Abstract
Objective
α-cells are the second most prominent cell type in pancreatic islets and are responsible for producing glucagon to increase plasma glucose levels in times of fasting. α-cell dysfunction and inappropriate glucagon secretion occur in both type 1 and type 2 diabetes. Thus, there is growing interest in studying both normal function and pathophysiology of α-cells. However, tools to target gene ablation or activation specifically of α-cells have been limited, compared to those available for β-cells. Previous Glucagon-Cre and Glucagon-CreER transgenic mouse lines have suffered from transgene silencing, and the only available Glucagon-CreER “knock-in” mouse line results in glucagon haploinsufficiency, which can confound the interpretation of gene deletion analyses. Therefore, we sought to develop a Glucagon-CreERT2 mouse line that would maintain normal glucagon expression and would be less susceptible to transgene silencing.
Methods
We utilized CRISPR-Cas9 technology to insert an IRES-CreERT2 sequence into the 3′ UTR of the Glucagon (Gcg) locus in mouse embryonic stem cells (ESCs). Targeted ESC clones were then injected into mouse blastocysts to obtain Gcg-CreERT2 mice. Recombination efficiency in GCG+ pancreatic α-cells and glucagon-like peptide 1 positive (GLP1+) enteroendocrine L-cells was measured in Gcg-CreERT2;Rosa26-LSL-YFP mice injected with tamoxifen during fetal development and adulthood.
Results
Tamoxifen injection of Gcg-CreERT2;Rosa26-LSL-YFP mice induced high recombination efficiency of the Rosa26-LSL-YFP locus in perinatal and adult α-cells (88% and 95%, respectively), as well as in first-wave fetal α-cells (36%) and adult enteroendocrine L-cells (33%). Mice homozygous for the Gcg-CreERT2 allele were phenotypically normal.
Conclusions
We successfully derived a Gcg-CreERT2 mouse line that expresses CreERT2 in pancreatic α-cells and enteroendocrine L-cells without disrupting preproglucagon gene expression. These mice will be a useful tool for performing temporally controlled genetic manipulation specifically in these cell types.
Keywords: Islet, α-cell, Enteroendocrine L-cell, Glucagon, GLP1, CRISPR
Abbreviations: Cre, Cre recombinase; CreERT2, tamoxifen-inducible Cre recombinase-estrogen receptor fusion protein; CRISPR, clustered regularly interspaced short palindromic repeat; DAPI, 4′,6-diamidino-2-phenylindole; ESC, embryonic stem cell; FACS, fluorescence-activated cell sorting; GCG, glucagon; GLP1, glucagon-like peptide 1; IRES, internal ribosomal entry site; LSL, loxP-stop-loxP; gRNA, guide RNA; UTR, untranslated region; YFP, yellow fluorescent protein
Highlights
-
•
Developed a new Gcg-CreERT2 knock-in mouse line using CRISPR-Cas9 gene targeting.
-
•
Gcg-CreERT2 mice exhibit high recombination in α-cells and L-cells.
-
•
There is no evidence of preproglucagon haploinsufficiency in Gcg-CreERT2 mice.
1. Introduction
Pancreatic α-cells secrete glucagon to increase plasma glucose levels in times of fasting and in opposition to insulin action [1]. α-cells make up 9–31% of the endocrine cells within murine islets of Langerhans, while the composition of human islets is much more variable than that of rodents, and the α-cell fraction of human islet endocrine cells can range from 10 to 65% [2]. It is now evident that α-cell dysfunction contributes to dysglycemia in type 1 and type 2 diabetes mellitus. Hyperglucagonemia exacerbates fasting and post-prandial hyperglycemia in type 2 diabetes [3], [4], [5], while patients with type 1 diabetes exhibit inappropriately low glucagon levels during hypoglycemia [6] but inappropriately elevated glucagon levels during hyperglycemia [3], leading to broad and unpredictable glucose fluctuations. At present, the underlying etiologies of α-cell dysfunction in these disorders are unclear but do not seem to be completely attributable to insulin deficiency or resistance. Thus, the fields of diabetes and islet biology research could benefit from further specific investigation of α-cell physiology and pathophysiology. Additionally, several studies have shown that α-cells can transdifferentiate into β-cells [7], [8], [9], and there is much interest in targeting α-cells to directly stimulate such transdifferentiation as a means of generating new β-cells for diabetes therapy.
Currently, there are few mouse models available with which to genetically modify α-cells in order to specifically study this cell population, and most of these mouse models are less effective than initially thought. The first Glucagon-Cre transgenic mouse line for α-cell specific gene ablation of loxP-flanked gene targets was generated in 2000. This randomly inserted transgene contained a 1.6 kb region of the rat Glucagon promoter and was initially reported to have a recombination efficiency in α-cells of 100% [10]. A later report in 2009 demonstrated only 85% recombination in α-cells [11]. Since then, multiple colonies of these Glucagon-Cre transgenic mice distributed to different labs have shown only 30–45% α-cell recombination, which is often insufficient to uncover the physiologic consequences of gene ablation, as the majority of α-cells still contain the wild type version of the gene of interest [12], [13], [14]. In 2013, a Glucagon-CreERT2 gene replacement mouse line was developed but demonstrated only 50–70% recombination efficiency in α-cells, which we confirmed (data not shown). However, a significant disadvantage of this Glucagon-CreERT2 mouse line is that the gene targeting design replaced the endogenous preproglucagon coding sequence with the CreERT2 cDNA, thus making these mice haploinsufficient for preproglucagon, confounding the analysis of gene deletions on α-cell function.
In addition to pancreatic α-cells, the Preproglucagon transcript is also expressed in the L-cells of the intestine. L-cells are a subtype of enteroendocrine cells that share a common developmental transcriptional pattern with α-cells [15], [16] and are located within the epithelial lining of the small intestine and colon. In contrast to α-cells, which utilize prohormone convertase 2 to process Preproglucagon into Glucagon, the L-cells utilize prohormone convertases 1 and 3 to generate Glucagon-like peptide 1 (GLP1), GLP2, Oxyntomodulin, and Glicentin [17], [18]. GLP1 has many beneficial effects on β-cells and is thus a well-known diabetes therapeutic target. Interestingly, there is some evidence that α-cells and L-cells can alter their expression of prohormone convertase type, resulting in GLP1 production by α-cells or Glucagon production by L-cells. Thus, there is much interest in better understanding and harnessing L-cell biology.
Here, we describe a novel Glucagon-CreERT2 gene-addition mouse line that was generated via CRISPR-Cas9 assisted gene targeting, does not disrupt glucagon expression from the targeted allele, and recapitulates endogenous glucagon and GLP1 expression. This will be a useful tool for the scientific community to perform specific genetic manipulations in murine α-cells and enteroendocrine L-cells.
2. Materials and methods
2.1. Cloning
The 5′ homology arm (1,044 bp mapping to chr2:62,474,721–62,475,764 on the mm10 build of the mouse genome, including part of exon 5, all of intron 5 and exon 6, and part of the 3′ untranslated region (UTR) of the Gcg locus) and the 3′ homology arm (1,039 bp mapping to chr2:62,473,652–62,474,690, including part of the 3′ UTR of the Gcg locus and distal intergenic sequence) were PCR-amplified from mouse genomic DNA using the Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA, USA). IRES and CreERT2 sequences were PCR-amplified from existing plasmids. See Supplemental Table 1 for PCR primers. The In-Fusion HD Cloning Kit (Clontech, Mountain View, CA, USA) was used to sub-clone the PCR-amplified sequences into the pBS vector (Stratagene, La Jolla, CA, USA) that was linearized with XmaI and EcoRI, to generate the gene targeting vector.
A guide RNA (gRNA) targeting the ATTATCGCAGTCACAACACC sequence in the Gcg 3′ UTR was cloned into the pX335 vector containing Cas9, following the CRISPR Genome Engineering Toolbox Target Sequence Cloning Protocol (http://www.genome-engineering.org/crispr/).
2.2. ESC targeting
Mouse V6.5 embryonic stem cells (ESCs; C57BL/6 and 129/sv F1 hybrid) were co-electroporated with the linearized pBS-based repair template vector, the pX335-sgRNA vector, and the Oct4-eGFP-PGK-PuroR plasmid [19]. Transient transfectants were selected with 2 μg/mL puromycin for 72 h. 192 clones were expanded and genotyped using primers to detect the 5′ and 3′ targeted sites (Supplemental Table 1), which confirmed proper targeting of seven clones. These clones were karyotyped, and two of the clones with the highest percentage of cells with normal karyotype were injected into pseudo-pregnant C57BL/6 female mice, resulting in two and eight chimeric pups, respectively. The chimeric mice were mated to C57BL/6 mice to obtain germline transmission of the Gcg-CreERT2 gene-addition allele.
2.3. Mice
Genotyping for Gcg-CreERT2 and Rosa26-LSL-YFP was performed using GoTaq Green Master Mix (Promega, Madison, WI, USA) with the primers and PCR conditions listed in Supplemental Table 2. Heterozygous Gcg-CreERT2 mice were mated to homozygous Rosa26-LSL-YFP mice on the C57BL/6 background (The Jackson Laboratory, Bar Harbor, ME, USA, strain #006148) to obtain Gcg-CreERT2;Rosa26-LSL-YFP mice. 2–3 month old male and female Gcg-CreERT2;Rosa26-LSL-YFP mice were injected intraperitoneally with 100 μg/g body weight of tamoxifen (Sigma–Aldrich, St. Louis, MO, USA, catalog #T5648) in corn oil (20 mg/mL), once daily for 3 consecutive days. Mice injected with or without tamoxifen were housed separately. The mice were killed 2 days, 7 days, or 1 month after the final injection. For the analysis of gene ablation efficacy during fetal development, adult male heterozygous Gcg-CreERT2 mice were mated to female homozygous Rosa26-LSL-YFP mice, and the mice were separated when a vaginal plug was observed (embryonic day [e] 0.5). Pregnant females were injected with 100 μg of tamoxifen at e9.5, e13.5, or e18.5, then 2 days later the embryos or pups (postnatal day [P] 1) were killed, and the intestinal tracts including pancreata were harvested. All mouse protocols were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
2.4. Immunofluorescence staining
Tissues were fixed with 4% paraformaldehyde (PFA) at 4 °C overnight (adult) or for 1 h (embryonic, postnatal), then rinsed in phosphate-buffered saline (PBS), and either dehydrated for paraffin embedding or immersed in 30% sucrose overnight at 4 °C and embedded in optimum cutting temperature compound (OCT), then frozen for cryosectioning. Paraffin sections underwent antigen retrieval with R Buffer A (Electron Microscopy Sciences, Hatfield, PA, USA) in a pressure cooker (PickCell Laboratories, Agoura Hills, CA) for 1 h. Cre immunolabeling required cryosections. Sections were blocked with CAS-Block (ThermoFisher, Waltham, MA, USA). Primary antibodies including goat anti-GFP (1:200, Abcam ab6673 lot 142576), rabbit anti-glucagon (1:200, Santa Cruz sc-13091), guinea pig anti-insulin (1:500, Invitrogen 180067), and rabbit anti-Cre (1:10,000, Millipore 69050-3) were incubated with the sections overnight at 4 °C. After washing, donkey Cy2-, Cy3-, and Cy5-conjugated secondary antibodies (1:250–1:500, Jackson ImmunoResearch Laboratories) were incubated with sections at room temperature for 3 h. Sections were then incubated with DAPI (4′,6-diamidino-2-phenylindole) for 1 min at room temperature to counterstain DNA prior to mounting with coverslips. Images were obtained using a Nikon Eclipse 80i widefield microscope. The anti-GFP antibody cross-reacts with YFP, and the anti-glucagon antibody cross-reacts with GLP1.
2.5. Islet isolation and fluorescence-activated cell sorting (FACS)
Islets from adult Gcg-CreERT2;Rosa26-LSL-YFP mice at 3–4 months of age that had been injected with tamoxifen (n = 4) or without (n = 3) were isolated by collagenase digestion of pancreatic tissue followed by hand-picking, as previously described [20], [21], yielding 800 and 500 islets per group, respectively. The islets were immediately dissociated with 0.05% trypsin for 3 min at 37 °C, and the reaction was quenched with fetal bovine serum (FBS). Dissociated islet cells were incubated with Aqua Live/Dead (ThermoFisher, Waltham, MA, USA) per manufacturer's instructions, then fixed with 1% PFA for 10 min at room temperature, and quenched with 3 mM glycine. Cells were then incubated with goat anti-GFP and rabbit anti-glucagon primary antibodies at 1:200 dilution followed by donkey Cy2-conjugated anti-goat and Cy5-conjugated anti-rabbit secondary antibodies at 1:500 dilution, both steps in blocking buffer (1% BSA, 0.1% saponin, 1X PBS) for 20 min at room temperature. Cells were sorted using FACSAria II (BD Biosciences, San Jose, CA, USA).
2.6. Plasma glucagon measurements
Adult mice at 2–4 months of age were fasted overnight for 16 h, then anesthetized using continuous isoflurane inhalation. Plasma glucose was measured from the tail tip using a Breeze 2 glucometer (Bayer, Mishawaka, IN, USA). Whole blood was then collected by cardiac puncture and transferred to heparinized tubes (Sarstedt, Nümbrecht, Germany), which were temporarily stored on ice, then centrifuged at 2,000×g for 5 min at 4 °C, and the plasma supernatant was stored at −20 °C. Plasma glucagon levels were measured using the Glucagon ELISA – 10 μL kit (Mercodia, Uppsala, Sweden), following the manufacturer's instructions and using technical duplicates.
2.7. Pancreas harvest for RT-qPCR and protein
Whole pancreas was collected from adult mice after overnight fast as described above, then immediately immersed in 15 mL TRIzol Reagent (ThermoFisher, Waltham, MA, USA), homogenized, snap-frozen in liquid nitrogen, and stored at −80 °C. RNA was later extracted from the aqueous phase per manufacturer's instructions using 0.2X chloroform, followed by isopropanol precipitation. RNA quality and concentration were analyzed with the RNA 6000 Nano kit (Agilent, Santa Clara, CA, USA). cDNA was synthesized from 1 μg of total RNA using SuperScript II Reverse Transcriptase (ThermoFisher, Waltham, MA, USA). qPCR was performed using the intron-spanning primers listed in Supplemental Table 3 and 2X Brilliant III SYBR Green QPCR Master Mix (Agilent, Santa Clara, CA, USA), with technical triplicates.
DNA and protein were extracted from the organic phase of the Trizol homogenate, per manufacturer's instructions, with modified protein resuspension based on [22]. Total protein content was measured using the Bio-Rad Protein Assay standard procedure for microtiter plates (Bio-Rad, Hercules, CA, USA) using technical triplicates. Pancreatic glucagon content was measured using the Glucagon ELISA – 10 μL kit (Mercodia, Uppsala, Sweden), following the manufacturer's instructions and using technical duplicates.
2.8. Statistical analyses
Prism 6.0 (GraphPad Software, La Jolla, CA, USA) was used to make graphs and perform statistical analyses. RT-qPCR and ELISA results were analyzed using one-way ANOVA with Tukey's multiple comparisons test with a single pooled variance.
3. Results
3.1. Gcg-CreERT2 expression
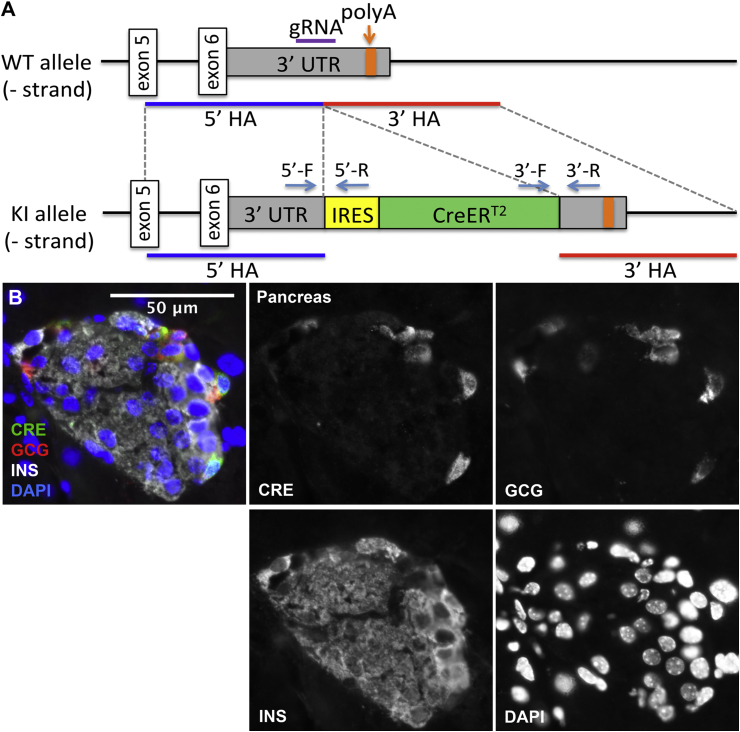
Mice with germline transmission of IRES-CreERT2 inserted in the 3′ UTR of the Gcg locus were derived from targeted mouse ESCs electroporated with vectors containing Cas9 nuclease with a gRNA designed to target the 3′ UTR of the Gcg allele, and a repair template containing the IRES-CreERT2 sequence flanked by 5′ and 3′ arms homologous to the Gcg locus (Figure 1A; for details see Materials and methods section). Pancreata from adult Gcg-CreERT2 mice showed Cre expression exclusively in α-cells (Figure 1B).
Figure 1.
Gcg-CreERT2targeting design and expression in the adult mouse pancreas. (A) The IRES-CreERT2 sequence was inserted into the 3′ UTR of the Gcg locus in murine embryonic stem cells by homology-directed repair. Note that the coding exons of Gcg were not altered after homologous recombination (HA: homology arm; 5′-F, 5′-R, 3′-F, and 3′-R denote the 5′ and 3′ Targeted Site primers, respectively). (B) Expression of CreERT2 in pancreas from adult Gcg-CreERT2 mice is restricted to α-cells, as detected by immunofluorescence staining for Cre protein (green) and glucagon (red), and is absent from β-cells, as detected by immunofluorescent labeling for insulin (white).
3.2. Gcg-CreERT2 recombination efficiency
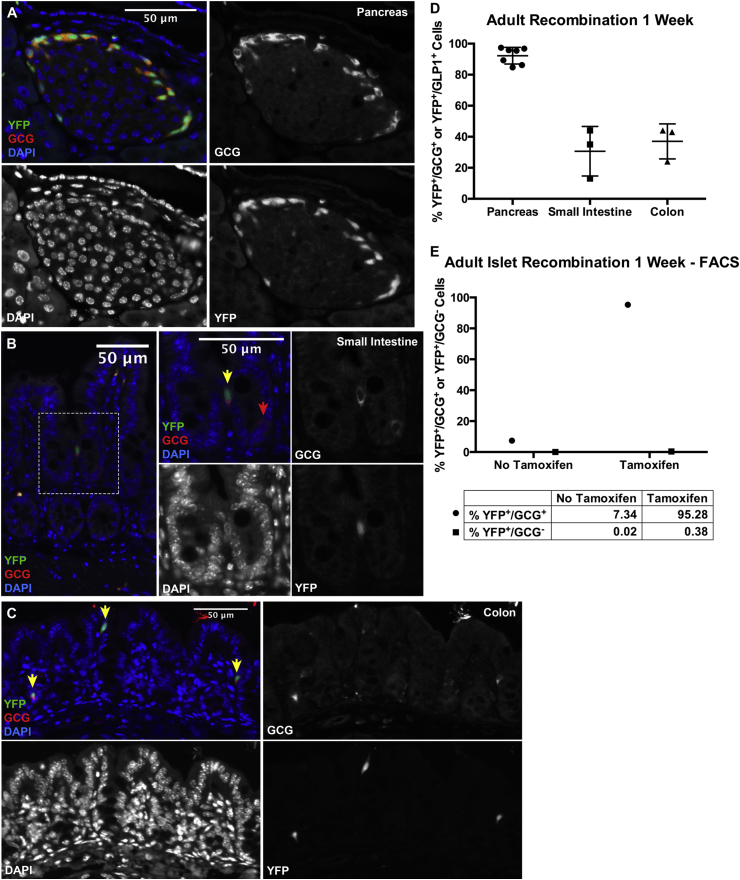
Adult Gcg-CreERT2;Rosa-LSL-YFP mice injected with tamoxifen and killed 7 days later exhibited YFP expression, indicative of CreERT2-mediated recombination of the Rosa-LSL-YFP locus, in 92% of GCG-positive pancreatic α-cells by histology (Figure 2A and D) and 95% by FACS (Figure 2E). YFP was also expressed in 31% of enteroendocrine L-cells in the small intestine (Figure 2B and D), and 37% of enteroendocrine L-cells in the colon (Figure 2C and D), respectively, by histology. A similar percentage of YFP+/GLP1+ cells were observed in the small intestine and colon of adult mice killed 2 days after the final tamoxifen injection (data not shown). YFP was very rarely identified in GCG-negative cells in islets (Figure 2A and E). However, some YFP+/GLP1− cells were observed in the intestine.
Figure 2.
Gcg-CreERT2efficiently recombines the Rosa-LSL-YFP locus in GCG-expressing α-cells in the adult mouse pancreas, and in GLP1-expressing L-cells in the adult mouse intestine. (A–C) Adult Gcg-CreERT2;Rosa-LSL-YFP mice injected with tamoxifen and killed 7 days later exhibited high recombination efficiency, indicated by YFP expression (green), in pancreatic α-cells (A) labeled with an anti-glucagon antibody (red), as well as in enteroendocrine L-cells marked by GLP1 expression (red) in the small intestine (B) and colon (C). YFP+/GLP1+ cells are indicated by yellow arrows, and YFP−/GLP1+ cells are indicated by red arrows. GLP1 was detected with a cross-reacting anti-glucagon antibody. Both the overlay and single channel immunofluorescence are shown to demonstrate the specificity of Cre activity in α- and enteroendocrine L-cells. (D) Quantification of percent YFP+/GCG+ cells in adult pancreas and YFP+/GLP1+ cells in adult small intestine and colon. All cells on at least 2 histological sections were counted for each mouse, resulting in at least 20 islets with approximately 150–650 GCG+ cells in the pancreas, 300–500 GLP1+ cells in the small intestine, or 900–2,000 GLP1+ cells in the colon, being counted per mouse. Data points for individual mice are shown. Error bars represent standard deviation. (E) Quantification of YFP+/GCG+ cells (circles) and YFP+/GCG− cells (squares) by FACS from adult islets of mice injected with or without tamoxifen. A total of 98,776 and 12,832 islet cells were counted, respectively.
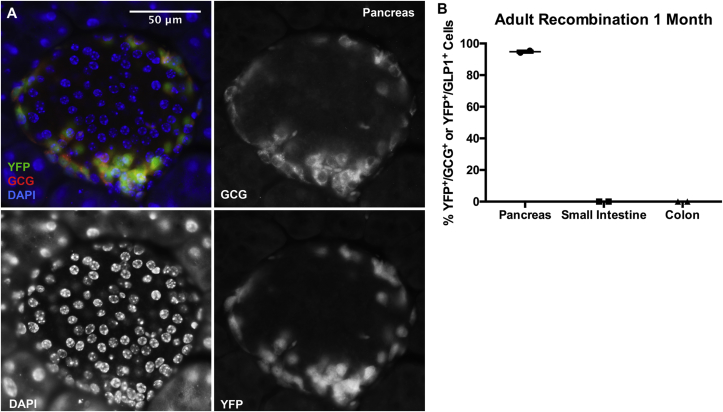
Furthermore, YFP expression persisted in 95% of pancreatic α-cells of adult Gcg-CreERT2;Rosa-LSL-YFP mice killed 1 month after tamoxifen injection (Figure 3A and B), consistent with the low turnover rate of these cells. In contrast, no enteroendocrine L-cells were found to express YFP 1 month after tamoxifen injection (Figure 3B), because enteroendocrine cells, like goblet cells and enterocytes, are replaced by new cells originating from intestinal crypts every three to five days, and because tamoxifen disappeared from the circulation.
Figure 3.
Gcg-CreERT2-induced α-cell lineage label is stable. (A) Adult Gcg-CreERT2;Rosa-LSL-YFP mice injected with tamoxifen and killed 1 month later exhibited persistent YFP expression in α-cells. (B) Quantification of percent YFP+/GCG+ cells in adult pancreas and YFP+/GLP1+ cells in the intestine, with data points for individual mice. 130–300 GCG+ cells in the pancreas, 200–400 GLP1+ cells in the small intestine, and 400–1,000 cells in the colon were counted per mouse. Error bars represent standard deviation.
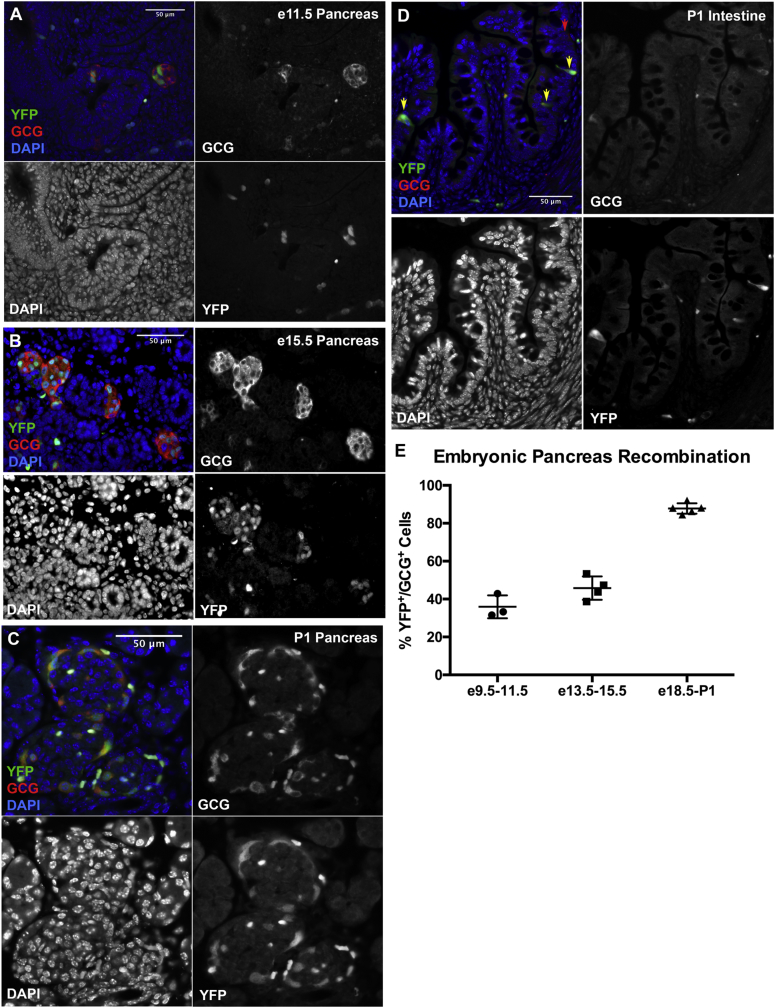
To evaluate CreERT2-mediated recombination efficiency during pancreas development, pregnant female mice were injected with tamoxifen at different embryonic stages. Expression of YFP was observed in 36% of first-wave GCG+ cells at e11.5 after low-dose tamoxifen injection at e9.5 (Figure 4A and E). Recombination was observed in 46% of early second-wave GCG+ cells at e15.5 after tamoxifen injection at e13.5 (Figure 4B and E), and in 88% of GCG+ cells at P1 after tamoxifen injection at e18.5 (Figure 4C and E). Among these time points, YFP and GLP1 expression were first identified in the intestine at P1, with a high percentage of YFP+/GLP1+ cells (Figure 4D, n = 5).
Figure 4.
Gcg-CreERT2drives recombination of loxP-flanked targets in embryogenesis in pancreatic α-cells and intestinal L-cells. (A–D) Low-dose tamoxifen administered to pregnant females induced YFP expression (green) in α-cells (red) of Gcg-CreERT2;Rosa-LSL-YFP embryos at e9.5-e11.5 (A), e13.5-e15.5 (B), and e18.5-P1 (C), as well as in enteroendocrine L-cells (red) at e18.5-P1 (D). (E) Quantification of percent YFP+/GCG+ cells in embryonic pancreas, with data points for individual mice. All sections containing e11.5 and e15.5 pancreas were used, resulting in 20–40 GCG+ cells being counted per pancreas for e11.5, and 120–880 GCG+ cells being counted per pancreas for e15.5. 70–400 GCG+ cells were counted per pancreas for P1. Error bars represent standard deviation.
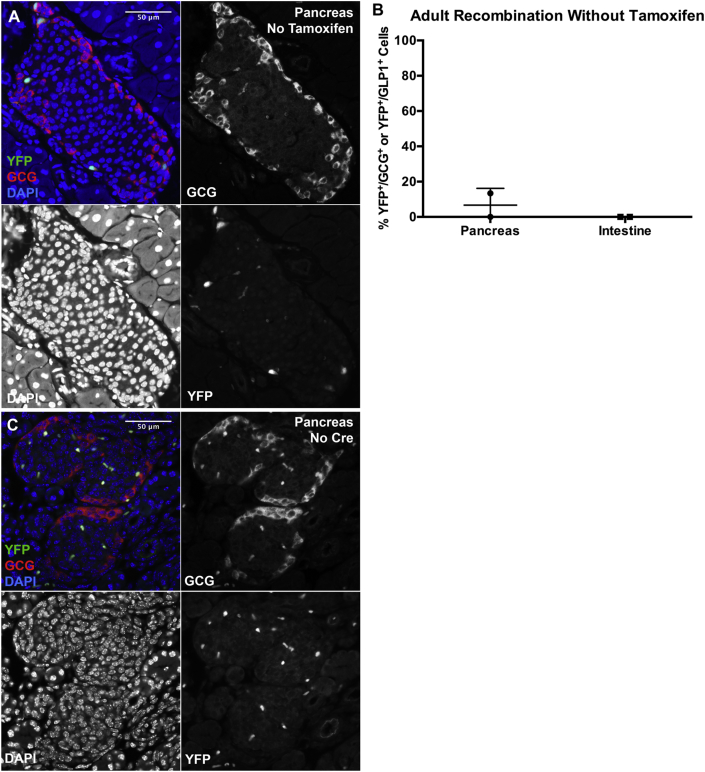
Notably, Gcg-CreERT2;Rosa-LSL-YFP mice exhibited some “leaky” recombination in the absence of tamoxifen, with 0–13% of adult α-cells expressing YFP without injection of tamoxifen by histology (Figure 5A and B) and an average of 7% by FACS (Figure 2E). There was no YFP expression observed in the intestine of Gcg-CreERT2;Rosa-LSL-YFP mice not injected with tamoxifen (Figure 5B) nor in pancreata from Rosa-LSL-YFP mice injected with tamoxifen (Figure 5C, n = 4). No YFP expression was detected in the midbrain (data not shown).
Figure 5.
The Gcg-CreERT2mouse line exhibits some leaky expression in the absence of tamoxifen administration. (A, B) YFP (green) is expressed at a low level in α-cells (A, B) but not enteroendocrine L-cells (B) of adult Gcg-CreERT2;Rosa-LSL-YFP mice not injected with tamoxifen. (C)Rosa-LSL-YFP P1 pups negative for the Gcg-CreERT2 allele show no YFP-positive cells 2 days after tamoxifen injection (the bright green and red spots are red blood cells).
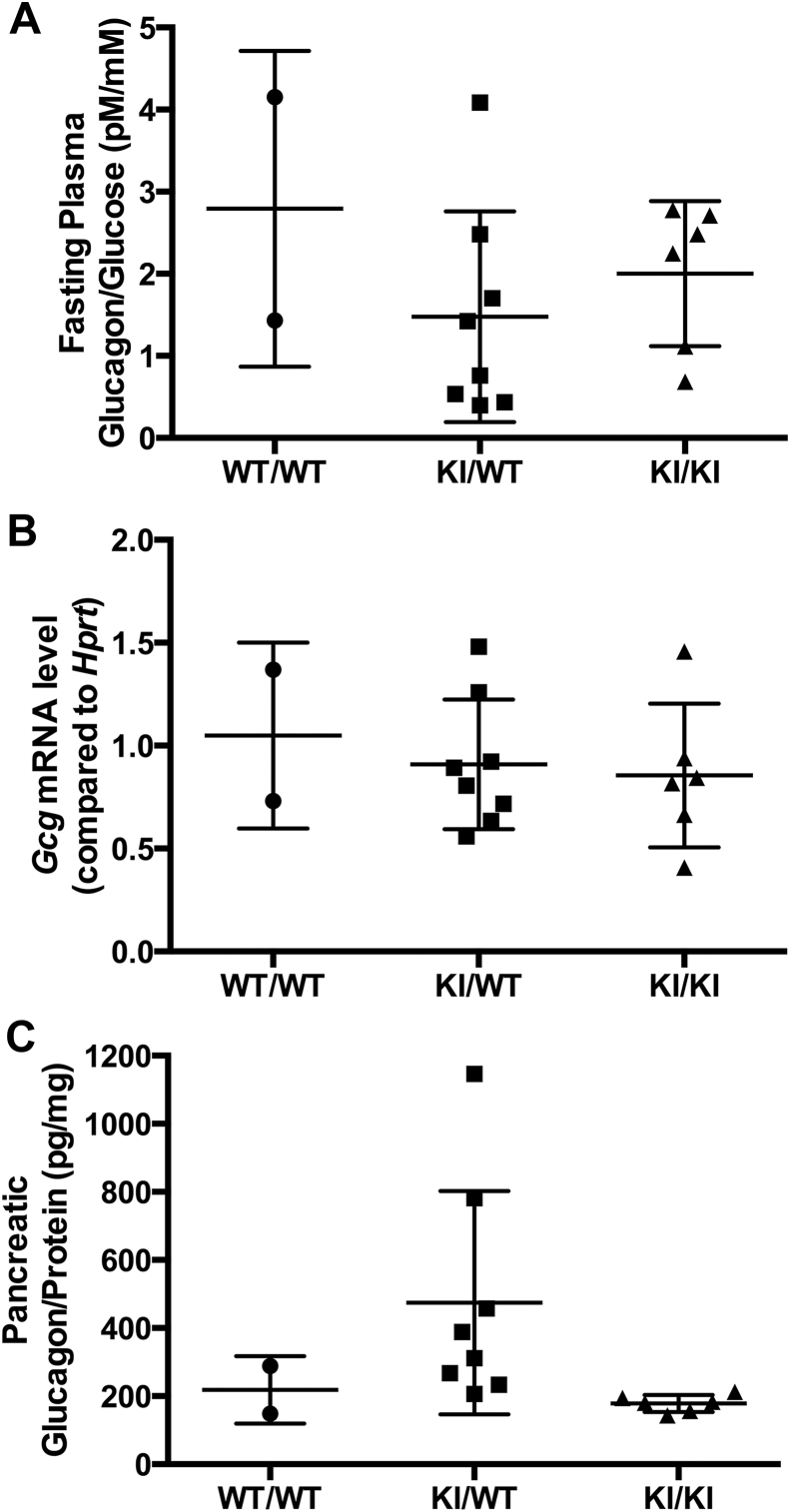
3.3. Glucagon expression
Gcg-CreERT2 heterozygous and homozygous mice were healthy and exhibited comparable fasting plasma glucagon levels as wild type littermates (Figure 6A). Likewise, Gcg mRNA and GCG protein levels in total pancreas homogenates were not significantly different in Gcg-CreERT2 heterozygous and homozygous mice compared to wild type littermate controls (Figure 6B and C). Thus, there is no indication of a deleterious effect of the Gcg-CreERT2 gene-addition allele described above.
Figure 6.
The Gcg-CreERT2allele does not alter glucagon expression or plasma glucagon levels even in the homozygous configuration. (A) Plasma glucagon levels were similar in wild type (WT) and Gcg-CreERT2 heterozygous (KI/WT) and homozygous (KI/KI) littermate mice after 16 h fast. Values were normalized to plasma glucose levels at the time of blood collection. Error bars represent standard deviation. (B)Glucagon mRNA levels in the whole pancreas were unchanged in Gcg-CreERT2 heterozygous and homozygous mice compared to wild type littermate controls. Delta-Ct values were obtained by comparing to the Hprt reference gene, and delta-delta-Ct values were obtained by normalizing to the wild type average. Error bars represent standard deviation. (C) Glucagon protein levels in the whole pancreas were unchanged in Gcg-CreERT2 heterozygous and homozygous mice compared to wild type littermate controls. Values were normalized to total pancreatic protein. Error bars represent standard deviation.
4. Discussion
Here, we derived an improved mouse model for conditional and inducible genetic manipulation in preproglucagon-expressing cell lineages including pancreatic α-cells and enteroendocrine L-cells. This system allows for time-controlled and cell type-specific gene ablation, gene mutation, or gene activation, depending on the design of the loxP-modified gene of interest. Because of the different kinetics of cell turnover between intestinal L-cells and pancreatic α-cells, it is even possible to obtain mice in which the target of interest is ablated only in α-cells, while in intestinal L-cells the original mutant cells have been replaced by wild type cells produced in the rapidly proliferating crypts. This can be achieved by simply waiting a few weeks following tamoxifen administration, for mutated L-cells to be replaced by wild type L-cells originating from the crypts.
The low level of Rosa-LSL-YFP recombination that was observed in α-cells in the absence of tamoxifen is similar to that described in other CreERT2 mouse models, including Pdx1-CreER mice [23], [24]. This leaky expression is likely due to a combination of high expression of CreERT2 from the preproglucagon locus, as well as the particularly easily-recombined nature of the Rosa locus in comparison to other loci.
Using CRISPR-Cas9 assisted homology-directed recombination, we inserted an IRES-CreERT2 sequence into the 3′ UTR of the Gcg locus in murine ESCs without disrupting the Gcg coding exons, allowing for co-expression of GCG and CreERT2. Because this mouse model utilizes the endogenous Gcg promoter to drive expression of CreERT2, expression of CreERT2 mirrors that of the endogenous preproglucagon gene, even at early developmental time points. Furthermore, CreERT2 expression in this mouse model is less likely to be silenced than in previous transgenic mouse models, as it is located in the native gene context.
5. Conclusions
We generated a new and improved Gcg-CreERT2 mouse line that expresses CreERT2 from the endogenous preproglucagon locus without disrupting preproglucagon expression. This mouse line exhibits high recombination efficiency in pancreatic α-cells and enteroendocrine L-cells without disrupting preproglucagon gene expression. We expect that because the CreERT2 gene utilizes an endogenous promoter, rather than a transgenic promoter, its expression will be less susceptible to gene silencing, as has affected other Gcg-CreER mouse lines. In summary, these Gcg-CreERT2 mice can be used to temporally manipulate genetics specifically in pancreatic α-cells and enteroendocrine L-cells.
Contributions
AMA designed experiments, collected data, analyzed data, drafted the article, revised the article, and approved the final version for publication. JZ conceived the study, designed experiments, revised the article, and approved the final version for publication. AH designed experiments and approved the final version for publication. AB collected and analyzed data and approved the final version for publication. KHK conceived the study, designed experiments, revised the article, and approved the final version for publication.
Acknowledgements
We thank the following core facilities at the University of Pennsylvania for their assistance: Gene Targeting Core and Laboratory, Transgenic and Chimeric Mouse Facility (supported by the NIDDK grants P30 DK019525 and P30 DK050306), Pancreatic Islet Cell Biology Core (supported by P30 DK19525), and Molecular Pathology and Imaging Core (supported by P30 DK050306 and P01 DK049210). We thank Tia Bernard-Banks for animal husbandry. This work was supported by the Juvenile Diabetes Research Foundation grant 3-PDF-2014-186-A-N to AMA, by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant UC4 DK104119 to KHK, and by the American Diabetes Association grant 7-12-MN-37 to KHK.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.01.003.
Contributor Information
Amanda M. Ackermann, Email: ackermanna@email.chop.edu.
Jia Zhang, Email: zhangjia@upenn.edu.
Aryel Heller, Email: aryelh@mail.med.upenn.edu.
Anna Briker, Email: abriker@villanova.edu.
Klaus H. Kaestner, Email: kaestner@mail.med.upenn.edu.
Conflicts of interest
The authors have no conflicts of interest to report.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Quesada I., Tuduri E., Ripoll C., Nadal A. Physiology of the pancreatic alpha-cell and glucagon secretion: role in glucose homeostasis and diabetes. The Journal of Endocrinology. 2008;199(1):5–19. doi: 10.1677/JOE-08-0290. [DOI] [PubMed] [Google Scholar]
- 2.Brissova M., Fowler M.J., Nicholson W.E., Chu A., Hirschberg B., Harlan D.M. Assessment of human pancreatic islet architecture and composition by laser scanning confocal microscopy. Journal of Histochemistry & Cytochemistry. 2005;53(9):1087–1097. doi: 10.1369/jhc.5C6684.2005. [DOI] [PubMed] [Google Scholar]
- 3.Unger R.H., Cherrington A.D. Glucagonocentric restructuring of diabetes: a pathophysiologic and therapeutic makeover. The Journal of Clinical Investigation. 2012;122(1):4–12. doi: 10.1172/JCI60016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knop F.K., Aaboe K., Vilsboll T., Volund A., Holst J.J., Krarup T. Impaired incretin effect and fasting hyperglucagonaemia characterizing type 2 diabetic subjects are early signs of dysmetabolism in obesity. Diabetes Obesity and Metabolism. 2012;14(6):500–510. doi: 10.1111/j.1463-1326.2011.01549.x. [DOI] [PubMed] [Google Scholar]
- 5.Michaliszyn S.F., Mari A., Lee S., Bacha F., Tfayli H., Farchoukh L. Beta-cell function, incretin effect, and incretin hormones in obese youth along the span of glucose tolerance from normal to prediabetes to type 2 diabetes. Diabetes. 2014;63(11):3846–3855. doi: 10.2337/db13-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rickels M.R., Fuller C., Dalton-Bakees C., Marksman E., Palanjian M., Cullison K. Restoration of glucose counterregulation by islet transplantation in long-standing type 1 diabetes. Diabetes. 2015;64(5):1713–1718. doi: 10.2337/db14-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thorel F., Nepote V., Avril I., Kohno K., Desgraz R., Chera S. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464(7292):1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung C.H., Hao E., Piran R., Keinan E., Levine F. Pancreatic beta-cell neogenesis by direct conversion from mature alpha-cells. Stem Cells. 2010;28(9):1630–1638. doi: 10.1002/stem.482. [DOI] [PubMed] [Google Scholar]
- 9.Courtney M., Gjernes E., Druelle N., Ravaud C., Vieira A., Ben-Othman N. The inactivation of Arx in pancreatic alpha-cells triggers their neogenesis and conversion into functional beta-like cells. PLoS Genetics. 2013;9(10):e1003934. doi: 10.1371/journal.pgen.1003934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrera P.L. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127(11):2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 11.Kawamori D., Kurd A.J., Hu J., Liew C.W., Shih J.L., Ford E.L. Insulin signaling in alpha cells modulates glucagon secretion in vivo. Cell Metabolism. 2009;9(4):350–361. doi: 10.1016/j.cmet.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tudurí E., Denroche H.C., Kara J.A., Asadi A., Fox J.K., Kieffer T.J. Partial ablation of leptin signaling in mouse pancreatic α-cells does not alter either glucose or lipid homeostasis. American Journal of Physiology – Endocrinology and Metabolism. 2014;306(7):E748–E755. doi: 10.1152/ajpendo.00681.2013. [DOI] [PubMed] [Google Scholar]
- 13.Solomou A., Meur G., Bellow E., Hodson D.J., Tomas A., Migrenne Li S. The zinc transporter Slc30a8/ZnT8 is required in a subpopulation of pancreatic α-cells for hypoglycemia-induced glucagon secretion. The Journal of Biological Chemistry. 2015;290(35):21432–21442. doi: 10.1074/jbc.M115.645291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun G., da Silva Xavier G., Gorman T., Priest C., Solomon A., Hodson D.J. LKB1 and AMPKα1 are required in pancreatic alpha cells for the normal regulation of glucagon secretion and responses to hypoglycemia. Molecular Metabolism. 2015;4(4):277–286. doi: 10.1016/j.molmet.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beucher A., Gjernes E., Collin C., Courtney M., Meunier A., Collombat P. The homeodomain-containing transcription factors Arx and Pax4 control enteroendocrine subtype specification in mice. PLoS One. 2012;7(5):e36449. doi: 10.1371/journal.pone.0036449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bramswig N.C., Kaestner K.H. Transcriptional regulation of alpha-cell differentiation. Diabetes Obesity and Metabolism. 2011;13(Suppl 1):13–20. doi: 10.1111/j.1463-1326.2011.01440.x. [DOI] [PubMed] [Google Scholar]
- 17.Drucker D.J. Evolving concepts and translational relevance of enteroendocrine cell biology. The Journal of Clinical Endocrinology and Metabolism. 2016;101(3):778–786. doi: 10.1210/jc.2015-3449. [DOI] [PubMed] [Google Scholar]
- 18.Habener J.F., Stanojevic V. Alpha cells come of age. Trends in Endocrinology and Metabolism. 2013;24(3):153–163. doi: 10.1016/j.tem.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P. Genetic engineering of human pluripotent cells using TALE nucleases. Nature Biotechnology. 2011;29(8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imai Y., Patel H.R., Dolby N.M., Matschinsky F.M., Tobias J.W., Ahima R.S. Analysis of gene expression in pancreatic islets from diet-induced obese mice. Physiological Genomics. 2008;36(1):43–51. doi: 10.1152/physiolgenomics.00050.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gotoh M., Maki T., Satomi S., Porter J., Bonner-Weir S., O'Hara C.J. Reproducible high yield of rat islets by stationary in vitro digestion following pancreatic ductal or portal venous collagenase injection. Transplantation. 1987;43(5):725–730. doi: 10.1097/00007890-198705000-00024. [DOI] [PubMed] [Google Scholar]
- 22.Simoes A.E., Pereira D.M., Amara J.D., Nunes A.F., Gomes S.E., Rodrigues P.M. Efficient recovery of proteins from multiple source samples after TRIzol((R)) or TRIzol((R))LS RNA extraction and long-term storage. BMC Genomics. 2013;14:181. doi: 10.1186/1471-2164-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandhu U., Cebula M., Behme S., Riemer P., Wodarczyk C., Metzger D. Strict control of transgene expression in a mouse model for sensitive biological applications based on RMCE compatible ES cells. Nucleic Acids Research. 2011;39(1):e1. doi: 10.1093/nar/gkq868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., Suckale J., Masjkur J., Magro M.G., Steffen A., Anastassiadis K. Tamoxifen-independent recombination in the RIP-CreER mouse. PLoS One. 2010;5(10):e13533. doi: 10.1371/journal.pone.0013533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.