Abstract
Epstein Barr virus (EBV)-encoded RNAs (EBER) in tumor tissue and cell-free plasma EBV-DNA (pEBVd) are detected in EBV-associated lymphomas. Studies have suggested that EBER+ peripheral T-cell lymphomas (PTCL) have worse prognosis, but the role of EBV in these neoplasms remains unclear. pEBVd is quantitative and more easily amenable to standardization than EBER, but frequency of pEBVd detection, clinical impact, and agreement with EBER status in PTCL are unknown. We retrospectively assessed frequency of detectable pre-treatment pEBVd, presence of EBER in tumor tissue, and outcomes in 61 of 135 EBV-assessable PTCL patients. Fifteen of 61 patients (24.5%, 95% CI: 14–37%) were pre-treatment pEBVd+, with no significant differences in baseline characteristics or treatment between pEBVd+ and pEBVd− patients. EBER-ISH was performed on 10 pEBVd+ and 35 pEBVd− tumors. All 10 pEBVd+ patients were EBER+, but 9 pEBVd− patients were also EBER+. With median follow up of 24 months (range 1–96), overall survival (OS) was shorter in pEBVd+ compared to pEBVd− patients (13 vs. 72 months; p=0.04). In this retrospective study, pre-treatment pEBVd was elevated in 25% of PTCL patients, was highly specific for EBER+ tumors, and was associated with shorter survival. pEBVd should be further explored as a prognostic variable and tumor biomarker in PTCL.
Keywords: peripheral T-cell lymphoma, PTCL, Epstein-Barr virus, EBV, cell-free DNA
INTRODUCTION
Peripheral T-cell lymphomas (PTCLs) are a heterogeneous family of lymphoid malignancies with four nodal subtypes representing more than 80% of the cases in the United States: peripheral T-cell lymphoma not otherwise specified (PTCL-NOS), angioimmunoblastic T-cell lymphoma (AITL), anaplastic lymphoma kinase (ALK) negative anaplastic large cell lymphoma (ALCL), and ALK positive ALCL. With the exception of ALK+ ALCL, PTCLs are characterized by poor prognosis (30% 5yr OS)1. Better therapies and improved tools for risk stratification and disease monitoring are urgently needed. Epstein-Barr virus (EBV) is a ubiquitous lymphotropic gammaherpesvirus associated with several lymphoma subtypes, including a subset of PTCLs.2 EBV-associated lymphomas are canonically defined by the presence of EBV-encoded RNAs (EBERs) in tumor tissue.3 While its contribution to disease development and progression in each lymphoma subtype is not fully understood, EBV has shown potential as a tumor biomarker, predictor of outcome, and therapeutic target.4–7 In some EBV-associated lymphomas, cell-free plasma EBV-DNA (pEBVd) can also be detected and quantified by quantitative real time (qRT) PCR, providing an easily attainable candidate biomarker to assess tumor’s linkage to EBV and disease burden. In some EBV-associated lymphomas the amount of pEBVd (colloquially known as viral load) has prognostic value.8, 9 Finally, therapies specifically targeting EBV in lymphoma are being introduced in the clinic with encouraging outcomes.4, 5, 10, 11
Multiple studies have looked for the presence of EBV (i.e. EBER) in PTCL tumor tissue with variable results.12–24 The incidence of EBV-associated PTCL differ substantially worldwide,25 and studies focused on North American and European populations are limited. Some studies have suggested a worse survival in EBER+ PTCL.2, 26, 27 However, the prognostic impact of pEBVd in PTCL has only been examined in patients with extranodal NK/T-cell lymphoma (ENKTL),28 and studies in North American patients or in other types of PTCL are lacking.
We conducted a retrospective study of pre-treatment pEBVd detection in PTCL patients. Given the histologic and molecular overlap between some PTCL subtypes29–31 we included all PTCL subtypes in our analysis. The objectives of this study were to: 1) estimate the frequency of pre-treatment pEBVd detection in our cohort of PTCL patients, 2) characterize agreement between pEBVd and EBER in tumor tissue, and 3) explore the prognostic or predictive implications of detectable pEBVd in PTCL. Secondarily, we measured plasma CMV-DNA (pCMVd) as an additional marker of impaired immune surveillance.
METHODS
Patient Selection & endpoints
Numerous instances of detectable pCMVd and/or pEBVd in patients with T-cell malignancies have prompted us to integrate the measurement of pEBVd and pCMVd via qRT-PCR in the Ohio State University Wexner Medical Center CLIA-approved molecular microbiology laboratory for patients with T-cell lymphoma (Supplemental Methods). To determine the clinical significance of pEBVd, we retrospectively reviewed data from the electronic medical record (EMR) and identified patients who had a pathologically confirmed diagnosis of PTCL according to WHO criteria29, were treated at the OSU James Cancer Hospital between 1/1/2004 and 12/31/2013, and had pre-treatment pEBVd measurement(s). Patients with ENKTL and cutaneous T-cell lymphoma (CTCL) were excluded. We reviewed patient characteristics, subtype, treatment history and outcomes. Stage was defined according to the Lugano modification of the Ann Arbor staging system.32 Patients treated prior to the revised Lugano classification were re-classified based on retrospective chart review. Date of diagnosis was defined as the date of the first biopsy showing a diagnosis of PTCL. We reviewed patient characteristics and outcomes including frequency and timing of detectable pEBVd. Response assessments were defined by the Lugano classification32. Dates of death were confirmed using our institutional EMR as well as social security death index. The OSU IRB approved this study (2013C0125). EBER in situ hybridization (ISH) was done at diagnosis in a prospective fashion. EBER positivity was defined as any amount of EBER positive cells on 200× field. In several cases, EBER-ISH was not done since it was felt unnecessary for diagnosis.
Statistical Analyses
EBER, pEBVd, and pCMVd status were defined as dichotomous variables of positive vs. negative (Supplemental Methods). Associations between pEBVd status (pEBVd+ vs. pEBVd−) and clinical characteristics were evaluated using Chi-square or Fisher’s exact tests for categorical variables and the Wilcoxon rank sum test for continuous variables. Agreement between EBER and pEBVd status was characterized using the kappa statistic33 and measures of sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). The primary clinical outcome of interest was overall survival (OS), calculated from the date of PTCL diagnosis to the date of death due to any cause, censoring patients who were alive at time of last follow-up. Kaplan-Meier analyses and log-rank tests were used to estimate and compare survival differences between pEBVd+ and pEBVd− groups. Given the limited number of deaths, multivariable analyses were restricted to two-covariate models and were seen as exploratory, where proportional hazards models were fit to evaluate the impact of pre-treatment pEBVd status on OS, controlling for one other prognostic factor as established by International Prognostic Index (IPI) and the Prognostic Index for PTCL (PIT), one at a time.29,34, 35 All tests were two-sided and statistical significance was declared for p<0.05. SAS statistical software Version 9.4 (SAS Institute Inc.) was used for these analyses.
RESULTS
Patient characteristics
We retrospectively identified 200 patients with PTCL seen at OSU between 2004 and 2013, not including patients with ENKTL and cutaneous T-cell lymphoma (CTCL). Of these 200 PTCL patients, 135 had at least one qRT-PCR pEBVd measurement. Out of the 135 PTCL patients with ≥1 pEBVd measurement, 61 had a pre-treatment pEBVd measurement. The clinical characteristics of the study cohort (pre-treatment pEBVd available; N=61) were not significantly different from those of the remaining patients (pre-treatment pEBVd not available; N=74) (Suppl. Table 1). While the selection bias cannot be eliminated, this comparison illustrates similarities between the two populations. The most common reason for a missing pre-treatment pEBVd measurement was that the patient’s first visit at OSU occurred after initiation of chemotherapy. Other reasons include lab error and sample not acquired, which occurred more frequently in patients diagnosed prior to 2010. To explore the prognostic impact of pEBVd detection in this population and to assess the agreement between pEBVd and EBER-ISH in tumor tissue, we focused our primary analyses on the 61 PTCL patients with a pre-treatment pEBVd measurement. (Figure 1)
Figure I. Schema of retrospectively identified PTCL patients with at least one plasma EBV-DNA (pEBVd) measurement.
*pEBVd+ defined as >2000 copies/mL by a clinical qRT-PCR
Table 1 shows the demographic and clinical features of the 61 PTCL patients and a comparison of the pEBVd+ and pEBVd− groups. Most patients had PTCL-NOS (N=28, 46%), while 15 had AITL (25%), 10 ALCL ALK− (17%), 4 ALCL ALK+ (6%), and 2 each had hepatosplenic T-cell lymphoma (HSTCL) and enteropathy associated T-cell lymphoma (EATL). Median age of diagnosis was 62 (range 19–85) with M:F ratio of 2:1. Most patients had stage III/IV disease (N=53, 87%), ECOG performance status 0–1 (N=34, 62%), elevated LDH (N=37, 63%), uninvolved bone marrow (N=39, 71%), and 0–1 sites of extranodal disease (N=37, 61%).
Table I.
Baseline characteristics and treatment for all patients with a pre-treatment plasma EBV-DNA measurement and by plasma EBV status
| All N=61 |
pEBVd+ N=15 |
pEBVd− N=46 |
P-value | |
|---|---|---|---|---|
| Median age, years (range) |
62 (19–85) | 62 (38–82) | 62 (19–85) | 0.90 |
| Males, no. (%) | 40 (66) | 12 (80) | 28 (61) | 0.18 |
| PTCL Subtype, no. (%) | ||||
| PTCL-NOS | 28 (46) | 7 (47) | 21 (46) | 0.06 |
| AITL | 15 (25) | 7 (47) | 8 (17) | |
| ALCL ALK+ | 4 (6) | 1 (6) | 3 (6) | |
| ALCL ALK− | 10 (17) | 0 (0) | 10 (22) | |
| Others | 4 (6) | 0 (0) | 4 (9) | |
| Stage, no. (%) | ||||
| I/ II | 8 (13) | 0 (0) | 8 (17) | 0.18 |
| III/ IV | 53 (87) | 15 (100) | 38 (83) | |
| ECOG PS >1, no. (%) | ||||
| Yes | 21(38) | 6 (46) | 15 (36) | 0.53 |
| No | 34(62) | 7 (54) | 27 (64) | |
| Missing | 6 | 2 | 4 | |
| Elevated LDH, no. (%) | ||||
| Yes | 37 (63) | 8 (57) | 29 (64) | 0.62 |
| No | 22 (37) | 6 (43) | 16 (36) | |
| Missing | 2 | 1 | 1 | |
| BM Involved, no. (%) | ||||
| Yes | 16 (29) | 5 (45) | 11 (25) | 0.27 |
| No | 39 (71) | 6 (55) | 33 (75) | |
| Missing | 6 | 4 | 2 | |
| >1 Extranodal Sites, no. (%) |
||||
| Yes | 24 (39) | 9 (60) | 15 (33) | 0.06 |
| No | 37 (61) | 6 (40) | 31 (67) | |
| IPI1, no. (%) | ||||
| 0–2 | 22(42) | 2 (17) | 20 (49) | 0.09 |
| 3–5 | 31(58) | 10 (83) | 21 (51) | |
| Missing | 8 | 3 | 5 | |
| Induction Therapy, no. (%) |
||||
| CHOP | 21 (36) | 3 (21) | 18 (41) | 0.25 |
| EPOCH | 21 (36) | 7 (50) | 14 (32) | |
| Others | 12(21) | 2 (14) | 10 (23) | |
| No Treatment | 4 (7) | 2 (14) | 2 (5) | |
| Missing | 3 | 1 | 2 | |
AITL=angioimmunoblastic T-cell lymphoma, ALCL=anaplastic large cell lymphoma, ALK=anaplastic lymphoma kinase; PS=performance status; LDH=lactate dehydrogenase; BM=bone marrow;
IPI=International Prognostic Index, 1 point for age>60yo, PS>1, LDH elevated, >1 extranodal site, stage III/IV; pEBVd=plasma EBV-DNA, PTCL-NOS=peripheral T-cell lymphoma not otherwise specified,
Among the 61 patients with a pre-treatment pEBVd measurement, 15 (24.5%, 95% CI: 14–37%) were pEBVd+. There were no significant differences in age, sex, race, stage, performance status, elevated LDH, or bone marrow involvement between the pEBVd+ and pEBVd− cohorts. There were moderate but not statistically significant associations between the pEBVd cohorts and stage of disease (p=0.18), international prognostic index (IPI; p=0.09), degree of extranodal involvement (p=0.06), and PTCL subtype (p=0.06). All pEBVd+ patients had stage III/IV disease compared with 83% of pEBVd− patients; likewise a higher proportion of pEBVd+ patients had more extranodal sites and higher IPI than pEBVd− patients. Notably, pEBVd+ patients were more likely to have AITL than pEBVd− patients (47% vs 17%) and less likely to have ALCL ALK− (0% vs 22%). There were no significant differences with regards to front line or salvage treatments between pEBVd cohorts (Table 1 & Suppl. Table 2). The most common front line regimens for all patients were EPOCH and CHOP (N=42; 72%).
Agreement between pEBVd and EBER
To determine the potential for pre-treatment pEBVd to serve as a surrogate biomarker for EBER detection in tumor tissue, we compared the frequency of EBER+ tumors using in-situ hybridization (Suppl. Methods). Overall, EBER-ISH testing was performed on 45 of the 61 PTCL patients with pre-treatment pEBVd testing. All 26 EBER− patients were pEBVd−. Of the 19 EBER+ patients, 10 were pEBVd+ and 9 were pEBVd−, resulting in an overall agreement rate of 80% (95% CI: 65–90%). The kappa statistic was 0.56 (95% CI: 0.33–0.79), corresponding to moderate but not substantial levels of agreement. The sensitivity of pEBVd as surrogate for EBER was 53% (95% CI: 29–76%) and specificity was 100% (95% CI: 87–100%). Importantly, positive predictive value (PPV) was 100% (95% CI: 69–100%) but negative predictive value (NPV) was only 74% (95% CI: 57–88%).
Impact of pre-treatment pEBVd and EBER on outcome
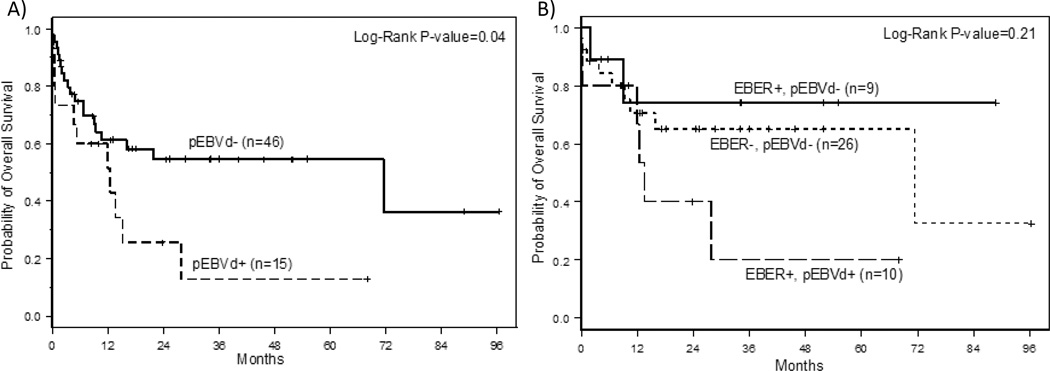
In addition to assessing agreement between pEBVd and EBER, one of the objectives of this study was to determine if pre-treatment pEBVd status (pEBVd+ vs. pEBVd−) was associated with survival. With a median follow up of 24 months, the pEBVd+ cohort had significantly shorter overall survival (OS) than the pEBVd− cohort (p=0.04) (Figure 2A). The estimated 2 year OS was 26% (95% CI: 6–51%) for pEBVd+ compared to 55% (95% CI: 37–69%) for pEBVd− patients. Unlike two previous studies26, 27, EBER+ status was not significantly associated with worse survival in our cohort (p=0.65). (Suppl. Figure 1) To further explore this association in the subset of patients for whom both EBER and pEBVd were available (N=45), EBER+/pEBVd+ patients had the lowest 2-year OS estimate (40%; 95% CI: 10–70%), whereas 2-year OS estimates for the EBER+/pEBVd− and EBER−/pEBVd− patients were 74% (95% CI: 29–93%) and 65% (95% CI: 42–81%), respectively (Figure 2B). Due to small patient numbers this data is only hypothesis generating but suggests that detection of pEBVd irrespective of EBER status identifies a subset of PTCL patients with worse prognosis.
Figure II. Overall survival by EBV status prior to treatment.
A) Overall survival by pEBVd status prior to therapy. pEBVd+ defined as >2000 copies/mL by a clinical qRT-PCR. B) Overall survival according to EBER and pEBVd status. There were no EBER− pEBVd+ patients
Due to the small number of deaths (n=30), adjustment for other potentially confounding variables via multivariable modeling was quite limited. However, separate bivariate models were fit, including pEBVd status and controlling for single known prognostic factors, one at a time (Table 2). The impact of pEBVd status on OS changed little when controlling for variables such as age, performance status, and elevated LDH. Since ALK+ ALCL patients carry a better prognosis than other types of PTCL we controlled for this factor and found that the worse survival in pEBVd+ patients was not altered by ALK status (HR 2.2; p=0.05). When controlling for AITL we found that pre-treatment pEBVd+ was associated with significantly worse overall survival (HR 2.9; p=0.008). Alternatively, when controlling for pEBVd+, a diagnosis of AITL was associated with significantly better prognosis (HR 0.38; p=0.04). However, when controlling for non-tumor specific factors such as stage, extranodal disease, bone marrow involvement, or IPI score, pEBVd status did not provide significant independent prognostic information in this cohort (p>0.05).
Table II.
Bivariate Cox proportional hazards models for overall survival
| Bivariate Models | Hazard Ratio | 95% CI | P-value |
|---|---|---|---|
| Pre-treatment pEBVd+ | 2.04 | 0.96–4.36 | 0.06 |
| Age ≥60 | 2.22 | 1.01–4.88 | 0.05 |
| Pre-treatment pEBVd+ | 1.71 | 0.80–3.69 | 0.17 |
| Stage IV | 3.20 | 1.28–7.99 | 0.01 |
| Pre-treatment pEBVd+ | 2.18 | 0.94–5.05 | 0.07 |
| ECOG PS >1 | 2.85 | 1.28–6.37 | 0.01 |
| Pre-treatment pEBVd+ | 2.11 | 0.97–4.61 | 0.06 |
| Elevated LDH | 2.18 | 0.92–5.16 | 0.08 |
| Pre-treatment pEBVd+ | 1.34 | 0.59–3.05 | 0.48 |
| >1 Extranodal Site | 3.11 | 1.37–7.05 | 0.007 |
| Pre-treatment pEBVd+ | 1.55 | 0.64–3.73 | 0.33 |
| BM Involvement | 2.44 | 1.11–5.36 | 0.03 |
| Pre-treatment pEBVd+ | 1.47 | 0.61–3.57 | 0.39 |
| IPI 3–5 | 2.68 | 0.98–7.37 | 0.06 |
| Pre-treatment pEBVd+ | 2.17 | 1.01–4.67 | 0.05 |
| ALCL ALK+ | 0.98 | 0.23–4.24 | 0.98 |
| Pre-treatment pEBVd+ | 2.91 | 1.33–6.40 | 0.008 |
| AITL | 0.38 | 0.15–0.97 | 0.04 |
pEBVd=plasma EBV-DNA; PS=performance status; LDH=lactate dehydrogenase; BM=bone marrow; IPI=International Prognostic Index, 1 point for age>60yo, PS>1, LDH elevated, >1 extranodal site, stage III/IV; ALCL=anaplastic large cell lymphoma, ALK=anaplastic lymphoma kinase;
Longitudinal measurement of pEBVd
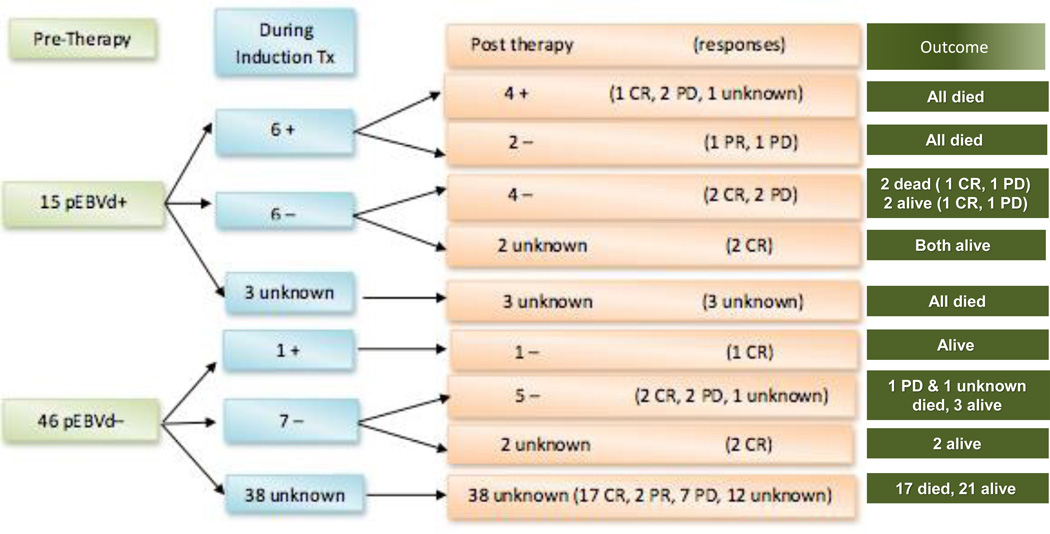
We hypothesized that, when detectable at diagnosis, pre-treatment pEBVd levels may serve as a tumor biomarker in PTCL, decreasing or normalizing with response to therapy and rising with disease relapse or progression. While the retrospective nature and limited sample size of this study only allow for descriptive observations, 12 of 15 pEBVd+ PTCL patients had more than one pEBVd measurement. Eight of 46 pEBVd− patients had pEBVd measurements after initiation of therapy. Figure 3 shows pEBVd measurements during and after therapy with corresponding outcomes. Six of the 12 pEBVd+ patients continued to have detectable pEBVd during therapy and 4 were positive at the end of therapy (including 1 patient in CR by Lugano criteria). All 6 patients with detectable pEBVd during therapy died of progressive disease. The other 6 pEBVd+ patients had undetectable pEBVd during therapy, and 4 remained pEBVd− after therapy (no data for the other 2). Only 2 of these 6 patients died of progressive disease. Of the 8 pEBVd− patients with longitudinal pEBVd measurements, 1 had detectable pEBVd during therapy but was negative at the end. Six of these 8 patients remain alive at last follow-up, 4 of which are alive at least 34 months since diagnosis.
Figure III. Serial pEBVd measurements before, during, and at the end of induction treatment with corresponding induction response and clinical outcomes.
pEBVd measurement reported as a dichotomous variable where “+” indicated pEBVd >2000 copies/mL. Response at end of front line therapy measured in accordance with Lugano classification. pEBVd was measured at the end of induction at the same time as imaging assessment. PR=partial response, CR=complete response, PD progressive disease
Frequency of pCMVd detection
Finally, we performed serial measurements of pCMVd in parallel with pEBVd via qRT-PCR as a surrogate for impaired immune surveillance. pCMVd was measured at least once in 55/61 (90%) patients. Forty-eight of 55 had pCMVd measured at diagnosis and 33 of 48 were measured at least once more after initiation of treatment. In total, 7 patients (13%; 7/55) had elevated pCMVd either at diagnosis or after initial treatment. Three were pCMVd+ at diagnosis, 2 of which continued to have elevated pCMVd after initiation of therapy. The other 4 individuals were only pCMVd+ after initiation of treatment. Positivity of pEBVd tended to be associated with positivity of pCMVd, where 29% of 14 pEBVd+ patients were also pCMVd+ compared with only 7% of 41 pEBVd− patients (p=0.06).
DISCUSSION
Although EBV is best known as a transforming virus for mature B-cells, a number of retrospective studies have shown that EBV infected cells, identified by the expression of non-coding EBV RNAs (EBER), are easily detected in a subset of lymphoid tumor biopsies from immune competent patients with mature T/NK-cell neoplasms, including PTCLs.3, 36–38 Some of the same studies have also suggested that EBER+ PTCL-NOS, defined as having at least one EBER+ cell per high power field (HPF), represent a relatively large subset (30–41%) of all cases and may have a worse outcome than EBER negative PTCL-NOS, although the age group affected differed among studies.3, 36–38 This would be in line with the negative prognostic impact conferred by EBV on diffuse large B-cell lymphoma (DLBCL) and classical Hodgkin lymphoma (cHL).9, 39
With the exception of ENKTL, where EBV uniformly infects NK cells, the lineage of the EBER+ cells in PTCL has generally been left unaddressed; mostly due to technical challenges and the poor resolution afforded by IHC-ISH double staining, especially when the number of EBER+ cells is small. However, EBER+ large, atypical T-cells have been described in PTCL, sometimes in large numbers2, 23, suggesting that EBV can infect neoplastic T-cells; therefore, raising questions about the mechanism of viral entry (T-cells do not typically express CD21, the canonical receptor for EBV) and the potential oncogenic role of the virus in these neoplasms. Regardless of whether it is found in neoplastic T-cells or in tumor-infiltrating B-cells in the nodal microenvironment, as in AITL40, EBV may affect tumor behavior41 and clinical outcomes in patients with PTCL through its pleiotropic effects - in cis and trans - on cell growth, apoptosis, DNA methylation, chromatin remodeling, and immune responses. Detection of EBER+ cells in PTCL tumor tissue, especially in large numbers, may therefore identify a subset of patients with a distinct biology, either because of virus-induced activation of oncogenic pathways in the neoplastic T-cells or because of ineffective anti-tumor responses in a “permissive” microenvironment (signaled by EBV reactivation in B-cells). Since in some cases, EBV can infect both T-cells42 and B-cells, these mechanisms may coexist. And, while clarifying the lineage of the EBV infected cells is essential to propose mechanistic hypotheses about direct oncogenic effects of the virus in PTCL, the impact of EBV as a prognostic variable or a marker of disease burden may not necessarily depend on which cells are infected. Thus, a blood- or plasma-based surrogate for EBER, such as pEBVd, could be utilized to pursue studies addressing the impact of EBV on key clinical endpoints in PTCL, even without knowledge of the cell type infected.
The selection of plasma vs. PBMC to measure EBV-DNA levels is supported by studies showing that the former correlates well with EBER-ISH, disease burden, or prognosis in other EBV-associated neoplasms.8, 9, 43 In a large prospective clinical trial in advanced cHL, pEBVd ≥60 copies/uL was found to predict failure free survival when measured pre-treatment and 6 months after treatment.9 Likewise, in ENKTL, detectable pEBVd after initiation of therapy was associated with relapsed and refractory disease and poor prognosis.43 The data presented in this study, and previous work from our group in advanced stage CTCL38, are the first to assess the value of pEBVd as a surrogate for EBER in PTCL and to suggest its potential as a prognostic biomarker comparable to previous studies2, 26, 27 that assessed EBER, rather than pEBVd. Overall we show that 25% of the assessed PTCL patients had elevated pre-treatment pEBVd levels, underlining the fact that EBV+ PTCL is not a rare occurrence. Furthermore, while preliminary, our data seem to confirm that EBV-association is not distributed equally across PTCL subtypes. One third (33%; N=14/43) of PTCL-NOS (N=7) and AITL (N=7) were pEBVd+ but only 1/14 CD30+ ALCL was pEBVd+. This difference suggests that detection of EBV is associated with different biological subsets of T-cell lymphoma and that either ALCL tumor cells are less prone to be infected by EBV or that the tumor microenvironment in CD30+ ALCL is less “permissive” to EBV reactivation. Although none of the associations with clinical characteristics reached statistical significance, patients who were pEBVd+ tended to have more advanced disease with a higher proportion of extranodal disease and IPI score.
One of the goals of this study was to assess the agreement between pEBVd and EBER. In this cohort of PTCL patients analyzed, elevated pEBVd at diagnosis was highly specific for EBER positivity in tumor tissue (100% specificity and PPV), but inadequately sensitive (53% sensitivity and 74% NPV). In other words, all patients who were pEBVd+ were also EBER+, but pEBVd failed to identify all EBER+ patients. The retrospective design and clinical qRT-PCR limit our ability to define a PCR cut-off that optimizes sensitivity and specificity. Questions regarding the optimal cut-off value can only be clarified using strategic prospective bio-specimen sampling and quantitative continuous measurements of both pEBVd and EBER. Future studies should compare, ideally prospectively, the agreement and prognostic implications of EBER and pEBVd in EBV+ PTCL. If EBER is the more robust outcome-predicting biomarker, our data suggest that pEBVd will not be an adequate surrogate. On the other hand, our preliminary data suggest that pEBVd rather than EBER may be the better prognostic marker. The high specificity and PPV, ease of attainment, reproducibility, and potential as an adaptable tumor biomarker for longitudinal surveillance (Figure 3), make pEBVd a strong alternative vis a vis EBER. Lastly, elevated pEBVd may simply identify the subset of EBV+ PTCL with larger numbers of EBER+ cells in tumor tissue. In advanced stage CTCL, we observed that higher pEBVd levels corresponded to samples with greater numbers of EBER+ cells (3–4+ vs 1–2+).38 Likewise, in ENKTL the level of pEBVd correlates with tumor burden.8
When we compared outcomes based on pEBVd status, pEBVd+ patients had worse 2-year OS, compared to pEBVd− patients (26 vs. 55%; p=0.04). We used bivariate modeling to control for previous independent prognostic factors.34, 35 While overall pEBVd+ was associated with worse overall survival, when controlling for non-tumor specific variables such as stage, extranodal disease, and IPI score, pEBVd did not provide independent prognostic information. When controlling for disease subtype (ALCL & AITL), pEBVd+ was independently prognostic. Within the AITL subtype patients who were pEBVd− tended to have better prognosis than pEBVd+. Similar to classifying and risk stratifying PTCL based on genetic alterations30, 44, pEBVd+ may indicate unique disease biology compared to EBV−, even within PTCL subtypes as suggested by Swerdlow et al45. Larger, prospective studies will be required to better determine the relative contribution of these findings. In addition, prognostic information might be gained by analyzing pEBVd as a continuous variable rather than a dichotomized pEBVd variable. Thus, we remain cautious to make one-sided conclusions in light of the study weaknesses. However, this is the first study analyzing the clinical significance of pEBVd across all PTCL subtypes. Unlike prognostic factors that entail a level of subjectivity, pEBVd is a quantifiable tumor specific variable that may provide additional clarity regarding PTCL heterogeneity and should be further investigated.
As mentioned, previous studies have assessed the prognostic impact of EBER status in T-cell lymphomas. The Danish study group (LYFO) reported worse survival in EBER+ T-cell lymphomas of various histologies in a registry-based population (7-year OS of 33% vs. 14%; p = .01).2 More recent studies assessed the prognostic impact of EBER within various PTCL subtypes, defined according to the WHO classification. A GELA study assessed the significance of EBER status on a cohort of prospectively treated PTCL-NOS patients (N=110) and showed that EBER+ patients over the age of 60 had worse survival (21 vs. 31% at 5 years)27; whereas, the International Peripheral T-cell Lymphoma (IPTCL) Project (N=340), a registry-based study, found that EBER positivity was associated with adverse survival in PTCL-NOS patients <60 years old (p=0.007)26. The IPTCL Project reported no significant differences in outcome between EBER+ and EBER− patients within the AITL subtype.46 The GELA and IPTCL Project evaluated the number of EBER+ cells semiquantitatively by counting the number of cells and averaging number per high power field. The IPTCL identified >10 EBER+ cells per 200× field as prognostic, while the GELA study found any amount of positivity prognostic. In our study, we assessed EBER as a dichotomous variable and found no significant difference in survival between EBER+ vs. EBER− patients (p=0.65). Differences in defining EBER+ tumors likely contribute to the inconsistent results across studies and demonstrate a need to further study EBV-association across all subtypes using more quantitative methods.
In our study, of the patients with pEBVd measurements during induction, half in the pEBVd+ cohort continued to have elevated pEBVd, all of which subsequently died. The other half of the pEBVd+ patients had undetectable levels during induction and 66% remain alive. Only one pEBVd− patient had elevated pEBVd after initiation of therapy. While conclusions are limited by small numbers and incomplete data sets, the results raise the question as to whether elevated pEBVd after initiation of therapy predicts worse outcomes.
A number of weaknesses, such as retrospective data collection, small sample size, and inability to further characterize EBV+ cells limit the impact of these observations to hypothesis generation. However, our data show that pEBVd is detectable in a large subset of PTCL, and that detection of pEBVd always implies the presence of an EBER+ tumor, thus alerting the practicing physician to the need to order EBER-ISH on the patient’s biopsy. Finally, pEBVd+ may have a role as a prognostic biomarker in EBV+ PTCL and should be prospectively studied as an outcome variable.
Supplementary Material
Novelty and Impact.
Discovering easily attainable prognostic markers that simultaneously identify PTCL patients who may benefit from specific therapies is a priority. The frequency of plasma EBV-DNA (pEBVd) detection, its clinical significance, and agreement with EBER status in PTCL are unknown. We demonstrated that pEBVd was detected in 25% of PTCL patients, was highly specific for EBER+ tumors, and associated with worse survival. This supports further investigation of pEBVd as a prognostic and predictive biomarker in PTCL.
Acknowledgments
The study was supported by a Pelotonia Idea Grant and philanthropic funding (PP, RAB). Dr. Haverkos was supported by an NIH/NCI T32 (T32CA165998) at the time the study was conducted.
Footnotes
None of the authors have a relevant conflict of interest to report.
REFERENCES
- 1.Vose J, Armitage J, Weisenburger D. International TCLP: International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;26:4124–4130. doi: 10.1200/JCO.2008.16.4558. [DOI] [PubMed] [Google Scholar]
- 2.d'Amore F, Johansen P, Houmand A, Weisenburger DD, Mortensen LS. Epstein-Barr virus genome in non-Hodgkin's lymphomas occurring in immunocompetent patients: highest prevalence in nonlymphoblastic T-cell lymphoma and correlation with a poor prognosis. Danish Lymphoma Study Group, LYFO. Blood. 1996;87:1045–1055. [PubMed] [Google Scholar]
- 3.Rezk SA, Weiss LM. Epstein-Barr virus-associated lymphoproliferative disorders. Human pathology. 2007;38:1293–1304. doi: 10.1016/j.humpath.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 4.Bollard CM, Gottschalk S, Torrano V, Diouf O, Ku S, Hazrat Y, Carrum G, Ramos C, Fayad L, Shpall EJ, Pro B, Liu H, Wu MF, Lee D, Sheehan AM, Zu Y, Gee AP, Brenner MK, Heslop HE, Rooney CM. Sustained Complete Responses in Patients With Lymphoma Receiving Autologous Cytotoxic T Lymphocytes Targeting Epstein-Barr Virus Latent Membrane Proteins. J Clin Oncol. 2014;32:798–808. doi: 10.1200/JCO.2013.51.5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roychowdhury S, Peng R, Baiocchi RA, Bhatt D, Vourganti S, Grecula J, Gupta N, Eisenbeis CF, Nuovo GJ, Yang W, Schmalbrock P, Ferketich A, Moeschberger M, Porcu P, Barth RF, Caligiuri MA. Experimental treatment of Epstein-Barr virus-associated primary central nervous system lymphoma. Cancer research. 2003;63:965–971. [PubMed] [Google Scholar]
- 6.Suzuki R, Yamaguchi M, Izutsu K, Yamamoto G, Takada K, Harabuchi Y, Isobe Y, Gomyo H, Koike T, Okamoto M, Hyo R, Suzumiya J, Nakamura S, Kawa K, Oshimi K Group NK-cTS. Prospective measurement of Epstein-Barr virus-DNA in plasma and peripheral blood mononuclear cells of extranodal NK/T-cell lymphoma, nasal type. Blood. 2011;118:6018–6022. doi: 10.1182/blood-2011-05-354142. [DOI] [PubMed] [Google Scholar]
- 7.Au WY, Pang A, Choy C, Chim CS, Kwong YL. Quantification of circulating Epstein-Barr virus (EBV) DNA in the diagnosis and monitoring of natural killer cell and EBV-positive lymphomas in immunocompetent patients. Blood. 2004;104:243–249. doi: 10.1182/blood-2003-12-4197. [DOI] [PubMed] [Google Scholar]
- 8.Wang ZY, Liu QF, Wang H, Jin J, Wang WH, Wang SL, Song YW, Liu YP, Fang H, Ren H, Wu RY, Chen B, Zhang XM, Lu NN, Zhou LQ, Li YX. Clinical implications of plasma Epstein-Barr virus DNA in early-stage extranodal nasal-type NK/T-cell lymphoma patients receiving primary radiotherapy. Blood. 2012;120:2003–2010. doi: 10.1182/blood-2012-06-435024. [DOI] [PubMed] [Google Scholar]
- 9.Kanakry JA, Li H, Gellert LL, Lemas MV, Hsieh WS, Hong F, Tan KL, Gascoyne RD, Gordon LI, Fisher RI, Bartlett NL, Stiff P, Cheson BD, Advani R, Miller TP, Kahl BS, Horning SJ, Ambinder RF. Plasma Epstein-Barr virus DNA predicts outcome in advanced Hodgkin lymphoma: correlative analysis from a large North American cooperative group trial. Blood. 2013;121:3547–3553. doi: 10.1182/blood-2012-09-454694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porcu P, Eisenbeis CF, Pelletier RP, Davies EA, Baiocchi RA, Roychowdhury S, Vourganti S, Nuovo GJ, Marsh WL, Ferketich AK, Henry ML, Ferguson RM, Caligiuri MA. Successful treatment of posttransplantation lymphoproliferative disorder (PTLD) following renal allografting is associated with sustained CD8(+) T-cell restoration. Blood. 2002;100:2341–2348. doi: 10.1182/blood-2002-01-0210. [DOI] [PubMed] [Google Scholar]
- 11.Alinari L, Mahasenan KV, Yan F, Karkhanis V, Chung JH, Smith EM, Quinion C, Smith PL, Kim L, Patton JT, Lapalombella R, Yu B, Wu Y, Roy S, De Leo A, Pileri S, Agostinelli C, Ayers L, Bradner JE, Chen-Kiang S, Elemento O, Motiwala T, Majumder S, Byrd JC, Jacob S, Sif S, Li C, Baiocchi RA. Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood. 2015;125:2530–2543. doi: 10.1182/blood-2014-12-619783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anagnostopoulos I, Hummel M, Stein H. Frequent presence of latent Epstein-Barr virus infection in peripheral T cell lymphomas. A review. Leukemia & lymphoma. 1995;19:1–12. doi: 10.3109/10428199509059657. [DOI] [PubMed] [Google Scholar]
- 13.Jones JF, Shurin S, Abramowsky C, Tubbs RR, Sciotto CG, Wahl R, Sands J, Gottman D, Katz BZ, Sklar J. T-cell lymphomas containing Epstein-Barr viral DNA in patients with chronic Epstein-Barr virus infections. The New England journal of medicine. 1988;318:733–741. doi: 10.1056/NEJM198803243181203. [DOI] [PubMed] [Google Scholar]
- 14.Ho JW, Ho FC, Chan AC, Liang RH, Srivastava G. Frequent detection of Epstein-Barr virus-infected B cells in peripheral T-cell lymphomas. The Journal of pathology. 1998;185:79–85. doi: 10.1002/(SICI)1096-9896(199805)185:1<79::AID-PATH52>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Chen YP, Jones D, Chen TY, Chang KC. Epstein-Barr virus present in T cells or B cells shows differential effects on hemophagocytic symptoms associated with outcome in T-cell lymphomas. Leukemia & lymphoma. 2014;55:2038–2047. doi: 10.3109/10428194.2013.861068. [DOI] [PubMed] [Google Scholar]
- 16.Zhou XG, Hamilton-Dutoit SJ, Yan QH, Pallesen G. High frequency of Epstein-Barr virus in Chinese peripheral T-cell lymphoma. Histopathology. 1994;24:115–122. doi: 10.1111/j.1365-2559.1994.tb01289.x. [DOI] [PubMed] [Google Scholar]
- 17.Ohshima K, Kikuchi M, Eguchi F, Masuda Y, Sumiyoshi Y, Mohtai H, Takeshita M, Kimura N. Analysis of Epstein-Barr viral genomes in lymphoid malignancy using Southern blotting, polymerase chain reaction and in situ hybridization. Virchows Archiv B, Cell pathology including molecular pathology. 1990;59:383–390. doi: 10.1007/BF02899428. [DOI] [PubMed] [Google Scholar]
- 18.Teramoto N, Sarker AB, Tonoyama Y, Yoshino T, Hayashi K, Takahashi K, Akagi T. Epstein-Barr virus infection in the neoplastic and nonneoplastic cells of lymphoid malignancies. Cancer. 1996;77:2339–2347. doi: 10.1002/(SICI)1097-0142(19960601)77:11<2339::AID-CNCR24>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 19.Huh J, Cho K, Heo DS, Kim JE, Kim CW. Detection of Epstein-Barr virus in Korean peripheral T-cell lymphoma. American journal of hematology. 1999;60:205–214. doi: 10.1002/(sici)1096-8652(199903)60:3<205::aid-ajh7>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 20.Hirose Y, Masaki Y, Sawaki T, Shimoyama K, Karasawa H, Kawabata H, Fukushima T, Ogawa N, Wano Y, Umehara H. Association of Epstein-Barr virus with human immunodeficiency virus-negative peripheral T-cell lymphomas in Japan. European journal of haematology. 2006;76:109–118. doi: 10.1111/j.0902-4441.2005.00575.x. [DOI] [PubMed] [Google Scholar]
- 21.De Bruin PC, Jiwa NM, Van der Valk P, Van Heerde P, Gordijn R, Ossenkoppele GJ, Walboomers JM, Meijer CJ. Detection of Epstein-Barr virus nucleic acid sequences and protein in nodal T-cell lymphomas: relation between latent membrane protein-1 positivity and clinical course. Histopathology. 1993;23:509–518. doi: 10.1111/j.1365-2559.1993.tb01236.x. [DOI] [PubMed] [Google Scholar]
- 22.Hamilton-Dutoit SJ, Pallesen G. A survey of Epstein-Barr virus gene expression in sporadic non-Hodgkin's lymphomas. Detection of Epstein-Barr virus in a subset of peripheral T-cell lymphomas. The American journal of pathology. 1992;140:1315–1325. [PMC free article] [PubMed] [Google Scholar]
- 23.Korbjuhn P, Anagnostopoulos I, Hummel M, Tiemann M, Dallenbach F, Parwaresch MR, Stein H. Frequent latent Epstein-Barr virus infection of neoplastic T cells and bystander B cells in human immunodeficiency virus-negative European peripheral pleomorphic T-cell lymphomas. Blood. 1993;82:217–223. [PubMed] [Google Scholar]
- 24.Ott G, Ott MM, Feller AC, Seidl S, Muller-Hermelink HK. Prevalence of Epstein-Barr virus DNA in different T-cell lymphoma entities in a European population. International journal of cancer Journal international du cancer. 1992;51:562–567. doi: 10.1002/ijc.2910510410. [DOI] [PubMed] [Google Scholar]
- 25.Yachie A, Kanegane H, Kasahara Y. Epstein-Barr virus-associated T-/natural killer cell lymphoproliferative diseases. Seminars in hematology. 2003;40:124–132. doi: 10.1053/shem.2003.50012. [DOI] [PubMed] [Google Scholar]
- 26.Weisenburger DD, Savage KJ, Harris NL, Gascoyne RD, Jaffe ES, MacLennan KA, Rudiger T, Pileri S, Nakamura S, Nathwani B, Campo E, Berger F, Coiffier B, Kim WS, Holte H, Federico M, Au WY, Tobinai K, Armitage JO, Vose JM International Peripheral TcLP. Peripheral T-cell lymphoma, not otherwise specified: a report of 340 cases from the International Peripheral T-cell Lymphoma Project. Blood. 2011;117:3402–3408. doi: 10.1182/blood-2010-09-310342. [DOI] [PubMed] [Google Scholar]
- 27.Dupuis J, Emile JF, Mounier N, Gisselbrecht C, Martin-Garcia N, Petrella T, Bouabdallah R, Berger F, Delmer A, Coiffier B, Reyes F, Gaulard P Groupe d'Etude des Lymphomes de l’Adulte (GELA) Prognostic significance of Epstein-Barr virus in nodal peripheral T-cell lymphoma, unspecified: A Groupe d'Etude des Lymphomes de l'Adulte (GELA) study. Blood. 2006;108:4163–4169. doi: 10.1182/blood-2006-04-017632. [DOI] [PubMed] [Google Scholar]
- 28.Suwiwat S, Pradutkanchana J, Ishida T, Mitarnun W. Quantitative analysis of cell-free Epstein-Barr virus DNA in the plasma of patients with peripheral T-cell and NK-cell lymphomas and peripheral T-cell proliferative diseases. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2007;40:277–283. doi: 10.1016/j.jcv.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Hematopoietic and Lymphoid Tissues. 4th. Lyon France: IARC; 2008. [Google Scholar]
- 30.Iqbal J, Wright G, Wang C, Rosenwald A, Gascoyne RD, Weisenburger DD, Greiner TC, Smith L, Guo S, Wilcox RA, Teh BT, Lim ST, Tan SY, Rimsza LM, Jaffe ES, Campo E, Martinez A, Delabie J, Braziel RM, Cook JR, Tubbs RR, Ott G, Geissinger E, Gaulard P, Piccaluga PP, Pileri SA, Au WY, Nakamura S, Seto M, Berger F, de Leval L, Connors JM, Armitage J, Vose J, Chan WC, Staudt LM. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood. 2014;123:2915–2923. doi: 10.1182/blood-2013-11-536359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Leval L, Rickman DS, Thielen C, Reynies A, Huang YL, Delsol G, Lamant L, Leroy K, Briere J, Molina T, Berger F, Gisselbrecht C, Xerri L, Gaulard P. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007;109:4952–4963. doi: 10.1182/blood-2006-10-055145. [DOI] [PubMed] [Google Scholar]
- 32.Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E, Lister TA. Recommendations for Initial Evaluation, Staging, and Response Assessment of Hodgkin and Non-Hodgkin Lymphoma: The Lugano Classification. J Clin Oncol. 2014 doi: 10.1200/JCO.2013.54.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 34.Gallamini A, Stelitano C, Calvi R, Bellei M, Mattei D, Vitolo U, Morabito F, Martelli M, Brusamolino E, Iannitto E, Zaja F, Cortelazzo S, Rigacci L, Devizzi L, Todeschini G, Santini G, Brugiatelli M, Federico M. Peripheral T-cell lymphoma unspecified (PTCL-U): a new prognostic model from a retrospective multicentric clinical study. Blood. 2004;103:2474–2479. doi: 10.1182/blood-2003-09-3080. [DOI] [PubMed] [Google Scholar]
- 35.A predictive model for aggressive non-Hodgkin's lymphoma. The International Non-Hodgkin's Lymphoma Prognostic Factors Project. The New England journal of medicine. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 36.Gru AA, Haverkos BH, Freud AG, Hastings J, Nowacki NB, Barrionuevo C, Vigil CE, Rochford R, Natkunam Y, Baiocchi RA, Porcu P. The Epstein-Barr Virus (EBV) in T Cell and NK Cell Lymphomas: Time for a Reassessment. Current hematologic malignancy reports. 2015 doi: 10.1007/s11899-015-0292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura H, Ito Y, Kawabe S, Gotoh K, Takahashi Y, Kojima S, Naoe T, Esaki S, Kikuta A, Sawada A, Kawa K, Ohshima K, Nakamura S. EBV-associated T/NK-cell lymphoproliferative diseases in nonimmunocompromised hosts: prospective analysis of 108 cases. Blood. 2012;119:673–686. doi: 10.1182/blood-2011-10-381921. [DOI] [PubMed] [Google Scholar]
- 38.Haverkos BM, et al. Increased Levels of Plasma Epstein Barr Virus DNA Identify a Poor-Risk Subset of Patients With Advanced Stage Cutaneous T-Cell Lymphoma. Clinical lymphoma, myeloma, & leukemia. 2016;16(Suppl):S181.e184–S190.e184. doi: 10.1016/j.clml.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tisi MC, Cupelli E, Santangelo R, Maiolo E, Alma E, Giachelia M, Martini M, Bellesi S, D'Alo F, Voso MT, Pompili M, Leone G, Larocca LM, Hohaus S. Whole blood EBV-DNA predicts outcome in diffuse large B-cell lymphoma. Leukemia & lymphoma. 2016;57:628–634. doi: 10.3109/10428194.2015.1072766. [DOI] [PubMed] [Google Scholar]
- 40.Weiss LM, Jaffe ES, Liu XF, Chen YY, Shibata D, Medeiros LJ. Detection and localization of Epstein-Barr viral genomes in angioimmunoblastic lymphadenopathy and angioimmunoblastic lymphadenopathy-like lymphoma. Blood. 1992;79:1789–1795. [PubMed] [Google Scholar]
- 41.Ferrajoli A, Ivan C, Ciccone M, Shimizu M, Kita Y, Ohtsuka M, D'Abundo L, Qiang J, Lerner S, Nouraee N, Rabe KG, Rassenti LZ, Van Roosbroeck K, Manning JT, Yuan Y, Zhang X, Shanafelt TD, Wierda WG, Sabbioni S, Tarrand JJ, Estrov Z, Radovich M, Liang H, Negrini M, Kipps TJ, Kay NE, Keating M, Calin GA. Epstein-Barr Virus MicroRNAs are Expressed in Patients with Chronic Lymphocytic Leukemia and Correlate with Overall Survival. EBioMedicine. 2015;2:572–582. doi: 10.1016/j.ebiom.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coleman CB, Wohlford EM, Smith NA, King CA, Ritchie JA, Baresel PC, Kimura H, Rochford R. Epstein-Barr virus type 2 latently infects T cells, inducing an atypical activation characterized by expression of lymphotactic cytokines. Journal of virology. 2015;89:2301–2312. doi: 10.1128/JVI.03001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Wang H, Wang JH, Xia ZJ, Lu Y, Huang HQ, Jiang WQ, Zhang YJ. Post-treatment plasma EBV-DNA positivity predicts early relapse and poor prognosis for patients with extranodal NK/T cell lymphoma in the era of asparaginase. Oncotarget. 2015;6:30317–30326. doi: 10.18632/oncotarget.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parrilla Castellar ER, Jaffe ES, Said JW, Swerdlow SH, Ketterling RP, Knudson RA, Sidhu JS, Hsi ED, Karikehalli S, Jiang L, Vasmatzis G, Gibson SE, Ondrejka S, Nicolae A, Grogg KL, Allmer C, Ristow KM, Wilson WH, Macon WR, Law ME, Cerhan JR, Habermann TM, Ansell SM, Dogan A, Maurer MJ, Feldman AL. ALK-negative anaplastic large cell lymphoma is a genetically heterogeneous disease with widely disparate clinical outcomes. Blood. 2014;124:1473–1480. doi: 10.1182/blood-2014-04-571091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swerdlow SH, Jaffe ES, Brousset P, Chan JK, de Leval L, Gaulard P, Harris NL, Pileri S, Weiss LM. Cytotoxic T-cell and NK-cell lymphomas: current questions and controversies. The American journal of surgical pathology. 2014;38:e60–e71. doi: 10.1097/PAS.0000000000000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Federico M, Rudiger T, Bellei M, Nathwani BN, Luminari S, Coiffier B, Harris NL, Jaffe ES, Pileri SA, Savage KJ, Weisenburger DD, Armitage JO, Mounier N, Vose JM. Clinicopathologic characteristics of angioimmunoblastic T-cell lymphoma: analysis of the international peripheral T-cell lymphoma project. J Clin Oncol. 2013;31:240–246. doi: 10.1200/JCO.2011.37.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.