Abstract
Objective
Variants in Proprotein Convertase Subtilisin/Kexin Type 1 (PCSK1) may be causative for obesity as suggested by monogenic cases and association studies. Here we assessed the functional relevance in experimental studies and the clinical relevance through detailed metabolic phenotyping of newly identified and known PCSK1 variants in children.
Results
In 52 obese children selected for elevated proinsulin levels and/or impaired glucose tolerance, we found eight known variants and two novel heterozygous variants (c.1095 + 1G > A and p.S24C) by sequencing the PCSK1 gene. Patients with the new variants presented with extreme obesity, impaired glucose tolerance, and PCOS. Functionally, c.1095 + 1G > A caused skipping of exon8 translation and a complete loss of enzymatic activity. The protein was retained within the endoplasmic reticulum (ER) causing ER stress. The p.S24C variant had no functional effect on protein size, cell trafficking, or enzymatic activity. The known variants rs6230, rs35753085, and rs725522 in the 5′ end did not affect PCSK1 promoter activity.
In clinical association studies in 1673 lean and obese children, we confirmed associations of rs6232 and rs6234 with BMI-SDS and of rs725522 with glucose stimulated insulin secretion and Matsuda index. We did not find the new variants in any other subjects.
Conclusions
We identified and functionally characterized two rare novel PCSK1 variants of which c.1095 + 1G > A caused complete loss of protein function. In addition to confirming rs6232 and rs6234 in PCSK1 as polygenic risk variants for childhood obesity, we describe an association of rs725522 with insulin metabolism. Our results support the contribution of PCSK1 variants to obesity predisposition in children.
Keywords: PCSK1, PC1/3, Obesity, Children, Prohormone convertase 1/3
Graphical abstract
Highlights
-
•
We identified two novel variants in PCSK1 in severely obese adolescents.
-
•
The phenotype of these two heterozygous carriers is more severe than in “common childhood obesity”.
-
•
The ΔEx8 variant leads to a truncated protein with a complete loss of function, which is retained within the ER.
-
•
For common variant rs725522 detailed metabolic phenotyping revealed impaired glucose dynamics.
-
•
Overall, variants in PCSK1 are not only associated with childhood obesity, but a more severe phenotype than in BMI-matched controls.
1. Introduction
Variants in the proprotein convertase subtilisin kexin type 1 (PCSK1) gene encoding proprotein convertase 1/3 (PC1/3) contribute to polygenic obesity risk as shown by candidate gene approaches [1] and subsequent genetic association studies [2] as well as genome-wide association studies (GWAS) for BMI and proinsulin [2], [3], [4]. Unlike some other genes identified in hypothesis-free, genome-wide association studies, PCSK1 constitutes a reasonable candidate for functional relevance based on its biological function. The PCSK1 gene is located on chromosome 5q [5] in a region originally showing linkage with type 2 diabetes [6]. In addition to this, linkage peaks on chromosome 5q have been identified for obesity [7], [8].
PCSK1 is mainly expressed in neuroendocrine tissues [9], where it is involved in tissue-specific processing of prohormones and neuropeptide precursors such as proopiomelanocortin, proinsulin, proglucagon, and other known key regulators of energy metabolism [10], [11].
Common variants in/near PCSK1 gene have been associated with obesity risk, body mass index variation, birth weight in association with body mass index, and proinsulin levels in several populations [1], [2], [4], [12], [13]. The coding variants rs6232, rs6234, and rs6235 have functional consequences on PCSK1 activity [1], [14], [15], [16].
Rare mutations in PCSK1 have also been described in individuals with early onset monogenic obesity. Null mutations in the PCSK1 gene cause recessive, monogenic, early-onset morbid obesity as a part of a complex syndrome including hypoadrenalism, hypogonadism, intestinal dysfunction, hyperphagia, altered proinsulin to insulin ratio, postprandial hypoglycemia, and diabetes insipidus [11], [17], [18], [19], [20], [21]. Moreover, heterozygous nonsense mutation p.R80* was linked in one family with dominantly inherited obesity and impaired glucose tolerance, and rare heterozygous missense PCSK1 variants were found in 0.83% of extremely obese individuals [22], [23].
These genotype-phenotype links in both common and rare variants are making this gene one of the most relevant players in the etiology of obesity.
In the present study, we aimed to i) identify new variants, ii) investigate their clinical relevance with regard to obesity and glucose metabolism, and iii) assess their functional relevance on secretion, localization, enzymatic activity, and the ability to induce endoplasmic reticulum (ER) stress.
2. Materials and methods
2.1. Study subjects
Genetic analyses were performed in a cohort of 1673 children and adolescents of the Leipzig school children cohort [24] and Leipzig Obesity Childhood cohort as described previously [25].
The body mass index (BMI) was standardized referring to national reference data [26]. Children with a BMI-standard deviation score (SDS) ≥1.88 (=97th percentile) were classified obese.
For the case control setting, 684 obese children (322 boys and 362 girls; mean age 11.53 ± 3.36 years; mean BMI-SDS 2.41 ± 0.66) were compared to 989 lean children (467 boys and 522 girls; mean age 11.83 ± 2.78 years; mean BMI-SDS −0.14 ± 0.78).
Written consent was obtained from both children >12 years and parents. The study was approved by the ethics committee of the University of Leipzig (NTC 02208141).
2.2. Genetic analyses
Sanger sequencing was performed using the Big Dye Terminator (Applied Biosystems, Foster City, CA) on an automated DNA capillary sequencer (ABI PRISM 3100 Avant; Applied Biosystem). Sequence information for all oligonucleotide primers used for variant screening is available upon request.
Genotypes of rs6230, rs6232, rs6234, and p.S24C in PCSK1 were determined using pre- or custom-designed Taqman single-nucleotide polymorphism genotyping assays (ABI) on an ABI Prism 7500 platform. Call rate was >95%.
PCR-restriction fragment length polymorphism (RFLP) technique was used for rs6230, rs35753085, rs725522, and c.1095 + 1G > A with the specified restriction enzyme (Supplemental Table 4) and evaluation of fragment length by electrophoresis.
For the p.S24C variant, the DNA sequence conservation (GERP, and phyloP100way provided by UCSC Genome Browser (http://genome.ucsc.edu) [27], [28] and pathogenicity was tested using in silico analyses (SIFT [29], PROVEAN [30] (http://provean.jcvi.org), MutationAssesor (http://mutationassessor.org) [31], CADD (http://cadd.gs.washington.edu/score) [32], and SNPs&GO (http://snps.biofold.org) [33]. Sequence alignment of PCSK1 proteins was done at www.uniprot.org and by Signal-BLAST at sigpep.services.came.sbg.ac.at/signalblast.html.
2.3. Enzyme activity assay, immunofluorescence, and immunoblot
Activity studies were performed according to the previously published protocol [34]. The enzymatic activity was assayed using a fluorogenic substrate p-Glu-Arg-Thr-Arg-Arg-amino methylcoumarin (Peptides International Inc., Kentucky, USA) and normalized to the amount of enzyme determined by immunoblot analysis using Image Station 440 (Eastman Kodak Co., New Haven).
Chinese hamster ovary (CHO) cells transfected and grown on glass slides were fixed in 4% paraformaldehyde. After washing in PBS, endogenous peroxidase was quenched with NH4Cl and blocked with blocking reagent (Roche) and 0.2% triton. Cells were stained with anti-FlagM2 primary antibody (1:600; Sigma–Aldrich) followed by incubation with a donkey anti-rabbit Dy488 secondary antibody (1:200/Jackson Immuno, Suffolk, UK).
PCSK1 was co-localized with dsRed-ER (Clontech) labeled ER by laser-scanning microscopy with a plan-neofluar 100×/1.3 oil objective (Carl Zeiss AG, Jena, Germany).
For western blot analysis, cells were lysed (RIPA buffer), samples were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and detected by chemiluminescence. All used antibodies are listed in Supplemental Table 5.
2.4. Statistical analyses
Association analyses were performed with Statistica 7.1 software (StatSoft, Tulsa, OK). Differences in the genotypes distribution between cases and controls were tested using an additive model adjusting for sex, age, and pubertal stage. Associations of polymorphisms with quantitative obesity-related metabolic traits were assessed by generalized linear regression models.
Homozygotes for the major allele (MM), heterozygotes (Mm) and homozygotes for the risk allele (mm) were coded to a continuous numeric variable for genotype (as 0, 1 and 2, respectively) in the additive model. A dominant model for the risk allele was defined as contrasting genotyping groups MM versus Mm + mm. Statistical power was calculated using the Quanto software (USC Biostats, Los Angeles, CA). For the rs6230, rs35753085, rs725522, rs6232, and rs6234, we had a statistical power in 12.1%, 13.6%, 10.7%, 56.4%, and 58%, respectively, to detect the observed effects at α = 0.5 in our cohort (see ORs in Table 2). To reach the power of 80%, 13005, 10776, 16155, 1727, and 1662 obese individuals with a corresponding number of controls would be required for the studied variants.
Table 2.
Association of PCSK1 genetic variants with obesity.
| SNP | Controls BMI SDS <1.28 (N = 989) | Cases BMI SDS ≥1.28 (N = 681) | p Value (adjusted for sex, age and PH) | OR (95% CI) |
|---|---|---|---|---|
| rs6230 | 0.93 | |||
| TT | 69.0% | 70.3% | 0.349 | (0.77; 1.12) |
| TC | 27.3% | 27.0% | ||
| CC | 3.7% | 2.7% | ||
| MAF | 17.4% | 16.3% | ||
| rs35753085 | 0.71 | |||
| CC | 98.2% | 98.7% | 0.289 | (0.30; 1.67) |
| CT | 1.8% | 1.3% | ||
| TT | 0.0% | 0.0% | ||
| MAF | 0.9% | 0.6% | ||
| rs725522 | 1.16 | |||
| AA | 95.1% | 94.0% | 0.530 | (0.76; 1.79) |
| AG | 4.7% | 6.0% | ||
| GG | 0.2% | 0.0% | ||
| MAF | 2.6% | 3.0% | ||
| rs6232 | 1.39 | |||
| AA | 90.9% | 87.4% | 0.022 | (1.00; 1.93) |
| AG | 8.8% | 12.2% | ||
| GG | 0.3% | 0.4% | ||
| MAF | 4.7% | 6.5% | ||
| rs6234 | 1.19 | |||
| CC | 57.9% | 52.5% | 0.039 | (1.01; 1.42) |
| CG | 35.7% | 39.6% | ||
| GG | 6.4% | 7.9% | ||
| MAF | 24.3% | 27.8% | ||
p Value and odds ratio (OR) with 95% confidence intervals (CI) were calculated under logistic regression analyses in additive mode of inheritance, adjusted for sex, age, and PH. MAF – minor allele frequency; PH – pubertal stages on basis of pubic hair development.
Non-normally distributed parameters were logarithmically transformed prior to statistical analysis. Hardy–Weinberg equilibrium was tested using χ2 test. All p-values are reported without Bonferroni corrections for multiple testing, which would require p < 0.002 (assuming 6 traits and 5 genetic variants included in the analyses). Therefore, all associations with p-values <0.05 but >0.002 were considered as nominal.
Detailed information about cell culture, transfection, plasmid construction, reporter gene, and apoptosis assay can be find in the Supplemental material.
3. Results
3.1. Identification of variants in PCSK1
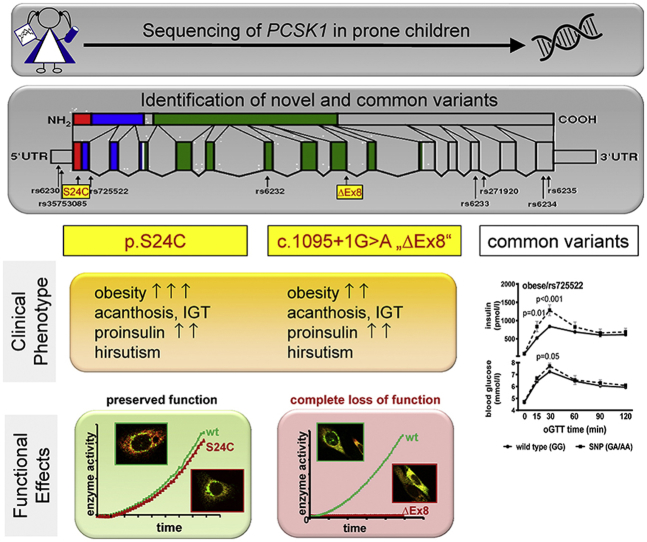
To identify novel variants in PCSK1 with the utmost probability, we selected 52 obese children (BMI SDS 3.0 ± 0.7) with high proinsulin levels >90th centile, impaired glucose tolerance, or high proinsulin:insulin ratio from our Leipzig Obesity Childhood cohort. By sequencing all 14 exons (NM 000439) with intron/exon boundaries including 254 bp of the 5′ UTR, we identified eight known and two novel heterozygous variants. Two variants were located in the 5′ UTR, five in the coding region, and three in the intronic region (Table 1, Supplementary Figure 1). The novel variant p.S24C (c.70A < T; in Figures called S24C) is located within exon1 and predicts a missense mutation p.S24C within the signal peptide of PC1/3. The novel c.1095 + 1G > A variant (in Figures called ΔEx8) affects the donor splice site of intron 8.
Table 1.
List of variants identified by sequencing of the PCSK1 coding region and 5′ UTR in 52 obese children.
| dbSNP | SNP position | SNP localization | Amino-acid change | Chromosome 5 position | Carrier/Allele frequency |
|---|---|---|---|---|---|
| rs6230 | c.-101T > C | 5′ UTR | – | 95,768,847 | 13/– |
| rs35753085 | c.-96C > T | 5′ UTR | – | 95,768,842 | 1/– |
| Novel p.S24C | c.70A > T | Exon 1 | p.S24C | 95,768,676 | 1/– |
| rs725522 | c.180 + 37C > T | Intron 1 | – | 95,768,530 | 4/0.0188 |
| rs6232 | c.661A > G | Exon 6 | p.N221D | 95,751,785 | 12/0.0398 |
| Novel c.1095 + 1G > A | c.1095 + 1G > A | Intron 8 | Truncation | 95,746,477 | 1/– |
| rs6233 | c.1650C > T | Exon 12 | p.N550N | 95,733,112 | 24/0.3528 |
| rs271920 | c.1722 + 54A > G | Intron 12 | – | 95,732,992 | 24/0.000009 |
| rs6234 | c.1993C > G | Exon 14 | p.Q665E | 95,728,974 | 34/0.2688 |
| rs6235 | c.2069C > G | Exon 14 | p.S690T | 95,728,898 | 34/0.2653 |
The novel heterozygous variants are marked in bold. SNP positions refers to the translation initiation site (NM 000439). Chromosome 5 positions are based on GRCh37 (ENSG00000175426). Carrier means the number of patients with the respective mutation in the sequencing cohort (N = 52). Where available, allele frequencies based on ExAC consortium are given.
3.2. Clinical relevance of identified variants
To assess potential associations with obesity and metabolic traits, we genotyped these PCSK1 variants in 989 lean and 684 obese children (Supplementary Table 1). All genotype distributions were consistent with Hardy–Weinberg equilibrium (p > 0.05). The two novel variants were not found in any other obese or lean child in our cohort confirming a survey in large variant databases (1000 genomes Project, NHLBI GO Exome Sequencing Project (ESP), Exome Aggregation Consortium (ExAC); all were accessed through the UCSC Genome Browser January 2016).
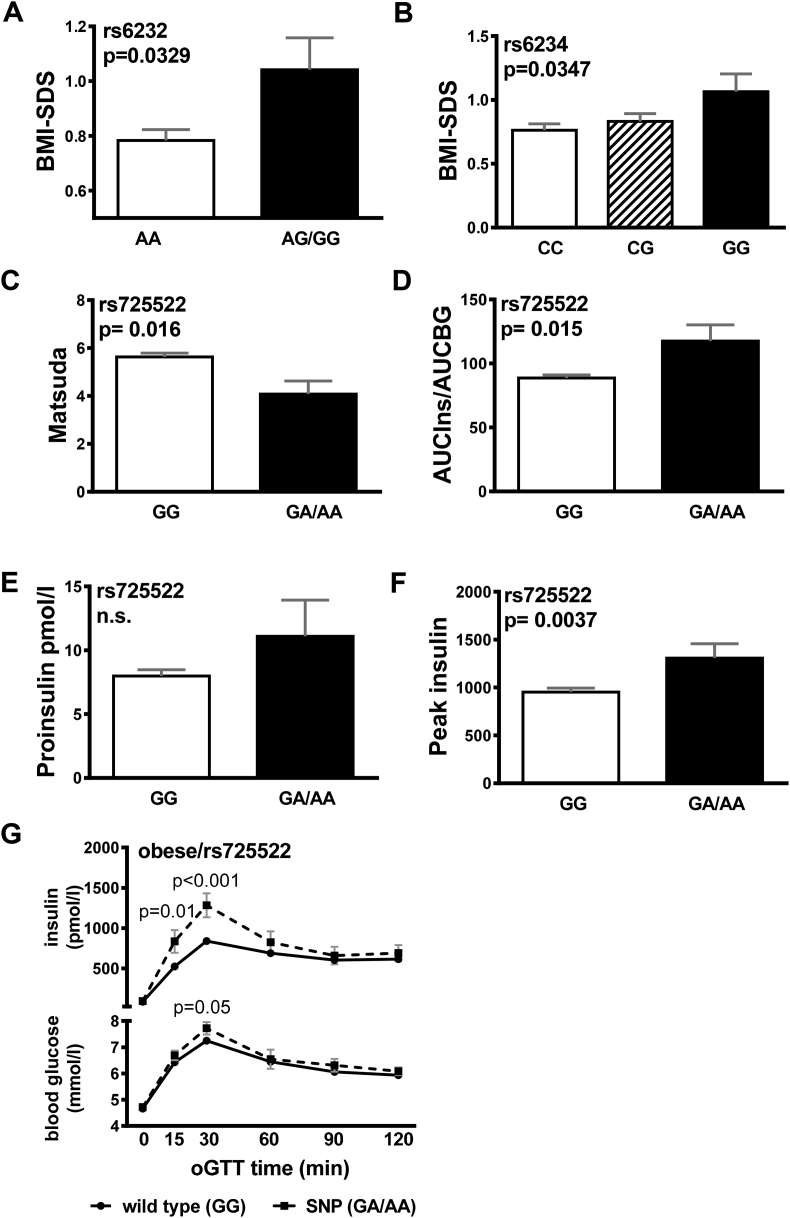
In the case control design, the minor alleles for rs6232 (p = 0.022) and rs6234 (p = 0.039) were significantly more prevalent in the obese group after adjustment for sex, age and pubertal stage (Table 2), conferring a higher risk of obesity for rs6232 (OR = 1.39, p = 0.022) and rs6234 (OR = 1.19, p = 0.039). Consistently, the minor alleles were nominally modestly associated with higher BMI-SDS (rs6232 p = 0.033, rs6234 p = 0.035; Figure 1A–B). In subsequent generalized linear regression analyses rs6232 and rs6234 contributed independently to BMI-SDS (Supplementary Table 2). Rs6230, rs35753085, and rs725522 were not associated with obesity.
Figure 1.
Association with obesity and metabolic factors. Carriers for rs6232 (A) and rs6234 (B) had higher BMI-SDS compared to wt. Carriers for rs725522 had higher (C) AUCIns/AUCBG and lower (D) Matsuda Insulin sensitivity indices (ISI) as compared to wt carriers. There were no significant differences in (E) proinsulin and (F) peak insulin levels. (G) Course of blood glucose and (H) insulin levels during oGTT in carriers of rs725522 wt (N = 473) and minor allele (N = 26). Data are given as mean ± SEM. AUC, area under the curve; wt, wild-type.
Furthermore, we assessed the association of PCSK1 variants with detailed traits related to glucose (including fasting blood glucose (BG), BG 120 min post oGTT, area under the curve BG), and insulin metabolism (including fasting plasma insulin, proinsulin, C-peptide, peak insulin, area under the curve insulin, Matsuda ISI, HOMA, QUICKI, Belfiore, and Stumvoll-index). Rs725522 was nominally significantly associated with Matsuda ISI (p = 0.016), AUC Ins-to-AUC BG ratio (p = 0.015), and peak insulin (p = 0.004) but not with proinsulin levels (Figure 1C–F). Furthermore, we found significant higher blood glucose 30min after glucose load and a significantly elevated first phase insulin response during oral glucose tolerance test (oGTT) in the group of the risk allele carriers (Figure 1G). In subsequent generalized linear regression analyses, rs725522 contributed independently from BMI-SDS and age to insulin secretion (Supplementary Table 3). The other variants were not associated with insulin traits (data not shown).
3.3. Functional characterization of PCSK1 variants
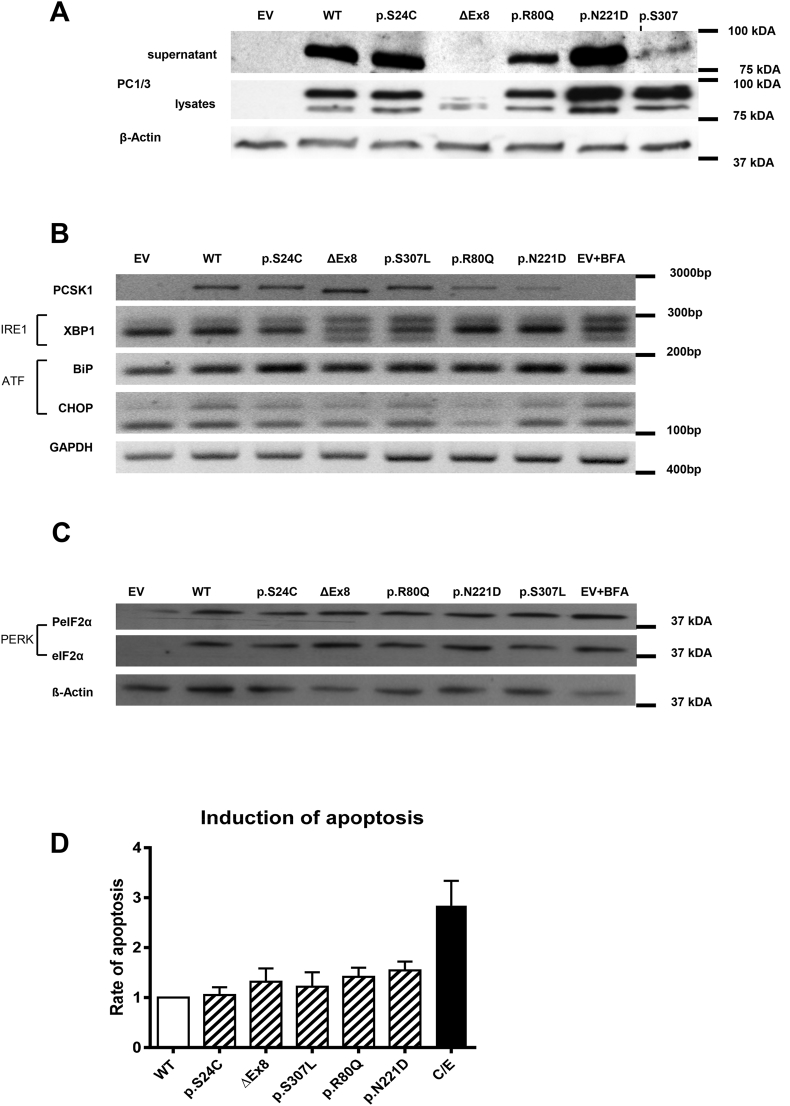
We first focused on the functional characterization of the two novel variants performing expression-, secretion-, and cellular localization studies and an enzyme activity assay. We also included the rs725522 variant since no functional investigations have been performed for this variant. However, we did not re-investigate rs6232 and rs6234 since similar investigations have already been published [1]. Second, we performed reporter gene assays to investigate all variants (rs35753085, rs6230, rs725522, p.S24C) that may affect PCSK1 promotor activity. Third, we investigated if variants can induce endoplasmic reticulum (ER) stress. For this, we preselected variants that were already described as variants with disturbed function (p.N221D [1], p.S307L [11], and p.R80Q [16] as well as our novel variants. Rs6234 and rs6235 were not investigated since former studies [1], [16] did not suggest an altered function of these variants.
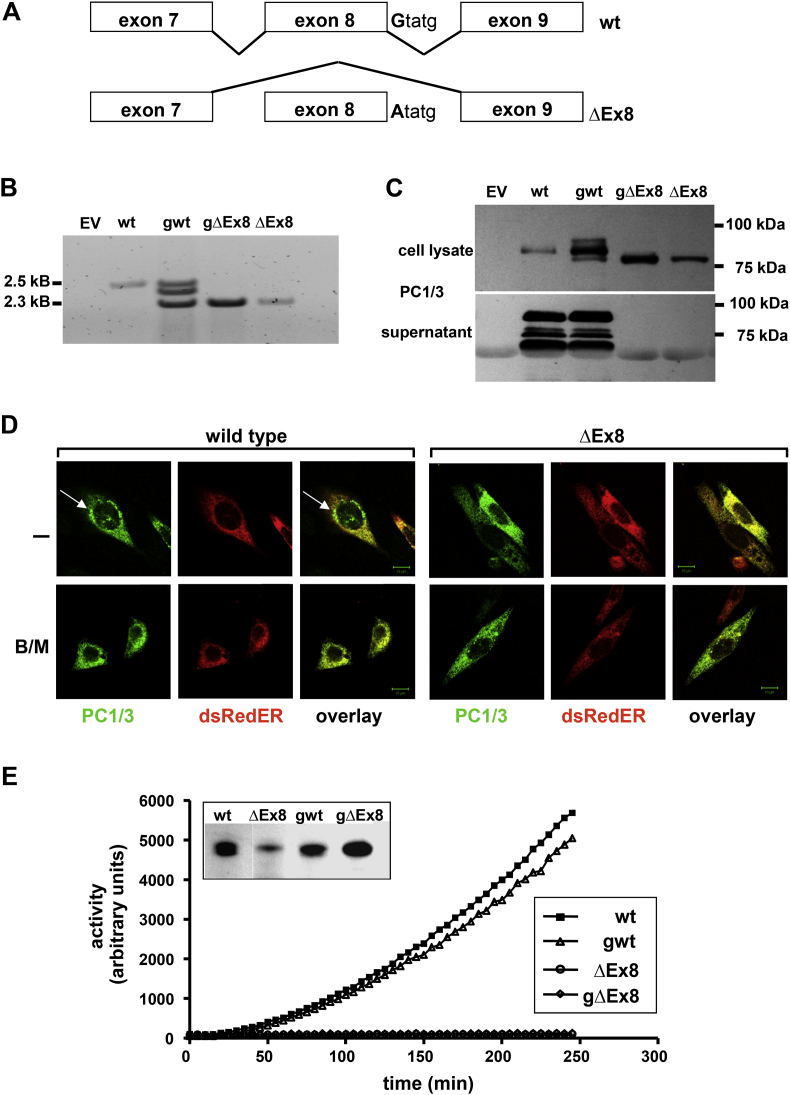
c.1095 + 1G > A variant. The occurrence of this variant in only one patient with morbid obesity, and the molecular localization with base-pair exchange of the first nucleotide of the intron 8 donor splice site suggested a potential functional relevance affecting splicing (Figure 2A).
Figure 2.
Functional characterization of the c.1095 + 1G > A mutation. The mutation c.1095 + 1G > A was named as ΔEx8. (A) Schematic representation of exon 8 skipping. Hek293 cells were transiently transfected with expression vectors encoding the Flag-tagged PC1/3 variants (wtcDNA; gwt; c.1095 + 1G > A (ΔEx8) cDNA; gc.1095 + 1G > A (ΔEx8)). (B) PCR products exons 1 to exon 14 showing different sized amplicons for gwt and c.1095 + 1G > A (ΔEx8) variants. EV, empty vector. (C) Immunoblot with antibodies to the Flag epitope showing protein in the cell lysates but not in the supernatants of c.1095 + 1G > A (ΔEx8) variants. (D) Confocal microscopy of CHO cells transfected with Flag-tagged PC1/3 variants wtcDNA (wild-type) and c.1095 + 1G > A cDNA (ΔEx8) as indicated and with dsRedER plasmids shows assembly of PC1/3 in the ER in wild-type and perinuclear localization (which was inhibited by Brefeldin/Monensin (B/M)) treatment. The c.1095 + 1G > A (ΔEx8) variant was detectable in the ER without assembly in perinuclear vesicles. (E) Enzymatic activity was completely abolished in Hek293 cells expressing the ΔEx8 variants compared to wildtype protein.
To verify the alternative splice effect, we constructed mini-genes (gwt, c.1095 + 1G > A (gΔEx8)) containing the complete introns 7 and 8 of PCSK1. These mini-gene constructs were compared with cDNA plasmids expressing wild-type (WT) cDNA (wtcDNA) and a c.1095 + 1G > A (ΔEx8) cDNA plasmid (c.1095 + 1G > A cDNA (ΔEx8)) (Figure 2A). The c.1095 + 1G > A (ΔEx8) coding plasmids generated mRNA with a molecular weight reduced by 213 bp compared to the WT constructs (Figure 2B; 2.5 kb versus 2.3 kb) predicted to result in a 71-amino-acids in-frame deletion (exon 8-skipping) within the catalytic domain of PC1/3. Cells transfected with the gwt-construct expressed both PCSK1 mRNA species (2.5 + 2.3 kb) and an additional third fragment of about 2.4 kb. We confirmed the identity of the WT and c.1095 + 1G > A (ΔEx8) fragments, and identified the third amplicon as a hybrid species generated during PCR reaction.
In concordance with mRNA expression, PC1/3 protein immunoblotting of cell lysates and supernatants revealed a 7 kDa smaller protein expressed from mutant cDNA and mini-gene constructs (Figure 2C). In contrast to the WT proteins, the mutant proteins were not secreted into the supernatant (Figure 2C). This cellular retention may be due to impaired cellular trafficking, which we subsequently investigated by immunofluorescence in transiently transfected CHO cells (Figure 2D). WT PC1/3 was readily detectable in the ER and in perinuclear vesicles; detection was lost after incubation with Golgi inhibitors BrefeldinA/Monensin (B/M) [35] as positive control, leading to accumulation of PC1/3 within the ER. The mutant protein was exclusively detected within the ER but not in perinuclear vesicles. Similar results were obtained in human embryonic kidney (Hek293) cells (data not shown). Finally, exon 8 deletion (c.1095 + 1G > A (ΔEx8)) led to a complete loss of enzymatic activity of PC1/3 compared to WT protein (Figure 2E).
Thus, the c.1095 + 1G > A (ΔEx8) variant leads to a truncated protein with a complete loss of function and which is retained within the ER.
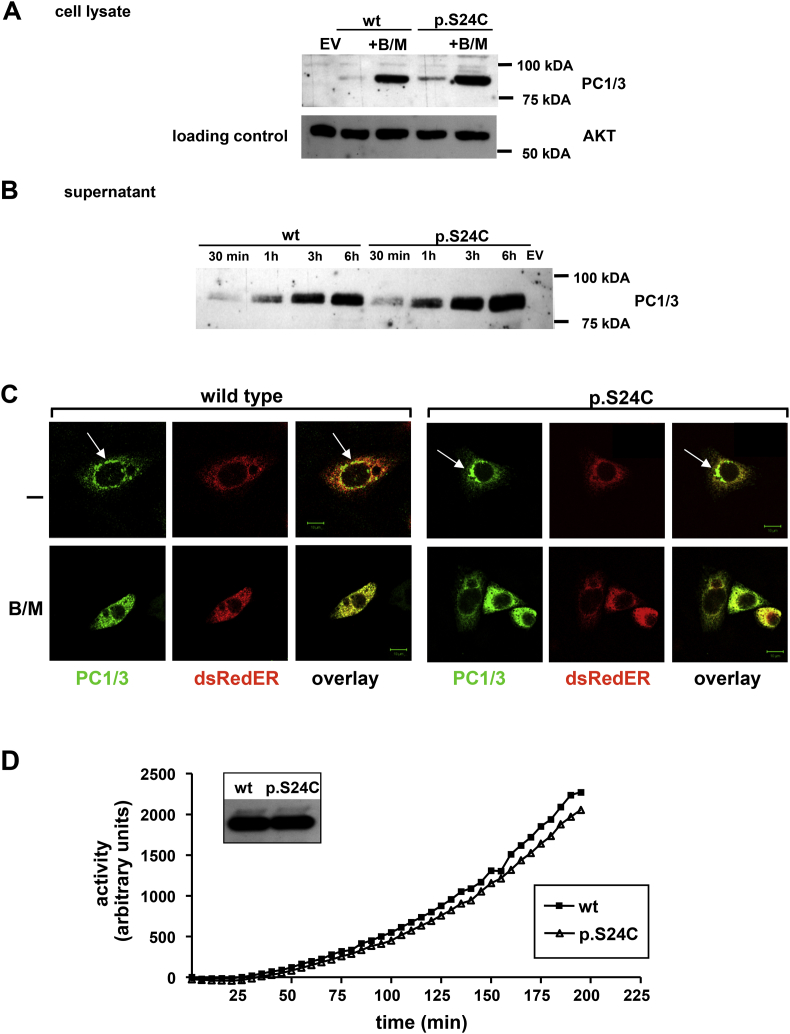
p.S24C variant. We similarly characterized potential functional consequences of the p.S24C variant on PC1/3 maturation, secretion, localization, and enzymatic function. The detection in whole cell lysates, release into supernatant, and the subcellular localization in the ER and perinuclear vesicles was identical to the WT protein (Figure 3A–C). The enzymatic activity of the p.S24C variant was not impaired in enzyme assays, providing evidence that enzymatic function is pertained when transfected into neuro2A (N2A) neuroblastoma (Figure 3D) or Hek293 cells (data not shown).
Figure 3.
Functional characterization of the p.S24C mutation. Immunoblot (A + B) with antibodies to the Flag epitope showing protein in the cell lysates and in the supernatants of wild-type (WT) and mutated variant. After Brefeldin/Monensin (B/M) treatment, proteins were retained in the cell. EV, empty vector. (C) Confocal microscopy of CHO cells transfected with Flag-tagged PC1/3 variants wtcDNA (wild-type) and p.S24CcDNA (p.S24C) as indicated and with dsRedER plasmids shows equally subcellular assembly of PC1/3 wt and variants in the ER and perinuclear localization (which was inhibited by Brefeldin/Monensin (B/M) treatment). (D) Enzymatic activity of PC1/3 was retained in p.S24C variant similar to the wild-type.
In in silico analyses, p.S24C was found tolerated/benign (MutationAssessor – Low impact, Provean – neutral, score −1.52, SIFT – tolerated, score 0.056, SNPs&GO: neutral polymorphism CADD: 13.37, PredictSNP – neutral with 89% accuracy) and not particularly conserved (phyloP100way: −0.115669, and GERP 0.168). According to the Uniprot, the first 27 amino acids are forming a signal peptide. Hence, p.S24C does not alter PC1/3 protein maturation and function.
rs725522 variant. To assess whether the nucleotide exchange in intron 1 may affect mRNA splicing, we again constructed mini-genes (gintron1wt and gintron1-37), containing the complete cDNA plus the first 218bp and the last 502bp of intron 1. PCR analysis revealed a product of predicted size for both the WT and variant (Supplementary Figure 4A) and hence no effect on PCSK1 mRNA splicing or PC1/3 maturation and secretion.
Promoter variants. We designed reporter constructs including −779bp to +109bp relative to the transcription start site (Supplementary Figure 5) to assess whether variants in 5′UTR or intron 1 affect promoter activity in Hek293 (non-neuroendocrine origin) and mouse insulinoma cells (βTC3; neuroendocrine origin). We included the p.S24C variant, for which we did not expect regulatory effects as an unregulated control. Compared to the WT construct we did not observe regulatory effects for any of the studied variants (rs35753085, rs6230, and p.S24C) (Supplementary Figure 5B).
As the presence of introns could elevate mRNA levels [36] by increasing the synthesis of mRNA, we analyzed the effects of rs725522 located in intron 1 on PCSK1 expression.
In a second promoter construct containing the exon 1 + 197bp of intron 1 (Supplementary Figure 5A), the presence of WT intronic sequence (fullex1, Supplementary Figure 5A) doubled luciferase activity in Hek293 and βTC3 cells compared to the WT construct (Supplementary Figure 5C). The insertion of the SNP rs725522, however, did not affect luciferase activity.
Overall, the analyzed variants did not affect promoter activity of PCSK1 in our cell systems.
Effects of PCSK1 variants on ER-stress. The retention of misfolded proteins in the ER can lead to ER stress and unfolded protein response (UPR) activation [37], [38] and eventually apoptosis [39], [40]. We, therefore, evaluated whether non-synonymous variants c.1095 + 1G > A and p.S24C and common variants p.R80Q (rs1799904), p.S307L and p.N221D (rs6232) in PCSK1 may lead to ER stress and the three branches of UPR activation (Inositol-Requiring Enzyme 1 (IRE1), Activating Transcription Factor 6 (ATF6), and PRKR-Like Endoplasmic Reticulum Kinase (PERK) [37].
The mutated p.S24C, p.R80Q, and p.N221D were readily detectable in cell lysates and supernatants, whereas release of c.1095 + 1G > A and p.S307L was blocked (Figure 4A).
Figure 4.
Endoplasmic reticulum stress. (A) Influence of PCSK1 variants on expression/maturation and release of PC1/3 wild-type (WT) and different variants as indicated by immunoblot analysis (EV = empty vector). (B) UPR activation was assessed via PCR for the IRE1 (XBP1) and ATF (BiP and CHOP) pathway (BFA = BrefeldinA [1 μg/ml]). (C) For analysis of PERK pathway activation, phosphorylated (PeIF2α) and unphosphorylated eIF2α was analyzed by western blotting. (D) Apoptosis was assessed by FACS identification of Annexin-V/Propidium Iodide stained cells. Results were normalized to the apoptosis rate of the wild-type transfected cells (C/E = camptothecin [2 μM]/etoposide [85 μM]).
Brefeldin was employed as positive control for inducing ER stress [41]. To assess the activation of the IRE1 pathway, we analyzed the processing of X-Box Binding Protein (XBP) 1 mRNA in an alternatively spliced variant [42] as described [43]. Expression of c.1095 + 1G > A and p.S307L resulted in an alternative XBP1 splicing pattern comparable to that of the Brefeldin control (Figure 4B), whereas the other variants did not affect XBP1 mRNA splicing. To evaluate activation of the ATF6 pathway we analyzed BIP (HSPA5) and CHOP (DDIT3) mRNA expression [42]. We did not detect an activation of the ATF6 pathway in any investigated variant. Thirdly, we examined the phosphorylation of the PERK target eIF2α but, again, did not find significant changes for the different variants (Figure 4C).
Finally, since persistently activated UPR can cause apoptosis, we analyzed the apoptosis and did not find an increased apoptosis rate after transfection with any of the variants, whereas camptothecin/etoposide as positive controls exerted the expected increase.
In summary, c.1095 + 1G > A and p.S307L cause an activation of the IRE1 pathway in Hek293 cells, indicating ER stress, whereas ATF6 and PERK pathway as well as apoptosis were not affected by PCSK1 variants.
3.4. Clinical phenotype of patients carrying novel variants
The patient carrying the c.1095 + 1G > A variant was a 15.4 year old girl with severe obesity (BMI-SDS 2.95 to 3.25 in sequential visits), acanthosis nigricans, and signs of hirsutism, hypertension, and polycystic ovary (PCO) syndrome. Metabolically, she had impaired glucose tolerance with hyperinsulinemia upon glucose provocation (Supplementary Figure 2A,C) and elevated proinsulin levels >4× upper reference limit (between 43.2 and 85.5 pmol/L). Both parents of the patient were obese (BMIs mother: 35.5 kg/m2, father: 34.7 kg/m2) but were, unfortunately, not available for further analysis.
The girl carrying the p.S24C variant had a similar phenotype with complete metabolic syndrome including severe obesity (BMI-SDS 3.45 to 3.64), acanthosis, hypertension, and PCO syndrome. She also had impaired fasting glucose and impaired glucose tolerance with delayed although elevated insulin response in oral glucose tolerance test (oGTT; Supplementary Figure 2B,D) and elevated basal proinsulin levels (42.8–71.2 pmol/L).
To assess whether those phenotypes differ significantly from “common obesity,” we matched controls, applying the criteria same sex (female), completed pubertal development, and similar age (between 14.4 and 18 years), which was fulfilled by 73 subjects from our cohort. Since the patient with p.S24C was also heterozygous for rs725522 and the patient with c.1095 + 1G > A did not carry any other variants, we also tried to match the genotype of the controls which left us with 4 controls for the p.S24C patient and 28 for the c.1095 + 1G > A patient. The phenotype of the carriers of novel variants appeared to be more severe, particularly with respect to insulin secretion and insulin resistance, compared to matched obese controls (Supplementary Figure 3).
4. Discussion
In this study, we characterized two novel and 8 known variants that had been identified by sequencing of the PCSK1 gene in a subset of obese patients, which we regarded as prone for PCSK1 mutations. In functional studies, we confirmed that the c.1095 + 1G > A (ΔEx8) variant caused a complete loss of enzymatic activity of the protein and cellular retention leading to ER stress. The other novel variant p.S24C did not have an effect on protein size, secretion, or enzymatic activity. Neither this nor the known variants rs6230, rs35753085, and rs725522 located in the 5´end of the gene affected PCSK1 promoter activity. Moreover, we performed detailed clinical studies for association with glucose or insulin metabolism, in which we confirmed association of rs6232 and rs6234 with obesity and describe a nominal association for rs725522 with insulin metabolism.
4.1. Novel rare PCSK1 variants
There are several reports of patients with monogenic obesity due to homozygous or compound heterozygous PCSK1 mutations [11], [17], [18], [19], [20], [21], [34] and on the heterozygous mutation carriers [22], [23]. Some of the previously reported heterozygous PCSK1 mutations have not co-segregated with the obesity phenotype in all of the patients [22], [23]. Functionally, Our newly identified c.1095 + 1G > A variant leads to abnormal splicing and subsequent skipping of exon 8 in the PCSK1 mRNA. The resulting truncated protein lacks 71 amino acids in the catalytic domain and, not surprisingly, has a complete loss of enzymatic function in vitro. In addition, it shows abnormal cellular localization. This retention of the mutated protein in the ER may be caused by binding to chaperones [44], as previously reported for other PC1/3 mutants [34], and lead to the induction of IRE1, an early ER stress marker. The activation of ER stress by PCSK1 variants, recently also shown by others [14], may present an additional impairment of the cells expressing PC1/3. In conclusion, our in vitro data provide strong evidence for a complete loss of function of the c.1095 + 1G > A mutation. Previously described rare heterozygous PCSK1 mutations were linked to obesity [22] or obesity and impaired glucose tolerance [23] but were not reported to present any other phenotypic features described in homozygous mutation carriers [11]. Nevertheless, our proband has some additional clinical features including PCO syndrome, acanthosis nigricans, and hypertension. Moreover, her proinsulin levels were 4-fold higher than in similarly obese controls; this is potentially caused by some degree of dysfunction of PC1/3 [34], which plays a major role in processing proinsulin in PC1/3-null mice [45]. The overall phenotype of these PC1/3-null mice with growth retardation but not obesity, however was not comparable with our patient [45]. Similarly, another mouse model with complete deletion of Pcsk1 was embryonically lethal and the heterozygous mice also had retarded growth [46]. Interestingly, however, a mouse model with a single nucleotide variation (N222D) showed a similar phenotype [47] as the initially reported first patient with hyperphagic severe early onset obesity with PC1/2 deficiency including hyperproinsulinemia [17].
On the other hand, it has been shown that human carriers with heterozygous mutations for PC1/3 do not exhibit significantly impaired proinsulin processing [34]. Moreover, because of high total insulin levels and apparent signs of insulin resistance, major defects in the insulin secretion seem rather unlikely in our patient. The phenotype of the c.1095 + 1G > A carrier including obesity, hypertension, impaired glucose tolerance, and PCO syndrome was found in less than <5% of obese females aged 16–18 years in our Leipzig childhood obesity cohort. Nevertheless, the proband carrying p.S24C variant presented a similar the phenotype, although this variant had no effect on the protein function in functional and in in silico analyses. The first 27 amino acids of the protein are forming a signal peptide, and hence the mutation may affect cell trafficking. As we showed that the intracellular localization of the p.S24C protein is normal even in homozygous experimentally mutated protein (release into supernatants, ER and Golgi localization, and, finally, enzymatic function), the mutation does apparently not affect protein function, particularly in the heterozygous state.
One major limitation of our study is that DNA or a more detailed metabolic phenotype of the parents was not available to verify the co-segregation of the novel variant with obesity in the family. As both parents were obese, the prevalence and the phenotype of the obesity in the affected family may be influenced by an obesogenic environment, as was the case in the p.R80* family [23]. We have not found any other patient with the c.1095 + 1G > A variant in our sample nor is there any case report in the literature that would corroborate our findings. Hence, we cannot speculate whether the phenotype of our patient is affected by the heterozygous variant, which would have deleterious effects, however, in the homozygous state.
4.2. Known common PCSK1 variants and their contribution to childhood onset polygenic obesity
PCSK1 variants contribute also to the polygenic type of obesity [2], [3]. Children present earlier stages of the disease and are less biased by co-morbidities. Hence, studies in children may allow a better discrimination between primary and secondary associations. In fact, recent reports indicated that the common variants in PCSK1 have a stronger effect on the obesity phenotype in children than in adults [2].
We confirmed the reported association of the common variants rs6232 and rs6234 with obesity and BMI-SDS in our cohort [1] but not the associations of these variants with proinsulin conversion and glucose homeostasis [48]. Our findings that both variants are only mild predictors for BMI-SDS or weight suggest that they are not major contributors to common obesity, which is in line with previous findings [49], [50]. Interestingly, we found that the PCSK1 rs725522 minor allele was significantly associated with parameters of insulin resistance (Matsuda ISI, peak insulin, AUCIns/AUCBG). These results suggest declined insulin sensitivity in the carriers. Albeit we did not find a significant association with proinsulin, the insulin levels were significantly higher after 15 and 30min of oGTT in the obese carrier group compared to non-carriers. This may reflect a difference in kinetics of insulin secretion in carriers and non-carriers at least under the conditions of a glucose load. Associations of obesity risk variants in PCSK1 with insulin sensitivity have so far only been described for the PCSK1 variants rs6232 [48] and rs3811951 [51].
Additionally, we investigated the potential functional relevance of the promoter variants. rs6230 and rs35753085 did not have significant effects on PCSK1 promoter activity in our cell systems. This is in line with results that cAMP-responsive elements [52] and thyroid hormone response element binding sites [53] were not affected by the investigated SNPs. The insertion of an intronic sequence per se into the reporter constructs increased the promoter activity. This enhancing effect of intronic sequences on transcription efficiencies is well documented in plants [36] and mammals [54]. Enhancement is perhaps mediated by the predicted binding sites for SP1, AP2α, or TCF-4E. The putative AP2α binding site is lost after insertion of rs725522, and recent publications indicated suppressive effects by AP2α on gene expression [55]. This may explain the slightly but not significantly increased promoter activity by rs725522 in Hek293 cells. We did not see evidence for effects of rs725522 on PCSK1 mRNA splicing and protein maturation.
There are several limitations of our study on common variants. First, a subset of variants was genotyped using RFLP experiments. This method may be prone to be subjective in the evaluation of PCR products and thereby prone to methodological bias. However, we objected to using pre-designed TaqMan assays due to poor discrimination quality. In addition, we confirmed suspicious samples by Sanger sequencing. Another limitation is that our sample size is small (<60% statistical power) regarding all of the analyzed common variants. We also report nominal p-values and Bonferroni correction would likely weaken some associations of common variants (including in rs725522). Nevertheless, the associations between rs6232 and rs6234 and obesity confirm recent findings.
Overall, our and previous publications coherently report an association of variants in PCSK1 with early onset obesity along a continuum on the severity of phenotype. The continuum ranges from extreme hyperphagic obesity with additional neuroendocrine defects in compound heterozygous individuals [17], [20] to dominant monogenic form of obesity in patients with heterozygous loss-of-function mutations [23] or oligogenic forms of obesity lacking severe additional phenotypes in patients with partial PCSK1 deficiency [22] to common variants reliably associated with obesity and elevated proinsulin [2], although not to the extent as described in monogenic forms of PCSK1 deficiency.
Our findings of the novel heterozygous loss of function PCSK1 variant c.1095 + 1G > A in a proband with severe early onset obesity, PCO syndrome, hypertension, and increased proinsulin levels, and associations of PCSK1 SNPs rs6232 and rs6234 with BMI-SDS and rs725522 with insulin resistance in a large pediatric cohort, support the role of PCSK1 variants contributing to obesity in children.
Funding
This work was supported by grants from by German Research Foundation (DFG) within the CRC Obesity Mechanisms, CRC1052 project C05, the European Community's Seventh Framework Programme (FP7/2007-2013) project Beta-JUDO under grant agreement no 279153, and the LIFE Child (Leipzig Research Center for Civilization Diseases, Universität Leipzig) funded by the European Union, by the European Regional Development Fund (ERDF) by means of the Free State of Saxony within the framework of the excellence initiative, the European Society for Paediatric Endocrinology (to DL), by the Federal Ministry of Education and Research (BMBF), Germany, IFB AdiposityDiseases FKZ: 01EO1501, and the German Diabetes Association (to AK and DL), J.S. was supported by the ESPE (European Society for Pediatric Endocrinology) Research Fellowship.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
We thank all children who participated in the studies. We very gratefully appreciate the help of the study nurses and physicians of our obesity outpatient clinic and the technical assistants (R. Tauscher, A. Berthold).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.12.002.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Benzinou M., Creemers J.W., Choquet H., Lobbens S., Dina C., Durand E. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nature Genetics. 2008;40:943–945. doi: 10.1038/ng.177. [DOI] [PubMed] [Google Scholar]
- 2.Nead K.T., Li A., Wehner M.R., Neupane B., Gustafsson S., Butterworth A. Contribution of common non-synonymous variants in PCSK1 to body mass index variation and risk of obesity: a systematic review and meta-analysis with evidence from up to 331 175 individuals. Human Molecular Genetics. 2015;24:3582–3594. doi: 10.1093/hmg/ddv097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stijnen P., Tuand K., Varga T.V., Franks P.W., Aertgeerts B., Creemers J.W. The association of common variants in PCSK1 with obesity: a HuGE review and meta-analysis. American Journal of Epidemiology. 2014;180:1051–1065. doi: 10.1093/aje/kwu237. [DOI] [PubMed] [Google Scholar]
- 4.Strawbridge R.J., Dupuis J., Prokopenko I., Barker A., Ahlqvist E., Rybin D. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seidah N.G., Mattei M.G., Gaspar L., Benjannet S., Mbikay M., Chretien M. Chromosomal assignments of the genes for neuroendocrine convertase PC1 (NEC1) to human 5q15-21, neuroendocrine convertase PC2 (NEC2) to human 20p11.1-11.2, and furin (mouse 7[D1-E2] region) Genomics. 1991;11:103–107. doi: 10.1016/0888-7543(91)90106-o. [DOI] [PubMed] [Google Scholar]
- 6.Martin L.J., Comuzzie A.G., Dupont S., Vionnet N., Dina C., Gallina S. A quantitative trait locus influencing type 2 diabetes susceptibility maps to a region on 5q in an extended French family. Diabetes. 2002;51:3568–3572. doi: 10.2337/diabetes.51.12.3568. [DOI] [PubMed] [Google Scholar]
- 7.Vaughan L.K., Wiener H.W., Aslibekyan S., Allison D.B., Havel P.J., Stanhope K.L. Linkage and association analysis of obesity traits reveals novel loci and interactions with dietary n-3 fatty acids in an Alaska Native (Yup'ik) population. Metabolism. 2015;64:689–697. doi: 10.1016/j.metabol.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barat-Houari M., Clement K., Vatin V., Dina C., Bonhomme G., Vasseur F. Positional candidate gene analysis of Lim domain homeobox gene (Isl-1) on chromosome 5q11-q13 in a French morbidly obese population suggests indication for association with type 2 diabetes. Diabetes. 2002;51:1640–1643. doi: 10.2337/diabetes.51.5.1640. [DOI] [PubMed] [Google Scholar]
- 9.Ramos-Molina B., Martin M.G., Lindberg I. PCSK1 variants and human obesity. Progress in Molecular Biology and Translational Science. 2016;140:47–74. doi: 10.1016/bs.pmbts.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stijnen P., Ramos-Molina B., O'Rahilly S., Creemers J.W. PCSK1 mutations and human endocrinopathies: from obesity to gastrointestinal disorders. Endocrine Reviews. 2016;37:347–371. doi: 10.1210/er.2015-1117. [DOI] [PubMed] [Google Scholar]
- 11.Farooqi I.S., Volders K., Stanhope R., Heuschkel R., White A., Lank E. Hyperphagia and early-onset obesity due to a novel homozygous missense mutation in prohormone convertase 1/3. The Journal of Clinical Endocrinology & Metabolism. 2007;92:3369–3373. doi: 10.1210/jc.2007-0687. [DOI] [PubMed] [Google Scholar]
- 12.Wen W., Cho Y.S., Zheng W., Dorajoo R., Kato N., Qi L. Meta-analysis identifies common variants associated with body mass index in East Asians. Nature Genetics. 2012;44:307–311. doi: 10.1038/ng.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruiz-Narvaez E.A., Haddad S.A., Rosenberg L., Palmer J.R. Birth weight modifies the association between central nervous system gene variation and adult body mass index. Journal of Human Genetics. 2016;61:193–198. doi: 10.1038/jhg.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco E.H., Ramos-Molina B., Lindberg I. Revisiting PC1/3 mutants: dominant-negative effect of endoplasmic reticulum-retained mutants. Endocrinology. 2015;156:3625–3637. doi: 10.1210/en.2015-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mbikay M., Sirois F., Nkongolo K.K., Basak A., Chretien M. Effects of rs6234/rs6235 and rs6232/rs6234/rs6235 PCSK1 single-nucleotide polymorphism clusters on proprotein convertase 1/3 biosynthesis and activity. Molecular Genetics and Metabolism. 2011;104:682–687. doi: 10.1016/j.ymgme.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 16.Pickett L.A., Yourshaw M., Albornoz V., Chen Z., Solorzano-Vargas R.S., Nelson S.F. Functional consequences of a novel variant of PCSK1. PLoS One. 2013;8:e55065. doi: 10.1371/journal.pone.0055065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson R.S., Creemers J.W., Ohagi S., Raffin-Sanson M.L., Sanders L., Montague C.T. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nature Genetics. 1997;16:303–306. doi: 10.1038/ng0797-303. [DOI] [PubMed] [Google Scholar]
- 18.Harter B., Fuchs I., Muller T., Akbulut U.E., Cakir M., Janecke A.R. Early clinical diagnosis of PC1/3 deficiency in a patient with a novel homozygous PCSK1 splice-site mutation. Journal of Pediatric Gastroenterology and Nutrition. 2016;62:577–580. doi: 10.1097/MPG.0000000000001018. [DOI] [PubMed] [Google Scholar]
- 19.Frank G.R., Fox J., Candela N., Jovanovic Z., Bochukova E., Levine J. Severe obesity and diabetes insipidus in a patient with PCSK1 deficiency. Molecular Genetics and Metabolism. 2013;110:191–194. doi: 10.1016/j.ymgme.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson R.S., Creemers J.W., Farooqi I.S., Raffin-Sanson M.L., Varro A., Dockray G.J. Small-intestinal dysfunction accompanies the complex endocrinopathy of human proprotein convertase 1 deficiency. Journal of Clinical Investigation. 2003;112:1550–1560. doi: 10.1172/JCI18784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin M.G., Lindberg I., Solorzano-Vargas R.S., Wang J., Avitzur Y., Bandsma R. Congenital proprotein convertase 1/3 deficiency causes malabsorptive diarrhea and other endocrinopathies in a pediatric cohort. Gastroenterology. 2013;145:138–148. doi: 10.1053/j.gastro.2013.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Creemers J.W., Choquet H., Stijnen P., Vatin V., Pigeyre M., Beckers S. Heterozygous mutations causing partial prohormone convertase 1 deficiency contribute to human obesity. Diabetes. 2012;61:383–390. doi: 10.2337/db11-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philippe J., Stijnen P., Meyre D., De Graeve F., Thuillier D., Delplanque J. A nonsense loss-of-function mutation in PCSK1 contributes to dominantly inherited human obesity. International Journal of Obesity (London) 2015;39:295–302. doi: 10.1038/ijo.2014.96. [DOI] [PubMed] [Google Scholar]
- 24.Reich A., Müller G., Gelbrich G., Deutscher K., Godicke R., Kiess W. Obesity and blood pressure-results from the examination of 2365 schoolchildren in Germany. International Journal of Obesity and Related Metabolic Disorders. 2003;27:1459–1464. doi: 10.1038/sj.ijo.0802462. [DOI] [PubMed] [Google Scholar]
- 25.Landgraf K., Friebe D., Ullrich T., Kratzsch J., Dittrich K., Herberth G. Chemerin as a mediator between obesity and vascular inflammation in children. The Journal of Clinical Endocrinology & Metabolism. 2012;97:E556–E564. doi: 10.1210/jc.2011-2937. [DOI] [PubMed] [Google Scholar]
- 26.Kromeyer-Hauschild K., Wabitsch M., Kunze D., Geller F., Geiß H.C., Hesse V. Perzentile für den Body-mass-Index für das Kindes- und Jugendalter unter Heranziehung verschiedener deutscher Stichproben. Monatsschrift Kinderheilkunde. 2001;149:807–818. [Google Scholar]
- 27.Rosenbloom K.R., Armstrong J., Barber G.P., Casper J., Clawson H., Diekhans M. The UCSC genome browser database: 2015 update. Nucleic Acids Research. 2015;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Computational Biology. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 30.Choi Y., Chan A.P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reva B., Antipin Y., Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biology. 2007;8:R232. doi: 10.1186/gb-2007-8-11-r232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Capriotti E., Calabrese R., Fariselli P., Martelli P.L., Altman R.B., Casadio R. WS-SNPs&GO: a web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genomics. 2013;14(Suppl. 3):S6. doi: 10.1186/1471-2164-14-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hazel M., Cooksey R.C., Jones D., Parker G., Neidigh J.L., Witherbee B. Activation of the hexosamine signaling pathway in adipose tissue results in decreased serum adiponectin and skeletal muscle insulin resistance. Endocrinology. 2003 doi: 10.1210/en.2003-0812. [DOI] [PubMed] [Google Scholar]
- 35.Orci L., Tagaya M., Amherdt M., Perrelet A., Donaldson J.G., Lippincott-Schwartz J. Brefeldin A, a drug that blocks secretion, prevents the assembly of non-clathrin-coated buds on Golgi cisternae. Cell. 1991;64:1183–1195. doi: 10.1016/0092-8674(91)90273-2. [DOI] [PubMed] [Google Scholar]
- 36.Rose A.B. Intron-mediated regulation of gene expression. Current Topics in Microbiology and Immunology. 2008;326:277–290. doi: 10.1007/978-3-540-76776-3_15. [DOI] [PubMed] [Google Scholar]
- 37.Back S.H., Kaufman R.J. Endoplasmic reticulum stress and type 2 diabetes. Annual Review of Biochemistry. 2012;81:767–793. doi: 10.1146/annurev-biochem-072909-095555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woehlbier U., Hetz C. Modulating stress responses by the UPRosome: a matter of life and death. Trends in Biochemical Sciences. 2011;36:329–337. doi: 10.1016/j.tibs.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Ma Y., Brewer J.W., Diehl J.A., Hendershot L.M. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. Journal of Molecular Biology. 2002;318:1351–1365. doi: 10.1016/s0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Negishi M. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6alpha and 6beta that activates the mammalian unfolded protein response. Molecular Cell Biology. 2001;21:1239–1248. doi: 10.1128/MCB.21.4.1239-1248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin W.C., Chuang Y.C., Chang Y.S., Lai M.D., Teng Y.N., Su I.J. Endoplasmic reticulum stress stimulates p53 expression through NF-kappaB activation. PLoS One. 2012;7:e39120. doi: 10.1371/journal.pone.0039120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter P., Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 43.Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fewell S.W., Travers K.J., Weissman J.S., Brodsky J.L. The action of molecular chaperones in the early secretory pathway. Annual Review of Genetics. 2001;35:149–191. doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- 45.Zhu X., Zhou A., Dey A., Norrbom C., Carroll R., Zhang C. Disruption of PC1/3 expression in mice causes dwarfism and multiple neuroendocrine peptide processing defects. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:10293–10298. doi: 10.1073/pnas.162352599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mbikay M., Croissandeau G., Sirois F., Anini Y., Mayne J., Seidah N.G. A targeted deletion/insertion in the mouse Pcsk1 locus is associated with homozygous embryo preimplantation lethality, mutant allele preferential transmission and heterozygous female susceptibility to dietary fat. Developmental Biology. 2007;306:584–598. doi: 10.1016/j.ydbio.2007.03.523. [DOI] [PubMed] [Google Scholar]
- 47.Lloyd D.J., Bohan S., Gekakis N. Obesity, hyperphagia and increased metabolic efficiency in Pc1 mutant mice. Human Molecular Genetics. 2006;15:1884–1893. doi: 10.1093/hmg/ddl111. [DOI] [PubMed] [Google Scholar]
- 48.Heni M., Haupt A., Schafer S.A., Ketterer C., Thamer C., Machicao F. Association of obesity risk SNPs in PCSK1 with insulin sensitivity and proinsulin conversion. BMC Medical Genetics. 2010;11:86. doi: 10.1186/1471-2350-11-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kilpelainen T.O., Bingham S.A., Khaw K.T., Wareham N.J., Loos R.J. Association of variants in the PCSK1 gene with obesity in the EPIC-Norfolk study. Human Molecular Genetics. 2009;18:3496–3501. doi: 10.1093/hmg/ddp280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qi Q., Li H., Loos R.J., Liu C., Hu F.B., Wu H. Association of PCSK1 rs6234 with obesity and related traits in a Chinese Han population. PLoS One. 2010;5:e10590. doi: 10.1371/journal.pone.0010590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang Y.C., Chiu Y.F., Shih K.C., Lin M.W., Sheu W.H., Donlon T. Common PCSK1 haplotypes are associated with obesity in the Chinese population. Obesity (Silver Spring) 2010;18:1404–1409. doi: 10.1038/oby.2009.390. [DOI] [PubMed] [Google Scholar]
- 52.Jansen E., Ayoubi T.A., Meulemans S.M., Van de Ven W.J. Neuroendocrine-specific expression of the human prohormone convertase 1 gene. Hormonal regulation of transcription through distinct cAMP response elements. Journal of Biological Chemistry. 1995;270:15391–15397. doi: 10.1074/jbc.270.25.15391. [DOI] [PubMed] [Google Scholar]
- 53.Shen X., Li Q.L., Brent G.A., Friedman T.C. Thyroid hormone regulation of prohormone convertase 1 (PC1): regional expression in rat brain and in vitro characterization of negative thyroid hormone response elements. Journal of molecular endocrinology. 2004;33:21–33. doi: 10.1677/jme.0.0330021. [DOI] [PubMed] [Google Scholar]
- 54.Bianchi M., Crinelli R., Giacomini E., Carloni E., Magnani M. A potent enhancer element in the 5′-UTR intron is crucial for transcriptional regulation of the human ubiquitin C gene. Gene. 2009;448:88–101. doi: 10.1016/j.gene.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 55.Bennett K.L., Romigh T., Arab K., Teresi R.E., Tada Y., Eng C. Activator protein 2 alpha (AP2alpha) suppresses 42 kDa C/CAAT enhancer binding protein alpha (p42(C/EBPalpha)) in head and neck squamous cell carcinoma. International Journal of Cancer. 2009;124:1285–1292. doi: 10.1002/ijc.24087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.