Abstract
The foo operon encodes F1651 fimbriae that belong to the P-regulatory family and are synthesized by septicemic Escherichia coli. Using an Lrp-deficient host and the lrp gene cloned under the arabinose pBAD promoter, we demonstrated that foo was transcribed proportionally to the amount of Lrp synthesized. l-Leucine and l-alanine decreased drastically the steady-state transcription of foo and modified phase variation, independently of the presence of FooI. Specific mutations in the C-terminal region of Lrp reduced or abolished the repressive effect of these amino acids, indicating that they modulate F1651 by affecting Lrp.
Virulence of Escherichia coli O115:KV165 strains associated with septicemia in young pigs involves synthesis of F1651 fimbriae, encoded by the foo gene cluster and belonging to the P fimbrial family (15). The regulatory region of the foo cluster contains two divergent open reading frames coding for the fooI and fooB genes, an arrangement similar to that of the pap operon (1, 14) (Fig. 1). The intercistronic region between the two promoters in both operons contains two GATC sites (GATC-I and GATC-II) separated by 102 bp (2).
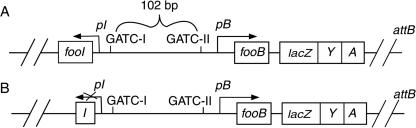
FIG. 1.
Schematic representations of fooIB-lacZYA and fooB-lacZYA fusions. (A) Strain FB1001 and derivatives bearing the fooIB-lacZYA fusion, including the fooI gene. (B) Strain FB2001 and derivatives bearing the fooB-lacZYA fusion, without the complete fooI gene. These fusions have been inserted at the attB site into the chromosome of E. coli CT4A by using the λRS45 derivative phages (λMT01 and λMT02). Both transcriptional fusions contained the foo regulatory region, including the two GATC sites. The arrows show the orientation of transcription, and p indicates foo promoters.
As is seen with many other virulence determinants, fimbriae are not expressed constitutively, and their synthesis is tightly regulated (20). Expression of F1651 is controlled at the transcriptional level by two mechanisms which modulate, on one hand, the percentage of cells producing fimbriae (ON cells) and not producing fimbriae (OFF cells) in a single colony (phase variation) and, on the other hand, the amount of antigen at the surface of a single cell (basal transcription) (8, 14). These two types of regulation are superimposed so that transcription in ON cells may be more or less active, leading to more or less fimbrial synthesis. Pap phase variation is regulated by a complex epigenetic mechanism that specifies the formation of specific DNA methylation patterns of two GATC sites (2, 16). Regulation of F1651 expression is equally complex, being mediated by Lrp (leucine-responsive regulatory protein), by Dam methylation, and by FooI and FooB, homologues of PapI and PapB, respectively (8).
The presence of Lrp is required for expression of F1651 and Pap (2, 8). It was shown that for some operons regulated by Lrp, exogenous leucine and alanine antagonize the effect of Lrp (4, 24). F1651 synthesis is strongly repressed by the presence of exogenous l-leucine or l-alanine (8), whereas the addition of amino acids doesn't affect the expression of Pap (2). However, expression of three other fimbriae is controlled by a mechanism involving Lrp and is regulated by exogenous leucine or alanine. Among them, CS31A belongs to the P-regulatory family (21), whereas K99 and type 1 fimbriae do not (2, 11).
Lrp was first identified as a global regulator that regulates the expression of 35 to 75 genes in E. coli (including many operons involved in the metabolism of amino acids and in the formation of fimbriae) by binding to specific DNA sequences and affecting DNA conformation (22). A microarray analysis has shown that more than 400 genes are significantly Lrp responsive and that most of them are involved in stationary-phase metabolism (26). The mechanism by which Lrp acts is now thought to involve different associations of Lrp monomers. Recently, it was shown that at micromolar concentrations Lrp self-associates to hexadecamers and to octamers instead of the dimer conformation previously suggested (5). The presence of leucine induces a transition of the hexadecamers or the octamers into leucine-bound octamers (5). The C-terminal domain of Lrp is primordial for this autoassociation (5). Whatever the mechanism, l-leucine modulates the effect of Lrp, antagonizing it or intensifying it, or in some cases not affecting it at all (2, 4, 11, 13, 24). There is much less knowledge about alanine as a regulator of gene expression, and such an effect might be mediated through Lrp (28). Mutations in Lrp have been isolated on the basis of their effect on ilvIH, an operon positively regulated by Lrp (24). Analysis of these mutations suggested the existence of three functional domains: an N-terminal domain containing a helix-turn-helix motif implicated in DNA binding, a middle domain responsible for transcriptional activation, and a C-terminal domain that is required for the response to leucine. The objective of this study was to determine, by introducing specific mutations in the C-terminal region of Lrp, whether leucine or alanine affects the interaction of Lrp with the regulatory region of foo.
Influence of leucine and alanine on Lrp-induced foo expression.
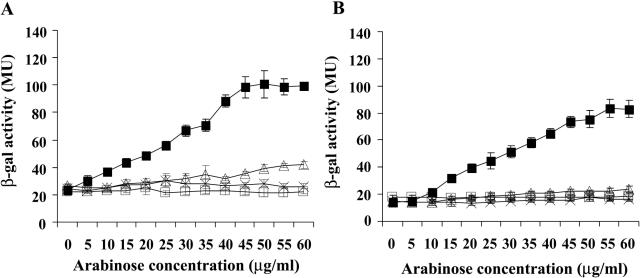
The lrp gene was introduced into pBAD22CM under the control of the araBAD promoter, which is inducible by exogenous arabinose (4). This vector was introduced into an lrp-deficient host strain (FB1001) containing a single chromosomal fooIBp-lacZ fusion with intact fooI and fooB genes and the whole foo intercistronic region, leading to strain FB1002 (Fig. 1; Table 1). The activity of the fooB promoter was monitored by assaying the β-galactosidase activity of FB1002 cells grown in NIV minimal medium with arabinose concentrations ranging from 0 to 60 μg/ml (4). The NIV medium is composed of 0.01 M potassium phosphate (pH 6.4), 0.2% ammonium sulfate, 0.001% CaCl2, and 0.02% MgSO4 neutralized with NaOH (pH 7.0) (23). Supplements were used at the following concentrations: 0.2% (wt/vol) glycerol, 50 μg of l-isoleucine/ml, 50 μg of valine/ml, and 2 mg of serine/ml. The fooB promoter activity increased proportionally to external arabinose concentrations up to a saturation level of 45 μg/ml (Fig. 2A). In contrast, foo expression was abolished in the Lrp-deficient strain FB1001 grown under the same conditions (Fig. 2A). Some residual β-galactosidase activity was detected in the absence of arabinose, probably due to the leakiness of the arabinose promoter (12).
TABLE 1.
Bacterial strains and bacteriophages used in this study
| Strain or phage | Description | Amino acids changed in Lrp | References |
|---|---|---|---|
| E. coli strains | |||
| CU1008 | ilvA | 27 | |
| MEW1 | CU1008 Δlac | 23 | |
| CT4A | ilvA lac ara lrp::Tn10 | 27 | |
| CV1008 | ilvI′-lacZ lrp-35::Tn10 | 25 | |
| FB1001 | CT4A fooIB-lacZ | This study | |
| FB1002 | FB1001 pBAD22CM-lrp | This study | |
| FB1003 | FB1001 pBAD22CM-lrp-1 | LrpS41P | This study |
| FB1004 | FB1001 pBAD22CM-lrp-2 | LrpL131R | This study |
| FB1005 | FB1001 pBAD22CM-lrp-3 | LrpL131A | This study |
| FB1006 | FB1001 pBAD22CM-lrp-4 | LrpL136R | This study |
| FB1007 | FB1001 pBAD22CM-lrp-5 | LrpV149A | This study |
| FB2001 | CT4A fooB-lacZ | This study | |
| FB2002 | FB2001 pBAD22CM-lrp | This study | |
| FB2006 | FB2001 pBAD22CM-lrp-4 | LrpL136R | This study |
| FB3002 | CV1008 pBAD22CM-lrp | This study | |
| FB3003 | CV1008 pBAD22CM-lrp-1 | LrpS41P | This study |
| FB3004 | CV1008 pBAD22CM-lrp-2 | LrpL131R | This study |
| FB3005 | CV1008 pBAD22CM-lrp-3 | LrpL131A | This study |
| FB3006 | CV1008 pBAD22CM-lrp-4 | LrpL136R | This study |
| FB3007 | CV1008 pBAD22CM-lrp-5 | LrpV149A | This study |
| Bacteriophages | |||
| λMT01 | λRS45 containing fooIB-lacZ | M. C. Tessier, C. Martin, and J. Harel (unpublished results) | |
| λMT02 | λRS45 containing fooB-lacZ | M. C. Tessier, C. Martin, and J. Harel (unpublished results) |
FIG. 2.
Influence of Lrp mutations and effects of leucine and alanine at 1.5 mM on the expression of fooBp. (A) fooIB-lacZ derivatives strain FB1002, containing wild-type Lrp (solid lines, black squares), and strain FB1001, deficient in Lrp (dotted line, empty squares). (B) fooB-lacZ derivatives FB2002, containing wild-type Lrp (solid lines, black squares), and FB2001, deficient in Lrp (dotted line, empty squares). The curves obtained in minimal medium without the addition of amino acids are represented by black squares, with the addition of alanine shown by empty triangles and the addition of leucine indicated by an X. The Durnett test was used to compare the effect of different Lrp concentrations on the foo operon (9). MU, Miller units.
In contrast to P fimbriae, the expression of foo in wild-type strains is repressed by the presence of exogenous leucine or alanine at 1.5 mM (14). At this concentration, both leucine and alanine repressed foo expression at all the arabinose concentrations tested (Fig. 2A). Leucine at 0.75 mM has been reported to repress expression of ilvIH (24), but alanine was not tested until now. Using the same concentration of leucine as Platko and Calvo, we observed a significant reduction of the ilvIH promoter activity of strain FB3002 (ilvI′-lacZ, pBAD-lrp) grown with arabinose at 50 μg/ml (Table 2). However, addition of alanine at 0.75 or 1.5 mM did not lower significantly the ilvIH expression (Table 2). Thus, ilvIH expression appears to be alanine insensitive, in contrast to foo expression.
TABLE 2.
β-Galactosidase activity of the different CV1008 derivatives (ilvI′-lacZ)a
| Strain | β-Galactosidase activity (%)
|
|
|---|---|---|
| Without amino acid | +Leucine, 1.5 mM | |
| FB3002 (Lrp) | 100.0 ± 19.2 | 17.4 ± 4.2 |
| FB3003 (LrpS41P) | 20.4 ± 5.9 | 18.0 ± 3.6 |
| FB3004 (LrpL131R) | 218.5 ± 37.7 | 174.3 ± 29.3 |
| FB3005 (LrpL131A) | 126.9 ± 2.4 | 161.6 ± 27.5 |
| FB3006 (LrpL136R) | 102.4 ± 18.6 | 83.8 ± 16.2 |
| FB3007 (LrpV149A) | 107.2 ± 15.6 | 48.0 ± 7.2 |
The arabinose concentration used was 50 μg/ml. Specific activities obtained were normalized to that of FB3002.
Effects of specific Lrp mutations on the repression of foo by leucine and alanine.
A study realized with the ilvIH promoter showed that the C-terminal third of Lrp is responsible for the response to l-leucine (24). In addition, in a recent study Ettema et al. (10) surveyed the structure of all known prokaryote Lrp-like molecules. They found a ligand-binding domain, RAM (regulation of amino acid metabolism) fused to the DNA-binding domain of Lrps. In the E. coli Lrp, this domain is located between residues 73 and 149. Following the procedure described by Labrie et al. (19) and by using the USE mutagenesis kit purchased from Amersham Pharmacia Biotech, we created mutations in the RAM domain of Lrp at those sites known to result in leucine insensitivity on the ilvIH promoter (L136R and V149A) and at an additional site in its C-terminal region based on the properties of some residues (L131R and L131A) (Table 1). As a negative control, a mutation in the N-terminal region was also introduced (S41P). This latter substitution abolished the fooIB-lacZ expression, whatever the arabinose concentration added (Table 3).
TABLE 3.
β-Galactosidase activity of the different FB1001 and FB2001 derivativesa
| Strain | β-Galactosidase activity (Miller units)
|
||
|---|---|---|---|
| Without amino acids | +Leucine, 1.5 mM | +Alanine, 1.5 mM | |
| FB1002 (Lrp) | 107.7 ± 9.8 | 28.3 ± 2.4 | 34.2 ± 2.7 |
| FB1003 (LrpS41P) | 12.2 ± 3.2 | 14.0 ± 3.7 | 14.8 ± 3.3 |
| FB1004 (LrpL131R) | 132.5 ± 2.8 | 147.2 ± 5.2 | 135.9 ± 2.6 |
| FB1005 (LrpL131A) | 109.0 ± 13.7 | 92.4 ± 8.5 | 121.6 ± 19.6 |
| FB1006 (LrpL136R) | 89.4 ± 2.2 | 105.1 ± 2.0 | 98.9 ± 4.0 |
| FB1007 (LrpV149A) | 93.8 ± 2.1 | 66.0 ± 5.3 | 74.3 ± 2.9 |
| FB1008 (LrpE133G) | 17.4 ± 3.8 | 14.1 ± 1.2 | 17.6 ± 4.1 |
| FB2001 (Lrp) | 75.5 ± 6.6 | 17.6 ± 2.7 | 22.0 ± 0.9 |
| FB2006 (LrpL136R) | 61.7 ± 8.7 | 62.5 ± 6.3 | 63.5 ± 5.7 |
The arabinose concentration used was 50 μg/ml. The post-hoc test of Tukey was used to analyze the effect of the medium on the expression of foo (9).
Strain FB1006 (L136R) and FB1004 (L131R) grown in glycerol minimal medium showed a similar fooIB-lacZ expression in response to arabinose as the parental strain carrying wild-type Lrp. However, neither leucine nor alanine repressed foo expression in these strains (Table 3). Thus, the L136R and L131R mutations, which caused leucine insensitivity at the ilvIH promoter, caused both leucine and alanine insensitivity at fooBp. A weak level of repression of fooB-lacZ expression due to leucine or alanine was also observed for strain FB1007 (V149A) at each arabinose concentration tested (Table 3). If these observations demonstrated an intrinsic characteristic of Lrp, they should be seen at other Lrp-regulated promoters. We therefore introduced the Lrp mutations in strains harboring an ilvI′-lacZ fusion. Transcription of ilvIH was shown to be leucine independent by using LrpL136R and LrpV149A, as also observed by Platko and Calvo, and by using LrpL131R (Table 2). Therefore, these results indicate that the residues at positions 131, 136, and 149 in Lrp are essential for the l-leucine and l-alanine responses. Because the Lrp mutants presented similar responses towards both amino acids, the mechanism of repression by alanine on foo appears to be similar to that of leucine. Among the operons regulated by Lrp and leucine, only a few are also regulated by alanine. Why alanine represses only a subset of leucine-responsive regulated operons remains to be elucidated. This might imply that another protein(s) could interact with Lrp to control the promoter activity, as was suggested for the Lrp-like LysM regulator in Sulfolobus solfataricus (3).
The Lrp leucine residues 131 and 136 were shown to be essential for the amino acid response. This hydrophobic residue was replaced at these positions by a hydrophilic one, arginine, to investigate whether the amino acid binding to Lrp involves a hydrophobic interaction. These Lrp mutants were unresponsive to leucine and alanine (Table 3). However, the lack of response did not seem to be the consequence of the interruption of a hydrophobic bond, since a hydrophobic residue conversion (LrpL131A) did not restore the repression.
Interestingly, Lrp carrying the L131R mutation differed in function from wild-type Lrp even in the absence of leucine and alanine. FB1004, which harbors this Lrp mutation, produced a significantly higher foo activity (about 1.2- to 1.7-fold) in minimal glycerol medium compared to FB1002 containing wild-type Lrp (Table 3). Thus, this mutation seems to intensify the activation by Lrp of foo expression in addition to causing a leucine- and alanine-nonresponsive phenotype. As described by Chen and Calvo (5), at micromolar concentrations Lrp self-associates primarily to hexadecamers, a combination of two octamers. The L136 residue, located in proximity to the L131 residue, is important for this autoassociation. Thus, the L131R mutation could be responsible for the formation of more-stable octamers. In this way, a higher number of LrpL131R octamers could bind the regulatory region of foo and intensify the activation of the operon. In contrast, this amplification of expression was not observed with the L131A substitution, indicating that the two Lrp molecules were quite different in terms of stability.
In addition to the C-terminal region of Lrp, it was already shown that residues A126, E133, and Thr134 are necessary for the activation of the pap operon in opposition to the ilvIH operon (18). The region containing these residues was considered pap specific. In our study, the same E133G mutation was introduced into Lrp to obtain strain FB1008. As observed for the pap operon, the expression of foo was abolished with this Lrp mutant.
Influence of FooI on leucine- or alanine-mediated foo repression.
Since FooI is involved in foo expression and phase variation (6), we wondered if the effect of leucine and/or alanine might be mediated by FooI. The fooBp activity was significantly lower in the absence of fooI than in its presence, at all the concentrations of arabinose tested (Fig. 1 and 2B). Thus, like PapI (17), FooI appears to be an activator of the foo operon. Moreover, we clearly demonstrated that the sensitivity of foo to leucine and alanine is independent of FooI, since the same degree of repression was obtained in the presence or in the absence of the fooI gene (Fig. 2A versus B; Table 3, FB1002 and FB2002). In addition, the mutant strains producing LrpL136R presented the same leucine- and alanine-nonresponsive phenotype independently of the presence of FooI (Table 3, FB1006 versus FB2006). Taken together, these results showed that FooI is not necessary for the repressive effect exerted by the amino acids on foo expression.
Effects of leucine and alanine on phase variation and on basal transcription of the foo operon.
To investigate whether leucine and/or alanine affected foo phase variation, the Lac phenotype of fooB-lacZ cultures was analyzed. A single FB1002 colony in the OFF or the ON state was picked from a Luria-Bertani plate and cultured overnight in Luria-Bertani broth at 37°C. This culture was diluted 50-fold in NIV broth containing 0.1% glycerol and 0.1% d-glucose and incubated overnight at 37°C. This culture was diluted in NIV-glycerol containing 50 μg of arabinose/ml with or without l-leucine or l-alanine and incubated at 37°C until an optical density at 600 nm between 0.4 and 0.5 was reached. Then, the phase variation assay was performed as previously described (7) on NIV 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside-agar plates containing 50 μg of arabinose/ml with or without amino acids by counting the number of blue and white colonies (Table 4). Phase variation occurred in strain FB1002, leading to a majority of colonies in the on phase using ON inoculating colonies, whereas a majority of colonies in the OFF phase was observed using OFF inoculating colonies (Table 4). When using ON inoculating colonies, without amino acids, 78.05% of cells were in the ON phase. In the presence of leucine or alanine, 2.1 and 4.35% of cells, respectively, were in the ON phase. A decrease in the number of ON colonies was also observed in the presence of both amino acids when using OFF inoculating colonies. Thus, the phase variation favored the OFF phenotype when alanine or leucine were added to the medium. Moreover, in the absence of amino acids, the blue colonies were dark blue or very dark blue. The very dark blue and dark blue colonies were excised from the plates and suspended separately in 1 ml of phosphate-buffered saline buffer for direct β-galactosidase activity measurement. The β-galactosidase activity representing the average of three very dark blue and dark blue colonies was 1,619 and 611 Miller units, respectively. In contrast, the pale blue cells grown with alanine or leucine demonstrated low β-galactosidase activities (305 and 381 Miller units, respectively). These results indicated that the amino acids had a drastic effect on the basal transcription of foo and on the phase variation control. For the strain expressing the mutated LrpL136R, neither the phase variation nor the basal transcription of foo was affected by the presence of the amino acids (data not shown). Taken together, our results suggest that the interaction between the C-terminal region of Lrp and the amino acids is mainly responsible for the reduction of foo basal transcription and phase variation.
TABLE 4.
foo phase variation in strain FB1002
| Medium | Starting colony phenotype | % Lac+ coloniesa | Avg % Lac+ colonies | Switch frequencyb
|
||
|---|---|---|---|---|---|---|
| ON to OFF | OFF to ON | Avg | ||||
| NIV | 1st blue | 82.6 ± 3.6 | 78.05 | 6.9 × 10−3 | 8.45 × 10−3 | |
| 2nd blue | 73.5 ± 5.9 | 1 × 10−2 | ||||
| 1st white | 45.3 ± 1.5 | 23.65 | 1.8 × 10−2 | 9.4 × 10−3 | ||
| 2nd white | 2 ± 0.5 | 8 × 10−4 | ||||
| NIV-leu | 1st blue | 1.5 ± 1 | 2.1 | 3.9 × 10−2 | 3.85 × 10−2 | |
| 2nd blue | 2.7 ± 0.7 | 3.8 × 10−2 | ||||
| 1st white | 0.027 ± 0.02 | 0.013 | 1 × 10−5 | 5 × 10−6 | ||
| 2nd white | 0 | 0 | ||||
| NIV-ala | 1st blue | 5 ± 2 | 4.25 | 3.7 × 10−2 | 3.75 × 10−2 | |
| 2nd blue | 3.5 ± 0.3 | 3.8 × 10−2 | ||||
| 1st white | 0.2 ± 0.2 | 0.2 | 8.5 × 10−5 | 7.4 × 10−5 | ||
| 2nd white | 0.2 ± 0.1 | 6.3 × 10−5 | ||||
Mean of at least two experiments.
Strains were inoculated onto NIV-glycerol-arabinose-X-Gal plates supplemented or not with l-leucine or l-alanine. Colonies showing a uniform phenotype were suspended in NIV salts. Appropriate dilutions were spread onto the same medium as the parent colony. After growth, the colonies were scored for a white or a blue phenotype. The switch frequencies were calculated using the formula (M/N)/g, where M is the number of cells that underwent phase transition, N is the total number of cells evaluated, and g is the total number of generations, estimated to be 27; that gave rise to the colony (7). A minimum of 2,000 colonies were screened for each test, and three independent tests were done to calculate the percentage of Lac+ colonies.
Conclusions.
In summary, this study demonstrates that the foo operon is transcribed proportionally to the amount of Lrp synthesized. We showed that the repressive effect of exogenous leucine and alanine on foo basal transcription and phase variation is mediated by Lrp, since specific mutations in the C-terminal region of Lrp totally abolished their action. In addition, we showed that FooI is not required for the repressive effect of both amino acids.
Acknowledgments
This work was supported in part by Fonds pour la Formation des Chercheurs et l’Aide à la Recherche (FCAR) grant 0214 and by the Natural Sciences Engineering Research Council of Canada (NSERC) grant 225155 to J.H. F.B. was funded by a scholarship from FCAR.
We thank Marie-Catherine Tessier for the creation of the γRS45 derivatives and Joseph M. Calvo for strain CV1008.
REFERENCES
- 1.Blyn, L. B., B. A. Braaten, and D. A. Low. 1990. Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 9:4045-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braaten, B. A., J. V. Platko, M. W. van der Woude, B. H. Simons, F. K. de Graaf, J. M. Calvo, and D. A. Low. 1992. Leucine-responsive regulatory protein controls the expression of both the pap and fan pili operons in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:4250-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinkman, A. B., S. D. Bell, R. J. Lebbink, W. M. de Vos, and J. van der Oost. 2002. The Sulfolobus solfataricus Lrp-like protein LysM regulates lysine biosynthesis in response to lysine availability. J. Biol. Chem. 277:29537-29549. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C., and E. B. Newman. 1998. Comparison of the sensitivities of two Escherichia coli genes to in vivo variation of Lrp concentration. J. Bacteriol. 180:655-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, S., and J. M. Calvo. 2002. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J. Mol. Biol. 318:1031-1042. [DOI] [PubMed] [Google Scholar]
- 6.Crost, C., J. Harel, F. Berthiaume, A. Garrivier, M. C. Tessier, H. Harivony Rakotoarivonina, and C. Martin. 2004. Influence of environmental cues on the transcriptional regulation of foo and clp coding for F1651 and CS31A adhesins in Escherichia coli. Res. Microbiol. 155:475-482. [DOI] [PubMed] [Google Scholar]
- 7.Crost, C., A. Garrivier, J. Harel, and C. Martin. 2003. Leucine-responsive regulatory protein-mediated repression of clp (encoding CS31A) expression by l-leucine and l-alanine in Escherichia coli. J. Bacteriol. 185:1886-1894. (Erratum, 185:2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daigle, F., C. Forget, C. Martin, M. Drolet, M. C. Tessier, H. Dezfulian, and J. Harel. 2000. Effects of global regulatory proteins and environmental conditions on fimbrial gene expression of F1651 and F1652 produced by Escherichia coli causing septicaemia in pigs. Res. Microbiol. 151:563-574. [DOI] [PubMed] [Google Scholar]
- 9.Dawson, B., and R. G. Trapp. 2001. Basic and clinical biostatistics. Lange Medical Books, New York, N.Y.
- 10.Ettema, T. J., A. B. Brinkman, T. H. Tani, J. B. Rafferty, and J. Van Der Oost. 2002. A novel ligand-binding domain involved in regulation of amino acid metabolism in prokaryotes. J. Biol. Chem. 277:37464-37468. [DOI] [PubMed] [Google Scholar]
- 11.Gally, D. L., J. A. Bogan, B. I. Eisenstein, and I. C. Blomfield. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 175:6186-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haney, S. A., J. V. Platko, D. L. Oxender, and J. M. Calvo. 1992. Lrp, a leucine-responsive protein, regulates branched-chain amino acid transport genes in Escherichia coli. J. Bacteriol. 174:108-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harel, J., F. Daigle, C. Forget, M. C. Tessier, C. Crost, and C. Martin. 2000. Phase variation of F1651 (Prs-like) fimbriae from Escherichia coli causing septicaemia in animals. Can. J. Microbiol. 46:1101-1107. [DOI] [PubMed] [Google Scholar]
- 15.Harel, J., F. Daigle, S. Maiti, C. Desautels, A. Labigne, and J. M. Fairbrother. 1991. Occurrence of pap-, sfa-, and afa-related sequences among F165-positive Escherichia coli from diseased animals. FEMS Microbiol. Lett. 66:177-182. [DOI] [PubMed] [Google Scholar]
- 16.Hernday, A., M. Krabbe, B. Braaten, and D. Low. 2002. Self-perpetuating epigenetic pili switches in bacteria. Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16470-16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernday, A. D., B. A. Braaten, and D. A. Low. 2003. The mechanism by which DNA adenine methylase and PapI activate the pap epigenetic switch. Mol. Cell 12:947-957. [DOI] [PubMed] [Google Scholar]
- 18.Kaltenbach, L., B. Braaten, J. Tucker, M. Krabbe, and D. Low. 1998. Use of a two-color genetic screen to identify a domain of the global regulator Lrp that is specifically required for pap phase variation. J. Bacteriol. 180:1224-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labrie, V., H. E. Beausoleil, J. Harel, and J. D. Dubreuil. 2001. Binding to sulfatide and enterotoxicity of various Escherichia coli STb mutants. Microbiology 147:3141-3148. [DOI] [PubMed] [Google Scholar]
- 20.Low, D., E. N. Robinson, Jr., Z. A. McGee, and S. Falkow. 1987. The frequency of expression of pyelonephritis-associated pili is under regulatory control. Mol. Microbiol. 1:335-346. [DOI] [PubMed] [Google Scholar]
- 21.Martin, C. 1996. The clp (CS31A) operon is negatively controlled by Lrp, ClpB, and l-alanine at the transcriptional level. Mol. Microbiol. 21:281-292. [DOI] [PubMed] [Google Scholar]
- 22.Newman, E. B., R. D'Ari, and R. T. Lin. 1992. The leucine-Lrp regulon in E. coli: a global response in search of a raison d'etre. Cell 68:617-619. [DOI] [PubMed] [Google Scholar]
- 23.Newman, E. B., B. Miller, L. D. Colebrook, and C. Walker. 1985. A mutation in Escherichia coli K-12 results in a requirement for thiamine and a decrease in l-serine deaminase activity. J. Bacteriol. 161:272-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Platko, J. V., and J. M. Calvo. 1993. Mutations affecting the ability of Escherichia coli Lrp to bind DNA, activate transcription, or respond to leucine. J. Bacteriol. 175:1110-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platko, J. V., D. A. Willins, and J. M. Calvo. 1990. The ilvIH operon of Escherichia coli is positively regulated. J. Bacteriol. 172:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthews. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuan, L. R., R. D'Ari, and E. B. Newman. 1990. The leucine regulon of Escherichia coli K-12: a mutation in rblA alters expression of l-leucine-dependent metabolic operons. J. Bacteriol. 172:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhi, J., E. Mathew, and M. Freundlich. 1999. Lrp binds to two regions in the dadAX promoter region of Escherichia coli to repress and activate transcription directly. Mol. Microbiol. 32:29-40. [DOI] [PubMed] [Google Scholar]