Abstract
Discovery of clinical and genetic predictors of exemestane pharmacokinetics was attempted in 246 post-menopausal patients with breast cancer enrolled on a prospective clinical study. A sample was collected two hours after exemestane dosing at a 1 or 3 month study visit to measure drug concentration. The primary hypothesis was that patients carrying the low-activity CYP3A4*22 (rs35599367) SNP would have greater exemestane concentration. Additional SNPs in genes relevant to exemestane metabolism (CYP1A1/2, CYP1B1, CYP3A4, CYP4A11, AKR1C3/4, AKR7A2) were screened in secondary analyses and adjusted for clinical covariates. CYP3A4*22 was associated with a 54% increase in exemestane concentration (p<0.01). Concentration was greater in patients who reported White race, had elevated aminotransferases, renal insufficiency, lower body mass index, and had not received chemotherapy (all p<0.05), and CYP3A4*22 maintained significance after adjustment for covariates (p<0.01). These genetic and clinical predictors of exemestane concentration may be useful for treatment individualization in patients with breast cancer.
Keywords: Pharmacogenetic, CYP3A4*22, exemestane, pharmacokinetic
Introduction
Exemestane is a second generation steroidal aromatase inhibitor (AI) used for the treatment of estrogen receptor (ER) positive breast cancer in post-menopausal women(1) or with ovarian suppression in pre-menopausal patients(2). Many patients receiving AI therapy in the adjuvant setting experience toxicity, most notably musculoskeletal arthralgias and myalgias that cause early treatment discontinuation in an estimated 25% of patients(3, 4). Patients who are unable to tolerate one AI medication are sometimes able to tolerate a different one, despite very similar mechanisms of action(3). One possible explanation is intra-patient differences in AI metabolism that result in differing levels of drug exposure. In addition, although 5 years of AI treatment improves disease-free and overall survival compared to 5 years of tamoxifen in post-menopausal patients with early stage breast cancer, approximately 19.1% of patients will experience breast cancer recurrence and 14.2% will die of breast cancer within 10 years(5). Although many of these cases are likely due to intrinsic or acquired tumor resistance, some may be due to insufficient circulating drug concentrations.
Clinical predictors of increased exemestane concentration have been reported, including diminished hepatic and renal function(6). Many patients treated with AI have previously received chemotherapy that is hepatotoxic(7), which may have a long-term effect on hepatic enzyme activity(8). Exemestane is primarily eliminated via CYP3A4 mediated metabolism, with a minor contribution from alternative metabolic pathways including CYP4A11, CYP1A1/2, aldo-keto reductases(9) and renal elimination(6). We hypothesized that genetic variability in exemestane’s metabolic pathways would affect drug concentration during treatment. We were particularly interested in the intronic CYP3A4*22 single nucleotide polymorphism (SNP) (rs35599367) found at an approximate frequency of 6% in Caucasians. This SNP confers reduced enzymatic activity in cancer patients, as measured by the probe substrate midazolam(10), and has been associated with decreased metabolism of several drugs(11) including some cancer agents(12, 13).
We previously conducted a prospective randomized clinical trial to evaluate the pharmacogenetics of two AIs, the non-steroidal aromatase inhibitor letrozole, and the steroid exemestane, in post-menopausal women with early stage, ER positive breast cancer (the Estrogen and Letrozole Pharmacogenetics clinical trial (ELPh)). An analysis of letrozole-treated patients from the ELPh study identified genetic variation in CYP2A6 and clinical variables including body-mass index (BMI) and age that were associated with letrozole concentrations(14). The primary objective of this analysis was to investigate the effect of CYP3A4*22 on steady-state exemestane concentrations in exemestane-treated patients, and secondarily, to examine associations with additional genetic and clinical factors.
Materials and Methods
Patient cohort and sample collection
This pharmacogenetic secondary analysis of steady state exemestane concentration was carried out in patients enrolled in a prospective, open-label, clinical trial conducted by the Consortium on Breast Cancer Pharmacogenomics (COBRA), details of which have been previously published (4, 15). Briefly, post-menopausal women diagnosed with stage I-III hormone receptor positive breast cancer considering AI therapy upfront or following tamoxifen were enrolled at three cancer centers (Indiana University Cancer Center, Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University, and the University of Michigan Comprehensive Cancer Center) from August 2005–July 2009. Local breast cancer therapy including surgery and/or radiation therapy as well as systemic chemotherapy were completed prior to enrollment. The protocol was approved by the Institutional Review Boards of each participating site and all enrolled patients provided written informed consent.
Patients were stratified based on previous chemotherapy, tamoxifen, and bisphosphonate therapy and randomized to receive oral exemestane 25 mg/day or letrozole 2.5 mg/day for 2 years. Samples were collected for pharmacokinetic analysis at 3 months after initiation of AI treatment, or after 1 month of treatment in patients who crossed-over to the alternative arm due to treatment-related toxicity, as defined by the clinical judgement of the treating oncologist. Patients were instructed to take their daily AI dose two hours before blood draw for analysis of an approximate concentration maximum (Cmax).
Measurement of exemestane concentration
Samples collected in heparinized tubes were centrifuged at 1600 g for 10 minutes at 4°C for plasma separation. Plasma exemestane concentration was measured by mass spectrometry using the following protocol. 500 μL of patient plasma was spiked with 500 μL of Glycine-NaOH Buffer (pH = 11.3) and 25 μL of internal standard (100 ng/mL nevirapine in methanol). 6 mL of ethyl acetate was used to extract the mixture followed by 5 minutes of centrifugation. After evaporation the residue of the organic supernatant was reconstituted with 100 μL of 1:1 acetonitrile and formic acid. After vortexing for 4 minutes, 50 μL was injected into a Shimadzu HPLC system (Columbia, MD) that was coupled with an Applied Biosystems API 2 000 triple-quadruple mass spectrometer (Foster City, CA) which controlled by Analyst software version 1.4.2 and equipped with a positive ionization turbo ion spray. An Agilent Zorbax Column SBC-18 (150 × 4.6 mm; 3.5 μm) was used for separation with the mobile phase composed of 3:2 acetonitrile and formic acid at a flow rate of 0.3 mL/min. The precursor to product-ion fragmentation were m/z 297.2 –> 121.0 for exemestane and m/z 267.1 –> 226.1 for nevirapine. Known exemestane concentrations spiked into plasma were used to construct a calibration curves with appropriate quality control. The lower-limit of quantification (LLOQ) of the exemestane assay was 2.5 ng/mL, with <15% inter- and intra-day coefficients of variation.
Genotyping
A whole blood sample was collected at enrollment for DNA isolation and germline genetic assessment. DNA was extracted prior to genotyping using the Qiamp DNA Blood Maxi Kit-Spin Protocol in accordance with the manufacturer’s instructions (Qiagen, Valencia, CA). Genotype determination was completed by the University of Michigan Medical School’s DNA Sequencing Core using the Sequenom MassARRAY iPLEX platform on previously validated multiplex assays(16). The SNPs included in these multiplex assays were selected for their relevance to a variety of ongoing and planned pharmacogenetic analyses in cohorts of cancer patients. Multiplex assays were designed using the Sequenom software (both online Assay Design Suite tools and desktop Assay Design 4.0) and performed according to manufacturer’s standard protocols. Results were processed to generate SNP calls automatically, using Sequenom TyperAnalyzer software, and manually reviewed to validate allele calls. Automatic SNP calls with questionable spectra, based on the judgement of the genotyping technician, were removed.
The primary and secondary analyses were defined prior to initiation of any analysis. The association of one SNP (CYP3A4*22, rs35599367) with exemestane concentration was selected as the primary hypothesis. Samples were genotyped on the multiplex panel that included the CYP3A4*22 SNP. An additional 11 putatively functionally consequential SNPs in genes relevant to drug metabolism (CYP3A4, CYP1A1/2, CYP1B1, CYP4A11, AKR1C3/4, and AKR7A2) were selected for secondary screening. The remaining 47 SNPs genotyped on the multiplex assay were excluded prior to analysis for lack of relevance to exemestane pharmacokinetics. Genotype quality control was conducted for these 12 SNPs including assessment of call rate (call rate < 90%), minor allele frequency (MAF < 5%), and departure from Hardy Weinberg Equilibrium (HWE) (p<0.004, 0.05/12). SNPs that departed significantly from HWE in the entire cohort were assessed stratified by race to differentiate likely genotyping failure from the effects of population admixture.
Statistical analysis
The primary endpoint for this analysis was the first exemestane concentration available (either at 1 or 3 months). Patients with exemestane concentrations below 2.5 ng/mL (the LLOQ) were left-censored at 2.5 ng/mL. Tobit regression was used in all analyses in which exemestane concentration was the outcome since it allows for the modeling of censored dependent variables(17). Exemestane concentration was log-transformed to address non-normality of residuals, and appropriate checks were performed to assess the validity of assumptions regarding residual variance and normality(18).
Each SNP was tested for an independent association with exemestane concentration, assuming an additive genetic model. CYP3A4*22 was selected a priori as the primary hypothesis and tested at the typical significance threshold of p<0.05. All other SNPs were screened in a hypothesis-generating secondary analysis that was uncorrected for multiple comparisons (p<0.05). Associations between exemestane concentration and clinical covariates including prior chemotherapy (yes vs. no), renal impairment (tertiles of creatinine clearance (CrCl) estimated using the Cockcroft-Gault equation(19) with baseline serum creatinine measurement), elevated aminotransferases (baseline AST or ALT >40), age, BMI, time of exemestane sample collection (1 vs. 3 months) and self-reported race (White vs. Black) were tested independently. Clinical variables with significant associations (p<0.05) were included in multivariable models to quantify the adjusted effect of each SNP of interest.
Results
Patients and exemestane concentration data
Of the 500 patients enrolled on the parent clinical trial, 246 had exemestane concentrations measured either after randomization (n=225) or crossover (n=21) and 231 of these 246 patients had DNA available for genotyping. Demographic and treatment information for the 246 patients can be found in Table 1. Overall the exemestane concentration data was log-normally distributed with an untransformed median of 7.7 ng/mL and a range after censoring of 2.5–72.0 ng/mL. Twenty-six (10.6%) patients with concentration <LLOQ were censored at 2.5 ng/mL. One patient had a concentration that was an extreme outlier (72.0 ng/mL). This patient was included in all analyses and analyses were re-run excluding this outlier to assess sensitivity of the findings (range: 2.5–46.7 ng/mL).
Table 1.
Demographic data for all patients included in the analysis.
| Characteristic | n=246 | |
|---|---|---|
| Self-Reported Race | White | 220 (89.4%) |
| Black | 20 (8.1%) | |
| Other/Unknown | 6 (2.4%) | |
| Age | At enrollment | 59 (35–83) |
| Size | Body Mass Index (kg/m2) | 29.3 (20.3–53.4) |
| Renal Function | 1st Tertile (CrCl > 108.5 mL/min) | 47 (19.1%) |
| 2nd Tertile (83.0 < CrCl ≤ 108.5) | 47 (19.1%) | |
| 3rd Tertile (CrCl ≤ 83.0 mL/min) | 47 (19.1%) | |
| Unknown | 105 (42.7%) | |
| Hepatic Function | Impaired (AST or ALT > 40) | 19 (7.7%) |
| Normal (AST and ALT ≤ 40) | 225 (91.5%) | |
| Unknown | 2 (0.1%) | |
| Prior Chemotherapy Treatment | Yes | 109 (44.3%) |
| No | 137 (55.7%) | |
| Sample collection time (months on exemestane) | 1 | 41 (16.7%) |
| 3 | 205 (83.3%) | |
Data reported as count (percentage) or median (range)
Genetic and clinical predictors of exemestane concentration
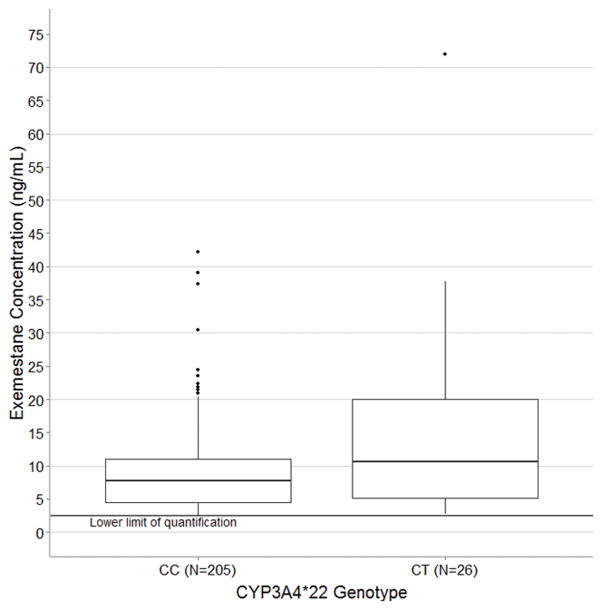
Discovery of predictors of steady-state exemestane concentration was conducted in all patients with clinical (n=246) and genetic (n=231) data, though some information was missing for some patients in these analyses. Of the 12 SNPs genotyped, one SNP failed genotyping as all patients were called heterozygous (CYP1A2*1C, rs2069514) and one was eliminated during quality control for low call rate and minor allele frequency (AKR1C3 E77G, rs11551177) (Table 2). In the primary analysis of 231 patients with genetic data the 26 patients carrying one copy of the CYP3A4*22 variant (MAF=0.06) had greater median exemestane concentration (10.7 vs. 7.7 ng/mL, relative difference = 54%, 95% Confidence interval: 14%–109%, p<0.01, Figure 1). In this cohort no patients were homozygous for the CYP3A4*22 allele. Exclusion of the outlier concentration, who was a carrier of the *22 allele, had a minor effect on the statistical association (10.5 vs. 7.7 ng/mL, difference = 43%, 95% CI: 5%–94%, p=0.02), as expected given the use of medians in the analysis, which are relatively insensitive to outliers. None of the other SNPs assessed in a secondary screening analysis were associated with exemestane concentration (all p>0.05, Table 3).
Table 2.
SNP Information and Genotyping Quality Control (n=231)
| Gene | Common Allele Name | rsID | Call Rate | MAF in this cohort | HWE p-value | Included in analysis |
|---|---|---|---|---|---|---|
| CYP3A4 | *22 | rs35599367 | 1.0 | 0.06 | 0.36 | Yes-1° |
| *1G | rs2242480 | 1.0 | 0.14 | <0.0011 | Yes-2° | |
| CYP4A11 | −845A/G | rs9332978 | 1.0 | 0.07 | 0.27 | Yes-2° |
| AKR1C3 | E77G | rs11551177 | 0.93 | 0.0022 | 0.97 | No |
| AKR1C4 | Leu311Val | rs17134592 | 1.0 | 0.14 | 0.16 | Yes-2° |
| AKR7A2 | A142T | rs1043657 | 1.0 | 0.09 | 0.88 | Yes-2° |
| CYP1A1 | 15:747248354 | rs2606345 | 1.0 | 0.37 | 0.51 | Yes-2° |
| *2 | rs4646903 | 1.0 | 0.11 | 0.67 | Yes-2° | |
| CYP1A2 | *1C | rs2069514 | 1.0 | 0.50 | <0.0013 | No |
| CYP1B1 | Arg48Gly | rs10012 | 0.99 | 0.30 | 0.57 | Yes-2° |
| Ala119Ser | rs1056827 | 1.0 | 0.30 | 0.47 | Yes-2° | |
| 2:380803674 | rs162555 | 1.0 | 0.19 | 0.30 | Yes-2° |
SNP was in HWE when assessed in each individual race (White: n=212, p=0.20, Black: n=19, p=1.0)
SNP excluded for low minor allele frequency
SNP failed genotyping (all patients called heterozygous)
Chromosomal location (Chromosome : position) using genome build GRCh38.p2 annotation release 107 from dbSNP)
1° Primary hypothesis
2° Secondary hypothesis-generating analysis
Figure 1.
Exemestane concentration stratified by CYP3A4 genotype comparing patients homozygous (CC, n=206) and heterozygous (CT, n=25) for the wild-type C allele. No patients included in this analysis were homozygous for the variant T allele. Concentrations below the LLOQ of exemestane (2.5 ng/mL) were censored at that value. Patients carrying the CYP3A4*22 allele had significantly greater median exemestane concentration (10.7 ng/mL vs. 7.7 ng/mL, p<0.01). In the box-and-whisker plot the middle line represents the median, the box represents the inter-quartile range (IQR), and the whiskers extend to 1.5 × IQR.
Table 3.
Pharmacogenetic association with exemestane concentration (n=231)
| Gene | Common Allele Name |
rsID | Homozygous Wild-type | Heterozygous | Homozygous Variant | Relative % change per variant allele (95% CI) |
Univariate additive p-value |
Adjusted1 additive p-value |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Median Concentration (range) |
N | Median Concentration (range) |
N | Median Concentration (range) |
||||||
| CYP3A4 | *22 | rs35599367 | 205 | 7.7 (2.5 – 42.2) | 26 | 10.7 (2.8 – 72.0) | 0 | -- | +54% (14%, 109%) | <0.01 | <0.01 |
| *1G | rs2242480 | 176 | 7.8 (2.5 – 72.0) | 43 | 8.1 (2.5 – 30.5) | 11 | 9.1 (2.5 – 21.8) | −4% (−20%, 15%) | 0.65 | 0.37 | |
| CYP4A11 | −845A/G | rs9332978 | 200 | 7.7 (2.5 – 72.0) | 31 | 9.8 (2.5 – 21.3) | 0 | -- | +14% (−15%, 51%) | 0.38 | 0.59 |
| AKR1C3 | E77G | rs11551177 | Excluded from analysis due to minor allele frequency = 0.002 | ||||||||
| AKR1C4 | Leu311Val | rs17134592 | 167 | 8.1 (2.5 – 72.0) | 61 | 7.5 (2.5 – 22.4) | 2 | 4.9 (3.5 – 6.3) | −8% (−25%, 13%) | 0.42 | 0.10 |
| AKR7A2 | A142T | rs1043657 | 192 | 7.7 (2.5 – 72.0) | 37 | 8.0 (2.5 – 27.5) | 2 | 22.4 (2.5 – 42.2) | −1% (−22%, 26%) | 0.94 | 0.97 |
| CYP1A1 | 15:747248352 | rs2606345 | 94 | 7.5 (2.5 – 39.0) | 103 | 8.3 (2.5 – 72.0) | 34 | 7.7 (2.5 – 24.4) | 0% (−13%, 15%) | 1.0 | 0.31 |
| *2 | rs4646903 | 183 | 7.5 (2.5 – 72.0) | 45 | 8.3 (2.5 – 30.5) | 2 | 12.3 (12.2 – 12.4) | +16% (−7%, 46%) | 0.20 | 0.24 | |
| CYP1A2 | *1C | rs2069514 | Excluded from analysis due to failed genotyping (all calls heterozygous) | ||||||||
| CYP1B1 | Arg48Gly | rs10012 | 110 | 7.8 (2.5 – 42.2) | 100 | 7.9 (2.5 – 72.0) | 19 | 7.5 (2.5 – 13.7) | −4% (−18%, 12%) | 0.59 | 0.54 |
| Ala119Ser | rs1056827 | 109 | 7.8 (2.5 – 42.2) | 102 | 7.8 (2.5 – 72.0) | 19 | 9.0 (2.5 – 13.7) | −1% (−15%, 16%) | 0.91 | 0.73 | |
| 2:380803672 | rs162555 | 153 | 7.5 (2.5 – 72.0) | 67 | 9.1 (2.5 – 37.8) | 11 | 3.7 (2.5 – 27.1) | 6% (−11%, 26%) | 0.51 | 0.57 | |
Adjustment for race, elevated aminotransferases, prior chemotherapy, and BMI in multivariable model.
Chromosomal location (Chromosome : position) using genome build GRCh38.p2 annotation release 107 from dbSNP)
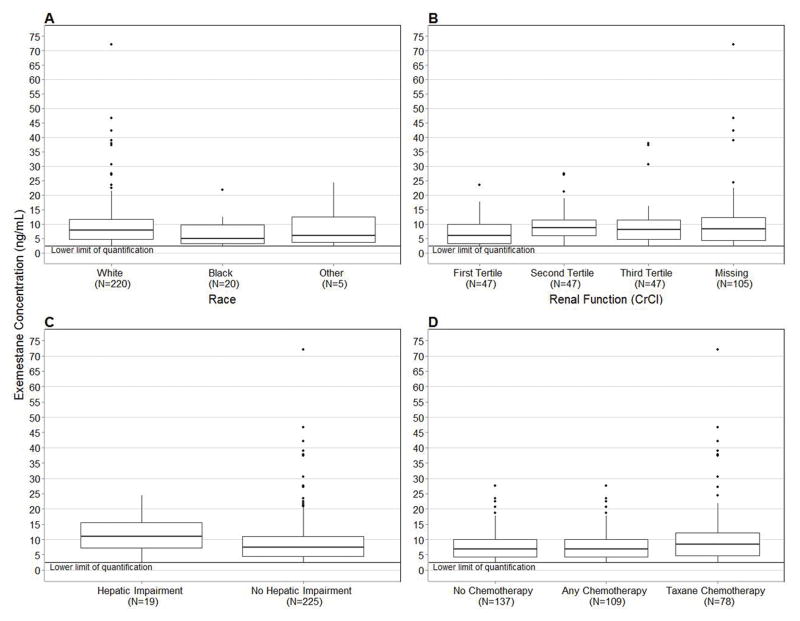
In analyses of baseline clinical variables performed in 246 patients, higher median exemestane concentrations were found in White vs. Black patients (7.9 ng/mL vs. 5.0 ng/mL, p=0.03, Figure 2A), patients in the second and third tertiles of estimated creatinine clearance (5.90 ng/mL vs. 8.60 ng/mL vs. 8.00 ng/mL, p=0.01, Figure 2B), patients who had elevated aminotransferases (11.0 ng/mL vs. 7.5 ng/mL, p=0.046, Figure 2C), patients who had not received prior chemotherapy (8.7 ng/mL vs. 6.9 ng/mL, p=0.03, Figure 2D,) and patients with lower body-mass index (1.5% decrease per unit increase in BMI, p=0.04, Table 4). For visual clarity Figure 2 excludes the high concentration outlier, a similar figure with the outlier included can be found in Supplemental Figure 1. All associations except for body-mass index (p=0.06) maintained significance in sensitivity analyses excluding the outlier.
Figure 2.
Clinical variables with significant associations with exemestane concentration. Concentrations below the LLOQ of exemestane (2.5 ng/mL) were censored at that value. 2A) Patients who self-reported as White had greater exemestane concentration compared with self-reported Black patients (7.9 ng/mL vs. 5.0 ng/mL, p=0.03). 2B) Patients in the first tertile of creatinine clearance (CrCl > 108.5 mL/min) had the lowest exemestane concentration (5.9 ng/mL vs. 8.6 ng/mL vs. 8.0 ng/mL, p=0.01). 2C) Patients with elevated aminotransferase levels (AST or ALT>40) had greater exemestane concentration (11.0 ng/mL vs. 7.5 ng/mL, p=0.05). 2D) Patients who received prior chemotherapy had lower exemestane concentration (6.9 vs. 8.7, p=0.03). In all box-and-whisker plots the middle line represents the median, the box represents the inter-quartile range (IQR), and the whiskers extend to 1.5 × IQR.
Table 4.
Association of clinical factors with exemestane concentration
| Clinical Variable | Category | Median Exemestane Concentration | Relative % Change Compared with Reference (95% Confidence Interval) | Univariate p-value | Adjusted p-value1 |
|---|---|---|---|---|---|
| Self-Reported Race | White | 7.9 ng/mL | Reference | 0.03 | 0.03 |
| Black | 5.0 ng/mL | −32.2% (−52.1%–−4.0%) | |||
| Renal Impairment | 1st Tertile (CrCl > 108.5 mL/min) | 5.9 ng/mL | Reference | 0.01 | NA |
| 2nd Tertile (83.0 < CrCl ≤ 108.5) | 8.6 ng/mL | +42.9% (8.3%–88.5%) | |||
| 3rd Tertile (CrCl ≤ 83.0 mL/min) | 8.0 ng/mL | +51.1% (14.5%–99.5%) | |||
| Elevated aminotransferases | No (AST and ALT ≤ 40) | 7.5 ng/mL | Reference | 0.05 | 0.08 |
| Yes (AST or ALT > 40) | 11.0 ng/mL | +42.9% (0.6%, 103%) | |||
| Prior chemotherapy | No | 8.7 ng/mL | Reference | 0.03 | 0.02 |
| Yes | 6.9 ng/mL | −18.6% (−33%, 2%) | |||
| BMI | Per unit increase | NA | −1.5% (−3%, 0%) | 0.04 | 0.27 |
| Age | Per year increase | NA | +0.8% (−0.4%, 2%) | 0.18 | NA |
| Exemestane Sample Collection Time | 1 month | 6.0 ng/mL | Reference | 0.07 | NA |
| 3 month | 8.0 ng/mL | −21.0% (−39%, 2%) |
Multivariable model of association for CYP3A4*22 (rs35599367) adjusted for race, elevated aminotransferases, prior chemotherapy, and BMI.
In the multivariable model, CYP3A4*22 maintained significance (p<0.01) after adjustment for significant clinical covariates (race, elevated aminotransferases, BMI, and prior chemotherapy). Estimated creatinine clearance could not be adjusted for due to substantial missing data (n=105). In the final model the only clinical variables that maintained independent significance were race and prior chemotherapy. The other 9 SNPs included in the secondary screen were not associated with exemestane concentration and were therefore not tested in multivariable models.
Discussion
In this large cohort of patients with ER positive breast cancer treated with exemestane we found that patients carrying the CYP3A4*22 SNP had significantly higher steady-state exemestane concentration 2 hours after dosing. Exemestane concentration was also higher in White patients, those with lower BMI, elevated aminotransferases and lower estimated renal function, and was, unexpectedly, lower in patients who had received prior chemotherapy. This is the first report to our knowledge of a pharmacogenetic variant that influences exemestane exposure. If exemestane concentration is predictive of treatment efficacy and/or toxicity these clinical and genetic factors could be useful for guiding individualized dosing in patients with breast cancer. As with all biomarker findings, particularly pharmacogenetic findings, this association should be replicated in independent patient cohorts to validate the association.
The intronic CYP3A4*22 (rs35599367, MAF≈5–7% in Caucasian cohorts (11)) SNP decreases CYP3A4 expression and enzymatic activity as measured by CYP3A4 probe substrates(10), and has been associated with the pharmacokinetics of several drugs(11). Due to the huge number of drugs that rely on CYP3A4-mediated metabolism, CYP3A4*22 could be clinically useful for individualizing treatment in many patients. However, this SNP only partially explains the inter-patient variability in CYP3A4 activity, which is likely dictated by a multitude of other factors including additional genetic variants (i.e. the less common inactive CYP3A4*20, MAF<1%)(20, 21), enzyme inhibition or induction due to concomitant medication, or clinical factors, such as hepatic dysfunction or age(22).
The effects of some of these additional factors on exemestane pharmacokinetics were assessed in secondary covariate analyses. Similar to previous reports, patients with elevated aminotransferases and diminished creatinine clearance at baseline had greater exemestane concentration(6). The prior study estimated that patients with hepatic impairment based on Child-Pugh grade had 2–3 fold greater exemestane exposure, as compared to our modest 20% concentration increase based on baseline elevation of AST or ALT. The patients in our study have much less severe hepatic damage than those included in the previous publication; however, our results suggest that even modest degrees of hepatic impairment can affect exemestane concentration. The previously referenced study also reported 2–3 fold greater exemestane exposure in patients with renal impairment. We again detected a more modest increase of approximately 30% in exemestane concentration for patients in the bottom two tertiles of creatinine clearance (CrCl ≤ 108.5 mL/min). Unfortunately, we could only estimate creatinine clearance for patients treated at one of the enrolling centers, leading to a substantial amount of missing data (43% of patients did not have baseline serum creatinine values), which limits the interpretability of this finding and prevents inclusion of estimated creatinine clearance in multivariate models.
Self-reported Black patients had significantly lower exemestane concentrations than White patients. The CYP3A4*22 allele has not been identified in the African cohort in the 1000 genomes project (http://browser.1000genomes.org/Homo_sapiens/Variation/Population?r=7:159138663-159138663;source=dbSNP;v=rs35599367;vdb=variation;vf=11871791). In a post-hoc secondary analysis including only self-reported White patients the CYP3A4*22 variant maintained its significant association with exemestane concentration (13.5 vs. 7.8 ng/mL, p<0.01), so this pharmacogenetic association is not due to racial confounding. Black patients could carry other genetic variants that modulate CYP3A activity that were not included in our multiplex genotyping platform. The lower concentrations in Black patients could reflect additional metabolic contribution of CYP3A5, as the wild-type CYP3A5*1 expresser genotype is more common in African-Americans than Caucasians(23), leading to greater CYP3A activity and decreased concentrations of some CYP3A substrates(24). Unfortunately, the CYP3A5*3 variant was not genotyped on this multiplex assay. Additional genotyping and further investigation of CYP3A genetic variation are planned based on these preliminary results.
It is possible that this racial difference is confounded by another variable, such as treatment adherence which may be lower in minority patients(25), or BMI which is greater in Black women(26). Patients with higher BMI had lower letrozole concentrations in our previous publication(14), as expected for drugs that are not dosed based on patient size. The clinical importance of the racial difference is also unclear. In the MA.17 trial there was no benefit of letrozole treatment compared to placebo in 352 minority patients (disease free survival hazard ratio=1.39, p=0.53), and these patients experienced significantly less toxicity than Caucasians, both of which would be consistent with subtherapeutic drug concentrations(27). The relatively small number of minority patients included in AI clinical trials and the myriad differences in patient and tumor characteristics confound the interpretation of all inter-race comparisons.
Finally, we found that patients who had received prior chemotherapy unexpectedly had lower exemestane concentration. Chemotherapy has been reported to diminish CYP2C19 metabolic activity(8), which we hypothesized was caused by hepatotoxicity that would affect all liver enzymes. In secondary exploratory analyses we did not detect an association between prior chemotherapy and any other clinical covariate that explains our finding of an effect in the opposite direction. It is conceivable that chemotherapy diminishes activity of some enzymes (i.e. CYP2C19) while inducing activity of others (i.e. CYP3A4), perhaps due to activation of enzyme-specific transcription factors such as PXR(28). However, enzyme induction is not expected to persist indefinitely, and these patients had completed chemotherapy prior to enrollment on this study, so this speculative hypothesis requires replication in independent patient cohorts and additional mechanistic validation.
The clinical relevance of discovering individualized predictors of exemestane concentration remains unclear. The FDA-approved exemestane package insert does not recommend dose adjustment for patients with hepatic or renal impairment or patients receiving concomitant CYP3A4 inhibitors, despite known increases in drug exposure. However, it is possible that long-term treatment with elevated concentration could have harmful effects that are not currently appreciated. Furthermore, the package insert does recommend dose escalation in patients on CYP3A4 inducers due to decreased drug concentrations. Patients who have inadequate exemestane exposure may not derive adequate benefit from treatment at standard doses, similar to that documented for the AI anastrozole(29). We are conducting similar analyses in this cohort to determine whether exemestane concentration is associated with estradiol depletion, treatment related toxicity, and other phenotypes that were collected on this prospective study. If any relationships are identified it will be of great interest to determine whether the clinical and genetic predictors of drug exposure identified in this analysis are predictive of these downstream clinical endpoints, and may be clinically useful for treatment individualization.
Due to the lack of available pharmacokinetic data, and the obvious clinical relevance of the phenotypes, many groups have attempted to discover genetic variants that predict efficacy and toxicity of AI treatment(30–37). Unfortunately, the vast majority of variants that have been discovered have not been successfully replicated. Our analysis used a more sensitive phenotype, drug concentration 2 hours after steady-state dosing, to interrogate SNPs with known functional consequence found in genes relevant to exemestane metabolism. Though prediction of drug exposure may not be as immediately clinically useful, this approach has been used to discover pharmacogenetic associations that can be successfully replicated prior to clinical translation(38, 39). Indeed, the clinical and genetic variables discovered in this analysis explained less than 10% of the overall variability in exemestane concentration, highlighting the sensitivity of this approach to pharmacogenetic discovery. It is possible that a minimum concentration measured just prior to dosing would have provided a more accurate phenotype to discover clinical and genetic predictors of drug elimination. Future replication efforts could consider using a trough concentration and enrolling larger numbers of patients to get a more accurate estimate of the magnitude of the association between CYP3A4*22 and exemestane pharmacokinetics, and to determine whether homozygous patients have even lower drug clearance than carriers of the low-activity allele. Furthermore, additional clinical and genetic predictors not included in this analysis may partially explain the residual variability in exemestane concentration. For example, our multiplex platform did not genotype for a deletion in UGT2B17 that has been reported to affect in vitro exemestane metabolism(40), or CYP3A5*3 discussed earlier, and we could not perform meaningful analyses of drug interactions given the small number of patients taking concomitant CYP3A4 inhibitors (n=8) or inducers (n=3).
In conclusion, post-menopausal patients with breast cancer who carry CYP3A4*22 have greater steady-state exemestane concentration. Exemestane concentration is also increased in patients with elevated aminotransferases, lower creatinine clearance, lower BMI, and possibly those who have not received prior chemotherapy. It is imperative that further research determines whether exemestane concentration is associated with efficacy or toxicity of exemestane treatment, including analysis of sensitive surrogate markers such as effective estrogen depletion, to ascertain whether the genetic and clinical predictors of drug concentration discovered in this study may have a role in individualization of AI therapy.
Supplementary Material
Acknowledgments
This research was supported by Pharmacogenetics Research Network Grant No. U-01 GM61373 (D.A.F.) and Clinical Pharmacology Training Grant No. 5T32-GM08425 (D.A.F.) from the National Institute of General Medical Sciences, National Institutes of Health (NIH), from Grants No. M01-RR000042 (University of Michigan), M01-RR00750 (Indiana University), and M01-RR00052 (Johns Hopkins University) from the National Center for Research Resources (NCRR), a component of the NIH, the Breast Cancer Research Foundation (BCRF) (N003173 to JMR and DFH), the National Cancer Institute (5T32CA083654), the National Institute of General Medical Sciences (GM099143 to J.M.R.) and the National Institutes of Health through the University of Michigan’s Cancer Center Support Grant (P30 CA046592) by the use of the following Cancer Center Core: University of Michigan DNA Sequencing Core. In addition, these studies were supported by grants from Pfizer (D.F.H.), Novartis Pharma AG (D.F.H.), the Fashion Footwear Association of New York/QVC Presents Shoes on Sale (D.F.H.). Drugs were supplied by Novartis and Pfizer.
Footnotes
This work was presented in part at the 2015 San Antonio Breast Cancer Symposium.
Conflict of Interest
This work was supported in part by Pfizer and Novartis Pharma AG. Dr. Stearns has received research funding from Abbvie, Celgene, Merck, Novartis, Medimmune, Pfizer, and Puma.
References
- 1.Coombes RC, Hall E, Gibson LJ, Paridaens R, Jassem J, Delozier T, et al. A Randomized Trial of Exemestane after Two to Three Years of Tamoxifen Therapy in Postmenopausal Women with Primary Breast Cancer. N Engl J Med. 2004 Mar 11;350(11):1081–92. doi: 10.1056/NEJMoa040331. 2015/03. [DOI] [PubMed] [Google Scholar]
- 2.Pagani O, Regan MM, Walley BA, Fleming GF, Colleoni M, Láng I, et al. Adjuvant Exemestane with Ovarian Suppression in Premenopausal Breast Cancer. N Engl J Med. 2014 Jul 10;371(2):107–18. doi: 10.1056/NEJMoa1404037. 2015/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henry NL, Azzouz F, Desta Z, Li L, Nguyen AT, Lemler S, et al. Predictors of aromatase inhibitor discontinuation as a result of treatment-emergent symptoms in early-stage breast cancer. J Clin Oncol. 2012 Mar 20;30(9):936–42. doi: 10.1200/JCO.2011.38.0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henry NL, Giles J, Ang D, Mohan M, Dadabhoy D, Robarge J, et al. Prospective characterization of musculoskeletal symptoms in early stage breast cancer patients treated with aromatase inhibitors. Breast Cancer Res Treat. 2008 Sep 01;111(2):365–72. doi: 10.1007/s10549-007-9774-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Dowsett M, Forbes JF, Bradley R, Ingle J, Aihara T, et al. Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet. 2015 Oct 3;386(10001):1341–52. doi: 10.1016/S0140-6736(15)61074-1. [DOI] [PubMed] [Google Scholar]
- 6.Jannuzzo MG, Poggesi I, Spinelli R, Rocchetti M, Cicioni P, Buchan P. The effects of degree of hepatic or renal impairment on the pharmacokinetics of exemestane in postmenopausal women. Cancer Chemother Pharmacol. 2004 Jun;53(6):475–81. doi: 10.1007/s00280-004-0774-5. [DOI] [PubMed] [Google Scholar]
- 7.King PD, Perry MC. Hepatotoxicity of Chemotherapy. The Oncologist. 2001 Apr 01;6(2):162–76. doi: 10.1634/theoncologist.6-2-162. [DOI] [PubMed] [Google Scholar]
- 8.Helsby NA, Lo WY, Sharples K, Riley G, Murray M, Spells K, et al. CYP2C19 pharmacogenetics in advanced cancer: compromised function independent of genotype. Br J Cancer. 2008 Oct 21;99(8):1251–5. doi: 10.1038/sj.bjc.6604699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamdem LK, Flockhart DA, Desta Z. In vitro cytochrome P450-mediated metabolism of exemestane. Drug Metab Dispos. 2011 Jan;39(1):98–105. doi: 10.1124/dmd.110.032276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elens L, Nieuweboer A, Clarke SJ, Charles KA, de Graan AJ, Haufroid V, et al. CYP3A4 intron 6 C>T SNP (CYP3A4*22) encodes lower CYP3A4 activity in cancer patients, as measured with probes midazolam and erythromycin. Pharmacogenomics. 2013 Jan;14(2):137–49. doi: 10.2217/pgs.12.202. [DOI] [PubMed] [Google Scholar]
- 11.Elens L, van Gelder T, Hesselink DA, Haufroid V, van Schaik RH. CYP3A4*22: promising newly identified CYP3A4 variant allele for personalizing pharmacotherapy. Pharmacogenomics. 2013 Jan;14(1):47–62. doi: 10.2217/pgs.12.187. [DOI] [PubMed] [Google Scholar]
- 12.Diekstra MH, Klumpen HJ, Lolkema MP, Yu H, Kloth JS, Gelderblom H, et al. Association analysis of genetic polymorphisms in genes related to sunitinib pharmacokinetics, specifically clearance of sunitinib and SU12662. Clin Pharmacol Ther. 2014 Jul;96(1):81–9. doi: 10.1038/clpt.2014.47. [DOI] [PubMed] [Google Scholar]
- 13.Teft WA, Gong IY, Dingle B, Potvin K, Younus J, Vandenberg TA, et al. CYP3A4 and seasonal variation in vitamin D status in addition to CYP2D6 contribute to therapeutic endoxifen level during tamoxifen therapy. Breast Cancer Res Treat. 2013 May;139(1):95–105. doi: 10.1007/s10549-013-2511-4. [DOI] [PubMed] [Google Scholar]
- 14.Desta Z, Kreutz Y, Nguyen AT, Li L, Skaar T, Kamdem LK, et al. Plasma letrozole concentrations in postmenopausal women with breast cancer are associated with CYP2A6 genetic variants, body mass index, and age. Clin Pharmacol Ther. 2011 Nov;90(5):693–700. doi: 10.1038/clpt.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henry NL, Jacobson JA, Banerjee M, Hayden J, Smerage JB, Van Poznak C, et al. A prospective study of aromatase inhibitor-associated musculoskeletal symptoms and abnormalities on serial high-resolution wrist ultrasonography. Cancer. 2010 Sep 15;116(18):4360–7. doi: 10.1002/cncr.25385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hertz DL, Kidwell KM, Thibert JN, Gersch C, Regan MM, Skaar TC, et al. Genotyping concordance in DNA extracted from formalin-fixed paraffin embedded (FFPE) breast tumor and whole blood for pharmacogenetic analyses. Mol Oncol. 2015 Nov;9(9):1868–76. doi: 10.1016/j.molonc.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tobin J. Estimation of Relationships for Limited Dependent Variables. Econometrica. 1958 Jan 01;26(1):24–36. [Google Scholar]
- 18.Hillis SL. Residual plots for the censored data linear regression model. Stat Med. 1995 Sep 30;14(18):2023–36. doi: 10.1002/sim.4780141808. [DOI] [PubMed] [Google Scholar]
- 19.Cockcroft DW, Gault MH. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16(1):31–41. doi: 10.1159/000180580. [DOI] [PubMed] [Google Scholar]
- 20.Werk AN, Cascorbi I. Functional gene variants of CYP3A4. Clin Pharmacol Ther. 2014 Sep;96(3):340–8. doi: 10.1038/clpt.2014.129. [DOI] [PubMed] [Google Scholar]
- 21.Westlind-Johnsson A, Hermann R, Huennemeyer A, Hauns B, Lahu G, Nassr N, et al. Identification and characterization of CYP3A4*20, a novel rare CYP3A4 allele without functional activity. Clin Pharmacol Ther. 2006 Apr;79(4):339–49. doi: 10.1016/j.clpt.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 22.Wilkinson GR. Drug Metabolism and Variability among Patients in Drug Response. N Engl J Med. 2005 May 26;352(21):2211–21. doi: 10.1056/NEJMra032424. 2015/10. [DOI] [PubMed] [Google Scholar]
- 23.Lee S, Goldstein JA. Functionally defective or altered CYP3A4 and CYP3A5 single nucleotide polymorphisms and their detection with genotyping tests. Pharmacogenomics. 2005 Jul 01;6(4):357–71. doi: 10.1517/14622416.6.4.357. 2013/01. [DOI] [PubMed] [Google Scholar]
- 24.Sanghavi K, Brundage RC, Miller MB, Schladt DP, Israni AK, Guan W, et al. Genotype-guided tacrolimus dosing in African-American kidney transplant recipients. Pharmacogenomics J. 2015 Dec 15; doi: 10.1038/tpj.2015.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawesi S, Carpenter JS, Jones J. Reasons for nonadherence to tamoxifen and aromatase inhibitors for the treatment of breast cancer: a literature review. Clin J Oncol Nurs. 2014 Jun;18(3):E50–7. doi: 10.1188/14.CJON.E50-E57. [DOI] [PubMed] [Google Scholar]
- 26.Flegal KM, Carroll MD, Ogden CL, Johnson CL. Prevalence and trends in obesity among US adults, 1999–2000. JAMA. 2002 Oct 9;288(14):1723–7. doi: 10.1001/jama.288.14.1723. [DOI] [PubMed] [Google Scholar]
- 27.Moy B, Tu D, Pater JL, Ingle JN, Shepherd LE, Whelan TJ, et al. Clinical outcomes of ethnic minority women in MA.17: a trial of letrozole after 5 years of tamoxifen in postmenopausal women with early stage breast cancer. Ann Oncol. 2006 Nov;17(11):1637–43. doi: 10.1093/annonc/mdl177. [DOI] [PubMed] [Google Scholar]
- 28.Harmsen S, Meijerman I, Beijnen JH, Schellens JHM. Nuclear receptor mediated induction of cytochrome P450 3A4 by anticancer drugs: a key role for the pregnane X receptor. Cancer Chemother Pharmacol. 2009 Jun 01;64(1):35–43. doi: 10.1007/s00280-008-0842-3. [DOI] [PubMed] [Google Scholar]
- 29.Ingle JN, Kalari KR, Buzdar AU, Robson ME, Goetz MP, Desta Z, et al. Estrogens and their precursors in postmenopausal women with early breast cancer receiving anastrozole. Steroids. 2015 Jul;99(Pt A):32–8. doi: 10.1016/j.steroids.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ingle JN, Schaid DJ, Goss PE, Liu M, Mushiroda T, Chapman JA, et al. Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. J Clin Oncol. 2010 Nov 1;28(31):4674–82. doi: 10.1200/JCO.2010.28.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry NL, Skaar TC, Dantzer J, Li L, Kidwell K, Gersch C, et al. Genetic associations with toxicity-related discontinuation of aromatase inhibitor therapy for breast cancer. Breast Cancer Res Treat. 2013 Apr;138(3):807–16. doi: 10.1007/s10549-013-2504-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontein DB, Houtsma D, Nortier JW, Baak-Pablo RF, Kranenbarg EM, van der Straaten TR, et al. Germline variants in the CYP19A1 gene are related to specific adverse events in aromatase inhibitor users: a substudy of Dutch patients in the TEAM trial. Breast Cancer Res Treat. 2014 Apr;144(3):599–606. doi: 10.1007/s10549-014-2873-2. [DOI] [PubMed] [Google Scholar]
- 33.Colomer R, Monzo M, Tusquets I, Rifa J, Baena JM, Barnadas A, et al. A Single-Nucleotide Polymorphism in the Aromatase Gene Is Associated with the Efficacy of the Aromatase Inhibitor Letrozole in Advanced Breast Carcinoma. Clin Cancer Res. 2008 Feb 1;14(3):811–6. doi: 10.1158/1078-0432.CCR-07-1923. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Casado Z, Guerrero-Zotano A, Llombart-Cussac A, Calatrava A, Fernandez-Serra A, Ruiz-Simon A, et al. A polymorphism at the 3′-UTR region of the aromatase gene defines a subgroup of postmenopausal breast cancer patients with poor response to neoadjuvant letrozole. BMC Cancer. 2010 Feb 9;10:36. doi: 10.1186/1471-2407-10-36. 2407-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferraldeschi R, Arnedos M, Hadfield KD, A’Hern R, Drury S, Wardley A, et al. Polymorphisms of CYP19A1 and response to aromatase inhibitors in metastatic breast cancer patients. Breast Cancer Res Treat. 2012 Jun;133(3):1191–8. doi: 10.1007/s10549-012-2010-z. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Bai YX, Zhou JH, Sun XW, Sui H, Zhang WJ, et al. A polymorphism at the 3′-UTR region of the aromatase gene is associated with the efficacy of the aromatase inhibitor, anastrozole, in metastatic breast carcinoma. Int J Mol Sci. 2013 Sep 13;14(9):18973–88. doi: 10.3390/ijms140918973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Ellsworth KA, Moon I, Pelleymounter LL, Eckloff BW, Martin YN, et al. Functional genetic polymorphisms in the aromatase gene CYP19 vary the response of breast cancer patients to neoadjuvant therapy with aromatase inhibitors. Cancer Res. 2010 Jan 1;70(1):319–28. doi: 10.1158/0008-5472.CAN-09-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramsey LB, Panetta JC, Smith C, Yang W, Fan Y, Winick NJ, et al. Genome-wide study of methotrexate clearance replicates SLCO1B1. Blood. 2013 Feb 07;121(6):898–904. doi: 10.1182/blood-2012-08-452839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hesselink DA, van Schaik RH, van der Heiden IP, van der Werf M, Gregoor PJ, Lindemans J, et al. Genetic polymorphisms of the CYP3A4, CYP3A5, and MDR-1 genes and pharmacokinetics of the calcineurin inhibitors cyclosporine and tacrolimus. Clin Pharmacol Ther. 2003 Sep;74(3):245–54. doi: 10.1016/S0009-9236(03)00168-1. [DOI] [PubMed] [Google Scholar]
- 40.Sun D, Chen G, Dellinger RW, Sharma AK, Lazarus P. Characterization of 17-dihydroexemestane glucuronidation: potential role of the UGT2B17 deletion in exemestane pharmacogenetics. Pharmacogenet Genomics. 2010 Oct;20(10):575–85. doi: 10.1097/FPC.0b013e32833b04af. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.