Abstract
AIM
To evaluate the role of biofilm formation on the resistance of Helicobacter pylori (H. pylori) to commonly prescribed antibiotics, the expression rates of resistance genes in biofilm-forming and planktonic cells were compared.
METHODS
A collection of 33 H. pylori isolates from children and adult patients with chronic infection were taken for the present study. The isolates were screened for biofilm formation ability, as well as for polymerase chain reaction (PCR) reaction with HP1165 and hp1165 efflux pump genes. Susceptibilities of the selected strains to antibiotic and differences between susceptibilities of planktonic and biofilm-forming cell populations were determined. Quantitative real-time PCR (qPCR) analysis was performed using 16S rRNA gene as a H. pylori-specific primer, and two efflux pumps-specific primers, hp1165 and hefA.
RESULTS
The strains were resistant to amoxicillin, metronidazole, and erythromycin, except for one strain, but they were all susceptible to tetracycline. Minimum bactericidal concentrations of antibiotics in the biofilm-forming cells were significantly higher than those of planktonic cells. qPCR demonstrated that the expression of efflux pump genes was significantly higher in the biofilm-forming cells as compared to the planktonic ones.
CONCLUSION
The present work demonstrated an association between H. pylori biofilm formation and decreased susceptibility to all the antibiotics tested. This decreased susceptibility to antibiotics was associated with enhanced functional activity of two efflux pumps: hp1165 and hefA.
Keywords: Helicobacter pylori, Biofilm, Antibiotic resistance, Efflux genes, hp1165, hefA
Core tip: The current study has addressed the co-incidence of biofilm formation by Helicobacter pylori (H. pylori) and decreased susceptibility to antibiotics. The results demonstrated a significantly higher expression of two efflux pump genes, hp1165 and hefA (involved in the specific resistance to tetracycline and multidrug resistance, respectively), in the biofilm-forming cells as compared to the planktonic cells. There was also association between H. pylori biofilm formation and decreased susceptibility to antibiotics. This event would probably be involved in the failure of H. pylori eradication and might be beneficial for developing new therapeutic approaches for this infection.
INTRODUCTION
Helicobacter pylori (H. pylori) colonizes the stomach of approximately half of the world’s population[1]. Gastritis may be the first manifestation of stomach colonization by H. pylori that can progress to the fatal states such as peptic ulceration and gastric adenocarcinoma. Although a low number of H. pylori-infected individuals can develop the serious symptoms of infection, in all cases non-treatment with appropriate antibiotics may result in a persistent infection, which can favor progression to the fatal outcomes[2].
The current treatment regimen of H. pylori infection includes a proton pump inhibitor in combination with two out of the following antibiotics: amoxicillin (AMX), metronidazole (MTZ), clarithromycin and tetracycline (TET)[3]. However, data from various geographical areas indicate that resistance to these antibiotics debilitates the treatment of H. pylori infections[3,4].
Bacterial biofilms are communities of individual cells that form microcolonies, grow in the third dimension and develop the phenotypic and biochemical properties which may differ from those of the corresponding freely existing (planktonic) cells[5,6]. Gene expression profiles in biofilms may, therefore, also be different from those of the planktonic cells[7]. The investigators have observed that biofilm formation by numerous pathogenic bacteria may be involved in increased rate of antibiotic resistance[8].
In practice, biofilm-forming cells may be observed on the surfaces of tissues and biomaterials at the site of persistent infections, where they can be protected from the killing effects of antibiotics[9]. Hence, during long-term infection, biofilm formation by pathogenic bacteria may play an important role in the emergence of resistant bacteria and can increase the rates of antibiotic resistance.
To date, the mechanisms of higher resistance to antibiotics in biofilm-forming cells are not fully understood. However, poor antibiotic penetration, adaptive stress responses due to expression of specific genes in the biofilm, slow growth rate, and formation of persister cells (which form multi-layers) may be the causes of higher resistance rates in biofilms[10,11].
Resistance mechanisms of H. pylori to macrolides, nitroimidazole, β-lactams, and TET have been reported to occur via mutations in the peptidyltransferase region encoded in 23S rRNA, different mutations involving the rdxA gene, multiple mutations in the penicillin binding protein “pbp gene” as well as production of β-lactamase, and mutations in the 16S rRNA gene, respectively[12]. Involvement of the efflux pumps may also be one of the important reasons for decreased susceptibility of H. pylori to various antibiotics[13-17]. Numerous and structurally different efflux pumps have been identified in the H. pylori 26695 genome, capable of acting either on the structurally non-related antibiotics or more specifically on one antibiotic[13-15]. For this purpose, investigators have studied the roles of a few putative efflux pumps in the resistance of H. pylori to antibiotics[16]. Liu et al[17], for example, have observed the role of hefA on the multidrug resistance of H. pylori and have demonstrated the role of Hp1165 in TET resistance.
To gain a better understanding of biofilm formation’s role in the resistance of H. pylori to commonly prescribed antibiotics, in the present study, the resistance profiles of biofilm-forming cells and their planktonic counterparts were evaluated and the expression of the two efflux pumps genes, hp1165 and hefA, were compared between biofilm-forming and planktonic cells.
MATERIALS AND METHODS
H. pylori strains and culture condition
A collection of 33 clinical H. pylori strains isolated from children and adult patients with chronic infections were examined for biofilm formation ability. The protocols, under which the biopsies for culture had been obtained, were in accordance with the Helsinki Declaration of 1975. The isolated strains were cultured at 37 °C under microaerobic atmosphere on modified Campy blood agar (MCBA) plates containing Brucella agar base (Merck, Germany), supplemented with 7% defibrinated sheep blood and antibiotics (polymyxin B, amphotericin B, vancomycin) for 3-7 d. The resulting colonies were identified by Gram staining, biochemical tests and polymerase chain reaction (PCR) using H. pylori-specific primers for glmM, as previously described by Espinoza et al[18].
Screening of the strains for biofilm formation ability
The strains were screened for biofilm formation ability according to our previously described protocol[19]. Briefly, bacteria were inoculated in Brucella broth (Biolife, Italy) supplemented with fetal calf serum (2%) and glucose (0.3%) (Merck). Suspensions were incubated at 37 °C under microaerobic atmosphere with shaking (100 rpm), to an optical density (OD) of 0.2 at 600 nm (A600) equivalent to approximately 5-8 × 103 CFU/mL. Portions of these cultures were inoculated into the wells of 96-well flat-bottomed tissue culture plates (BIOFIL, Jet Bio-Filtration Products Co., Ltd, China) and were incubated at 37 °C under microaerobic conditions for 6 d. The wells were vigorously washed (3 times) with sterile phosphate buffered saline (PBS). Tightly attached bacteria were fixed with 99% ethanol (200 μL per well) for 20 min, and air-dried. Plates were then stained with 1% crystal violet (200 μL per well) for 5 min and the excess of stain was rinsed off with running tap water. Dried plates were treated with 33% glacial acetic acid (160 μL per well) and the OD of the wells was measured at 505 nm using an ELISA reader (SCO GmbH, Germany). A culture medium (Brucella broth) without bacterial cells was used as negative control.
Antibiotics
Four classes of the antibiotics were selected for present study: MTZ as a nitromidazole (Sigma, United States), ERY as a macrolide (Sigma), AMX as a β-lactam (Shafa Pharmaceutical Co., Iran), and TET as a tetracycline (Sigma).
The stock solutions of MTZ (128 μg/mL) and AMX (200 μg/mL) were prepared by addition of acetic acid and dimethyl sulfoxide, respectively; those of TET (128 μg/mL) and ERY (128 μg/mL) were obtained by addition of ethanol.
Antibiotic susceptibility testing
The minimal inhibitory concentration (MIC) was determined using agar dilution method in accordance with the guidelines established by the Clinical and Laboratory Standards Institute[20]. For this purpose, Mueller-Hinton agar (MH) plates containing 7% fresh sheep blood were supplemented with increasing concentration of antibiotics. A bacterial suspension adjusted to number 2 of McFarland standard, corresponding to 108-2 × 108 CFU/mL was inoculated onto the agar dilution plates. After spreading the bacteria, the plates were incubated under microaerobic conditions for 72 h. Quality control for the antibiotics was ensured using Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 strains. MIC breakpoints for resistance were defined as follows: MTZ: 8 mg/mL; ERY: 1.0 mg/mL; AMX: > 0.5 mg/mL; TET: 8 mg/mL[21,22].
Comparison of antibiotic susceptibilities between biofilm-forming and planktonic cell populations
The susceptibilities of biofilm-forming cells (group B) and planktonic cells (group P) to the antibiotics were evaluated by comparing the minimal bactericidal concentration (MBC). For this purpose, the 96-well culture plates were inoculated with 100 μL of bacterial suspension containing 5-8 × 103 CFU/mL according to the protocol described for biofilm formation. After biofilm formation, the planktonic cells were removed for further MBC determination. The remaining biofilm forming cells were washed (3 times) and exposed to 2-fold increasing concentrations of the antibiotics (from 0.5 μg/mL to 64 μg/mL) for 24 h. After the incubation period, the cells were removed via an ultrasonic bath for 7 min (Elmasonic S 60/ (H); Elma Schmidbauer GmbH, Germany) using ultrasonic frequency of 37 kHz, and cultured on MCBA plates[10]. To determine the susceptibility of planktonic cells, similar concentrations of the antibiotics were added to the 96-well plates containing 5-8 × 103 CFU/mL planktonic bacteria (previously removed from the plates after biofilm formation) and incubated for 24 h. Their antibiotic susceptibility was determined by culturing a small amount (10 μL) of bacterial suspension on the MCBA plates. The plates were then incubated at 37 °C under microaerobic conditions for > 72 h incubation. Test controls were similar culture plates containing bacteria but without antibiotics. The results presented for this analysis correspond to the mean of 3 independent tests.
Determination of minimal biofilm inhibitory concentration
By this assay, minimal concentration of the antibiotics which inhibited formation of the biofilms on plates was determined. For this purpose, similar serial dilutions of the antibiotics noted above were made in the culture medium. The microplates were seeded with these serial dilutions of antibiotics and were inoculated with 100 μL of bacterial suspension containing 108-2 × 108 CFU/mL. After incubation, the plates were vigorously washed (3 times) with sterile PBS, in order to remove all non-adherent bacteria, and then incubated with Brucella broth supplemented with 0.05% (w/v) of 2,3,5-triphenyl tetrazolium chloride (TTC; Merck) at 37 °C under microaerobic conditions for 24 h. The control assay was performed with the same medium but without bacteria. After incubation, the broth was removed, the wells were air-dried and the bound TTC dyes were dissolved using 20% acetone/80% ethanol. To quantify the biofilm formation, absorbance (OD) of the wells was measured at 505 nm[23].
Detection of 16S rRNA, hp1165, and hefA genes by PCR
H. pylori specific primers for 16S rRNA and glmM, were designed according to the previously described protocol[24]. Efflux pump-specific primers for hp1165 was designed according to hp1165 gene sequences using Primer3 software online, and efflux-specific primers for hefA were designed according to Zhang et al[25]. All primer sequences, including that of glmM, that were used for bacterial identification process are listed in Table 1.
Table 1.
Polymerase chain reaction primers
| Primer name | Sequence | Product size, bp |
| glm-F | 5' GGATAAGCTTTTAGGGGTGTTAGGGG 3' | 140 |
| glm-R | 5' GCATTCACAAACTTATCCCCAATC 3' | |
| 16s rRNA-F | 5' GAAGATAATGACGGTATCTAAC 3' | 139 |
| 16s rRNA-R | 5' ATTTCACACCTGACTGACTAT 3' | |
| hp1165-F | 5' F; TACAACCCCCACGCTAAAAG 3' | 117 |
| hp1165-R | 5' GGATTTGATGAGCCGAAAAA 3' | |
| hefA-F | 5' CTCGCTCGCATGATCGC 3' | 162 |
| hefA-R | 5' CGTATTCGCTCAAATTCCCT 3' |
DNA was extracted using boiling method according to the previously described protocol[26]. PCR reactions were performed in a 25 mL mixture containing 0.5 μg of extracted DNA, 0.2 mmol (each) deoxyribonucleoside triphosphates, 0.2-0.4 μmol/L (each) primer, 1.5-2 mmol/L MgCl2, and 5 U of Taq polymerase in PCR buffer (SinaClon BioScience, Iran). Following denaturation at 94 °C (2 min), the fragments were amplified through 35 cycles at 94 °C (30 s). Annealing for 16S rRNA gene, hp1165 and hefA was performed at 58 °C, 59 °C and 59.5 °C respectively. Reactions were continued at 72 °C for 1 min and 72 °C for 10 min.
Quantitative real-time reverse transcription-PCR analysis
Biofilms were removed via an ultrasonic bath for 7 min. RNA was extracted from the planktonic and biofilm-forming cell cultures. For this purpose and following bacterial wash with PBS (× 3), total RNA extraction was carried out using a commercial Cinna Pure RNA kit (SinaClon BioScience). The RNA samples were then treated with DNase I (Roche Diagnostics GmbH, Germany) according to the manufacturer's recommendations. Reverse transcription (RT) was carried out with an RT-PCR Kit (Vivantis Technologies, Malaysia) according to the company's recommendations for using 2 ng RNA sample. Real-time RT-PCR was performed for the cDNA samples using 16S rRNA-specific primers and efflux pump-specific primers, including hp1165 and hefA[25]. Real-time PCR was performed with the QuantiFast SYBR Green PCR Kit (Qiagen, Germany) in a StepOne Real-time PCR system (Applied Biosystems, United States). The PCR reaction was held at 95 °C for 5 min, followed by 2- step cycling consisting of 40 cycles at 95 °C for 10 s and 60 °C for 30 s.
The final results were expressed as the amount of expression for each efflux pump gene relative to that of the 16S rRNA gene. We selected 16S rRNA for control of the quantitative real-time RT-PCR (qPCR) analysis since its expression does not change in biofilm-forming and planktonic cells in the RNA samples.
Statistical analyses were carried out using the ANOVA one-way test with Minitab 17 statistical software, and probability levels of < 0.05 were considered as statistically significant.
RESULTS
Selection of strains
Screening of clinical isolates for biofilm production demonstrated their variable ability for biofilm formation, such that their biofilm formation abilities could be classified as high (one strain), moderate (23 strains), and low (six strains). The strains were also screened for detection of two efflux genes, hp1165 and hefA, by PCR. The PCR results were then analyzed according to production of the sharp bands on gels. To select the most suitable strains for further experiments, beside higher biofilm formation ability and PCR result, their subculture states (none more than three laboratory passages) were also taken into account. PCR results from three selected strains (Hp141, Hp932 and Hp70) for hp1165, hefA, and 16Sr RNA genes which demonstrated moderate biofilm formation ability are shown in Figure 1.
Figure 1.
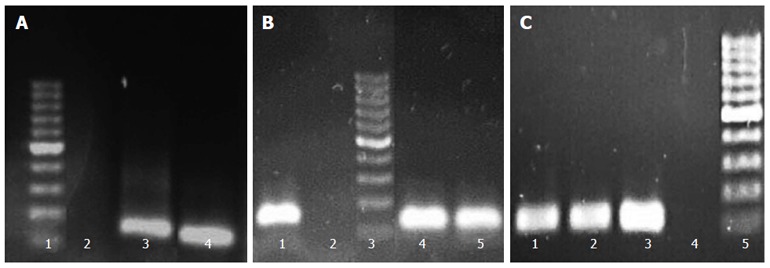
Polymerase chain reaction products of hefA (162 bp), hp1165 (117 bp) and 16S rRNA genes on 1.5% agarose gel. A: Detection of hefA gene: 1: 100 bp size marker, 2: Control, 3: Hp932 strain, 4: Hp141 strain; B: Detection of hp1165 gene: 1: HP70 strain, 2: Control, 3: Size marker, 4: Hp932 strain, 5: Hp141 strain; C: Detection of 16S rRNA gene: 1: Hp932 strain, 2: Hp141 strain, 3: Hp70 strain, 4: Control, 5: Size marker.
Determination of MIC by agar dilution
All three selected isolates were resistant to AMX, MTZ and ERY, except one. All three were susceptible to TET. MICs of AMX for Hp932, Hp141 and Hp70 were 2, 2 and 1 μg/mL, respectively. The MIC of TET was 4 μg/mL for all the three isolates. MICs of ERY for Hp141, Hp932 and Hp70 were 2, 2 and 1 μg/mL, respectively. MICs of MTZ for Hp932, Hp141 and Hp70 were 16, 8 and 8 μg/mL, respectively.
Comparison of antibiotic resistance between biofilm-forming and planktonic cell populations
For this comparison, MBCs were selected as the better criteria. The results of comparison between MBC of biofilm-forming cells and their planktonic counterparts are demonstrated in Table 2. We noted that the MBCs to all the antibiotics were increased significantly in biofilm-forming cells as compared to those of planktonic ones.
Table 2.
Minimal bactericidal concentration of selected antibiotics in biofilm-forming cells and their planktonic counterparts
| Strain |
MBC, μg/mL |
|||||||
|
AMX |
TET |
ERY |
MTZ |
|||||
| MBC-P | MBC-B | MBC-P | MBC-B | MBC-P | MBC-B | MBC-P | MBC-B | |
| Hp932 | 2 | 8 | 8 | 16 | 4 | 4 | 16 | 32 |
| Hp141 | 2 | 4 | 4 | 16 | 2 | 4 | 16 | 32 |
| Hp70 | 1 | 4 | 4 | 8 | 2 | 4 | 8 | 16 |
MBC: Minimal bactericidal concentration; AMX: Amoxicillin; TET: Tetracycline; MTZ: Metronidazole; B: Biofilm-forming; P: Planktonic counterparts.
Determination of minimal biofilm inhibitory concentration
We observed that the minimal biofilm inhibitory concentrations (MBICs) of AMX required for preventing biofilm formation was similar to that of the MICs for all three isolates. However, MBICs of TET, ERY and MTZ were at least 2-folds lower than the MICs of the isolates determined in agar dilution (Table 3).
Table 3.
Minimal biofilm inhibitory concentration
| Strain |
MBIC, μg/mL |
|||
| AMX | TET | ERY | MTZ | |
| Hp932 | 2 | 2 | 1 | 8 |
| Hp141 | 2 | 2 | 1 | 4 |
| Hp70 | 1 | 1 | 0.5 | 4 |
MBIC: Minimal biofilm inhibitory concentration; AMX: Amoxicillin; TET: Tetracycline; MTZ: Metronidazole.
Expression of hp1165 and hefA in biofilm-forming and planktonic cells
The differences in transcription levels of hefA and hp1165 efflux genes between biofilm-forming and planktonic cells, using hp16S rRNA as internal control, were compared. For this purpose, the quantities of cDNA corresponding to these genes were determined by real-time qPCR and their amounts were normalized using 16S rRNA gene in each unique reaction. Each experiment was repeated 3 times with at least duplicate samples from independently isolated RNA preparations. Data are expressed as the means of all experiments ± standard error. The results of qPCR products for hp1165 (117 bp), hefA (162 bp), and 16S rRNA (136 bp) genes in biofilm-forming (group B) and planktonic (group P) populations on 1.5% agarose gel are demonstrated in Figure 2. The results displaying the amounts of expression for each efflux pump gene relative to that of the 16S rRNA gene are demonstrated in Figure 3. We noted that expression of these genes was significantly higher in the biofilm-forming cells as compared to their planktonic counterparts.
Figure 2.
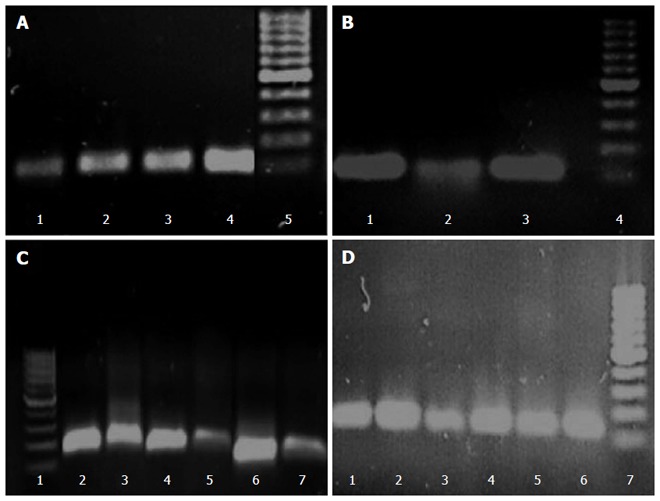
Real-time reverse transcription-polymerase chain reaction products of hp1165 (117 bp), hefA (162 bp), and 16S rRNA (136 bp) genes in biofilm-forming (B) and planktonic (P) populations on 1.5% agarose gel. A: Detection of hp1165 gene: 1: Hp141 strain (B), 2: Hp141 strain (P), 3: Hp932 strain (B), 4: Hp932 strain (P), 5: 100 bp size marker; B: Detection of hp1165 gene: 1-2: Hp70 strain (B), 3: Hp70 strain (P), 4: Size marker; C: Detection of hefA gene: 1: Size marker, 2: Hp932 strain (B), 3: Hp932 strain (P), 4: Hp141 strain (B), Hp141 strain (P), 5: Hp70 strain (B), 6: Hp70 strain (P); D: Detection of 16S rRNA gene: 1: Hp932 strain (B), 2: Hp932 strain (P), 3: Hp141 strain (B), 4: Hp141 strain (P), 5: Hp70 strain (B), 6: Hp70 strain (P), 7: Size marker.
Figure 3.
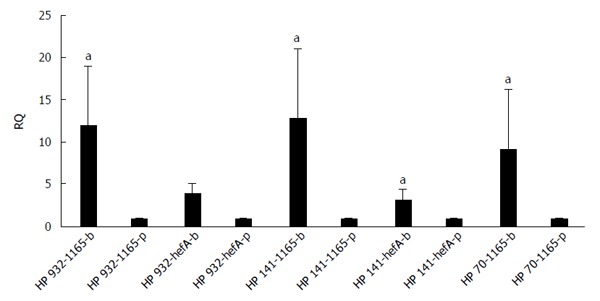
Expression rates of Helicobacter pylori efflux pump genes, hefA and hp1165. Data are expressed as the means of multiple experiments ± standard deviation. Significantly different (aP ≤ 0.05) relative to the mRNA expression level (planktonic vs biofilm-forming).
DISCUSSION
Long-term infection of humans by H. pylori may favor the possibilities of biofilm formation in the stomach, which might be a barrier for antibiotic capture, thereby increasing resistance to antibiotic[11].
Several investigators have reported that H. pylori can form the biofilm in vitro and most probably in vivo[27-31]. In light of these investigations, a few scientific observations, including that of Yonezawa et al[29], have reported in vitro biofilm formation by H. pylori strains.
In a previous study, we investigated the role of biofilm formation on H. pylori colonization of the mouse model[19]. In the present work, we investigated its role in the resistance to commonly used antibiotics by comparing the expression rates of two efflux pumps between the biofilm-forming cells and their planktonic counterparts.
To observe the effectiveness of antibiotics against planktonic and biofilm-forming cells, we compared their MBCs, instead of MICs, since it was demonstrated that planktonic cells are more susceptible to antibiotics and the eventual presence of planktonic cells (even at low density) in established biofilms may not result in a fair MIC comparison between biofilm-forming and planktonic cells[32]. Furthermore, the possibility exists that in such an evaluation, instead of MIC comparison, the MIC of planktonic cells be compared with the MBIC of biofilm-forming cells. This comparison may result in an erroneous conclusion since MBIC and MIC may not be similar for the majority of the antibiotics. In agreement with this situation, we found that MBIC of selected H. pylori isolates for TET, ERY and MTZ were at least 2-folds lower than their MIC (Table 3). Using MBC as a criterion of comparison, the MBC of biofilm-forming cells were at least 2-folds higher than their planktonic counterparts (Table 2).
Expression analysis of mRNA for two efflux pump genes, hp1165 and hefA, involved in the specific resistance to TET and multidrug resistance, respectively, showed that they were active in both the biofilm-forming and planktonic cell populations (Figure 2). By analyzing the amount of expression, we observed that both efflux genes had significantly increased expression in biofilm-forming cells, compared to their planktonic counterparts (Figure 3).
Impact of biofilm formation by H. pylori on antibiotic resistance was also studied by Yonezawa and coworkers[33], who found a decreased susceptibility to clarithromycin via increased frequency of mutations in the biofilm-forming cells.
Although the actual treatment regimen for eradication of H. pylori infection may be effective in some cases, the failure of this approach is well documented in many cases[34]. The main reasons for unsuccessful eradication appear to be the emergence of resistant strains or resistant populations to selected antibiotics. Biofilm-forming populations may develop multiple functional genes which favor more resistance to antibiotics, while planktonic cells do not. Involvement of the efflux pumps may be one of the important reasons for reduced susceptibility of these resistant populations to the various antibiotics. Situations triggering an overexpression of the efflux pump genes could also have several effects on biofilm formation through an increase in the interchange of quorum sensing molecules[35].
The present research provides a new finding about the roles of biofilm on the increased expression of two efflux pumps, hefA and hp1165, in the clinical isolates of H. pylori. Future investigations based upon these findings may reveal the nature of this mechanism and illuminate the role of biofilm formation in the antibiotic resistance of H. pylori in vivo.
As prevalence of H. pylori infection is high in West Asia, particularly in Iran[3], the need exists for improving our knowledge about the mechanism(s) of antibiotic resistance during chronic infection by H. pylori. Furthermore, investigations about the role and the nature of biofilms may help select better eradication regiments capable of acting on biofilm-forming cells as well.
The results of the present work demonstrate the existence of an association between H. pylori biofilm formation and decreased susceptibility to various antibiotics via increased functional activity of two efflux pumps, hp1165 and hefA, in biofilm-forming cells.
ACKNOWLEDGMENTS
We thank Alzahra University for supporting this work. We cordially thank Dr. Sara Gharavi and Miss Zahra Landarani from Alzahra University, Tehran, Iran for helping in this work.
COMMENTS
Background
In this study, the authors investigated the role of biofilm formation on the resistance of Helicobacter pylori (H. pylori) to commonly prescribed antibiotics and examined expression of two efflux pump genes, hp1165 and hefA, involved in the specific resistance to tetracycline and multidrug resistance, respectively, in the biofilm-forming and planktonic cells. In addition, these studies can provide insights into the way for development and application of the new therapeutic approaches in the case of H. pylori infection.
Research frontiers
The authors investigated the importance of biofilm formation in the antibiotic resistance of clinical isolates of H. pylori. This study will provide new approaches towards the treatment of H. pylori-related diseases.
Innovations and breakthroughs
The current research showed significantly increased expression levels of hefA and hp1165 efflux pumps genes in the biofilm-producing H. pylori cell populations. In addition, the results showed that the efflux pumps may be one of the important reasons for reduced susceptibility of these resistant populations to various antibiotics.
Applications
One of the fatal outcomes of chronic H. pylori infection may be production of gastric cancer. So, the next step of this research would be to understand the mechanism and modality of antibiotic resistance in biofilm formation during chronic infection by H. pylori. Another application would be the development of novel therapeutic strategies to overcome biofilm formation in vivo.
Terminology
In bacteria, the biofilm is a well-organized assembly of the cells clustered together to form microcolonies. These colonies attach to surfaces and take up different characteristics from the free-floating or planktonic bacteria. Gram-negative bacteria are able to exclude drugs by efflux pumps, which may represent the mechanism by which they can be protected from toxic effects of non-desirable compounds. In general, the efflux pumps of Gram-negative bacteria consist of an inner-membrane protein, a periplasmic adaptor protein and an outer-membrane protein in which the first one acts with the second and third ones. These efficiently structured complexes are involved in specific resistance as well as multi-resistance to antibiotics.
Peer-review
This is an interesting manuscript about up-regulation of two efflux pumps in biofilm-forming cell populations. Also, the manuscript provides valuable and interesting information about H. pylori.
Footnotes
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Iran
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Institutional review board statement: The H. pylori strains used for this study were isolated from the patients who were admitted to Medical Centers for their persistent upper gastrointestinal problems, and for this reason they underwent endoscopy for biopsy. All routine biopsy specimens from the patients were taken after informed consent and the protocols, under which the biopsies for culture were obtained, were in accordance with the Helsinki Declaration of 1975.
Conflict-of-interest statement: The authors have no conflict of interest.
Data sharing statement: No additional data are available.
Peer-review started: September 25, 2016
First decision: October 20, 2016
Article in press: January 4, 2017
P- Reviewer: Chai FY, March-Rossello GA, Yakoob J, Yamaoka Y S- Editor: Qi Y L- Editor: Filipodia E- Editor: Zhang FF
References
- 1.Andersen LP, Rasmussen L. Helicobacter pylori-coccoid forms and biofilm formation. FEMS Immunol Med Microbiol. 2009;56:112–115. doi: 10.1111/j.1574-695X.2009.00556.x. [DOI] [PubMed] [Google Scholar]
- 2.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fakheri H, Bari Z, Aarabi M, Malekzadeh R. Helicobacter pylori eradication in West Asia: a review. World J Gastroenterol. 2014;20:10355–10367. doi: 10.3748/wjg.v20.i30.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Francesco V, Giorgio F, Hassan C, Manes G, Vannella L, Panella C, Ierardi E, Zullo A. Worldwide H. pylori antibiotic resistance: a systematic review. J Gastrointestin Liver Dis. 2010;19:409–414. [PubMed] [Google Scholar]
- 5.Cole SP, Harwood J, Lee R, She R, Guiney DG. Characterization of monospecies biofilm formation by Helicobacter pylori. J Bacteriol. 2004;186:3124–3132. doi: 10.1128/JB.186.10.3124-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cellini L, Grande R, Di Campli E, Di Bartolomeo S, Di Giulio M, Traini T, Trubiani O. Characterization of an Helicobacter pylori environmental strain. J Appl Microbiol. 2008;105:761–769. doi: 10.1111/j.1365-2672.2008.03808.x. [DOI] [PubMed] [Google Scholar]
- 7.Resch A, Rosenstein R, Nerz C, Götz F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol. 2005;71:2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cammarota G, Sanguinetti M, Gallo A, Posteraro B. Review article: biofilm formation by Helicobacter pylori as a target for eradication of resistant infection. Aliment Pharmacol Ther. 2012;36:222–230. doi: 10.1111/j.1365-2036.2012.05165.x. [DOI] [PubMed] [Google Scholar]
- 9.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 10.Beaudoin T, Zhang L, Hinz AJ, Parr CJ, Mah TF. The biofilm-specific antibiotic resistance gene ndvB is important for expression of ethanol oxidation genes in Pseudomonas aeruginosa biofilms. J Bacteriol. 2012;194:3128–3136. doi: 10.1128/JB.06178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mah TF. Biofilm-specific antibiotic resistance. Future Microbiol. 2012;7:1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- 12.Nishizawa T, Suzuki H. Mechanisms of Helicobacter pylori antibiotic resistance and molecular testing. Front Mol Biosci. 2014;1:19. doi: 10.3389/fmolb.2014.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwamoto A, Tanahashi T, Okada R, Yoshida Y, Kikuchi K, Keida Y, Murakami Y, Yang L, Yamamoto K, Nishiumi S, et al. Whole-genome sequencing of clarithromycin resistant Helicobacter pylori characterizes unidentified variants of multidrug resistant efflux pump genes. Gut Pathog. 2014;6:27. doi: 10.1186/1757-4749-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francesco VD, Zullo A, Hassan C, Giorgio F, Rosania R, Ierardi E. Mechanisms of Helicobacter pylori antibiotic resistance: An updated appraisal. World J Gastrointest Pathophysiol. 2011;2:35–41. doi: 10.4291/wjgp.v2.i3.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kutschke A, de Jonge BL. Compound efflux in Helicobacter pylori. Antimicrob Agents Chemother. 2005;49:3009–3010. doi: 10.1128/AAC.49.7.3009-3010.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bina JE, Alm RA, Uria-Nickelsen M, Thomas SR, Trust TJ, Hancock RE. Helicobacter pylori uptake and efflux: basis for intrinsic susceptibility to antibiotics in vitro. Antimicrob Agents Chemother. 2000;44:248–254. doi: 10.1128/aac.44.2.248-254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu ZQ, Zheng PY, Yang PC. Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J Gastroenterol. 2008;14:5217–5222. doi: 10.3748/wjg.14.5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espinoza MG, Vazquez RG, Mendez IM, Vargas CR, Cerezo SG. Detection of the glmM gene in Helicobacter pylori isolates with a novel primer by PCR. J Clin Microbiol. 2011;49:1650–1652. doi: 10.1128/JCM.00461-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Attaran B, Falsafi T, Moghaddam AN. Study of biofilm formation in C57Bl/6J mice by clinical isolates of Helicobacter pylori. Saudi J Gastroenterol. 2016;22:161–168. doi: 10.4103/1319-3767.178529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franklin R, Cockerill III M. Performance Standards for Antimicrobial Susceptibility Testing, Twenty-First Informational Supplement M100-S21. Clinical and Laboratory Standard Institute: 2011. pp. 68–80. [Google Scholar]
- 21.Gerrits MM, van Vliet AH, Kuipers EJ, Kusters JG. Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis. 2006;6:699–709. doi: 10.1016/S1473-3099(06)70627-2. [DOI] [PubMed] [Google Scholar]
- 22.Grignon B, Tankovic J, Mégraud F, Glupczynski Y, Husson MO, Conroy MC, Emond JP, Loulergue J, Raymond J, Fauchère JL. Validation of diffusion methods for macrolide susceptibility testing of Helicobacter pylori. Microb Drug Resist. 2002;8:61–66. doi: 10.1089/10766290252913773. [DOI] [PubMed] [Google Scholar]
- 23.Brown HL, van Vliet AH, Betts RP, Reuter M. Tetrazolium reduction allows assessment of biofilm formation by Campylobacter jejuni in a food matrix model. J Appl Microbiol. 2013;115:1212–1221. doi: 10.1111/jam.12316. [DOI] [PubMed] [Google Scholar]
- 24.Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97:1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Liu ZQ, Zheng PY, Tang FA, Yang PC. Influence of efflux pump inhibitors on the multidrug resistance of Helicobacter pylori. World J Gastroenterol. 2010;16:1279–1284. doi: 10.3748/wjg.v16.i10.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Queipo-Ortuño MI, De Dios Colmenero J, Macias M, Bravo MJ, Morata P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin Vaccine Immunol. 2008;15:293–296. doi: 10.1128/CVI.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stark RM, Gerwig GJ, Pitman RS, Potts LF, Williams NA, Greenman J, Weinzweig IP, Hirst TR, Millar MR. Biofilm formation by Helicobacter pylori. Lett Appl Microbiol. 1999;28:121–126. doi: 10.1046/j.1365-2672.1999.00481.x. [DOI] [PubMed] [Google Scholar]
- 28.Cellini L, Grande R, Traini T, Di Campli E, Di Bartolomeo S, Di Iorio D, Caputi S. Biofilm formation and modulation of luxS and rpoD expression by Helicobacter pylori. Biofilms. 2005;2:1–9. [Google Scholar]
- 29.Yonezawa H, Osaki T, Kurata S, Zaman C, Hanawa T, Kamiya S. Assessment of in vitro biofilm formation by Helicobacter pylori. J Gastroenterol Hepatol. 2010;25 Suppl 1:S90–S94. doi: 10.1111/j.1440-1746.2009.06213.x. [DOI] [PubMed] [Google Scholar]
- 30.Carron MA, Tran VR, Sugawa C, Coticchia JM. Identification of Helicobacter pylori biofilms in human gastric mucosa. J Gastrointest Surg. 2006;10:712–717. doi: 10.1016/j.gassur.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 31.Coticchia JM, Sugawa C, Tran VR, Gurrola J, Kowalski E, Carron MA. Presence and density of Helicobacter pylori biofilms in human gastric mucosa in patients with peptic ulcer disease. J Gastrointest Surg. 2006;10:883–889. doi: 10.1016/j.gassur.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Cerca N, Martins S, Cerca F, Jefferson KK, Pier GB, Oliveira R, Azeredo J. Comparative assessment of antibiotic susceptibility of coagulase-negative staphylococci in biofilm versus planktonic culture as assessed by bacterial enumeration or rapid XTT colorimetry. J Antimicrob Chemother. 2005;56:331–336. doi: 10.1093/jac/dki217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yonezawa H, Osaki T, Hanawa T, Kurata S, Ochiai K, Kamiya S. Impact of Helicobacter pylori biofilm formation on clarithromycin susceptibility and generation of resistance mutations. PLoS One. 2013;8:e73301. doi: 10.1371/journal.pone.0073301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’’Morain C, Smith S. Helicobacter pylori Treatment Failure: The Rationale for Alternative Antibiotics. Digestion. 2016;93:309–310. doi: 10.1159/000444362. [DOI] [PubMed] [Google Scholar]
- 35.Soto SM. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence. 2013;4:223–229. doi: 10.4161/viru.23724. [DOI] [PMC free article] [PubMed] [Google Scholar]


