<0.0001) (Figure 2Bc) and subcutaneous (6.85 ± 4.44 vs 1.12 ± 0.22, p<0.05) (Figure 2Cc) of SO+D+ compared to SO−D−. There was also a significant increase in the ratio of TREM-1/TREM-2 transcripts in the liver (6.88 ± 3.38 vs 2.81 ± 2.07, p<0.05) (Figure 2Ac) and omentum (9.19 ± 3.60 vs 4.75 ± 2.21, p<0.01) (Figure 2Bc) tissues of SO+D+ compared to SO+D−. The expression of other classical inflammatory genes, such as TNF-α, IL-5, IL-6, IL-1β, CD64 and CRP, is shown in Figure 3.
Increased levels of sTREM-1 in SO+D+ group
sTREM-1 and sTREM-2 follow the trend shown in the tissue biopsy samples. sTREM-1 levels were significantly higher in SO+D+ compared to SO+D− (587.14 ± 290.91 vs 255.79 ± 151.71 pg/ml, p<0.05) and SO−D− groups (587.14 ± 290.91 vs 98.12 ± 12.60 pg/ml, p<0.01) (Figure 2Da). At the same time sTREM-2 levels were significantly decreased in the SO+D+ compared to SO+D− (77.83 ± 19.42 vs 114.33 ± 32.55 pg/ml, p<0.01) and SO−D− (77.83 ± 19.42 vs 132.85 ± 25.02 pg/ml, p<0.001) (Figure 2Db). The sTREM-1/sTREM-2 ratio also followed the trend of sTREM-1 and sTREM-2 expression. We found significantly increased serum sTREM-1/sTREM-2 ratio in SO+D+ compared to SO+D− (10.23 ± 4.36 vs 3.50 ± 2.38, p<0.001) and SO−D− (10.23 ± 4.36 vs 1.02 ± 0.18, p<0.001) (Figure 2Dc).
Increased expression of TREM-1 protein levels in the tissues of SO+D+ group
The TREM-1 protein levels was significantly higher in SO+D+ of liver (Figure 2Ea), omentum (Figure 2Fa), and subcutaneous (Figure 2Ga) tissue biopsies compared to SO+D− and SO−D− groups. At the same time, TREM-2 levels were significantly decreased in the SO+D+ compared to SO−D−. TREM-2 was significantly decreased in SO+D+ of liver (Figure 2Eb), and subcutaneous (Figure 2Gb), tissue biopsies compared to SO−D−. The significant downregulation of TREM-2 was also observed only in the omentum (Figure 2Fb) of SO+D− compared to SO−D− group. The ratio of TREM-1/TREM-2 transcripts followed the trend of TREM-1 protein expression in liver (Figure 2Ec), omentum (Figure 2Fc), and subcutaneous (Figure 2Gc) tissues of SO+D+ group. The TREM-1/TREM-2 ratio was significantly higher in the omentum of SO+D− compared to SO−D− group.
High immunoreactivity of TREM-1 in the tissue biopsies of SO+D+ group
The dual immunofluorescence study showed a significant increase in TREM-1 levels and a decrease in TREM-2 immunoreactivity in the liver (Figure 4), omentum and subcutaneous (Figure 5) tissue of SO+D+ compared to SO+D− and SO−D− groups.
The immunostaining of TREM-1, as quantified by MFI, was significantly higher in SO+D+ of liver (15.64 ± 5.69 vs 4.92 ± 2.92, p<0.0001; 15.64 ± 5.69 vs 2.18 ± 0.42, p<0.0001) (Figure 4D), omentum (5.70 ± 1.22 vs 3.32 ± 1.02, p<0.001; 5.70 ± 1.22 vs 1.73 ± 0.31, p<0.0001) (Figure 5Am), and subcutaneous (4.16 ± 0.97 vs 1.77 ± 0.76, p<0.0001; 4.16 ± 0.97 vs 0.92 ± 0.20, p<0.0001) (Figure 5Bm) tissues compared to SO+D− and SO−D−, respectively. The MFI of TREM-1 was also found to be increased in omentum tissue biopsies of SO+D− compared to SO−D− group (3.32 ± 1.02 vs 1.73 ± 0.31, p<0.05) (Figure 5Am).
The MFI of TREM-2 was significantly down regulated in liver (2.47 ± 0.39 vs 3.62 ± 0.53, p<0.001) (Figure 4D), omentum (0.98 ± 0.35 vs 2.15 ± 0.24, p<0.0001) (Figure 5An), and subcutaneous (0.63 ± 0.16 vs 1.05 ± 0.34, p<0.01) (Figure 5Bn) tissues of SO+D+ compared to SO−D−. The significant downregulation in MFI of TREM-2 was also observed in the omentum (0.98 ± 0.35 vs 1.69 ± 0.36, p<0.001) (Figure 5Am) and subcutaneous tissue (0.63 ± 0.16 vs 1.02 ± 0.21, p<0.01) (Figure 5Bn) between SO+D+ and SO+D− groups.
Strong correlation between increased TREM-1/TREM-2 ratio and SO+D+ group
Higher number of patients with increased TREM-1 and TREM-1/TREM-2 ratio was found in both SO+D+ and SO+D− group compared to SO−D− group. Among SO+D+ and SO+D− group, SO+D+ had a significantly higher number of patients with increased expression of liver and serum TREM-1 (100% vs 57.14%, r=0.582, p<0.05) compared to SO+D−. Similar trend was observed with TREM-1/TREM-2 ratios (Table 2). The association graphs for TREM-1, TREM-2 and TREM-1/TREM-2 ratio in liver, omentum and subcutaneous and serum samples between SO+D− and SO+D+ are also shown in Figure S4. The association analyses of TNF-α, IL-5, IL-6, IL-1β, CD64 and CRP in these samples between SO+D+ and SO+D− groups are shown in Table S6. On correlation analysis of liver TREM-1/TREM-2 ratio with co-morbid conditions, a significantly high correlation was found between TREM-1/TREM-2 ratio and obesity (r=0.582, p<0.05), hypertension (r=0.658, p<0.05) and SO+D+. This ratio also had a significantly positive association with hepatosteatosis (r=0.645, p<0.05) in SO+D+ group (Table 3; Figure S5).
Table 2.
Expression of TREM-1, TREM-2 and TREM-1/TREM-2 ratio in subjects with obesity compared to controls.
| TREM1 and TREM2 expression in subjects with obesity (22) |
SO+D− (7) | SO+D+ (15) | Correlation (R); P value |
||
|---|---|---|---|---|---|
| TREM1 (over- expression) |
Liver | 19 (86.4%) | 4 (57.14%) | 15 (100%)* | R=0.582; P=0.023 |
| Omentum | 21 (95.45%) | 6 (85.71%) | 15 (100%) | NS | |
| Subcutaneous | 16 (72.72%) | 4 (57.14%) | 12 (80%) | NS | |
| Serum | 19 (95.45%) | 4 (57.14%) | 15 (100%)* | R=0.582; P=0.023 | |
| TREM2 (under- expression) |
Liver | 17 (77.27%) | 4 (57.14%) | 13 (86.66%) | |
| Omentum | 21 (95.45%) | 6 (85.71%) | 15 (100%) | ||
| Subcutaneous | 15 (68.18%) | 3 (42.85%) | 12 (80%) | NS | |
| Serum | 20 (90.90%) | 5 (71.4%) | 15 (100%) | ||
| TREM1/ TREM2 ratio |
Liver | 19 (86.4%) | 4 (57.14%) | 15 (100%)* | R=0.582; P=0.023 |
| Omentum | 21 (95.45) | 6 (85.7%) | 15 (100%) | NS | |
| Subcutaneous | 16 (72.72%) | 4 (57.1%) | 12 (80%) | NS | |
| Serum | 19 (95.45%) | 4 (57.14%) | 15 (100%)* | R=0.582; P=0.023 | |
Higher number of subjects with overexpression of TREM-1, down regulation of TREM-2 and TREM-1/TREM-2 ratio were analyzed between SO+D− and SO+D+ using Fisher’s exact test or Pearson’s χ2 for categorical variables. Increased in folds of TREM-1 or TREM-2 expressions were regarded as over expression, likewise decrease in folds as under-expression. To describe the balance between pro and anti-inflammatory state, we analyzed TREM-1/TREM-2 ratio in all the study subjects. Data show number of subjects having higher values of these compared to controls (SO−D−). Values show number of subjects (% subjects of total), not significant (NS). Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
p<0.05.
Table 3.
Association of liver and serum TREM-1/TERM2 ratio with co-morbid conditions and biochemical profile of study subjects
| Co-morbid conditions and biochemical markers |
SO+D− (7) | SO+D+ (15) | Correlation (R); P value |
||
|---|---|---|---|---|---|
|
| |||||
| n | TREM-1/ TREM-2 ratio |
n | TREM-1/ TREM-2 ratio |
||
| Liver TREM-1/TREM-2 ratio association with co-morbid conditions | |||||
| Obesity | 7 | 4/7 (57/1%) | 15 | 15/15 (100%) * | R=0.582; P=0.023 |
| Hypertension | 4 | 2/4 (50%) | 13 | 13/13 (100%) * | R=0.658; P=0.044 |
| Sleep apnea | 2 | 2/2 (100%) | 9 | 9/9 (100%) | NS |
| Hyperlipidemia | 1 | 1/1 (100%) | 6 | 6/6 (100%) | NS |
| Former Smokers | 4 | 3/4 (75%) | 7 | 7/7 (100%) | NS |
| Liver TREM-1/TREM-2 ratio association with fatty liver grading | |||||
| Inflammation | 3 | 3/3 (100%) | 8 | 8/8 (100%) | NS |
| Hepato-steatosis | 6 | 3/6 (50%) | 15 | 15/15 (100%) * | R=0.645; P=0.015 |
| Fibrosis | 2 | 2/2 (100%) | 6 | 6/6 (100%) | NS |
| Serum TREM-1/TREM-2 ratio association with biochemical markers | |||||
| Cholesterol (mg/dl), >200 | 3 | 1/3 (33.3%) | 3 | 3/3 (100%) | NS |
| Triglycerides(mg/dl),>149 | 4 | 2/4 (50%) | 13 | 13/13 (100%) * | R=0.658; P=0.044 |
| FFA (μM/ml), >0.65, | 5 | 2/5 (100%) | 15 | 15/15 100%) ** | R=0.728; P=0.009 |
| VLDL (mg/dl), >30, | 4 | 2/4 (50%) | 10 | 10/10 (100%) | |
| HDL (mg/dl), <40 | 3 | 2/3 (66.7%) | 10 | 10/10 (100%) | |
| LDL (mg/dl), >99 | 4 | 2/4 (50%) | 4 | 4/4 (100%) | |
| Cholesterol:HDL >4.4 | 3 | 2/3 (66.7%) | 5 | 5/5 (100%) | |
| LDL:HDL >3.2 | 2 | 1/2 (50%) | 3 | 3/3 (100%) | |
| CRP (mg/l ) > 10 | 5 | 4/5 (80%) | 13 | 13/13 (100%) | NS |
| HbA1c (%), >6 | 4 | 4/4 (100%) | 15 | 15/15 (100%) | |
| Glucose (mg/dl) >100 | 4 | 4/4 (100%) | 15 | 15/15 (100%) | |
| Insulin (μIU/mL) >8.4 | 4 | 4/4 (100%) | 15 | 15/15 (100%) | |
| HOMA-IR, >2 | 4 | 4/4 (100%) | 15 | 15/15 (100%) | |
| HOMA-β%, >100 | 7 | 4/7 (57.1%) | 11 | 11/11 (100%) | |
| TNF-α (pg/ml) >Control | 4 | 4/4 (100%) | 12 | 12/12 (100%) | |
| IL-1β (pg/ml) >Control | 4 | 3/4 (75%) | 11 | 11/11 (100%) | |
| IL-6 (pg/ml) >Control | 4 | 3/4 (75%) | 10 | 10/10 (100%) | |
Subject’s with strong association between liver TREM-1/TREM-2 ratio and co-morbid conditions or fatty liver grading; strong association between sTREM-1/sTREM-2 ratio and biochemical profiles were analysed among SO+D− and SO+D+ groups using Fisher’s exact test or Pearson’s χ2 for categorical variables. Data shows number of subjects (% of total subjects) having high TREM-1/TREM-2 ratio in association with co-morbid conditions in the biopsy samples; high sTREM-1/ sTREM-2 level in association with biochemical profiles. Number (n), not significant (NS). Abbreviations: SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
p<0.05,
p<0.01.
High association between sTREM-1/sTREM-2 ratio and biochemical profile in SO+D+ group
The sTREM-1/sTREM-2 ratio showed a significantly positive correlation between high FFA (r=0.728, p<0.01) and triglycerides (r= 0.658, p<0.05) levels in SO+D+ compared to SO+D−. We also found positive association between glucose, insulin and HOMA-IR and high sTREM-1/sTREM-2 ratio in SO+D+ (15/15) and majority of SO+D− (4/7) group. It is noteworthy that sTREM-1/sTREM-2 ratio and sTREM-1 were not increased in SO+D− (3/7) with normal HOMA-IR index (Table 3). Association graph between sTREM-1/sTREM-2 ratio and biochemical profiles are shown in Figure S6.
Discussion
Our results demonstrate significantly increased expression of TREM-1 in tissue biopsies and circulating neutrophils and monocytes, and increased levels of sTREM-1 in SO+D+ group. These findings suggest that TREM-1 could play a critical role in the underlying pathophysiology of obesity-induced chronic inflammation. TREM-1 activates monocytes and neutrophils through DAP12 (DNAX activation protein of 12kDa), a trans-membrane adaptor protein (13). TREM-2 is also expressed on macrophages and monocyte-derived dendritic cells and regulates innate immune cells via inhibitory signalling (14). Several cell surface receptors and intracellular kinases have been implicated in the pathogenesis of insulin resistance in the patients with obesity (15).
In acute phase of inflammation, pro-inflammatory cytokines activate TREM-1, resulting in cytokine secretion and activation of inflammatory cell surface markers, including MHC class I and CD markers on neutrophils and monocytes (12). We found that SO+D+ group had significantly increased expression of TREM-1 and under-expression of TREM-2 on neutrophils. FFA in subjects with obesity not only activate monocytes/macrophage lineage but also activate acute phage inflammation (16, 17). The findings in the current study further support the hypothesis of obesity-induced inflammation involving acute inflammatory components like neutrophils. We also found that TREM-1 expression is increased- on monocytes along with increased sTREM-1 in SO+D+ compared to SO+D− and SO−D− groups. These findings suggest that increased TREM-1 expression can potentially play a key role in chronic inflammation-induced insulin resistance (18, 19).
Chronic low grade inflammation at the tissue level results in insulin resistance (20). This could be secondary to adipose tissue macrophages (ATMs) as illustrated by several studies (21). Infiltration of ATMs and increased secretion of TNF-α and IL-6 have been implicated as the primary reason in the pathogenesis of adipose tissue-induced insulin resistance (22). Our results from mRNA and protein studies in biopsy tissues of SO+D+ showed significantly increased TREM-1 compared to the SO−D−, supporting their potential role in increased inflammation (23). Overall, we found that TREM-1 expression is increased in the fat tissue of subjects with obesity (omentum 21/22; 95.45%, subcutaneous 16/22; 72.72%) compared to control patients. These results could be secondary to the higher number of ATMs in omentum early in the development of insulin resistance (24). There was significantly increased expression of liver TREM-1 in SO+D+(15/15; 100%) compared to SO+D− (4/7; 57.1%), but not significant in omentum and subcutaneous tissues. Therefore, we believe that kupffer cells in the liver are involved early in the pathogenesis of insulin resistance and metabolic syndrome (20). The mRNA expression levels of other inflammatory genes (TNF-α, IL-5, IL-6, IL-1β, CD64 and CRP) were also increased at tissue levels in both SO+D+ and SO+D− groups compared to SO−D− group. However, higher numbers of patients with increased expression of these genes were not significantly associated between SO+D+ and SO+D− groups.
Accumulation of fatty acids (25, 26) can lead to increased oxidative stress and inflammatory activity (27). Our results showed that the majority of subjects with obesity (both SO+D+ and SO+D−) (21/22, 95.45%) had hepatosteatosis, various signs of inflammation (11/22, 50%) and early sign of fibrosis and cirrhosis (8/22, 36.36%). Kupffer cells in the liver are critical in the pathogenesis of hepatosteatosis and possibly insulin resistance (21, 28). Fatty liver grading showed inflammation in 50% (11/22) of subjects with obesity (SO+D−; 3/7 and SO+D+; 8/15); however, TREM-1 was increased in the liver of 86.4% (19/22) of subjects with obesity (SO+D−; 4/7 and SO+D+; 15/15). This finding suggests that increased expression of TREM-1 not only precedes the histological changes which could be delayed for several years, but also occurs only in patients who develop insulin resistance.
Our study found strong association between obesity, decreased insulin sensitivity and hypertension. The strong correlation between hypertension with increased TREM-1/TREM-2 ratio indicates the potential role of TREM-1 in the pathogenesis of insulin resistance in SO+D+ with hypertension. Additionally, we found strong association of increased sTREM-1/sTREM-2 ratio with higher FFA and triglycerides levels in SO+D+ compared to SO+D−. Higher FFA and triglyceride levels have been associated with insulin resistance (26, 29) and increased expression of sTREM-1 in our patients suggest a possible role of TREM-1 in the induction of insulin resistance in SO+D+. In our study, HOMA-IR, a marker for insulin resistance, was found to be higher in all SO+D+ and 57% (4/7) SO+D− compared to SO−D−. Although the 4 SO+D− were not clinically diabetic, they did have features of pre-diabetes. These findings demonstrated that sTREM-1 is significantly increased in SO+D− and in SO+D+ with HOMA-IR index >2.
The results of this study further support our hypothesis that subjects with diabetes and pre-diabetes have increased sTREM-1 suggesting that the increased TREM-1 expression can precede in subjects with pre-diabetes earlier than changes in the serum levels of other inflammatory markers. Higher sTREM-1 in subjects with T2DM supports that obesity is an inflammatory condition and sTREM-1 levels increase during inflammation (30). The routine clinical test being used to measure insulin resistance are insulin tolerance test, gold standard hyperinsulinemic euglycemic clamp test, steady state plasma glucose test, oral glucose tolerance test, intravenous glucose tolerance test, fasting insulin and derived indices (HOMA-IR), retinol binding protein 4 (RBP4) level, YLK-40, α-hydroxybutyrate and adiponectin level (31–34). However, these tests remain limited to clinical research due to time and cost constraints. It is evident from our study that serum TREM-1 level could fulfil this gap.
Conclusion
The findings in our study support an association between obesity-induced inflammation and TREM-1, suggesting the critical role of TREM-1 in the pathogenesis of insulin resistance in subjects with obesity (Figure S7). Significantly increased TREM-1 expression in subjects with obesity compared to control subjects indicates that these cell surface receptors are activated during the initiation and progression of DM-II. Thus, increased expression of TREM-1 surface receptor could be used as one of the biomarker for insulin insensitivity in SO+D+ group. However, future studies are warranted to confirm the role of TREM-1 as a potential marker for insulin resistance.
Supplementary Material
Figure S1 Isotype control for TREM-1 and TREM-2 expression in monocytes and neutrophils of the study subjects. Cell phenotype was performed using anti-human antibodies and isotypes for monocytes (CD45+, CD14+, HLA-DR+) represented in figure A and neutrophils (CD45+, CD16+) in figure B. A) The representative figures for isotype control for monocytes gating; live monocytes were gated from FSC/SSC gating. Then CD45+ were gated using the basal level of CD45 Isotype and analyzed for CD14 and HLA-DR expression with respective CD14 and HLA-DR Isotypes. The double positive populations (CD14+ HLA-DR+) were further gated for expression of TREM-1+ and TREM-2+ using the basal level of TREM-1 and TREM-2 Isotypes. B) The representative figures for Isotype control for neutrophils gating; live granulocytes were gated from FSC/SSC gating. Then CD45+ were gated using the basal level of CD45 Isotype and analyzed for CD16 expression using the basal level of CD16 Isotype. CD16+ populations were further gated for expression of TREM-1 and TREM-2 using the basal level of TREM-1 and TREM-2 Isotypes.
Figure S2 Hematoxylin and Eosin staining for fatty liver grading and inflammation. H & E in biopsy samples of SO−D−, SO+D− and SO+D+ groups respectively in liver (images Aa, Ab & Ac), omental fat (images Ba, Bb & Bc) and subcutaneous fat (images Ca, Cb & Cc). Liver biopsy samples showed steatosis in SO+D− and fibrosis and cirrhosis in SO+D+ groups. Size of the adipocyte was larger in subjects with obesity (images Bb, Bc, Cb, & Cc) and inflammation was increased in SO+D+ (images Bc & Cc) compared to SO−D− group (N= 5 SO−D−; 7 SO+D−; 15 SO+D+). Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Figure S3 TREM-1 and TREM-2 expression in monocytes and neutrophils of the study subjects. The protein expression of TREM-1 and TREM-2 in monocytes and neutrophils were compared between SO−D−, SO+D− and SO+D+ groups using One-way ANOVA for continuous variables. A) The expression of TREM1+ TREM2−, TREM1− TREM2+ and TREM1+ TREM2+ levels in monocytes (%). Data are shown as mean ± SD (N= 5 SO−D−; 7 SO+D−; 15 SO+D+); A) The expression of TREM1+ TREM2−, TREM1− TREM2+ and TREM1+ TREM2+ levels in neutrophils (%). Data are shown as mean ± SD (N= 5 SO−D−; 7 SO+D−; 15 SO+D+); *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Figure S4 Correlation of TREM-1, TREM-2 and TREM-1/TREM-2 ratio in liver, omentum and subcutaneous and serum samples between SO+D− and SO+D+ groups. A) Correlation analysis in Liver biopsy sample (Aa) TREM-1; (Ab) TREM-2; (Ac) TREM-1/TREM-2 ratio. B) Correlation analysis in Omentum biopsy sample (Ba) TREM-1; (Bb) TREM-2; (Bc) TREM-1/TREM-2 ratio. C) Correlation analysis in Subcutaneous biopsy sample (Ca) TREM-1; (Cb) TREM-2; (Cc) TREM-1/TREM-2 ratio. D) Correlation analysis in serum (Da) TREM-1; (Db) TREM-2; (Dc) TREM-1/TREM-2 ratio. Abbreviations: SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Figure S5 Graphical representation of the analysis of the liver TREM-1/TREM-2 ratio in association with co-morbid conditions (hypertension, hyperlipidemia, sleep apnea, and smoking) and fatty liver grading (inflammation, steatosis, and fibrosis) in liver biopsy samples (Table 3). Subject’s with strong correlation between liver TREM-1/TREM-2 ratio and co-morbid conditions or fatty liver grading were analyzed among SO+D− and SO+D+ groups. Analysis showed strong association of the increased TREM-1/TREM-2 ratio in subjects with SO+D+. A) Association between liver TREM-1/TREM-2 ratio with comorbid conditions; B) Association between liver TREM-1/TREM-2 ratio with fatty liver grading. Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Figure S6 Correlation of sTREM-1/sTERM2 ratio and biochemical profile of study subjects. SPSS scatter blot was used to analyze the association between sTREM-1/sTREM-2 ratio and biochemical profiles among SO+D− and SO+D+ groups. Abbreviations: SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Figure S7 Model of TREM-1 induction in obesity and insulin resistance. 1- Obesity results in deposition of white adipose tissue in liver, omentum and subcutaneous space, and results in metabolic syndrome with increased parameters like hypertension, hyperlipidemia, hyperinsulinemia and hyperglycemia. Obesity leads to obstructive sleep apnea, weight gain and increase in BMI. 2- Chronic inflammation in white adipose tissue leads to increased TREM-1 via secretion of proinflammatory cytokines like IL-6 and TNF-alpha, and decreased TREM-2 expression. Chronic inflammation further increases the levels of TREM-1. Detachment of TREM-1 protein from the cell surface increases sTREM-1. sTREM-1 increases more with chronic inflammation in SO+D+ group. 3- Over expression of TREM-1 leads to insulin resistance in obesity. Insulin resistance decreases the glucose transport from blood to liver resulting in further increase in the FFA, TG, glucose and lipoprotein levels by decreasing lipolysis and gluconeogenesis.
Table S1 Biochemical profile of subjects with obesity
Table S2 The criteria used for fatty liver grading and inflammation
Table S3 The fluorescent conjugated antibody panel used for determination of TREM-1 and TREM-2 in monocytes and neutrophils
Table S4 Primers used in this study for real time PCR
Table S5 Grading of Fatty liver in subjects with obesity
Table S6 Expression of TNF-α, IL-6, IL-1β, CRP, IL-5, and CD64 in subjects with obesity compared to controls.
Study Importance.
What is already known about this subject?
Chronic inflammation in obesity mediates the insulin resistance.
TREM-1 is an inflammatory marker.
What does this study add?
For the first time this study demonstrates the increased expression of TREM-1 in the liver and adipose tissue and in circulating neutrophils and monocytes in the subjects with obesity (with or without diabetes).
TREM-1 expression associates with increased insulin resistance in subjects with obesity.
Acknowledgements
This study was supported by a research grant to Dr. Kalyana C Nandipati from the LB692 Nebraska Tobacco Settlement Funds to Creighton University, and by the research grants R01HL116042 and R01HL128063 from the National Institutes of Health, USA to DK Agrawal. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbrebiations
- TREM
Triggering Receptor Expressed on Myeloid Cells
- DM-II
Diabetes Mellitus type-II
- ATMs
Adipose Tissue Macrophages
- FFA
Free Fatty Acids
- LDL
Low Density Lipoprotein
- HDL
High Density lipoprotein
- HOMA-IR
Homeostatic model assessment-Insulin Resistance
- DAP-12
DNAX Activation Protein-12KDa
- MCP-1
Monocyte Chemoattractant protein-1
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Author Contributions
SS- designing experiment, conducting experiments, acquiring and analyzing data, writing the manuscript; PKP- providing tissue samples; VR- conducting experiments, editing the manuscript; PS- analyzing the histological slides; DKA- concept of the study, designing experiments, providing reagents, analysis and critical evaluation of the data, editing the manuscript; KCN- concept of the study, designing experiments, providing samples and reagents, writing and editing the manuscript.
Disclosure: The authours declare no conflict of interest.
References
- 1.McArdle MA, Finucane OM, Connaughton RM, McMorrow AM, Roche HM. Mechanisms of obesity-induced inflammation and insulin resistance: insights into the emerging role of nutritional strategies. Front Endocrinol (Lausanne) 2013;4:52. doi: 10.3389/fendo.2013.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouloumié A, Curat CA, Sengenès C, Lolmède K, Miranville A, Busse R. Role of macrophage tissue infiltration in metabolic diseases. Curr Opin Clin Nutr Metab Care. 2005;8:347–54. doi: 10.1097/01.mco.0000172571.41149.52. [DOI] [PubMed] [Google Scholar]
- 3.Ota T. Obesity-induced inflammation and insulin resistance. Front Endocrinol (Lausanne) 2014;5:204. doi: 10.3389/fendo.2014.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serrano-Marco L, Chacón MR, Maymó-Masip E, et al. TNF-α inhibits PPARβ/δ activity and SIRT1 expression through NF-κB in human adipocytes. Biochim Biophys Acta. 2012;1821:1177–85. doi: 10.1016/j.bbalip.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Schenk M, Bouchon A, Seibold F, Mueller C. TREM-1--expressing intestinal macrophages crucially amplify chronic inflammation in experimental colitis and inflammatory bowel diseases. J Clin Invest. 2007;117:3097–106. doi: 10.1172/JCI30602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchon A, Dietrich J, Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 2000;164:4991–5. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- 7.Radsak MP, Taube C, Haselmayer P, et al. Soluble triggering receptor expressed on myeloid cells 1 is released in patients with stable chronic obstructive pulmonary disease. Clin Dev Immunol. 2007;2007:52040. doi: 10.1155/2007/52040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagler H, Sharif O, Haslinger I, et al. TREM-1 activation alters the dynamics of pulmonary IRAK-M expression in vivo and improves host defense during pneumococcal pneumonia. J Immunol. 2009;183:2027–36. doi: 10.4049/jimmunol.0803862. [DOI] [PubMed] [Google Scholar]
- 9.Zanzinger K, Schellack C, Nausch N, Cerwenka A. Regulation of triggering receptor expressed on myeloid cells 1 expression on mouse inflammatory monocytes. Immunology. 2009;128:185–95. doi: 10.1111/j.1365-2567.2009.03091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turnbull IR, Gilfillan S, Cella M, et al. Cutting edge: TREM-2 attenuates macrophage activation. J Immunol. 2006;177:3520–4. doi: 10.4049/jimmunol.177.6.3520. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K, Prinz M, Stagi M, Chechneva O, Neumann H. TREM2-transduced myeloid precursors mediate nervous tissue debris clearance and facilitate recovery in an animal model of multiple sclerosis. PLoS Med. 2007;4:e124. doi: 10.1371/journal.pmed.0040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genua M, Rutella S, Correale C, Danese S. The triggering receptor expressed on myeloid cells (TREM) in inflammatory bowel disease pathogenesis. J Transl Med. 2014;12:293. doi: 10.1186/s12967-014-0293-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Bremen T, Drömann D, Luitjens K, et al. Triggering receptor expressed on myeloid cells-1 (Trem-1) on blood neutrophils is associated with cytokine inducibility in human E. coli sepsis. Diagn Pathol. 2013;8:24. doi: 10.1186/1746-1596-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelham CJ, Agrawal DK. Emerging roles for triggering receptor expressed on myeloid cells receptor family signaling in inflammatory diseases. Expert Rev Clin Immunol. 2014;10:243–56. doi: 10.1586/1744666X.2014.866519. [DOI] [PubMed] [Google Scholar]
- 15.Ruderman NB, Carling D, Prentki M, Cacicedo JM. AMPK, insulin resistance, and the metabolic syndrome. J Clin Invest. 2013;123:2764–72. doi: 10.1172/JCI67227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dasu MR, Jialal I. Free fatty acids in the presence of high glucose amplify monocyte inflammation via Toll-like receptors. Am J Physiol Endocrinol Metab. 2011;300:E145–54. doi: 10.1152/ajpendo.00490.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chawla A, Nguyen KD, Goh YPS. Macrophage-mediated inflammation in metabolic disease. Nat Rev Immunol. 2011;11:738–49. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sartipy P, Loskutoff DJ. Monocyte chemoattractant protein 1 in obesity and insulin resistance. Proc Natl Acad Sci U S A. 2003;100:7265–70. doi: 10.1073/pnas.1133870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luck H, Tsai S, Chung J, et al. Regulation of obesity-related insulin resistance with gut anti-inflammatory agents. Cell Metab. 2015;21:527–42. doi: 10.1016/j.cmet.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Tilg H, Moschen AR. Insulin resistance, inflammation, and non-alcoholic fatty liver disease. Trends Endocrinol Metab. 2008;19:371–9. doi: 10.1016/j.tem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Cai D, Yuan M, Frantz DF, et al. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat Med. 2005;11:183–90. doi: 10.1038/nm1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniele G, Guardado Mendoza R, Winnier D, et al. The inflammatory status score including IL-6, TNF-α, osteopontin, fractalkine, MCP-1 and adiponectin underlies whole-body insulin resistance and hyperglycemia in type 2 diabetes mellitus. Acta Diabetol. 2014;51:123–31. doi: 10.1007/s00592-013-0543-1. [DOI] [PubMed] [Google Scholar]
- 23.Kim EJ, Choi M-R, Park H, et al. Dietary fat increases solid tumor growth and metastasis of 4T1 murine mammary carcinoma cells and mortality in obesity-resistant BALB/c mice. Breast Cancer Res. 2011;13:R78. doi: 10.1186/bcr2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bigornia SJ, Farb MG, Mott MM, et al. Relation of depot-specific adipose inflammation to insulin resistance in human obesity. Nutr Diabetes. 2012;2:e30. doi: 10.1038/nutd.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karpe F, Dickmann JR, Frayn KN. Fatty acids, obesity, and insulin resistance: time for a reevaluation. Diabetes. 2011;60:2441–9. doi: 10.2337/db11-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boden G. Obesity, insulin resistance and free fatty acids. Curr Opin Endocrinol Diabetes Obes. 2011;18:139–43. doi: 10.1097/MED.0b013e3283444b09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuzawa-Nagata N, Takamura T, Ando H, et al. Increased oxidative stress precedes the onset of high-fat diet-induced insulin resistance and obesity. Metabolism. 2008;57:1071–7. doi: 10.1016/j.metabol.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Huang W, Metlakunta A, Dedousis N, et al. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes. 2010;59:347–57. doi: 10.2337/db09-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiang J-K, Lai N-S, Chang J-K, Koo M. Predicting insulin resistance using the triglyceride-to-high-density lipoprotein cholesterol ratio in Taiwanese adults. Cardiovasc Diabetol. 2011;10:93. doi: 10.1186/1475-2840-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gómez-Piña V, Soares-Schanoski A, Rodríguez-Rojas A, et al. Metalloproteinases shed TREM-1 ectodomain from lipopolysaccharide-stimulated human monocytes. J Immunol. 2007;179:4065–73. doi: 10.4049/jimmunol.179.6.4065. [DOI] [PubMed] [Google Scholar]
- 31.Gall WE, Beebe K, Lawton KA, et al. alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One. 2010;5:e10883. doi: 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham TE, Yang Q, Blüher M, et al. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med. 2006;354:2552–63. doi: 10.1056/NEJMoa054862. [DOI] [PubMed] [Google Scholar]
- 33.Rathcke CN, Johansen JS, Vestergaard H. YKL-40, a biomarker of inflammation, is elevated in patients with type 2 diabetes and is related to insulin resistance. Inflamm Res. 2006;55:53–9. doi: 10.1007/s00011-005-0010-8. [DOI] [PubMed] [Google Scholar]
- 34.Kadowaki T, Yamauchi T, Kubota N, Hara K, Ueki K, Tobe K. Adiponectin and adiponectin receptors in insulin resistance, diabetes, and the metabolic syndrome. J Clin Invest. 2006;116:1784–92. doi: 10.1172/JCI29126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Isotype control for TREM-1 and TREM-2 expression in monocytes and neutrophils of the study subjects. Cell phenotype was performed using anti-human antibodies and isotypes for monocytes (CD45+, CD14+, HLA-DR+) represented in figure A and neutrophils (CD45+, CD16+) in figure B. A) The representative figures for isotype control for monocytes gating; live monocytes were gated from FSC/SSC gating. Then CD45+ were gated using the basal level of CD45 Isotype and analyzed for CD14 and HLA-DR expression with respective CD14 and HLA-DR Isotypes. The double positive populations (CD14+ HLA-DR+) were further gated for expression of TREM-1+ and TREM-2+ using the basal level of TREM-1 and TREM-2 Isotypes. B) The representative figures for Isotype control for neutrophils gating; live granulocytes were gated from FSC/SSC gating. Then CD45+ were gated using the basal level of CD45 Isotype and analyzed for CD16 expression using the basal level of CD16 Isotype. CD16+ populations were further gated for expression of TREM-1 and TREM-2 using the basal level of TREM-1 and TREM-2 Isotypes.
Figure S2 Hematoxylin and Eosin staining for fatty liver grading and inflammation. H & E in biopsy samples of SO−D−, SO+D− and SO+D+ groups respectively in liver (images Aa, Ab & Ac), omental fat (images Ba, Bb & Bc) and subcutaneous fat (images Ca, Cb & Cc). Liver biopsy samples showed steatosis in SO+D− and fibrosis and cirrhosis in SO+D+ groups. Size of the adipocyte was larger in subjects with obesity (images Bb, Bc, Cb, & Cc) and inflammation was increased in SO+D+ (images Bc & Cc) compared to SO−D− group (N= 5 SO−D−; 7 SO+D−; 15 SO+D+). Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Figure S3 TREM-1 and TREM-2 expression in monocytes and neutrophils of the study subjects. The protein expression of TREM-1 and TREM-2 in monocytes and neutrophils were compared between SO−D−, SO+D− and SO+D+ groups using One-way ANOVA for continuous variables. A) The expression of TREM1+ TREM2−, TREM1− TREM2+ and TREM1+ TREM2+ levels in monocytes (%). Data are shown as mean ± SD (N= 5 SO−D−; 7 SO+D−; 15 SO+D+); A) The expression of TREM1+ TREM2−, TREM1− TREM2+ and TREM1+ TREM2+ levels in neutrophils (%). Data are shown as mean ± SD (N= 5 SO−D−; 7 SO+D−; 15 SO+D+); *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001. Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Figure S4 Correlation of TREM-1, TREM-2 and TREM-1/TREM-2 ratio in liver, omentum and subcutaneous and serum samples between SO+D− and SO+D+ groups. A) Correlation analysis in Liver biopsy sample (Aa) TREM-1; (Ab) TREM-2; (Ac) TREM-1/TREM-2 ratio. B) Correlation analysis in Omentum biopsy sample (Ba) TREM-1; (Bb) TREM-2; (Bc) TREM-1/TREM-2 ratio. C) Correlation analysis in Subcutaneous biopsy sample (Ca) TREM-1; (Cb) TREM-2; (Cc) TREM-1/TREM-2 ratio. D) Correlation analysis in serum (Da) TREM-1; (Db) TREM-2; (Dc) TREM-1/TREM-2 ratio. Abbreviations: SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Figure S5 Graphical representation of the analysis of the liver TREM-1/TREM-2 ratio in association with co-morbid conditions (hypertension, hyperlipidemia, sleep apnea, and smoking) and fatty liver grading (inflammation, steatosis, and fibrosis) in liver biopsy samples (Table 3). Subject’s with strong correlation between liver TREM-1/TREM-2 ratio and co-morbid conditions or fatty liver grading were analyzed among SO+D− and SO+D+ groups. Analysis showed strong association of the increased TREM-1/TREM-2 ratio in subjects with SO+D+. A) Association between liver TREM-1/TREM-2 ratio with comorbid conditions; B) Association between liver TREM-1/TREM-2 ratio with fatty liver grading. Abbreviations: SO−D− = subjects without obesity and diabetes; SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Figure S6 Correlation of sTREM-1/sTERM2 ratio and biochemical profile of study subjects. SPSS scatter blot was used to analyze the association between sTREM-1/sTREM-2 ratio and biochemical profiles among SO+D− and SO+D+ groups. Abbreviations: SO+D− = subjects with obesity but not diabetes; SO+D+ = subjects with obesity and diabetes.
Figure S7 Model of TREM-1 induction in obesity and insulin resistance. 1- Obesity results in deposition of white adipose tissue in liver, omentum and subcutaneous space, and results in metabolic syndrome with increased parameters like hypertension, hyperlipidemia, hyperinsulinemia and hyperglycemia. Obesity leads to obstructive sleep apnea, weight gain and increase in BMI. 2- Chronic inflammation in white adipose tissue leads to increased TREM-1 via secretion of proinflammatory cytokines like IL-6 and TNF-alpha, and decreased TREM-2 expression. Chronic inflammation further increases the levels of TREM-1. Detachment of TREM-1 protein from the cell surface increases sTREM-1. sTREM-1 increases more with chronic inflammation in SO+D+ group. 3- Over expression of TREM-1 leads to insulin resistance in obesity. Insulin resistance decreases the glucose transport from blood to liver resulting in further increase in the FFA, TG, glucose and lipoprotein levels by decreasing lipolysis and gluconeogenesis.
Table S1 Biochemical profile of subjects with obesity
Table S2 The criteria used for fatty liver grading and inflammation
Table S3 The fluorescent conjugated antibody panel used for determination of TREM-1 and TREM-2 in monocytes and neutrophils
Table S4 Primers used in this study for real time PCR
Table S5 Grading of Fatty liver in subjects with obesity
Table S6 Expression of TNF-α, IL-6, IL-1β, CRP, IL-5, and CD64 in subjects with obesity compared to controls.