Summary
The fungal pathogen Candida albicans can transition from budding to hyphal growth, which promotes biofilm formation and invasive growth into tissues. Stimulation of adenylyl cyclase to form cAMP induces hyphal morphogenesis. The failure of cells lacking adenylyl cyclase (cyr1Δ) to form hyphae has suggested that cAMP signaling is essential for hyphal growth. However, cyr1Δ mutants also grow slowly and have defects in morphogenesis, making it unclear whether hyphal inducers must stimulate cAMP, or if normal basal levels of cAMP are required to maintain cellular health needed for hyphal growth. Interestingly, supplementation of cyr1Δ cells with low levels of cAMP enabled them to form hyphae in response to the inducer N-acetylglucosamine (GlcNAc), suggesting that a basal level of cAMP is sufficient for stimulation. Furthermore, we isolated faster-growing cyr1Δ pseudorevertant strains that can be induced to form hyphae even though they lack cAMP. The pseudorevertant strains were not induced by CO2, consistent with reports that CO2 directly stimulates adenylyl cyclase. Mutational analysis showed that induction of hyphae in a pseudorevertant strain was independent of RAS1, but was dependent on the EFG1 transcription factor that acts downstream of protein kinase A. Thus, cAMP-independent signals contribute to the induction of hyphal responses.
Keywords: Candida albicans, hyphal morphogenesis, adenylyl cyclase, cAMP, pseudorevertants
Abbreviated Summary
The cAMP signaling pathway induces a switch from budding to hyphal morphogenesis that promotes biofilm formation and invasive growth by the human fungal pathogen Candida albicans. In this study we identified pseudorevertant strains that can form hyphae in the absence of adenylyl cyclase, which indicates that cAMP-independent pathways also contribute to this morphological switch.
Introduction
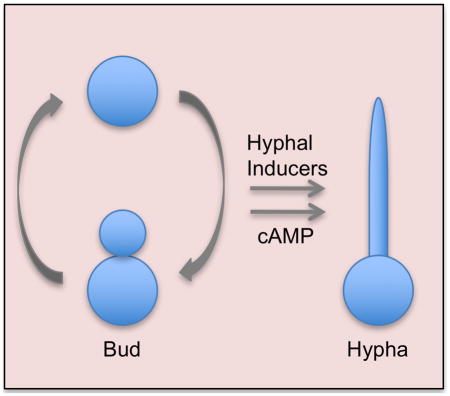
The human fungal pathogen Candida albicans is commonly found in the GI tract as a commensal organism (Odds, 1988, Heitman et al., 2006). Severe systemic infections can result from conditions that promote the overgrowth of C. albicans in the host, or compromise the immune system. Better therapies are needed to treat systemic candidiasis; despite advances in antifungal therapy there is about 40% attributable mortality (Pfaller & Diekema, 2010). A key factor for virulence is the ability of C. albicans to undergo a transition from growing as budding cells to instead forming long chains of hyphal or pseudohyphal cells that grow invasively into tissues (Sudbery, 2011). This change in the pattern of morphogenesis to filamentous growth is also important for C. albicans to form biofilms on catheters and medical devices (Blankenship & Mitchell, 2006). Cells induced to form hyphae also show increased expression of virulence factors including adhesin proteins that promote biofilm formation and enzymes that protect the cells from oxidative attack by the immune system (Whiteway & Oberholzer, 2004, Kumamoto & Vinces, 2005, Blankenship & Mitchell, 2006).
Various stimuli induce C. albicans to undergo hyphal growth in vitro including serum, alkaline pH, CO2, bacterial peptidoglycan breakdown products, and N-acetylglucosamine (GlcNAc) (Biswas et al., 2007, Whiteway & Bachewich, 2007, Davis, 2009, Sudbery, 2011). Several signal transduction pathways contribute to induction of hyphal growth in C. albicans; however, the cAMP pathway has been considered the most important (Wang, 2013, Hogan & Muhlschlegel, 2011). The C. albicans adenylyl cyclase Cyr1 is thought to act as a key sensor that integrates information from the environment as it can be activated by the small GTP-binding protein Ras1, the Gα protein Gpa2, peptidoglycan breakdown products, and bicarbonate derived from CO2 (Wang, 2013, Hogan & Muhlschlegel, 2011, Rocha et al., 2001).
Activation of adenylyl cyclase to produce cAMP is thought to promote hyphal induction because addition of exogenous cAMP can stimulate hyphal growth (Rocha et al., 2001, Sabie & Gadd, 1992). Furthermore, mutants with higher basal levels of cAMP are hyperfilamentous, such as mutants that produce constitutively active forms of Ras1 or Cyr1 (Bahn et al., 2003, Davis-Hanna et al., 2008, Bai et al., 2011). Sustained elevation of cAMP levels does not appear to be necessary to maintain hyphal growth; some hyphal inducers have been reported to cause only a transient spike in cAMP levels (Maidan et al., 2005, Fang & Wang, 2006, Lu et al., 2014).
Elevated cAMP is thought to promote hyphal growth by activation of PKA (protein kinase A) (Maidan et al., 2005, Fang & Wang, 2006, Lu et al., 2014). The events downstream of PKA are not well understood, but hyphal morphogenesis is regulated in part by the Hgc1-Cdc28 cyclin-dependent kinase and phosphorylation of morphogenesis proteins (Sudbery, 2011, Wang, 2009). The PKA pathway also promotes activation of the Efg1 transcription factor, which induces the hyphal-specific genes (Lu et al., 2014, Carlisle & Kadosh, 2013, Doedt et al., 2004).
Cells lacking Cyr1 are devoid of cAMP and do not respond to a wide range of hyphal inducers (Rocha et al., 2001, Wang, 2013, Hogan & Muhlschlegel, 2011). However, it is not clear that cAMP signaling is necessary to induce hyphal growth. The cyr1Δ mutant has other phenotypes that could contribute to its defects in hyphal morphogenesis. For example, the cyr1Δ mutant grows about three-fold slower than wild-type cells, presumably due to altered expression of metabolic genes (Rocha et al., 2001, Harcus et al., 2004). The Cyr1 protein also complexes with actin and Srv2 in wild-type cells, indicating it may have roles in morphogenesis in addition to the production of cAMP (Zou et al., 2010). Thus, it is not clear that all hyphal inducers act specifically through cAMP, or if a normal basal level of cAMP is needed for cells to be competent to respond to hyphal inducers. To better define the role of cAMP signaling in C. albicans hyphal growth, we showed that supplementing cyr1Δ cells with low levels of a cell permeable cAMP analog enabled them to induce hyphal growth in response to GlcNAc, in agreement with similar results for cells induced with serum (Rocha et al., 2001). Additionally, we discovered spontaneous pseudorevertants of cyr1Δ that grew faster and could be induced to undergo hyphal growth. These results indicate that cAMP-independent pathways also promote the transition to filamentous hyphal growth.
Results
Role of Csc25, Ras1, and Cyr1 in hyphal induction
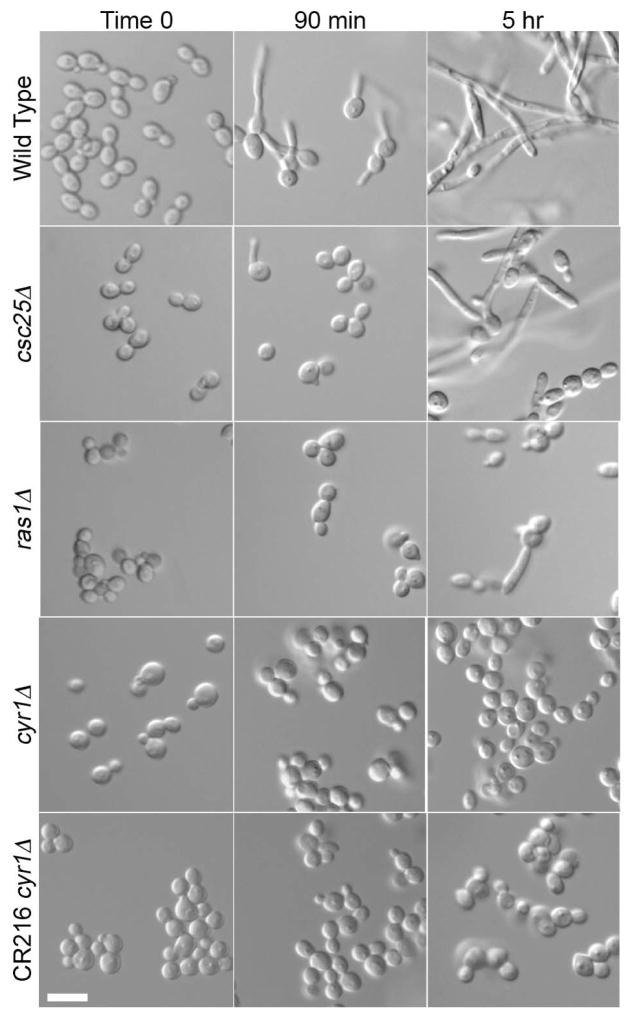
Our interests in GlcNAc signaling (Naseem & Konopka, 2015) led us to analyze the role of the cAMP pathway in inducing C. albicans hyphal growth. We examined the effects of deleting CYR1, RAS1, and CSC25, which encode adenylyl cyclase, its upstream activator Ras1, and the closest homolog of S. cerevisiae Cdc25 that acts as a guanine nucleotide exchange factor for Ras1. Although cyr1Δ and ras1Δ deletion mutants have been studied, to our knowledge only an insertional mutation of CSC25 was analyzed in previous studies (Enloe et al., 2000, Maidan et al., 2005). Therefore, we compared the effects of deleting these genes in a similar manner in the same strain background. The mutant cells were then induced with GlcNAc to determine if they could undergo hyphal morphogenesis. Wild-type cells induced in GlcNAc medium showed the expected formation of small hyphal outgrowths (germ tubes) by 90 min (Fig. 1). In contrast, the csc25Δ mutant showed limited germ tube formation at 90 min, and the ras1Δ and the cyr1Δ mutants showed strong defects in filamentous growth, similar to results for the previously described cyr1Δ strain CR216 (Rocha et al., 2001).
Figure 1. Deletion of cAMP pathway members results in hyphal defects.
Hyphal formation was induced by growing cells in synthetic medium containing 100 mM GlcNAc at 37°C for the indicated time. Deletion of CSC25 (csc25Δ) caused a mild defect in hyphal growth, deletion of RAS1 (ras1Δ) caused a stronger defect, and deletion of CYR1 (cyr1Δ) completely abrogated it. The wild type (DIC185), csc25Δ (SP53-1), ras1Δ (SP54-1), cyr1Δ (SP60-66) strains were derived from BWP17 and the CR216 cyr1Δ strain was derived from the CAI4 strain of C. albicans as described previously (Rocha et al., 2001). Bars, 10 μm.
After 5 h incubation, wild-type cells formed elongated chains of hyphal cells (Fig. 1). Interestingly, the csc25Δ cells also formed hyphae, indicating they could do so after a lag. The ras1Δ mutant also formed some filamentous cells after 5 h, but to a lesser degree. The cyr1Δ mutant strains failed to induce any filamentous growth forms even after 5 h. Filamentation at 5 h was not due to the effect of GlcNAc catabolism on extracellular pH, since buffering the media to pH 4 did not prevent filamentation. Previous studies have shown that growth of cells on GlcNAc medium raises the extracellular pH, which can also contribute to hyphal signaling, presumably because excess nitrogen is exported as ammonia (Naseem et al., 2015).
Synergy between GlcNAc and exogenously added cAMP
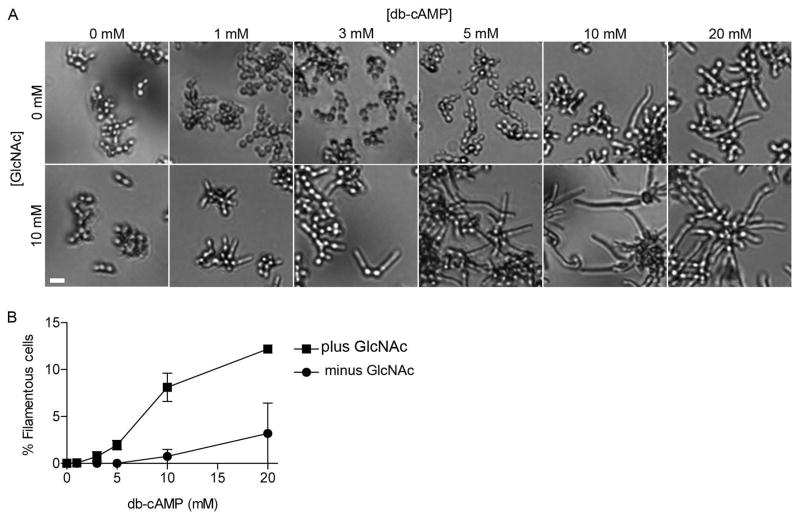
To more directly examine whether a sudden rise in cAMP or just a normal basal level of cAMP is important for hyphal growth, we restored intracellular cAMP levels by supplementing cyr1Δ cells with the cell permeable cAMP analog dibutyryl-cAMP (db-cAMP). Specifically, we cultured cells of strain CR216 cyr1Δ in a 96-well plate with various concentrations of db-cAMP. The cells were grown in medium containing galactose as a nutrient source, because it does not repress the expression of the GlcNAc-specific transporter Ngt1 as does glucose, thus allowing cells to grow normally and remain fully able to respond to GlcNAc (Alvarez & Konopka, 2007). After an initial incubation for 2 h at 37° C, GlcNAc was added to a final concentration of 10 mM to one set of cultures and then cells were incubated for an additional 3 h. In the absence of GlcNAc, filamentous growth was obvious at db-cAMP concentrations of 10 and 20 mM, but not at the lower concentrations (Figure 2A, top row). In contrast, GlcNAc induction of filamentous growth was detectable at the lowest concentration of db-cAMP tested (1 mM) and became more prevalent at higher concentrations of db-cAMP (Figure 2A, bottom row). This showed that restoring a basal level of cAMP enabled cells to respond to GlcNAc in the absence of adenylyl cyclase. Similar results have been reported that cyr1Δ cells treated with db-cAMP can be induced with serum to form hyphae (Rocha et al., 2001).
Figure 2. Addition of non-inducing levels of db-cAMP to cyr1Δ restores hyphal switching in response to GlcNAc.
A. Cells of cyr1Δ strain CR216 were cultured in rich YP medium containing 100 mM galactose plus the indicated amount of dibutyryl cAMP (db-cAMP). The cells were incubated at 37°C for 2 h, GlcNAc was added to a final concentration of 10 mM, and then the cells were incubated at 37°C for 3 h before being photographed. Bars, 10 μm.
B. Quantification of the average number of filamentous cells induced in the experiments described in panel A. Pre-incubation of cyr1Δ cells with non-inducing concentrations of db-cAMP restored the ability of GlcNAc to stimulate hyphal growth. The results represent the average of three different experiments, with at least 300 cells counted per experiment. Error bars indicate SD.
Pseudorevertants of cyr1Δ exhibit better growth and can be induced to form hyphae
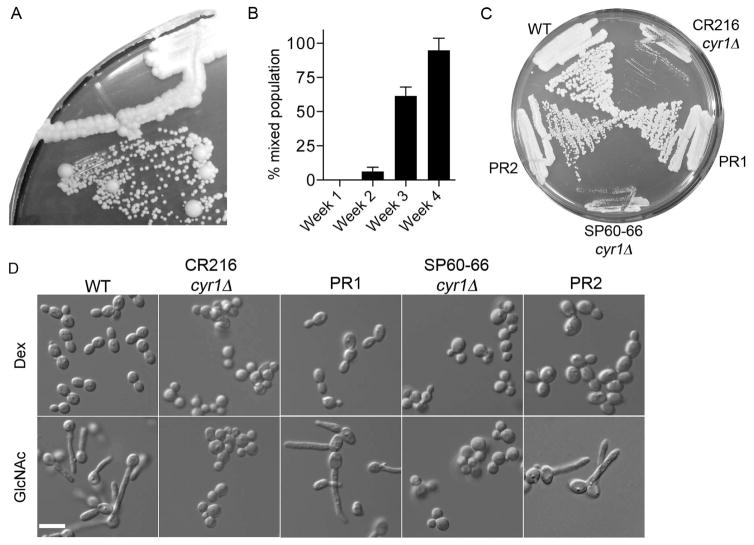
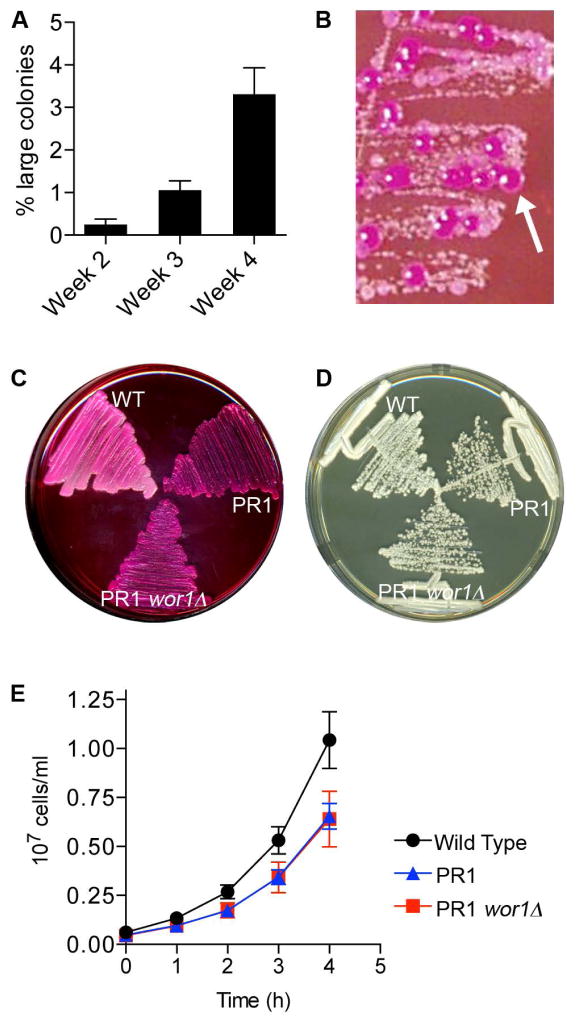
During the course of these studies, we observed that cyr1Δ mutants that had been maintained for a few weeks on agar medium exhibited two populations of cells when restreaked onto a fresh agar plate; the expected small colonies and also large colonies that grew similar to a wild type control (Fig. 3A). Further analysis showed that the larger colonies arose in a time-dependent manner (Fig. 3B). Only small colonies were detected when cyr1Δ strain CR216 was restreaked after incubation for one week on an agar plate at room temperature. However, after 2 weeks incubation larger colonies began to be detected in about 6% of the restreaked colonies, and by 4 weeks incubation 100% of the restreaked cyr1Δ CR216 cells showed at least one larger colony when restreaked onto fresh YPD medium. The larger colonies continued to grow faster than the parental cyr1Δ strain when restreaked onto fresh agar medium (Fig. 3C).
Figure 3. Faster growing pseudorevertants spontaneously arise from cyr1Δ cultures over time.
A. Cells streaked onto YPD agar plate that were derived from a representative colony of cyr1Δ (CR216) cells that had been incubated at room temperature for 3 weeks. At least two populations of cells are evident: small colonies similar to the cyr1Δ parental cells and large colonies of pseudorevertants.
B. Percent of cyr1Δ (CR216) colonies incubated for the indicated time that gave rise to mixed populations of large and small cells upon restreaking onto a fresh agar medium plate. Colonies that, when restreaked, gave at least 1 large colony were scored as positive. Results represent the average for 30 colonies assayed this way in each of 3 independent experiments. Error bars indicate SD.
C. Growth rate comparison between the parental cyr1Δ cells and pseudorevertants derived from them. Indicated strains were streaked on YPD and grown for 3 days at 30°C. Both the previously reported published cyr1Δ strain CR216 and a newly made cyr1Δ strain we constructed (SP60-66) gave rise to pseudorevertants that grew at approximately similar rates. Pseudorevertant PR1 was derived from CR216 while PR2 was derived from SP60-66.
D. GlcNAc induction of hyphal growth. Cells grown in SC-URA plus 100 mM dextrose medium, were washed and then resuspended in the same medium containing either 100 mM dextrose or 100 mM GlcNAc and cultured at 37°C for 90 min. As expected, wild-type cells formed hyphae but the cyr1Δ strains were completely defective. Interestingly, the faster growing pseudorevertants derived from both cyr1Δ strains were also stimulated to undergo filamentous growth in GlcNAc medium. Bars, 10 μm.
The faster growing cyr1Δ cells were then tested for ability to be induced with GlcNAc to form hyphae. Interestingly, the cells were readily induced by GlcNAc to form filamentous cells, even at an early 90 min time point (Figure 3D). The spontaneous emergence of faster growing cells was not limited to the previously constructed CR216 cyr1Δ, as faster growing cells were isolated from newly made cyr1Δ strains in the BWP17 strain background that had been incubated at room temperature for 3 weeks (Fig. 3C). The pseudorevertants that arose from the cyr1Δ mutant in the BWP17 strain background were also induced by GlcNAc to undergo filamentous growth (Fig. 3D). These results indicate that adenylyl cyclase is not needed for GlcNAc to induce hyphal growth.
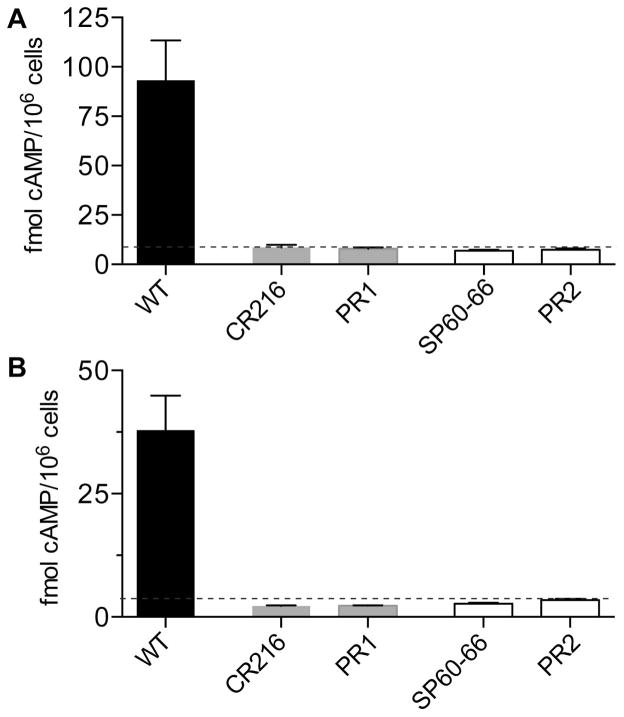
To ensure that the faster growing cells were not due to contamination, PCR analysis was carried out with 4 sets of primers designed to detect the catalytic domain of CYR1, all of which gave negative results (Supporting Information Fig. S1). As an independent way to confirm this, cell extracts were assayed for the presence of cAMP. Cells were cultured in rich media, frozen cell pellets were extracted, and then the levels of cAMP were assessed using an immunoassay kit (Fig. 4A). Wild type control strain DIC185 consistently showed detectable levels of cAMP that agreed well with previous studies (~1.8 pmol mg−1 dry weight) (Xu et al., 2008). In contrast, neither the cyr1Δ nor the pseudorevertants gave a signal above the limit of detection (Fig. 4A). Similar results were observed for both of the two independent cyr1Δ strains and the pseudorevertants derived from them. The cAMP assays were also performed on cells that were treated under conditions similar to those used to induce hyphal growth of the pseudorevertants. In brief, cells grown in synthetic medium were induced by addition of bovine calf serum for 30 min, a time previously reported to coincide with the spike in cAMP levels (Maidan et al., 2005). The cyr1Δ strains and the derived pseudorevertants did not show cAMP above the limit of detection for the assay (Fig. 4B). Wild-type cells consistently gave a detectable signal, although it was slightly lower for cells grown in synthetic medium than for the cells grown in rich YPD medium in Fig. 4A. These results confirm that the faster growing cells are cyr1Δ pseudorevertants that lack cAMP.
Figure 4. Pseudorevertants do not contain detectable cAMP.
A. Wild type, cyr1Δ, and derived pseudorevertant cells were grown overnight in rich YPD medium, cAMP was extracted, and then cAMP levels were assayed using a GE Amersham cAMP EIA-Immunoassay kit with the non-acetylation protocol. The pseudorevertant strains PR1 and PR2 showed low signals that were at or below the limit of detection, indicating that no cAMP could be detected, similar to their parental strains cyr1Δ strain CR216 and cyr1Δ strain SP60-66.
B. The strains described above were grown in synthetic SC-URA medium with 100 mM dextrose and the induced with 30% serum at 37° C for 30 min. cAMP was extracted as in panel A and assayed using the more sensitive acetylation protocol to detect cAMP. The cyr1Δ strains and their derived pseudorevertants contained no detectable cAMP under these conditions, similar to the results obtained with rich medium in panel A. The results for each strain represent the average cAMP value obtained from the analysis of at least four independent colonies. The dashed line indicates the limit of detection of the assay. Error bars indicate SD.
In preliminary studies we also observed fast-growing isolates of ras1Δ cells (not shown), demonstrating that care should be taken when working with cyr1Δ and ras1Δ cells to avoid enrichment for the faster growing pseudorevertants.
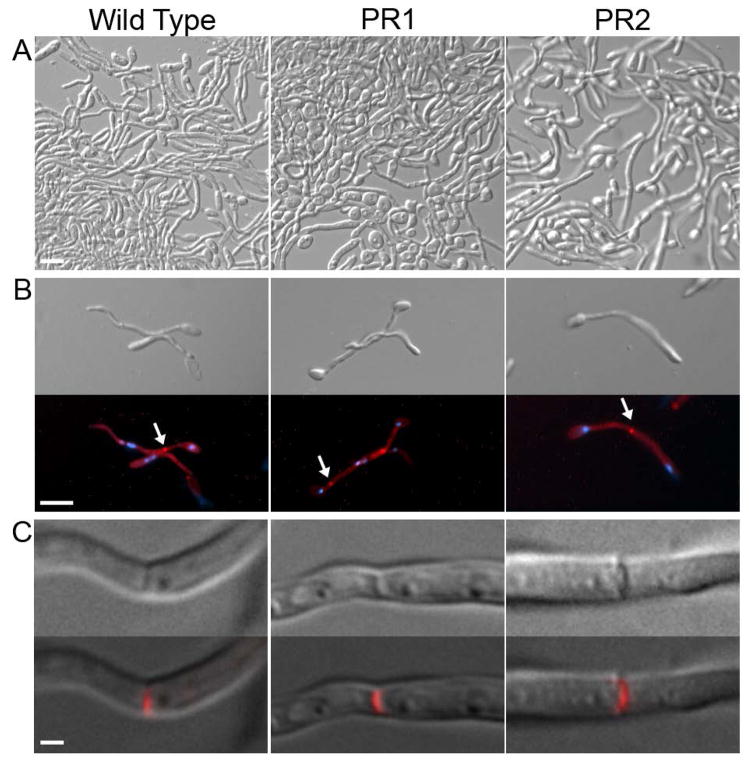
Pseudorevertant strains form true hyphae
The morphology of the filamentous cells formed by the pseudorevertants was examined further to determine if they were forming true hyphae. There are many types of filamentous growth, and not all are considered to be true hyphal growth. Hyphal growth is characterized by chains of elongated cells with parallel cell walls, a first septum distal to the bud neck, and multiple nuclei separated by septa (Sudbery et al., 2004). Pseudohyphal cells are characterized as chains of elongated buds with a first septum at the neck of the mother cell and indentations at subsequent septa. In addition, some mutants can form elongated cells that resemble hyphal filaments, such as elongated buds formed by septin mutants or in response to stress (Li et al., 2012, Shi et al., 2007, da Silva Dantas et al., 2010). To determine if cyr1Δ pseudorevertants form true hyphae, two independent pseudorevertant strains were induced with GlcNAc. Microscopic analysis at relatively low magnification revealed that germ tubes could extend into long hyphal cells for both the wild type and the pseudorevertants (Figure 5A). To further characterize whether these were true hyphae, the cells were stained with Pontamine Fast Scarlet 4B to detect cell wall chitin and with Hoechst 33342 to detect nuclear DNA. As expected for true hyphae, wild-type cells formed the first septum distal to the bud neck and subsequent nuclei were separated by septa (Figure 5B). Similar results were observed for two independently derived pseudorevertant strains. A higher magnification image of the septa indicated that the pseudorevertants were similar to the wild type in forming a flat zone of cell wall across the septum. These results indicate that the pseudorevertants are capable of forming true hyphae.
Figure 5. Pseudorevertants undergo true hyphal growth.
A. Wild-type cells (DIC185), and pseudorevertant cells derived from both cyr1Δ strains (CR216 and SP60-66) were cultured overnight in 100 mM GlcNAc at 37°C and then photographed at low magnification. Bar, 30 μm.
B. Hyphal cells from the cultures in panel A were stained with Hoechst 33342 to detect DNA (Blue) and Pontamine Fast Scarlet 34 (Red) to detect cell wall chitin. Hyphal cells from all three strains displayed characteristics of true hyphae including a first septum distal to the mother cell (arrow). Bar, 10 μm.
C. Upper panels show a close-up view of a single septum. Lower panels show the septum image merged with Red staining from Pontamine Fast Scarlet 34 to verify the presence of the septum. Bar, 1 μm.
Pseudorevertants induce hyphal morphology and gene expression in response to some but not all inducers
Activation of the cAMP pathway has been reported to play an important role in the induction of a specific set of genes in hyphae. It is thought that the hyphal repressor Nrg1 must be evicted from the promoters of hyphal-specific genes by a process that is activated by signaling through the cAMP pathway (Lu et al., 2011). To determine if hyphal genes are induced during hyphal growth in pseudorevertant strains, cells were engineered to express a reporter gene consisting of GFP under the control of the hyphal-specific promoter of HWP1. It is important to note that the cyr1Δ HWP1-GFP strain was constructed by first introducing the HWP1-GFP construct into a heterozygous strain containing one copy of CYR1, which was then deleted. This minimized the effects of the standard lithium acetate transformation procedure, which we found increased the frequency of cyr1Δ pseudorevertants. In contrast, deleting the remaining copy of CYR1 as the last step helped to ensure isolation of the expected slower growing cells rather than pseudorevertants. The slow growing cyr1Δ HWP1-GFP strain was then used to isolate a faster growing pseudorevertant by incubating for 3 weeks on solid media as described earlier.
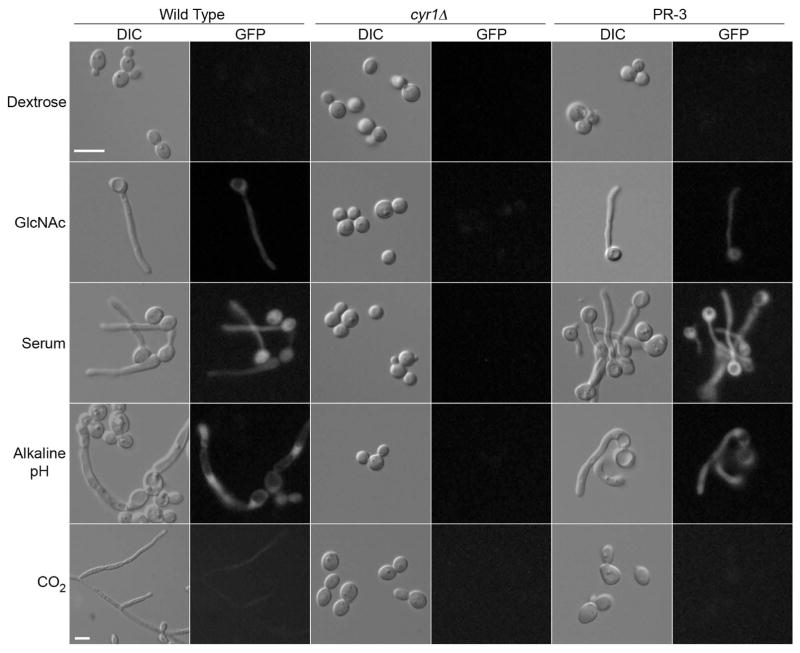
Strains carrying the HWP1-GFP reporter gene were treated with GlcNAc, serum, high pH, or CO2 to determine how they would respond to these different hyphal inducers (Fig. 6). As expected, wild-type cells formed hyphae and expressed GFP under all inducing conditions, whereas the cyr1Δ HWP1-GFP strain neither formed filamentous cells nor showed GFP expression. The pseudorevertant cyr1Δ HWP1-GFP strain formed hyphal cells and expressed GFP when induced with GlcNAc, serum, and high pH, providing additional evidence that they are stimulated to undergo typical hyphal morphogenesis.
Figure 6. Pseudorevertants induce hyphal morphology and gene expression in response to many inducers, but not CO2.
Strains carrying the HWP1-GFP reporter gene were incubated under the indicated hyphal inducing conditions and then photographed using light microscopy to detect cell morphology and fluorescence microscopy to determine if GFP was produced. As expected, all of the inducers stimulated the wild type control strain to form hyphae and produce GFP, whereas the cyr1Δ strain showed no detectable hyphae or GFP. In contrast, the pseudorevertant strain PR3 showed induction of filamentous cells and GFP levels similar to the wild type under all conditions except for CO2. All inductions were performed at 37°C. GlcNAc induction was performed using SC-URA medium with 100 mM GlcNAc and was visualized at 90 min. Serum induction was performed in SC-URA plus 100 mM dextrose and 30% bovine calf serum and was visualized after 90 min. Hyphal induction with alkaline pH was performed by culturing cells overnight in SC medium buffered to pH 8 with 150 mM HEPES. CO2 induction was performed by culturing cells overnight in a chamber with 5% CO2.
Interestingly, growing cells in an environment enriched in CO2 did not induce hyphae or the HWP1-GFP reporter gene in the pseudorevertant strain. This is consistent with previous reports that adenylyl cyclase is a sensor for CO2 in C. albicans and other organisms (Hall et al., 2010). After entering cells, CO2 is converted to bicarbonate ion, which is then thought to bind the catalytic lysine of adenylyl cyclase, increasing the specific activity of the cyclase and thereby increasing cAMP levels. These data therefore support the model that bicarbonate stimulates hyphal growth by activating adenylyl cyclase.
Pseudorevertants show some similarity to opaque cells, but are not dependent on WOR1
As described above, after 4 weeks incubation on solid agar medium essentially 100% of cyr1Δ colonies restreaked onto a fresh agar plate will give rise to a subset of cells that grow faster. We also observed that the relative proportion of large colonies increased with time. When 4-week-old colonies were restreaked onto fresh medium, the large pseudorevertant colonies accounted for about 3% of total colonies (Fig. 7A). This relatively high spontaneous rate of appearance of the pseudorevertants raised the possibility that the suppression of the slow growth phenotype may be due to an epigenetic change, rather than a mutation in the genome. The most commonly studied spontaneous epigenetic change in C. albicans is the White-Opaque switch (Soll, 2009, Scaduto & Bennett, 2015). Opaque cells are named for their distinctive colony morphology and have a very distinct pattern of gene expression compared to the White phase cells, including changes in expression of metabolic genes, (Lohse & Johnson, 2009). To determine if the pseudorevertants show similarity to Opaque cells, they were grown on medium containing Phloxine B; a dye that stains Opaque cells dark magenta. Interestingly, the large pseudorevertant colonies stained deep magenta and appeared somewhat flatter, as expected for Opaque cells. In contrast the smaller colonies were stained a lighter color, as expected for White phase cells (Fig. 7B).
Figure 7. Pseudorevertants show increased Phloxine B staining, but not other characteristics of Opaque cells.
A. The cyr1Δ strain CR216 was incubated at room temperature on YPD agar medium for the indicated time, restreaked onto fresh medium, and then the resulting colonies were assessed to determine the percent of faster-growing large colonies rather than the expected small colonies. The results represent the average for 6 colonies aged for two weeks, 59 colonies aged for three weeks, and 91 colonies aged for four weeks.
B. The cyr1Δ cells (CR216) were incubated 3 weeks on YPD agar plates and then streaked onto a YPD plate containing 45 μg ml−1 Phloxine B. Larger pseudorevertant colonies were observed to stain magenta (arrow), similar to that expected for cells in the Opaque phase.
C. Wild type (DIC185), a cyr1Δ pseudorevertant (PR1) derived from strain CR216, and its wor1Δ derivative (PR1-wor1Δ) were streaked onto YPD agar plate containing 45 μg ml−1 Phloxine B, showing that deletion of WOR1 did not change Phloxine B staining of the pseudorevertant.
D. The strains described in panel C were streaked for single colonies on YPD and incubated at 30° C for 2 days.
E. The strains described in panel C were grown in liquid YPD media at 30° C. Panels D and E show that deletion of WOR1 does not affect the growth rate of the pseudorevertant strain PR1.
To examine this further, the WOR1 gene was deleted in a pseudorevertant strain. WOR1 encodes a transcription factor that is considered to be the master regulator of the transition from White to Opaque phase (Zordan et al., 2007). However, deletion of WOR1 in a pseudorevertant strain had no effect on the Phloxine B staining, growth rate, or ability to induce hyphae (Figure 7C, D, E). In addition, out of a total of 35 pseudorevertant strains tested by PCR, 28 tested positive for the presence of both MTLa1 and MTLα1, including strains PR1, PR2, and PR3 that were used in this study. In the strain background used in these studies, MTLa1 or MTLα1 must be lost before a strain becomes capable of the White-Opaque switch (Miller & Johnson, 2002). Furthermore, although Opaque cells will switch back to the White phase at 37° C, incubating cyr1Δ colonies at 37° C did not prevent the appearance of pseudorevertants, and growth of cells at 37°C did not alter their ability to stain more darkly with Phloxine B. These results suggest that in spite of some similarities with Opaque cells, the cyr1Δ pseudorevertants arise through a distinct mechanism.
Hyphal signaling in pseudorevertants is independent of Ras1, but dependent on Efg1
Genome sequence analysis of three different pseudorevertant strains showed that they each contained an average of 131 polymorphisms relative to the cyr1Δ parental strain, but did not reveal an obvious mutation in common that could explain the basis for the faster growth rate (Supporting Information Table S1). For example, there were only three genes containing mutations in both alleles that were common to all three pseudorevertant strains, and these genes do not appear likely to account for the suppression of the cyr1Δ mutation. The three genes that were mutated are predicted to encode a GPI-anchored adhesin-like protein (IFF6), a nuclear importin (MTR10), and a protein of unknown function (CR_06260W) (Noble et al., 2010). Thus, it is not clear yet whether the pseudorevertants arise by an epigenetic mechanism or if further genome sequence analysis will be required to identify a genetic basis. However, it was significant that the analysis confirmed the absence of the CYR1 gene from the pseudorevertants, and that no mutations were detected in other components of the cAMP pathway, including the protein kinase A subunits (BCY1, TPK1, TPK2) or the EFG1 transcription factor.
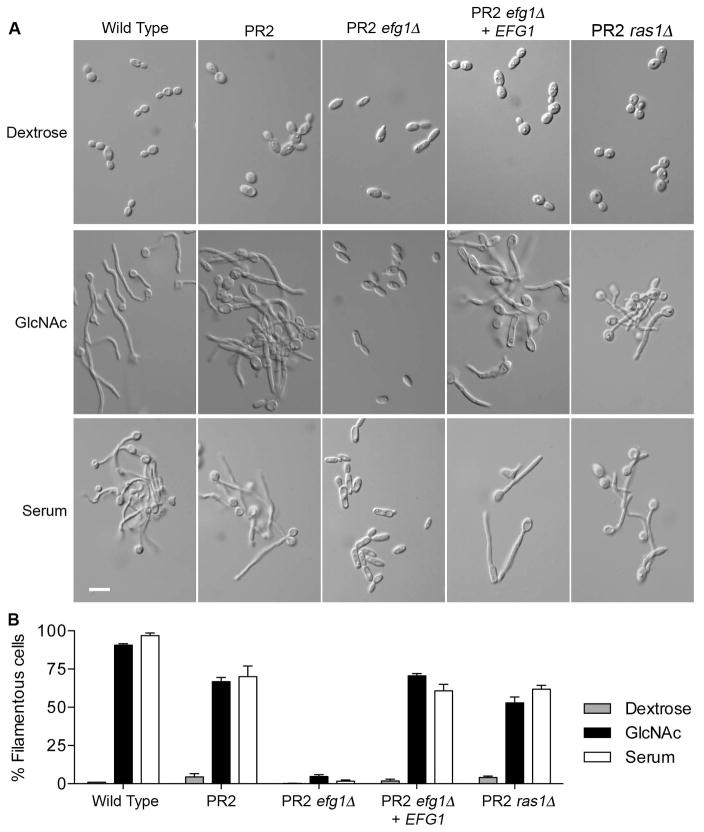
To gain further insight into the mechanisms that suppress the cyr1Δ mutation, we examined the effects of deleting RAS1 and EFG1 in the pseudorevertant strain PR2. A PR2 efg1Δ mutant failed to form hyphae in response to GlcNAc or serum, indicating that Efg1 is required for hyphal signaling in the PR2 mutant cells as it is in wild-type cells (Fig 8A). In contrast, a PR2 ras1Δ mutant was still capable of forming hyphae, albeit at slightly lower efficiency (Fig. 8A,B). These results indicate that the ability to suppress the hyphal defect of cyr1Δ cells is not due to activation of a Ras1 pathway that acts in parallel to adenylyl cyclase.
Figure 8. Pseudorevertant strain PR2 induces hyphal growth independently of RAS1, but dependent on EFG1.
A. Photographs of cells grown in dextrose medium or induced to undergo hyphal morphogenesis in medium containing 50 mM GlcNAc or 10% bovine serum. Cells were induced at 37°C with GlcNAc for 3h and with serum for 2 h.
B. Graph indicating the percent of filamentous cells after growth in conditions described in panel A. Values represent the average of three independent assays with at least 200 cells counted for each condition. The strains used included a wild type control (DIC185), pseudorevertant (PR2), PR2 with EFG1 deleted (PR2 efg1Δ; strain HS628), the PR2 efg1Δ strain with EFG1 restored (PR2 efg1Δ + EFG1; strain HS6281), and PR2 with RAS1 deleted (PR2 ras1Δ; strain HS631).
We also determined whether deletion of EFG1 or RAS1 from strain PR2 affected the suppression of the slow growth caused by cyr1Δ. Interestingly, the doubling times in rich YPD medium were similar for these strains (PR2 ~ 114.5 ± 5.5 m, PR2 efg1Δ 124.8 ± 10.4 m, and PR2 ras1Δ 117.9 ± 13.1 m). This contrasts with a doubling time of 68.6 ± 4.7 min for the wild type control strain and about 160.7 ± 2.5 min for the cyr1Δ mutant. This indicates that the suppression of cyr1Δ slow growth is independent of Efg1 and Ras1, even though Efg1 was needed for hyphal growth.
Discussion
Activation of adenylyl cyclase is sufficient to induce hyphal growth in C. albicans, since hyperactive adenylyl cyclase alleles or addition of exogenous cAMP can promote filamentous morphogenesis (Sabie & Gadd, 1992, Bai et al., 2011, Bahn et al., 2003, Davis-Hanna et al., 2008). Signaling through adenylyl cyclase has also been suggested to be necessary to respond to most hyphal inducers, because cyr1Δ mutants are defective in forming hyphae in response to a wide range inducing conditions (Wang, 2013, Hogan & Muhlschlegel, 2011). However, a complicating factor is that cAMP is not exclusively involved in hyphal signaling; it is also important for normal cellular function. The cyr1Δ cells show reduced expression of many metabolic genes, including those encoding ribosomal proteins, RNA polymerase subunits, and enzymes involved in the Krebs cycle, pyrimidine metabolism, heme and sterols (Harcus et al., 2004). Consistent with this, cyr1Δ cells have abnormal shape, size, and ~ three-fold slower growth rate (Rocha et al., 2001, Xu et al., 2008). This raised the question as to whether all hyphal inducers must act by stimulating adenylyl cyclase, or if a normal basal level of cAMP is required for cells to be competent to undergo hyphal morphogenesis. Our results indicate that both cAMP-dependent and cAMP-independent pathways promote hyphal growth.
Basal cAMP level is important for hyphal induction
The hyphal defects of the cyr1Δ, ras1Δ and csc25Δ mutants (Fig. 1) were proportional to their basal levels of cAMP (Maidan et al., 2005). To examine the role of the basal level of cAMP in hyphal signaling, cyr1Δ cells were exposed to different levels of the cell permeable analog db-cAMP and then examined for ability to undergo filamentous growth in response GlcNAc. Interestingly, GlcNAc induced hyphal growth at low levels of db-cAMP that were not sufficient to induce hyphae on their own (Fig. 2). Similar results were reported in other studies, which showed that db-cAMP supplementation restored the ability to form hyphae to a ras1Δ mutant induced with GlcNAc (Davis-Hanna et al., 2008) and a cyr1Δ mutant induced with serum (Rocha et al., 2001). These results indicate that the basal level of cAMP is important for cells to be competent to be induced by GlcNAc. In addition, these results suggest that there are cAMP-independent pathways that can function in the absence of adenylyl cyclase.
Pseudorevertants form hyphae in the absence of adenylyl cyclase or cAMP
To further examine cAMP-independent signaling we took advantage of the observation that cyr1Δ cells gave rise to faster growing pseudorevertant strains that could be stimulated to form hyphae even though they lacked adenylyl cyclase (Figs. 3,4,5). The pseudorevertants formed true hyphae (Fig. 5) and not just abnormal filamentous growth that has been reported for septin mutants or for cells exposed to oxidative or genotoxic stress (Li et al., 2012, Shi et al., 2007, da Silva Dantas et al., 2010). The pseudorevertants were also similar to the wild type in that they induced a hyphal-specific reporter gene, HWP1-GFP (Fig. 6). However, the pseudorevertants were distinct from wild-type cells in that they lacked cAMP (Fig. 4). Thus, there must be cAMP-independent pathways that can induce the cyr1Δ pseudorevertants to form hyphae in the absence of adenylyl cyclase.
Potential for synergy between hyphal inducing pathways
The ability of diverse stimuli including GlcNAc, serum, and alkaline pH to stimulate hyphal growth of the pseudorevertants in a cAMP-independent manner indicates that there are multiple inputs that can promote filamentous growth (Fig. 6). The existence of multiple pathways for hyphal induction is consistent with observations that different hyphal-inducing conditions stimulate distinct sets of overlapping genes, rather than inducing coordinate levels of expression of the same set of genes as would be expected if there was only a single main pathway (Martin et al., 2013, Naseem et al., 2015). Serum contains several components that can induce hyphae (Xu et al., 2008), suggesting it could be capable of inducing multiple signal pathways. GlcNAc is known to activate a cAMP-independent pathway to induce the genes needed for GlcNAc catabolism, since these genes can be induced in a cyr1Δ mutant (Gunasekera et al., 2010). These cAMP-independent pathways may also help explain the induction of hyphal growth at alkaline pH, which is mediated by the Rim101 pathway in a manner that does not obviously intersect with adenylyl cyclase (Davis, 2009). However, not all hyphal inducers may stimulate multiple pathways. The pseudorevertants were not induced by CO2 to form hyphae, consistent with previous reports that CO2 becomes converted into bicarbonate and acts directly on adenylyl cyclase to stimulate cAMP production and hyphal growth (Hall et al., 2010). The pseudorevertants will therefore provide a useful new tool to study hyphal induction pathways independently of the effects of adenylyl cyclase on cAMP levels.
The failure of cyr1Δ cells to form hyphae indicates that the cAMP-independent pathways must not be efficient in the absence of a normal basal level of cAMP. This could be due to the slow growth rate and other problems associated with the lack of adenylyl cyclase (Rocha et al., 2001, Xu et al., 2008). In addition, there may be synergy between the cAMP-dependent and cAMP-independent pathways that would be lost in a cyr1Δ mutant. For example, stimulation of adenylyl cyclase can increase the level of other signaling components, which could promote synergy between different pathways (Piispanen et al., 2013, Grahl et al., 2015). Thus, serum and GlcNAc may be potent inducers of hyphal growth because they can stimulate both cAMP-dependent and cAMP-independent pathways.
Hyphal growth of pseudorevertants is dependent on EFG1 but independent of RAS1
The high frequency with which the pseudorevertants arose (Figs. 3,7) suggested they may occur by an epigenetic mechanism (Scaduto & Bennett, 2015). However, the pseudorevertant phenotype was independent of the Wor1 transcription factor that regulates epigenetic White-Opaque switching in C. albicans (Fig. 7), indicating that some other mechanism must be involved. Genome sequence analysis was carried out to search for genetic changes, but the results did not reveal any obvious mutations in common for three independent pseudorevertants that could explain the suppression of the cyr1Δ phenotypes. In particular, there were no mutations detected in the cAMP pathway genes.
To gain better insight into the mechanism of suppression, the EFG1 and RAS1 genes were deleted from the cyr1Δ pseudorevertant strain PR2. Interestingly, although Ras1 has been suggested to have functions independent of its ability to activate Adenylyl Cyclase (Sudbery, 2011), deletion of RAS1 from a cyr1Δ pseudorevertant strain did not abrogate the ability to form hyphae (Fig. 8). In contrast, deletion of EFG1 did block hyphal growth, indicating that this transcription factor that acts downstream of protein kinase A is still required to induce hyphae in the pseudorevertant cells (Fig. 8). This suggests that downstream aspects of the cAMP pathway are activated in the pseudorevertants. However, neither EFG1 nor RAS1 were needed in pseudorevertant strain PR2 for the suppression of the slow growth rate due to cyr1Δ.
Adenylyl cyclase is important in diverse fungal pathogens
The results of this study have important implications for the study of other fungal pathogens, as the cAMP pathway has been reported to regulate virulence functions in a diverse set of pathogenic fungi. For example, cAMP levels govern the morphological change from filamentous growth to the pathogenic yeast form in the dimorphic fungus, Paracoccidioides brasiliensis (Chen et al., 2007), virulence functions in the mold Aspergillus fumigatus (Fuller et al., 2011), and in the budding yeast Cryptococcus neoformans, the cAMP pathway controls the formation of the polysaccharide capsule and deposition of melanin into the cell wall, both of which are key virulence functions (Kozubowski et al., 2009). Since the cAMP pathway also regulates growth properties in other fungi as it does in C. albicans, it will be similarly important to distinguish between a direct role of adenylyl cyclase in regulating virulence functions and an indirect effect due to more global effects on metabolism.
Experimental Procedures
Strains and media
The C. albicans strains used in this study are described in Table 1. C. albicans cells were grown in rich yeast extract-peptone-dextrose (YPD) medium or in complete synthetic medium (SC) made with yeast nitrogen base (YNB) and select amino acids or uridine (Ura) (Sherman, 2002). Homozygous gene deletion mutant strains were constructed either in strain BWP17 URA3+ or in the indicated parental strain by the sequential deletion of both copies of the targeted gene (C. albicans is diploid). BWP17 URA3+ was generated by transforming BWP17 (Wilson et al., 1999) with a URA3-containing fragment to complement the auxotrophy. The plasmid pBSK-URA3 was digested with the restriction enzymes Pst1 and Not1 to liberate the URA3-IRO1 sequence, which was then transformed into BWP17 and integrated by homologous recombination into the genome to restore URA3 at its native locus as well as the fragment of the IRO1 gene removed during the making of BWP17.
Table 1.
C. albicans strains used in this study.
| Strain | Parent | Genotype |
|---|---|---|
| BWP17 | SC5314 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| DIC185 | BWP17 | ura3Δ::λimm434/URA3 his1:hisG/HIS1 arg4::hisG/ARG4 |
| BWP17URA3+ | BWP17 | ura3Δ::λimm434/URA3 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| SP53-1 | BWP17 URA3+ | csc25Δ::SAT1-FLIP/csc25Δ::ARG4 ura3Δ::λimm434/URA3 his1::hisG/HIS1 arg4::hisG/arg4 |
| SP54-1 | BWP17 URA3+ | ras1Δ::SAT1-FLIP/ras1Δ::ARG4 ura3Δ::λimm434/URA3 his1::hisG/HIS1 arg4::hisG/arg4 |
| SP60-66 | BWP17 URA3+ | cyr1Δ::SAT1-FLIP/cyr1Δ::ARG4 ura3Δ::λimm434/URA3 his1::hisG/HIS1 arg4::hisG/arg4::hisG |
| PR2 | SP60-66 | cyr1Δ::SAT1-FLIP/cyr1Δ::ARG4 ura3Δ::λimm434/URA3 his1::hisG/HIS1 arg4::hisG/arg4::hisG |
| SP57-1 | BWP17 URA3+ | HWP1-GFP-HIS1 ura3Δ::λimm434/URA3 his1::hisG/his1::hisG arg4::hisG/ARG4 |
| SP62-88 | BWP17 URA3+ | HWP1-GFP-HIS1 cyr1Δ::SAT1-FLIP/cyr1Δ::ARG4 ura3Δ::λimm434/URA3 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| PR3 | SP62-88 | HWP1-GFP-HIS1 cyr1Δ::SAT1-FLIP/cyr1Δ::ARG4 ura3Δ::λimm434/URA3 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| CR216 | CAI4 | ura3::λimm434/ura3::1imm434 cyr1Δ::hisG-URA3-hisG/cyr1Δ::hisG |
| PR1 | CR216 | ura3::λimm434/ura3::λimm434 cyr1Δ::hisG-URA3-hisG/cyr1Δ::hisG |
| PR1-wor1Δ | PR1 | wor1Δ::FRT/wor1Δ::SAT1-FLIP ura3::λimm434/ura3::λimm434 cyr1Δ::hisG-URA3-hisG/cyr1Δ::hisG |
| HS628 | PR2 | cyr1Δ::FRT/cyr1Δ::ARG4 ura3Δ::λimm434/URA3 his1::hisG/HIS1 arg4::hisG/arg4 efg1Δ::FRT/efg1Δ::FRT |
| HS6281 | HS628 | cyr1Δ::FRT/cyr1Δ::ARG4 ura3Δ::λimm434/URA3 his1::hisG/HIS1 arg4::hisG/arg4 efg1Δ::FRT/efg1Δ::FRT EFG1::SAT1 |
| HS631 | PR2 | cyr1Δ::SAT1-FRT/cyr1Δ::ARG4 ura3Δ::λimm434/URA3 his1::hisG/HIS1 arg4::hisG/arg4 ras1Δ::SAT1/ras1Δ::FRT |
The csc25Δ (SP53-1), ras1Δ (SP54-1), and cyr1Δ (SP57-1) strains were constructed by deleting one copy of the indicated gene using the SAT1 flipper (Reuss et al., 2004). The SAT1 flipper was PCR amplified in a manner that added 80 bp on each end of DNA sequence that flanked the open reading frame of the targeted gene to promote homologous recombination. The SAT1 flipper contains a nourseothricin resistance marker CaSAT1, and a modified flippase gene, CaFLP. The appropriate mutants were identified by PCR analysis using a combination of primers outside the sites of cassette integration and internal primers. The second allele was deleted in a similar manner using a PCR amplified version of the ARG4 selectable marker with 80 bp of flanking homology to the targeted gene. The histidine auxotrophy was corrected either with a PCR product containing 2050 bp of HIS1 or a GFP-HIS1 cassette (Zhang & Konopka, 2010) targeted to the HWP1 locus to create the HWP1-GFP reporter gene in strains SP57-1 and SP62-88. Gene deletion was verified by PCR analysis using a combination of primers that detect the gene deletion cassettes at the locus as well as primers designed to detect the open reading frame. Strain PR1-wor1Δ was constructed from strain PR1 by sequential deletion of both copies of WOR1 using the SAT1 flipper method (Reuss et al., 2004).
Hyphal growth assays
Unless otherwise indicated, cells were grown overnight at 30°C at low density (<1×107 cells ml−1) in SC-Ura medium lacking uridine and containing 100 mM dextrose. The following morning, 1.5 ml aliquots were harvested by centrifugation, washed with 1 ml of YNB, inoculated into 7 ml of SC-Ura + 100 mM dextrose or 100 mM GlcNAc medium, and incubated at 37°C for the indicated time to induce hyphal morphogenesis. Serum induction was performed by inoculating cells into SC-Ura medium containing 100 mM dextrose and 30% bovine calf serum. Alkaline pH induction of hyphal growth was performed by modification of a previously described method (Davis et al., 2000) in which cells were cultured overnight in YPD at 30°C, washed, and then inoculated into SC containing 0.1% dextrose + 150 mM HEPES buffered to pH 8, and cultured overnight at 37°C before visualization. CO2 induction was performed by a modification of a previously described protocol (Klengel et al., 2005). Cells were inoculated into YNB medium containing 100 mM dextrose and 150 mM PIPES buffered to pH 7 in a total volume of 100 μl in a 96-well plate. The plate was covered with a breathable cover and incubated overnight at 37°C with 5% CO2, and then the next day the cells were removed from the wells and examined by microscopy.
To assess the production of GFP by the HWP1-GFP reporter gene, a hyphal growth induction was performed with the GFP-tagged strains and then live cells were harvested and examined using a fluorescence microscope.
Staining cells with Hoechst 33342 and Pontamine Fast Scarlet 4B
To visualize cell septa and nuclear DNA, cells were first stained with Hoechst 33342 (2 μg ml−1; Invitrogen, Grand Island, NY) to detect DNA and then stained with Pontamine Fast Scarlet 4B (0.3 μg ml−1; gift from Charles Sprecht, University of Massachusetts Medical School, Worcester, MA) to detect cell wall and septa. Pontamine Fast Scarlet 4B stains cells similar to Calcofluor White (Hoch et al., 2005) but it fluoresces at a distinct wavelength from Calcofluor White, making it possible to carry out double-staining analyses with Hoechst stain.
db-cAMP supplementation
Cells of strain CR216 cyr1Δ were grown overnight in YP medium containing 100 mM galactose. Galactose was used because, unlike dextrose, it does not repress the expression of the GlcNAc specific transporter NGT1 (Alvarez & Konopka, 2007). The next day, cells were washed and resuspended in the same medium with the indicated amount of db-cAMP at a density of 5 × 106 cells ml−1 in a total volume of 150 μl in a 96-well microplate with an air permeable cover. The cells were cultured for 2 hours at 37°C before addition of GlcNAc, then grown an additional 3 hours. Cells in the microplate were then imaged by microscopy. Quantification was performed by counting at least 300 cells per experiment in 3 separate experiments.
Pseudorevertant frequency assay
Strain CR216 was freshly streaked from a frozen stock and grown on YPD plates at 30°C. Four different colonies were then restreaked onto fresh plates and grown for 4 days at 30°C. The plates were then incubated at room temperature for 1, 2, 3, or 4 weeks. After incubation at room temperature, 32 different colonies were restreaked on YPD and incubated at 30°C. The number of large and small colonies was then counted. This assay was performed three independent times. Similar results were observed in similar experiments with cyr1Δ strain SP60-66.
Intracellular cAMP determination
cAMP was extracted using the protocol essentially as described previously (Davis-Hanna et al., 2008). A pellet of 1×108 cells was snap frozen in liquid nitrogen. The cAMP was then extracted by adding 1 ml of ice-cold 1 M formic acid saturated with N-butanol and then the sample was mixed until resuspended. Samples were kept on ice and mixed 3 more times over the course of 10 min. Cell debris was pelleted in a 4°C refrigerated microcentrifuge at 20,000 × g for 10 min. 150 μl of the supernatant was collected and dried under vacuum. The amount of cAMP was determined using a cAMP Direct Biotrak EIA immunoassay kit (GE Amersham, Pittsburgh, PA). The dried cAMP pellet was resuspended in 1 ml of sample buffer and assayed using either the non-acetylation or acetylation protocols as per the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank Dr. Malcolm Whiteway for sending strains, Dr. Charles Sprecht for the kind gift of Pontamine Scarlet 4B, and members of our lab for their helpful advice and suggestions on the manuscript. This research was supported by Public Health Service grants awarded to J.B.K. from the National Institutes of Health (RO1 GM087368 and RO1 GM116048). S.P. was supported in part by a training grant (NIH T32AI007539) from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
Footnotes
Author Contributions
SMP, HS, SN, KG, JG, and JBK contributed to the conception and design of the studies. SMP, HS, SN, KG, JG, and JBK contributed to the acquisition, analysis, and interpretation of the data. SMP, HS, and JBK contributed to the writing of the manuscript.
References
- Alvarez FJ, Konopka JB. Identification of an N-acetylglucosamine transporter that mediates hyphal induction in Candida albicans. Mol Biol Cell. 2007;18:965–975. doi: 10.1091/mbc.E06-10-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, Staab J, Sundstrom P. Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol Microbiol. 2003;50:391–409. doi: 10.1046/j.1365-2958.2003.03692.x. [DOI] [PubMed] [Google Scholar]
- Bai C, Xu XL, Wang HS, Wang YM, Chan FY, Wang Y. Characterization of a hyperactive Cyr1 mutant reveals new regulatory mechanisms for cellular cAMP levels in Candida albicans. Mol Microbiol. 2011;82:879–893. doi: 10.1111/j.1365-2958.2011.07859.x. [DOI] [PubMed] [Google Scholar]
- Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JR, Mitchell AP. How to build a biofilm: a fungal perspective. Curr Opin Microbiol. 2006 doi: 10.1016/j.mib.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Carlisle PL, Kadosh D. A genome-wide transcriptional analysis of morphology determination in Candida albicans. Mol Biol Cell. 2013;24:246–260. doi: 10.1091/mbc.E12-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Janganan TK, Chen G, Marques ER, Kress MR, Goldman GH, Walmsley AR, Borges-Walmsley MI. The cAMP pathway is important for controlling the morphological switch to the pathogenic yeast form of Paracoccidioides brasiliensis. Mol Microbiol. 2007;65:761–779. doi: 10.1111/j.1365-2958.2007.05824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Dantas A, Patterson MJ, Smith DA, Maccallum DM, Erwig LP, Morgan BA, Quinn J. Thioredoxin regulates multiple hydrogen peroxide-induced signaling pathways in Candida albicans. Mol Cell Biol. 2010;30:4550–4563. doi: 10.1128/MCB.00313-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D, Wilson RB, Mitchell AP. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20:971–978. doi: 10.1128/mcb.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DA. How human pathogenic fungi sense and adapt to pH: the link to virulence. Curr Opin Microbiol. 2009;12:365–370. doi: 10.1016/j.mib.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Davis-Hanna A, Piispanen AE, Stateva LI, Hogan DA. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol. 2008;67:47–62. doi: 10.1111/j.1365-2958.2007.06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doedt T, Krishnamurthy S, Bockmuhl DP, Tebarth B, Stempel C, Russell CL, Brown AJ, Ernst JF. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol Biol Cell. 2004;15:3167–3180. doi: 10.1091/10.1091/mbc.E03-11-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enloe B, Diamond A, Mitchell AP. A single-transformation gene function test in diploid Candida albicans. J Bacteriol. 2000;182:5730–5736. doi: 10.1128/jb.182.20.5730-5736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang HM, Wang Y. RA domain-mediated interaction of Cdc35 with Ras1 is essential for increasing cellular cAMP level for Candida albicans hyphal development. Mol Microbiol. 2006;61:484–496. doi: 10.1111/j.1365-2958.2006.05248.x. [DOI] [PubMed] [Google Scholar]
- Fuller KK, Richie DL, Feng X, Krishnan K, Stephens TJ, Wikenheiser-Brokamp KA, Askew DS, Rhodes JC. Divergent Protein Kinase A isoforms co-ordinately regulate conidial germination, carbohydrate metabolism and virulence in Aspergillus fumigatus. Mol Microbiol. 2011;79:1045–1062. doi: 10.1111/j.1365-2958.2010.07509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahl N, Demers EG, Lindsay AK, Harty CE, Willger SD, Piispanen AE, Hogan DA. Mitochondrial Activity and Cyr1 Are Key Regulators of Ras1 Activation of C. albicans Virulence Pathways. PLoS Pathog. 2015;11:e1005133. doi: 10.1371/journal.ppat.1005133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera A, Alvarez FJ, Douglas LM, Wang HX, Rosebrock AP, Konopka JB. Identification of GIG1, a GlcNAc-induced gene in Candida albicans needed for normal sensitivity to the chitin synthase inhibitor nikkomycin Z. Eukaryot. Cell. 2010;9:1476–1483. doi: 10.1128/EC.00178-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RA, De Sordi L, Maccallum DM, Topal H, Eaton R, Bloor JW, Robinson GK, Levin LR, Buck J, Wang Y, Gow NA, Steegborn C, Muhlschlegel FA. CO2 acts as a signalling molecule in populations of the fungal pathogen Candida albicans. PLoS Pathog. 2010;6:e1001193. doi: 10.1371/journal.ppat.1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcus D, Nantel A, Marcil A, Rigby T, Whiteway M. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol Biol Cell. 2004;15:4490–4499. doi: 10.1091/mbc.E04-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Filler SG, Edwards JEJ, Mitchell AP. Molecular Principles of Fungal Pathogenesis. ASM Press; Washington, DC, USA: 2006. [Google Scholar]
- Hoch HC, Galvani CD, Szarowski DH, Turner JN. Two new fluorescent dyes applicable for visualization of fungal cell walls. Mycologia. 2005;97:580–588. doi: 10.3852/mycologia.97.3.580. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Muhlschlegel FA. Candida albicans developmental regulation: adenylyl cyclase as a coincidence detector of parallel signals. Curr Opin Microbiol. 2011;14:682–686. doi: 10.1016/j.mib.2011.09.014. [DOI] [PubMed] [Google Scholar]
- Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schroppel K, Naglik JR, Eckert SE, Mogensen EG, Haynes K, Tuite MF, Levin LR, Buck J, Muhlschlegel FA. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol. 2005;15:2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L, Lee SC, Heitman J. Signalling pathways in the pathogenesis of Cryptococcus. Cell Microbiol. 2009;11:370–380. doi: 10.1111/j.1462-5822.2008.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD. Alternative Candida albicans Lifestyles: Growth on Surfaces. Annu Rev Microbiol. 2005 doi: 10.1146/annurev.micro.59.030804.121034. [DOI] [PubMed] [Google Scholar]
- Li L, Zhang C, Konopka JB. A Candida albicans Temperature-Sensitive cdc12-6 Mutant Identifies Roles for Septins in Selection of Sites of Germ Tube Formation and Hyphal Morphogenesis. Eukaryot Cell. 2012;11:1210–1218. doi: 10.1128/EC.00216-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Johnson AD. White-opaque switching in Candida albicans. Curr Opin Microbiol. 2009;12:650–654. doi: 10.1016/j.mib.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Su C, Liu H. Candida albicans hyphal initiation and elongation. Trends Microbiol. 2014;22:707–714. doi: 10.1016/j.tim.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Su C, Wang A, Liu H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 2011;9:e1001105. doi: 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidan MM, De Rop L, Serneels J, Exler S, Rupp S, Tournu H, Thevelein JM, Van Dijck P. The G Protein-coupled Receptor Gpr1 and the Ga Protein Gpa2 Act through the cAMP-PKA Pathway to Induce Morphogenesis in Candida albicans. Mol Biol Cell. 2005;16:1971–1986. doi: 10.1091/mbc.E04-09-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Albrecht-Eckardt D, Brunke S, Hube B, Hunniger K, Kurzai O. A core filamentation response network in Candida albicans is restricted to eight genes. PloS one. 2013;8:e58613. doi: 10.1371/journal.pone.0058613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- Naseem S, Araya E, Konopka JB. Hyphal growth in Candida albicans does not require induction of hyphal-specific gene expression. Mol Biol Cell. 2015;26:1174–1187. doi: 10.1091/mbc.E14-08-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseem S, Konopka JB. N-acetylglucosamine regulates virulence properties in microbial pathogens. PLoS Pathog. 2015;11:e1004947. doi: 10.1371/journal.ppat.1004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC. Candida and Candidosis. Bailliere Tindall; Philadelphia, PA, USA: 1988. [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol. 2010;36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- Piispanen AE, Grahl N, Hollomon JM, Hogan DA. Regulated proteolysis of Candida albicans Ras1 is involved in morphogenesis and quorum sensing regulation. Mol Microbiol. 2013;89:166–178. doi: 10.1111/mmi.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Rocha CR, Schroppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M, Leberer E. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell. 2001;12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabie FT, Gadd GM. Effect of nucleosides and nucleotides and the relationship between cellular adenosine 3′:5′-cyclic monophosphate (cyclic AMP) and germ tube formation in Candida albicans. Mycopathologia. 1992;119:147–156. doi: 10.1007/BF00448812. [DOI] [PubMed] [Google Scholar]
- Scaduto CM, Bennett RJ. Candida albicans the chameleon: transitions and interactions between multiple phenotypic states confer phenotypic plasticity. Curr Opin Microbiol. 2015;26:102–108. doi: 10.1016/j.mib.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Shi QM, Wang YM, Zheng XD, Lee RT, Wang Y. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol Biol Cell. 2007;18:815–826. doi: 10.1091/mbc.E06-05-0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll DR. Why does Candida albicans switch? FEMS Yeast Res. 2009;9:973–989. doi: 10.1111/j.1567-1364.2009.00562.x. [DOI] [PubMed] [Google Scholar]
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Wang Y. CDKs and the yeast-hyphal decision. Curr Opin Microbiol. 2009;12:644–649. doi: 10.1016/j.mib.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Wang Y. Fungal adenylyl cyclase acts as a signal sensor and integrator and plays a central role in interaction with bacteria. PLoS Pathog. 2013;9:e1003612. doi: 10.1371/journal.ppat.1003612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–553. doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M, Oberholzer U. Candida morphogenesis and host-pathogen interactions. Curr Opin Microbiol. 2004;7:350–357. doi: 10.1016/j.mib.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XL, Lee RT, Fang HM, Wang YM, Li R, Zou H, Zhu Y, Wang Y. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell host & microbe. 2008;4:28–39. doi: 10.1016/j.chom.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Zhang C, Konopka JB. A photostable green fluorescent protein variant for analysis of protein localization in Candida albicans. Eukaryot Cell. 2010;9:224–226. doi: 10.1128/EC.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Fang HM, Zhu Y, Wang Y. Candida albicans Cyr1, Cap1 and G-actin form a sensor/effector apparatus for activating cAMP synthesis in hyphal growth. Mol Microbiol. 2010;75:579–591. doi: 10.1111/j.1365-2958.2009.06980.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.