Abstract
Rhizobium leguminosarum bv. trifolii mutants unable to catabolize the methyl-pentose rhamnose are unable to compete effectively for nodule occupancy. In this work we show that the locus responsible for the transport and catabolism of rhamnose spans 10,959 bp. Mutations in this region were generated by transposon mutagenesis, and representative mutants were characterized. The locus contains genes coding for an ABC-type transporter, a putative dehydrogenase, a probable isomerase, and a sugar kinase necessary for the transport and subsequent catabolism of rhamnose. The regulation of these genes, which are inducible by rhamnose, is carried out in part by a DeoR-type negative regulator (RhaR) that is encoded within the same transcript as the ABC-type transporter but is separated from the structural genes encoding the transporter by a terminator-like sequence. RNA dot blot analysis demonstrated that this terminator-like sequence is correlated with transcript attenuation only under noninducing conditions. Transport assays utilizing tritiated rhamnose demonstrated that uptake of rhamnose was inducible and dependent upon the presence of the ABC transporter at this locus. Phenotypic analyses of representative mutants from this locus provide genetic evidence that the catabolism of rhamnose differs from previously described methyl-pentose catabolic pathways.
Rhizobium leguminosarum bv. trifolii is a gram-negative aerobic soil bacterium that can exist as a free-living heterotropic saprophyte or can form nitrogen-fixing nodules on various species of clovers (Trifolium spp.). The interaction of rhizobia with their hosts is a sequence of events that begins with an exchange of signals in the rhizosphere, followed by the invasion of the plant through infection threads and ultimately by release of the bacteria into plant cells, where they differentiate into nitrogen-fixing bacteroids. Through this symbiotic association, the plant provides the bacteria with energy for growth, and in return the Rhizobium provides the plant with fixed nitrogen. Although a great deal has been elucidated regarding the actual steps during nodulation (27, 56) and the metabolic pathways utilized while in the bacteroid state (39), comparatively little is known about what influences the growth of the bacteria prior to or during their interaction with plant roots.
The rhizosphere has been described as the region directly under the influence of secreted compounds from the plant root (12). Competition for nodule occupancy exists between strains of Rhizobium within the rhizosphere and has been well documented in the literature (16, 54). A large number of biotic and abiotic factors influence competition between strains for nodule occupancy. Some of the best-characterized strain-dependent factors include production of antibiotics and bacteriocins (36, 42, 57) and the ability to catabolize specific compounds present in the root environment (11, 13, 18, 22, 23, 35, 43, 49, 55), as well as the synthesis or utilization of specific vitamins (38, 50).
Rhamnose is a methyl-pentose sugar and is found as a constituent of pectin in the form of rhamnogalacturonan within the cell walls of dicotyledonous plants (28). It has also been found in the mucilage of a number of legume plants (26). R. leguminosarum mutants unable to utilize rhamnose as a sole carbon source were found to be significantly impaired in their nodulation competitiveness (35). The reason for the uncompetitive nature of these mutants is not understood. It was hypothesized that if the genetic region responsible for the catabolism of rhamnose were characterized, a better understanding of why these mutants were uncompetitive could be achieved. The objective of the present work, therefore, was to identify and characterize the genes in this region.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used and generated in this work are listed in Table 1. R. leguminosarum strains were routinely grown at 30°C on TY as a complex medium (8) and VMM (58) as a defined medium. Glycerol, glucose, or rhamnose (or a combination of two of these sugars) was used instead of mannitol as a carbon source in VMM, according to the needs of individual experiments. Carbon sources were filter sterilized and added to defined media to a final concentration of 15 mM or 0.4% (wt/vol). When required, antibiotics were added to solid or liquid media at the following concentrations: tetracycline (Tc), either 10 or 5 μg ml−1; neomycin (Nm), 200 μg ml−1; streptomycin, 200 μg ml−1; gentamicin, either 15 or 30 μg ml−1; ampicillin, 100 μg ml−1; kanamycin, 50 μg ml−1. Bacterial growth was monitored spectrophotometrically at 600 nm.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or phenotypea | Reference or source |
|---|---|---|
| R. leguminosarum strains | ||
| Rlt100 | W14-2, R. leguminosarum bv. trifolii streptomycin-resistant wild type | 6 |
| Rlt101 | W14-2 b, cured of plasmid a and d, deletion in plasmid c | 31 |
| Rlt105 | Rlt100 rhaD1::Tn5B20 | 35 |
| Rlt106 | Rlt100 rhaT2::Tn5B20 | 35 |
| Rlt112 | Rlt100 rhaS17::TnphoA | This work |
| Rlt115 | Rlt100 rhaP19::Tn5 | This work |
| Rlt117 | Rlt100 rhaR25::Tn5B20 | This work |
| Rlt118 | Rlt100 rhaD26::Tn5B20 | This work |
| Rlt121 | Rlt100 rhaS29::Tn5B20 | This work |
| Rlt124 | Rlt100 rhaI31::Tn5B20 | This work |
| Rlt125 | Rlt100 rhaI32::Tn5B20 | This work |
| Rlt128 | Rlt100 rhaP36::Tn5B20 | This work |
| Rlt130 | Rlt100 rhaI39::Tn5B20 | This work |
| Rlt131 | Rlt100 rhaK40::Tn5B20 | This work |
| Rlt144 | Rlt100 rhaK50::Tn5B20 | This work |
| Rlt146 | Rlt100 rhaK52::Tn5B20 | This work |
| Rlt151 | Rlt100 rhaQ38::Tn5B20 | This work |
| Rlt152 | Rlt100(pW3C1) | This work |
| Rlt153 | Rlt101(pMR73, contains rhaR25) | This work |
| Rlt154 | Rlt100(pMR73, contains rhaR25) | This work |
| Rlt197 | Rlt117(pMR53, contains rhaR+) | This work |
| Rlt211 | Rlt100 rhaD1::Tn5B20, rhaK58::pKNOCK-Tc | This work |
| Rlt212 | Rlt100 rhaI31:Tn5B20, rhaK58::pKNOCK-Tc | This work |
| E. coli strains | ||
| MM294A | pro-82 thi-1 hsdR17 supE44 | 17 |
| MT607 | MM294A recA56 | 17 |
| EcA101 | MT607 with a Tn5B20 insert in the chromosome | 14 |
| MT614 | MT607 with a Tn5 insert in the chromosome | 17 |
| MT621 | MT607 MM294A malF::TnPhoA | 61 |
| MT616 | MT607 (pRK600) | 17 |
| DH5α | endA hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA) U169 φ80dlacZΔM15 | B.R.L. Inc. |
| S17-1 | recA derivative of MM294A with RP4-2 (Tc::Mu::Km::Tn7) integrated into the chromosome | 48 |
| Plasmids | ||
| pUC19 | Cloning vector, ColE1 oriV, Apr | 60 |
| pBlueScript II SK | Cloning vector, ColE1 oriV, Apr | Stratagene |
| pJQ200 SK+ | Suicide vector for homogenotization; P15a ori, mob sacB, Gmr | 41 |
| pKNOCK-Tc | Suicide vector for insertional mutagenesis; R6K ori, RP4 orit, Tcr | 2 |
| pRK600 | pRK2013 npt::Tn9 Cmr Nm-Kms | 17 |
| pRK7813 | RK2 derivative carrying pUC9 polylinker and lambda cos site, Tcr | 24 |
| pPH1JI | IncP plasmid, Gmr | 9 |
| pW3A | pRK7813 cosmid from Rlt100 wild-type cosmid bank, complements Rlt101 for rhamnose utilization | 35 |
| pW3C1 | pRK7813 cosmid from Rlt100 wild-type cosmid bank, complements Rlt101 for rhamnose and sorbitol utilization | 35 |
| pMR3 | 12-kb HindIII fragment from pW3C1 in pBluescript | This work |
| pMR5 | EcoRI deletion of pMR3 in pBluescript | This work |
| pMR6 | 1.1-kb EcoRI subclone from pMR3 in pBluescript | This work |
| pMR28 | rhaK coding sequence in pBluescript | This work |
| pW3AR1 | pW3A rhaD1::Tn5B20 | 35 |
| pW3AR2 | pW3A rhaT2::Tn5B20 | 35 |
| pW3AR11 | pW3A rhaS11::TnphoA | This work |
| pW3AR44 | pW3A rhaI44::Tn5B20 | This work |
| pW3CR1B | pW3C1 rhaT12::Tn5 | 35 |
| pW3CR3A | pW3C1 rhaT13::Tn5 | 35 |
| pW3CR5 | pW3C1 rhaQ14::Tn5 | 35 |
| pW3CR7 | pW3C1 rhaK7::Tn5 | This work |
| pMR53 | pRK7813 with rhaR+ expressed from plac | This work |
| pMR65 | pW3C1 rhaS17::TnphoA | This work |
| pMR67 | pW3C1 rhaP19::Tn5 | This work |
| pMR70 | pW3C1 rhaK22::Tn5 | This work |
| pMR71 | pW3C1 rhaP23::Tn5 | This work |
| pMR73 | pW3C1 rhaR25::Tn5B20 | This work |
| pMR74 | pW3C1 rhaD26::Tn5B20 | This work |
| pMR77 | pW3C1 rhaS29::Tn5B20 | This work |
| pMR79 | pW3C1 rhaI31::Tn5B20 | This work |
| pMR80 | pW3C1 rhaI32::Tn5B20 | This work |
| pMR84 | pW3C1 rhaQ36::Tn5B20 | This work |
| pMR86 | pW3C1 rhaQ38::Tn5B20 | This work |
| pMR87 | pW3C1 rhaI39::Tn5B20 | This work |
| pMR88 | pW3C1 rhaK40::Tn5B20 | This work |
| pMR98 | pW3C1 rhaK50::Tn5B20 | This work |
| pMR100 | pW3C1 rhaK52::Tn5B20 | This work |
| pMR146 | pKNOCK-Tc with 465-bp EcoRI fragment from rhaK | This work |
Abbreviations for antibiotics are as follows: Ap, ampicillin; Cm, chloramphenicol; Gm, gentamicin; Km, kanamycin; Nm, neomycin; Sm, streptomycin; Tc, tetracycline.
DNA manipulations, Southern blot analysis, and constructions.
Standard techniques were used for plasmid isolation, restriction enzyme digestion, and agarose gel electrophoresis (46). For Southern analysis, genomic DNA was isolated using a slightly modified version of the protocol outlined by Meade et al. (29), as has been previously described (34).
Genomic DNA for Southern blot hybridizations was restricted to completion with SalI and electrophoresed through a 0.8% agarose gel using 1× Tris-acetate-EDTA buffer. DNA was transferred onto nylon membranes (Roche Diagnostics, Laval, Quebec, Canada) overnight by alkaline transfer (46), neutralized using 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and hybridized overnight with digoxigenin-labeled probes. Hybridization was done at 68°C overnight, and membranes were washed two times for 5 min with 2× SSC-0.1% sodium dodecyl sulfate (SDS) at room temperature and two times for 15 min at 68°C with 0.1% SSC-0.1% SDS. Detection was done utilizing CSPD reagent (Roche Diagnostics) as specified by the manufacturer.
Southern blotting probes were constructed by digesting a plasmid pW3C1 (isolated from Rlt100 genomic library) with EcoRI. The fragments used as probes were labeled with digoxigenin (Roche Diagnostics) as suggested by the manufacturer.
Construction of pMR53 was accomplished in two steps. First, PCR amplification of rhaR was achieved by using the primers RhaR5′ (5′-ATATCTGCAGAAGGAGTTGATCAATGCACGAACGCGAACGC-3′) and RhaR3′ (5′-ATATGAATTCCCCGGGTCATCGGACCGACGAGGA-3′) and pW3C1 as the template. The RhaR5′ primer contained a BclI site immediately preceding the predicted ATG start site and a ribosome binding site with optimal spacing from the ATG. The amplification product was cloned into pBluescript II SK using the PstI and EcoRI restriction sites that were incorporated into the primers, yielding pMR41. This construct was verified by nucleotide sequencing. pMR53 was subsequently generated by subcloning the PstI/EcoRI fragment from pMR41 into pRK7813.
Construction of pMR28 was carried out by amplifying the coding region of rhaK using the primers RhaK5′ (5′-ATAGGATCCATGACCGCCAGTTCCTATC-3′) and RhaK3′ (5′-ATAAAGCTTCTATGCCATCGCCGCGTA-3′) utilizing pW3C1 as a template. The amplification product was cloned as a BamHI/HindIII fragment into pBluescript II SK by utilizing the restriction sites that were introduced into the primers. The construct was verified by nucleotide sequencing.
Genetic manipulations.
Isolation of Tn5, Tn5B20, and TnphoA inserts was carried out by using strains MT614, EcA101, and MT620, respectively, as previously described (14, 17, 61). Briefly, pW3C1, which had previously been shown to complement Rlt101 for its inability to utilize rhamnose as a sole carbon source (35), was introduced into each of these strains and subsequently mobilized into Rlt101. Tcr and Nmr transconjugants were screened for the inability to complement. Transposon inserts of interest were subsequently subcloned into pBluescript II SK such that DNA flanking the insert could be sequenced by utilizing an IS50 primer (5′-TAGGAGGTCACATGGAAGTCAGAT-3′).
Gene replacement experiments were accomplished either by the use of pJQ200SK (41) or by the use of the incompatible plasmid pPH1JI (9) essentially as described by Ruvkun and Ausubel (45). Correct replacement of the wild-type gene was confirmed by Southern blot analysis of genomic DNA cut with SalI and probed with pW3C1.
The construction of rhaD/rhaK and rhaI/rhaK double mutants was accomplished by isolating the internal 465-bp EcoRI fragment from rhaK and ligating it into pKNOCK-Tc (2), yielding pMR146. pMR146 was subsequently conjugated into strains Rlt105 and Rlt124, which contained rhaD1 and rhaI31 alleles, respectively. Single crossovers of pMR146 in either Rlt105 or Rlt124 were selected on TY agar containing Nm and Tc. A number of colonies were purified and checked by Southern hybridization to ensure correct integration into rhaK. Two strains, Rlt211 and Rlt212, containing rhaD1/rhaK58 and rhaI31/rhaK58, respectively, were subsequently used.
DNA sequencing and sequence analysis.
The rhamnose catabolism region was sequenced by a combination of sequencing the DNA flanking Tn5 inserts in pW3C1 that had lost the ability to confer growth on rhamnose on strain Rlt101, as well as primer walking to fill in gaps. Sequencing reactions were done using dye terminators at the University of Calgary Core DNA Services by using an ABI automated sequencer. Sequence data were analyzed using DNASIS (Hitachi Software Engineering Co., San Bruno, Calif.). Database searches were done using the BLASTX program (3). Both strands were completely sequenced, and the sequence has been deposited in the GenBank database.
RNA isolation, RNA probe preparation, and RNA dot blots.
Rapid isolation of RNA was accomplished as previously described (4) with the following modifications: 500 ml of cells was used for the isolation, an additional phenol-chloroform-isoamyl alcohol (24:25:1) extraction was added, and the final resuspension contained 40 U of RNaseOUT (GibcoBRL, Life Technologies). The concentration of RNA for each preparation was determined by assaying an adequate dilution at 260 nm by using a Shimadzu MultiSpec-1501 instrument. The quality of the prep was also visualized by running the sample on a denaturing agarose gel.
To generate gene-specific RNA-labeled probes utilizing digoxigenin, an in vitro labeling kit utilizing either T3 or T7 polymerase was utilized as recommended by the manufacturer (Roche Diagnostics). Briefly, constructs containing the desired open reading frames (ORFs) were linearized with an appropriate enzyme and RNA was synthesized using either T3 or T7 RNA polymerase. To generate a rhaR-specific probe, pMR41 was linearized with PstI, and T7 RNA polymerase was used. pMR28 was linearized with BamHI, and T7 RNA polymerase was used to generate a rhaK RNA probe. Probes specific for rhaS and rhaT were generated by digesting pMR49 and pMR51 with SalI, and T3 RNA polymerase was used for both.
Dot blots were carried out by first quantitating the RNA (by the optical density at 260 nm [OD260]), adjusting the concentration of the isolated RNA such that all would be at an equivalent concentration, and serially diluting each sample (using diethyl pyrocarbonate-treated double-distilled H2O-20× SSC-formaldehyde in the ratio of 5:3:2). These were subsequently boiled for 5 min, quenched on ice, and spotted on a positively charged nylon membrane (Roche Diagnostics). Samples were applied such that equivalent volumes with identical concentrations of RNA were applied to the membranes. Samples were cross-linked using UV light, and hybridizations were carried out at 68°C overnight. Washes and chemiluminescent detection were carried out as suggested by the manufacturer (Roche Diagnostics). Results were obtained by autoradiography as well as chemiluminescent analysis (FluorChem version 0.2.01; Alpha Innotech Corporation). Signals were quantitated by using the linear portion of the phosphorimager data as well as by using NIH Image version 1.62 (National Institutes of Health).
β-Galactosidase activity.
Rhizobium cultures containing active fusions were assayed following an overnight induction with rhamnose. Cultures used for assays were first grown overnight in TY and then used to inoculate a fresh broth culture of defined medium supplemented with 15 mM glycerol and 15 mM rhamnose. Cells were grown overnight in VMM-glycerol-rhamnose broth to mid-log phase (OD600 of approximately 0.5) and were assayed as described by Miller (30). Cultures were assayed in duplicate, and a minimum of three independent cultures was used for each experiment. β-Galactosidase activity was shown to be linear over the duration of the assay (data not shown).
Rhamnose transport assay.
Radioactive [3H]rhamnose (5 Ci/mmol) was purchased from American Radiolabeled Chemicals Ltd. (St. Louis, Mo.). Strains were grown for 20 h in 5-ml cultures and then subcultured into 50 ml of VMM-glycerol-rhamnose. Cells were harvested by centrifugation (5,000 × g for 10 min) at mid-log phase (OD600 = 0.5 to 0.7), washed twice in VMM-glycerol, and resuspended in this medium to a final OD600 of ∼0.3 in a total volume of 10 ml. Transport assays were initiated by the addition of tritiated rhamnose to a final concentration of 2 μM (125,000 dpm), and aliquots of 0.5 ml were withdrawn at appropriate time points and rapidly filtered through a Millipore 0.45-μm Hv filter on a Millipore sampling manifold. Filtered cells were immediately washed with 5 ml of the defined salts medium, and the radioactivity that remained on the filter was determined using a liquid scintillation spectrophotometer (Beckman LS6500). Samples were taken every 20 s and continued for up to 2 min. Transport rates were generally linear over the first minute of the assay. Linearity, however, did vary slightly between experiments. To determine the Km of the ABC transporter, 15- and 30-s time points were used to determine initial rates.
Nucleotide sequence accession number.
The sequence data for the rhamnose region of R. leguminosarum bv. trifolii strain Rlt100 have been submitted to the GenBank database and assigned accession number AF085687.
RESULTS
Rhamnose locus contains catabolic and transporter genes.
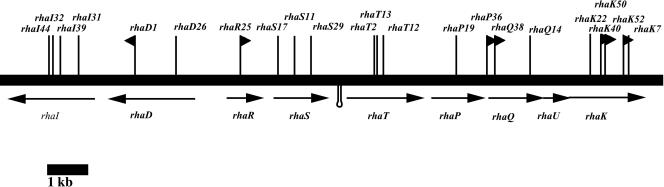
The cosmid pW3C1 was isolated from a cosmid bank constructed from the wild-type Rlt100 on the basis of its ability to complement Rlt101 for its inability to utilize rhamnose as a sole carbon source (35). To delineate the region necessary for the complementation phenotype, random transposon mutagenesis was carried out on pW3C1 utilizing Tn5, Tn5B20, and TnphoA, and a total of 56 independent mutants were isolated, mapped, and recombined into Rlt100 (Fig. 1). The entire region on pW3C1 was sequenced on both strands by a combination of sequencing the DNA flanking each of the transposon inserts as well as primer walking. The rhamnose region spans 10,959 bp. The DNA sequence revealed the presence of nine plausible ORFs apparently arranged as at least two divergently transcribed units. The genes rhaD and rhaI are transcribed in one direction, whereas the genes rhaR, rhaS, rhaT, rhaP, rhaQ, rhaU, and rhaK are divergently transcribed (Fig. 1).
FIG. 1.
Rhamnose region of R. leguminosarum bv. trifolii strain Rlt100. Heavy solid horizontal line, Rlt100 genomic DNA; lines with arrows, ORFs; vertical lines, transposon inserts; vertical lines with flags, Tn5B20 inserts that yielded lacZ fusions; stem-loop, position of a transcriptional attenuator. Gene names are under each respective ORF. The sequence data for the region were submitted to the GenBank database and assigned accession number AF085687.
The genes rhaD and rhaI code for a putative oxidoreductase and a putative sugar isomerase, respectively. RhaD is predicted to be a 698-amino-acid protein that has 87% identity with a similar hypothetical oxidoreductase in Agrobacterium tumefaciens. Analysis with the Conserved Domain Architecture Retrieval tool (21) of the translated peptide revealed that this protein contains two domains, a short-chain dehydrogenase domain as well as a type II aldolase domain. A putative NAD+ binding site was also predicted to exist.
A gap of 193 bp was found between the rhaD and rhaI genes. The RhaI protein is predicted to be a 430-amino-acid sugar isomerase showing 84% identity to the predicted product of a similar gene from A. tumefaciens. Transposon inserts yielding mutants unable to use rhamnose were not found beyond rhaI, and sequencing beyond rhaI did not reveal any further ORFs which might be associated with rhamnose utilization.
Genes rhaRSTPQUK were found to be oriented divergently from rhaDI (Fig. 1). The rhaR gene encodes a 270-amino-acid protein that displays 79% identity to a DeoR-type negative regulator from A. tumefaciens and 68% identity to Mesorhizobium loti and Sinorhizobium meliloti DeoR-type negative regulators. Following the regulator gene, separated by 46 bp, is rhaS. The gene rhaS encodes a potential periplasmic sugar binding protein. The genes rhaTPQ encode an ABC-type protein followed by two permeases, respectively. The genes rhaS and rhaT are separated by 73 bp, whereas the intergenic spaces between rhaT and rhaP and between rhaP and rhaQ are 9 and 36 bp, respectively. We note that the intergenic region between rhaS and rhaT appears to have a palindromic sequence that has the potential to form a hairpin loop consisting of a 10-bp GC-rich stem with a 4-bp loop. This structure has a predicted ΔG0 value of −24.4 kcal/mol and a predicted Tm of 126°C.
The genes rhaU and rhaK follow rhaQ. The start site of rhaU is predicted to overlap the stop codon of rhaQ (ATGA). BLASTX analysis of RhaU shows that this protein is similar to hypothetically conserved proteins that are found in a wide variety of Eubacteria. These genes are all classified as being similar to yulD from Bacillus subtilis. Interestingly, the gene yulD is found in a cluster of genes that resemble the rhamnose catabolic operon in Escherichia coli. There is no ascribed function to the gene product of yulD in any organism.
The final gene in this region is rhaK. RhaK is predicted to encode a 460-amino-acid protein that is 74% similar to a putative rhamnose kinase from A. tumefaciens. The rhaK predicted ORF overlaps the stop codon of rhaU (ATGA). No transposon inserts were isolated beyond rhaK that led to an inability of pW3C1 to complement Rlt101. BLASTX analysis of the nucleotide sequence following rhaK did not reveal any significant homologies, suggesting that rhaK was on a terminal end of the rhamnose-complementing region.
It is of interest that there are clusters of genes in both A. tumefaciens (59) and S. meliloti (19) that appear to correspond very closely to the rhamnose utilization gene cluster we have identified here. The order of the genes is precisely conserved, as is the presence of the stem-loop structure between rhaS and rhaT. However, the genetic context of the gene clusters is highly different in the three bacteria. In A. tumefaciens C58 they are located on the linear chromosome (Atu3484 to Atu3492) and are flanked by panCB and a predicted nonheme haloperoxidase, whereas in S. meliloti 1021 they are chromosomally encoded (Smc02321 to Smc03003) and located immediately adjacent to the chemotaxis operon. This is in contrast to both strains of R. leguminosarum, Rlt100 and 3841, in which the genes are located on a large plasmid (35). A comparison with the R. leguminosarum 3841 genomic sequence, currently being sequenced at the Sanger Center (http://www.sanger.ac.uk/Projects/R_leguminosarum/), reveals a similar organization of the rha genes to that reported here, and the predicted proteins encoded by the strain 3841 sequence are 88 to 96% identical to those from Rlt100. It is of note that the genes are found in an entirely different context in these three related members of the Rhizobiaceae family.
The rhamnose region contains two complementation groups.
Sequence analysis of the rhamnose region revealed several intergenic gaps that could contain putative promoters. To address the possibility that multiple transcripts were produced in this region, representative insertion mutants for each gene (carried on either pW3C1 or pW3A) were chosen and a complementation analysis was carried out (Table 2). The results clearly showed that although an intergenic gap of 193 bp exists between rhaD and rhaI, these two genes are in one transcriptional unit. Similarly, the 46- and 76-bp intergenic regions that exist between rhaR/rhaS and rhaS/rhaT, respectively, do not contain independent promoters, and all the genes transcribed divergently from rhaDI appear to constitute a single transcript (Table 2).
TABLE 2.
Complementation of rhamnose mutants
| Strain | Relevant genotype | Gene mutated | Growth with plasmida
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| pW3AR1 (rhaD1 dehydrogenase) | pMR79 (rhaI31 isomerase) | pMR74 (rhaR25 regulator) | pMR65 (rhaS17 PBP) | pW3AR2 (rhaT2 ABC) | pMR67 (rhaP19 permease) | pMR98 (rhaK50 kinase) | |||
| Rlt100 | Wild type | NAb | + | + | + | + | + | + | + |
| Rlt105 | rhaD1 | Dehydrogenase | − | − | + | + | + | + | + |
| Rlt124 | rhaI31 | Isomerase | − | − | + | + | + | + | + |
| Rlt117 | rhaR25 | Regulator | + | + | − | − | − | − | − |
| Rlt112 | rhaS17 | Binding protein | + | + | − | − | − | − | − |
| Rlt106 | rhaT2 | ABC protein | + | + | − | − | − | − | − |
| Rlt115 | rhaP19 | Permease | + | + | − | − | − | − | − |
| Rlt144 | rhaK50 | Kinase | + | + | − | − | − | − | − |
Designations are as follows: +, like wild-type growth; −, no growth.
NA, not applicable.
Regulation of rhaRSTPQUK.
Initial regulatory experiments have been previously carried out utilizing the rhaD1 allele either on a plasmid or as a single-copy chromosomal recombinant (35). Those data showed that rhaD1 was induced by rhamnose and clover root exudate but repressed by glucose (35). Since the rhaRSTPQUK transcript contained a regulatory gene and components that could be used for the transport and catabolism of rhamnose, it was of interest to examine the expression of this region by using representative Tn5B20 fusions that were spread out across the transcriptional unit (Table 3).
TABLE 3.
Effect of rhamnose and glucose on β-galactosidase fusions to genes in the rhamnose transport operon
| Strain | Relevant genotype | Glucosea | Rhamnose | Inductionb |
|---|---|---|---|---|
| Rlt117 | rhaR25::Tn5B20 | 2,824 (185)c | 10,451 (751)d | 3.7 |
| Rlt128 | rhaP36::Tn5B20 | 24 (1) | 660 (46)e | 28 |
| Rlt151 | rhaQ38::Tn5B20 | 33 (4) | 424 (41) | 13 |
| Rlt131 | rhaK40::Tn5B20 | 2 (1) | 5 (0) | 2 |
| Rlt144 | rhaK50::Tn5B20 | 2 (1) | 11 (2)e | 5 |
| Rlt146 | rhaK52::Tn5B20 | 4 (2) | 12 (1)e | 3 |
The values given represent β-galactosidase activity expressed in Miller units following overnight growth in defined media with either glucose-glycerol or rhamnose-glycerol as carbon sources. Values are the average of at least four independent replicates; values in parentheses represent standard error.
Ratio of rhamnose values to the glucose values.
Value represents seven independent replicates.
Value represents six independent replicates.
Value represents three independent replicates.
All the strains carrying fusions in this unit showed higher activity when grown in the presence of rhamnose than in the presence of glucose (Table 3). Strains Rlt128 and Rlt151, containing rhaP36::Tn5B20 and rhaQ38::Tn5B20, showed an induction of 28- and 13-fold, respectively. The results suggested that transcription of the operon was induced by the presence of rhamnose and that rhaPQ were essentially not transcribed under noninducing conditions.
Fusions to rhaK showed low levels of β-galactosidase activity. Initially, Rlt146 carrying rhaK52::Tn5B20 was assayed and showed a low induction. To ensure that induction was indeed occurring, two additional rhaK alleles, rhaK40::Tn5B20 and rhaK50::Tn5B20 in strains Rlt131 and Rlt144, were also assayed. Consistent with the results with Rlt146, these fusions also showed reproducible low fusion activity that showed between a two- and fivefold increase of β-galactosidase activity when grown in the presence of rhamnose (Table 3). Low β-galactosidase activity was also observed when these alleles were present on the multicopy plasmid pW3C1 in a Rlt100 background (data not shown).
In contrast to the activity found with fusions to either of the permease genes or the kinase gene, rhaR25::Tn5B20 in strain Rlt117 had very high activity under noninducing conditions (Table 3). However, this fusion also showed induction in the presence of rhamnose. The rhaR25 fusion showed the same high activity when grown on glucose regardless of whether it was integrated into the chromosome or present on a multicopy plasmid (Tables 3 and 4).
TABLE 4.
Effect of RhaR on rhaR expression
| Strain | Relevant genotype (chromosomal/plasmid) | Glucosea | Rhamnose | Inductionb |
|---|---|---|---|---|
| Rlt100 | Wild type | 17 (2) | 15 (2) | 1 |
| Rlt101 | Δrha/− | 28 (1) | 21 (1) | 1 |
| Rlt154c | Wild type/rhaR25 | 1,108 (84) | 2,408 (210) | 2.2 |
| Rlt153d | Δrha/rhaR25 | 2,650 (44) | 7,029 (288) | 2.7 |
| Rlt117 | rhaR25/− | 2,624 (70) | 7,780 (255) | 3.0 |
| Rlt197e | rhaR25/rhaR+ | 155 (13) | 600 (32) | 3.9 |
The values given represent β-galactosidase activity expressed in Miller units following overnight growth in defined media with either glucose-glycerol or rhamnose-glycerol as carbon sources. Values are the average of at least four independent replicates except for Rlt117, which is an average of three independent replicates; values in parentheses represent standard errors.
Induction was calculated by dividing activity in the presence of rhamnose by the activity in the presence of glucose.
Rlt154 = Rlt100(pMR73).
Rlt153 = Rlt101(pMR73).
Rlt197 = Rlt117(pMR53).
RhaR is a negative regulator.
Sequence analysis of rhaR suggested that this gene encodes a DeoR-type negative regulator (32). To show that RhaR acts as a negative regulator, it was reasoned that operon basal activity should increase if a strain lacking rhaR were constructed. To test this hypothesis, pMR73 carrying rhaR25::Tn5B20 was mobilized into both the wild-type Rlt100 as well as Rlt101 which was missing the entire rhamnose region. The results showed that in the absence of a functional copy of RhaR, fusion activity, as measured by rhaR25, increased 2.4-fold when the strain was grown on defined medium with glucose (Table 4, Rlt153 versus Rlt154). This is comparable to the level of induction seen when the wild-type Rlt100 carrying the fusion rhaR25 is grown in the presence of rhamnose (Table 4). The fusion activity of rhaR25 in a strain devoid of rhaR was still inducible and in fact appeared to induce to higher levels (Table 4). These data are also consistent with the levels observed for Rlt117, which is also lacking functional RhaR (Tables 3 and 4).
To provide additional evidence that RhaR is a negative regulator, it was reasoned that if RhaR were overexpressed in a rhaR25 background, the overall transcriptional activity should be reduced. To test this, the entire coding region of rhaR was PCR amplified and cloned into pRK7813, such that it would be transcribed constitutively from the plac present as part of the multiple cloning site. To ensure expression, a Shine-Dalgarno sequence was also added. The introduction of this construct, pMR53, into Rlt117 reduced the expression of a chromosomal copy of rhaR25 by approximately 17-fold when grown on glucose (Table 4, Rlt117 versus Rlt197). However, it was observed that even with the overexpression of RhaR in Rlt197, the rhaR25 fusion was still inducible by rhamnose. Together, these data suggest that the induction of this operon is independent of the negative regulation provided by RhaR (Table 4).
Transcription extends past rhaR but not beyond rhaS under noninducing conditions.
Analysis of the transcriptional fusion data suggested that the transcriptional pattern of rhaR was very different from that of the rest of the transcript (Table 3). However, mutations in rhaR were shown to be polar on rhaSTPQUK (Table 2). This suggests that a level of control must exist to allow differential transcription of the promoter-proximal genes relative to distal genes in this operon (Fig. 1). It was hypothesized that the putative hairpin loop that had a similarity to a transcriptional terminator might play a role in the regulation of this operon by preventing high levels of transcription beyond rhaS when the bacteria are grown under noninducing conditions.
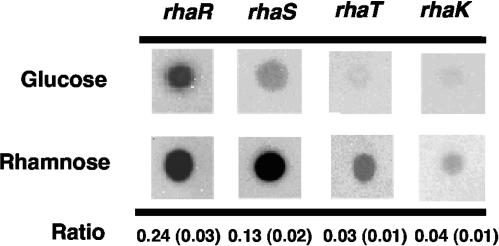
To provide direct physical evidence to support this hypothesis, transcriptional analysis was carried out using RNA dot blot analysis. Individual RNA probes corresponding to rhaR, rhaS, rhaT, and rhaK were generated and used to probe RNA isolated from both Rlt100 and Rlt100(pW3C1) grown on defined media with either glycerol-glucose or glycerol-rhamnose. The data from these experiments showed that the steady-state mRNA ratios under noninducing conditions compared to those for full induction for rhaR, rhaS, rhaT, and rhaK were 0.24 ± 0.03, 0.13 ± 0.02, 0.03 ± 0.01, and 0.04 ± 0.01, respectively (mean ± standard deviation). These data are consistent with the hypothesis that the hairpin loop found in the intergenic region between rhaS and rhaT plays a role in attenuating further transcription into the operon (Fig. 2). Moreover, RNA isolated from Rlt117 (containing rhaR25) grown in either the presence of glucose or rhamnose and probed with either rhaS, rhaT, or rhaK did not show any hybridization (data not shown). This also provides further evidence that all genes in this transcript are transcribed from a single promoter.
FIG. 2.
RNA dot blot experiments. Equal volumes and concentrations of total RNA (6 μg) from Rlt100 cultures grown in defined media with either glycerol-glucose or glycerol-rhamnose as carbon sources were spotted onto nylon charged membranes and probed with probes specific for rhaR, rhaS, rhaT, and rhaK as described in Materials and Methods. The figure shows the results of one representative experiment. Ratios represent the quantitated glucose values divided by the quantitated rhamnose values derived from three independent experiments. Values in parentheses represent the standard deviations.
Transport of rhamnose is inducible and is carried out by the ABC transporter.
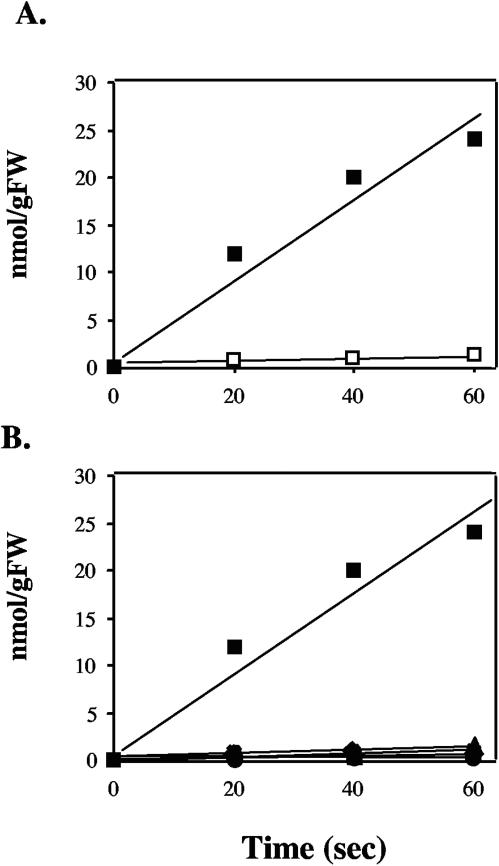
Sequence analysis suggested that rhaSTPQ might code for an ABC-type transporter. To show functionality, transport assays utilizing tritiated rhamnose were carried out. Strain Rlt100, when grown in the presence of rhamnose, was able to take up tritiated rhamnose from the culture medium (Fig. 3A). However, when Rlt100 was grown in the presence of glucose, the amount of tritiated rhamnose uptake was not above background levels.
FIG. 3.
Rhamnose transport experiments. Strains were grown to mid-log phase in defined medium containing either glucose-glycerol (open symbols) or rhamnose-glycerol (closed symbols), and initial uptake rates were determined using [3H]rhamnose. Symbols: squares, Rlt100 (wild-type); diamonds, Rlt117 (rhaR25); triangles, Rlt112 (rhaS17); circles, Rlt106; +, Rlt151 (rhaQ38); X, Rlt128 (rhaP36). (A) Comparison of rhamnose uptake by Rlt100 grown on rhamnose-glycerol or glucose-glycerol. (B) Comparison of uptake of induced wild-type and representative mutants. Note that symbols for the mutants are so low that they are superimposed. The data represent the averages of three independent experiments. Experiments in both panels were carried out at the same time. Error bars are smaller than the symbols in all cases.
To further characterize the rhamnose transport system, experiments were carried out to determine the Km for rhamnose transport. The Km for rhamnose uptake in Rlt100 was found to be approximately 13 ± 2 μM (n = 4) (data not shown).
Naturally occurring sugars are generally found in a d configuration. The exceptions to this are the naturally occurring sugars l-rhamnose, l-fucose (which is also a methyl-pentose), and l-arabinose. These sugars are also aldoses, which are found as constituents of plant cell walls (28). To determine specificity of the rhamnose transporter, a series of inhibition experiments was carried out using tritiated rhamnose and an excess of unlabeled rhamnose, fucose, or arabinose. It was determined that the transport of tritiated rhamnose was not inhibited by either l-fucose or l-arabinose, even if these were present at a 100-fold excess with respect to the tritiated rhamnose. The transport of tritiated rhamnose, however, was reduced to 13% with a 7.5-fold excess of unlabeled rhamnose and completely inhibited at a 75-fold excess (data not shown).
To determine if the proteins encoded by rhaSTPQ were responsible for the ability of Rlt100 to transport rhamnose, strains Rlt112, Rlt106, Rlt128, and Rlt151, corresponding to mutations in rhaS, rhaT, rhaP, and rhaQ, respectively, were tested for their ability to transport rhamnose. In all cases, the mutants were unable to take up the labeled rhamnose from the medium (Fig. 3B).
Additionally, Rlt117 as well as Rlt105 were assayed for their ability to transport rhamnose. It was found that Rlt117, which has an insertion upstream of rhaSTPQ, was unable to transport rhamnose (Fig. 3B), whereas Rlt105, which carries a mutation in the rhaDI transcript, had transport rates similar to that of wild-type Rlt100 (data not shown).
Catabolism of rhamnose follows a novel pathway.
The catabolism of rhamnose has been studied at both the biochemical and genetic levels in E. coli. The pathway has been shown to proceed by an isomerization of rhamnose to yield rhamnulose and then a phosphorylation to give rhamnulose-1-phosphate, followed by an aldolase reaction which cleaves the rhamnulose-1-phosphate to give dihydroxy-acetone phosphate and lactaldehyde (40, 47, 52, 53).
In E. coli, a biochemical lesion in a catabolic pathway following the synthesis of a phosphorylated sugar intermediate can give rise to a conditional lethal phenotype if the bacterium is grown on a medium that contains a noninducing carbon source that can be catabolized. This particular phenomenon was first shown with respect to galactose utilization in E. coli. Galactose utilization is carried out by the subsequent actions of three enzymes, GalK (galactose kinase), GalT (galactose-1-phosphate uridyl transferase), and GalE (UDP-galactose 4-epimerase), which convert galactose first to galactose-1-phosphate, then to UDP-galactose, and finally to glucose-1-phosphate. Whereas galK mutants are able to grow in the presence of glycerol and galactose, galE and galT mutants are sensitive to galactose in the presence of glycerol. This sensitivity is correlated with the accumulation of intermediates of this pathway: galactose-1-phosphate and/or UDP-galactose (1, 51). An analogous growth inhibition and accumulation of rhamnulose-1-phosphate have also been demonstrated to take place in the rhamnose catabolic pathway (40).
Since the gene order as well as the predicted functions of the ORFs in Rlt100 differed from those of the E. coli rhamnose operon, we wished to determine whether the catabolism of this methyl-pentose followed the same biochemical pathway. To determine this, R. leguminosarum rhaI, rhaD, and rhaK mutants were tested for their ability to grow on rhamnose, rhamnose-glycerol, and glycerol defined medium. The results showed that, as expected, rhaK mutants were able to grow on glycerol-rhamnose medium (Table 5). However, rhaD and rhaI mutants were unable to grow on glycerol-rhamnose (Table 5). Taken together, these data suggest that unlike E. coli, where the first step of the catabolic pathway is an isomerization, the first step in the catabolism of rhamnose in R. leguminosarum strain Rlt100 is a phosphorylation.
TABLE 5.
Growth of mutant Rhizobium strains with a representative in each of the on glycerol, rhamnose, and glycerol-rhamnose as carbon sourcesa
| Strain | Relevant genotype | Location of mutation(s) | Growth on:
|
||
|---|---|---|---|---|---|
| Glyc | Glyc-Rha | Rha | |||
| Rlt100 | Wild type | NA | + | + | + |
| Rlt106 | rhaT2 | ABC | + | + | − |
| Rlt117 | rhaR25 | Regulator | + | + | − |
| Rlt144 | rhaK50 | Kinase | + | + | − |
| Rlt151 | rhaQ38 | Permease | + | + | − |
| Rlt105 | rhaD1 | Dehydrogenase | + | − | − |
| Rlt124 | rhaI31 | Isomerase | + | − | − |
| Rlt211 | rhaD1 rhaK58 | Dehydrognase, kinase | + | + | − |
| Rlt212 | rhaI31 rhaK58 | Isomerase, kinase | + | + | − |
Designations are as follows: +, like wild-type growth; −, no growth. A 15 mM concentration of each carbon source was added. Glyc, glycerol; Rha, rhamnose; NA, not applicable.
It was hypothesized that if RhaK were indeed the first step of rhamnose catabolism, then either a rhaD/rhaK or a rhaI/rhaK double mutant should have the same phenotype on glycerol-rhamnose medium as a rhaK mutant. To test this, Rlt211 containing rhaD1/rhaK58 and Rlt212 containing rhaI31/rhaK58 were constructed. When these strains were tested on glycerol-rhamnose medium, the results showed that both of these strains were able to grow (Table 5). These data are consistent with the hypothesis that in R. leguminosarum, RhaK and not RhaI is the first biochemical step in the catabolism of rhamnose.
DISCUSSION
Genes involved in the transport and catabolism of rhamnose in R. leguminosarum bv. trifolii strain Rlt100 are plasmid encoded and are necessary for the bacterium to be competitive for nodule occupancy (6, 35). Sequence analysis of this region was consistent with the hypothesis that the locus contained two transcripts, rhaDI and rhaRSTPQUK. The analysis was ambiguous, however, because intergenic regions that could contain promoters existed within the putative rhaRSTPQUK transcript (Fig. 1), and β-galactosidase fusion analysis showed differential expression (Table 3). It was therefore of interest to determine whether these genes were in one transcript. Based on the isolation of transposon inserts in a negative regulator, RhaR (Fig. 1), the inability of Rlt117, which carries the polar rhaR25 allele, to transport rhamnose (Fig. 3), the complementation data (Table 2), and the lack of detectable mRNA for genes distal to rhaR25, we conclude that rhaRSTPQUK is transcribed as a single transcript.
The transcription of the rha::lacZ fusions in the rhaRSTPQUK operon can be roughly divided into three groups: the activity exhibited by rhaR25::Tn5B20 (Tables 3 and 4), the activity exhibited by rhaP36::Tn5B20 and rhaQ38::Tn5B20, and that of fusions in rhaK (Table 3). Stoichiometric balance between components of an ATP binding cassette transport system is often necessary for correct functioning. As such, it is not unexpected to find ABC transporter operons organized such that components that need to be found in higher concentrations are encoded by genes which are found at the 5′ end of an operon (10, 15).
The in vivo activity of RhaR is consistent with its sequence analysis as a DeoR-type negative regulator (Table 4). Transcriptional activity of the rhaR25 fusion responds to the absence or presence of RhaR with corresponding increases or decreases in assayed activity. It is of interest that although the basal activity of a strain carrying the rhaR25 fusion grown on glucose can be manipulated to increase or decrease with respect to the absence or presence of RhaR, this fusion is still inducible by rhamnose (Table 4). The complete absence of RhaR leads to a large increase in the rate of transcription of rhaR as measured by the rhaR25 fusion allele (Tables 3 and 4). This argues that the regulation of the rhaR transcript is under two levels of control, a negative regulator, RhaR, as well as another regulator that is not encoded on genes found within this region. It is also of note that induction in Rlt117 is occurring even though we are unable to detect the transport of rhamnose into the cell (Fig. 3).
The presence of hairpin attenuators has been previously described in a number of operons (5, 44). RNA dot blot experiments were carried out to address whether the hairpin-like sequence that is found in the intergenic region between rhaS and rhaT was correlated with the lack of activity of fusions found distal to rhaS when grown in the presence of glucose (Table 3). The data clearly show that transcription drops off dramatically between rhaS and rhaT (Fig. 2). The mechanism(s) by which antitermination might occur under inducing conditions in the rhaRSTPQUK operon is at present unknown.
The RNA dot blot data also show that the gene for the sugar binding component, rhaS, is also expressed when Rlt100 is grown in the presence of glucose. Assuming transcription is proportional to translation, this would indicate that under noninducing conditions RhaS is present in the periplasm. The presence of the periplasmic binding component under noninducing conditions suggests that this protein may play a role in addition to transport. Sugar binding proteins have been shown to participate in signal transduction cascades in chemotaxis (7, 20, 25, 33, 63) and in activation of other gene regulation pathways (37) in addition to being substrate recognition proteins for ABC transporters. Tests using a previously described swarm plate assay (62) to evaluate whether strains carrying a rhaS mutation had altered chemotaxis toward rhamnose found no difference between these strains and the wild type (data not shown).
The β-galactosidase fusion data from three independent rhaK alleles, either integrated into the chromosome (Table 3) or as plasmids in a wild-type background, yielded values that were consistently lower than for any other alleles in the operon. Dot blot data, however, clearly showed that induction of rhaK occurs when grown in the presence of rhamnose (Fig. 2). The data are consistent with a lower rate of transcription through rhaK, which in turn may play a role in limiting the amount of RhaK that is produced. It is possible that RhaK is a very stable or a very active protein, and therefore the gene does not need to be transcribed at the same level as the transporter. We have, however, shown that strains carrying rhaD and rhaI mutations have a sensitivity phenotype if they are grown on rhamnose-glycerol that is rhaK dependent (Table 5). The phenotype is presumably due to the buildup of a phosphorylated intermediate (1, 40, 51). Since it is highly unlikely that the bacterium would ever encounter a situation where it would entirely depend upon rhamnose as its sole source of carbon in nature, RhaK may need to be limiting to ensure that the metabolic pool of the phosphorylated intermediate is kept at a subinhibitory concentration.
Transport experiments using tritiated rhamnose show that the wild-type strain Rlt100 does not take up rhamnose when grown in the presence of glucose and that transport is inducible by the presence of rhamnose. The inducible nature of the transporter is consistent with the regulatory studies carried out with the genetic fusions as well as the RNA dot blot assays. The inability of representative mutants in the regulator and the components of the ABC transporter to take up labeled rhamnose demonstrate that the operon containing the ABC transporter is necessary for the transport of rhamnose and that a secondary transporter for this sugar is not present or is not expressed under the growth conditions being utilized. Mutants carrying lesions in rhaD or rhaI were able to transport rhamnose, although they were unable to grow on the sugar as a sole carbon source (data not shown).
The biochemical pathway for the catabolism of methyl-pentoses is relatively well understood in enteric bacteria, and this model has been assumed to be true for all organisms. The simple phenotypic testing of representative mutants in Rlt100 unable to grow using rhamnose as a sole carbon compound suggests that the pathway order, and hence the order of the biochemical reactions, may not be conserved. The simplest interpretation of our data suggests that following transport of rhamnose into the cell the first catabolic step is the phosphorylation of rhamnose and not an isomerization to yield a keto-sugar, as seen in E. coli. Although the position of a gene does not imply the order of a biochemical pathway or its function, it may be a noteworthy observation that the kinase is found in the same transcript as the sugar transporter, suggesting that the primary catabolic step may work in concert with the transport of the sugar.
Since the pathway appears to be initiated with the phosphorylation of rhamnose rather than rhamnulose, the presumed substrates of each of the subsequent enzymatic steps are currently unavailable commercially, making it difficult to demonstrate biochemical activity for either the isomerase or the dehydrogenase-aldolase. Nevertheless, we attempted to develop a coupled in vitro assay to demonstrate dehydrogenase-aldolase activity utilizing a rhamnose-induced cell extract, ATP, rhamnose, and NAD(P)+. The goal of such an assay would be to test strains carrying either a dehydrogenase or an isomerase mutation and thus be able to order the pathway. We were unsuccessful in our attempts. Presumably, either the quantity of the product from the kinase and/or the isomerase reaction may have been insufficient to act as a substrate for the dehydrogenase reaction, or a necessary cofactor was absent. Our work in this area is ongoing.
In this report we have characterized a locus necessary for rhamnose catabolism and determined that in R. leguminosarum bv. trifolii rhamnose transport is inducible and dependent on an ABC-type transporter that is regulated by a DeoR-type negative regulator. As well, a level of transcriptional regulation may be due to a hairpin loop located in the rhaRSTPQUK transcript that may aid in expression of genes needed for uptake and catabolism at levels proportional to the amount of each individual protein required. Additionally, we have shown that the catabolism of rhamnose follows a novel pathway that to our knowledge has not been previously described. We are currently focusing our work on understanding the molecular aspects of rhamnose catabolism and regulation in R. leguminosarum. By understanding these fundamental aspects, it is hoped that we can achieve a better understanding of how these mutants grow in the rhizosphere and why their inability to catabolize rhamnose leads to a noncompetitive nodulation phenotype.
Acknowledgments
This work was supported by NSERC grants to I.J.O. and M.F.H.
We thank D. Court for invaluable advice to carry out RNA dot blot and Northern blot analyses and Karen LoVetri and E. Worobec for advice on carrying out transport assays. We are also grateful to Josh Adam, who contributed technical assistance as an undergraduate project student in the lab of I.J.O. at the University of Manitoba.
REFERENCES
- 1.Adhya, S. L., and J. A. Shapiro. 1969. The galactose operon of E. coli K-12. I. Structural and pleiotropic mutations of the operon. Genetics 62:231-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeyev, M. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. BioTechniques 26:824-828. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M. R., R. E. Brent, D. D. Kingston, J. G. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. J. Wiley and Sons, New York, N.Y.
- 5.Bailey, M. J. A., C. Hughees, and V. Koronakis. 1997. RfaH and the ops element, components of a system controlling bacterial transcription elongation. Mol. Microbiol. 26:845-851. [DOI] [PubMed] [Google Scholar]
- 6.Baldani, J. I., R. W. Weaver, M. F. Hynes, and B. D. Eardly. 1992. Utilization of carbon substrates, electrophoretic enzyme patterns, and symbiotic performance of plasmid-cured rhizobia. Appl. Environ. Microbiol. 58:2308-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bastelaere, E. V., M. Lambrecht, H. Vermeiren, V. Keijers, P. Proost, and J. Vanderleyden. 1999. Characterization of a sugar-binding protein from Azospirllum basilense mediating chemotaxis to and uptake of sugars. Mol. Microbiol. 32:703-714. [DOI] [PubMed] [Google Scholar]
- 8.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 9.Beringer, J. E., J. L. Beynon, A. V. Buchanan-Wollason, and A. W. B. Johnston. 1978. Transfer of the drug resistance transposon Tn5 to Rhizobium. Nature 276:633-634. [Google Scholar]
- 10.Boos, W., and J. M. Lucht. 1996. Periplasmic binding protein-dependent ABC transporters, p. 1175-1209. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 11.Borthakur, D., M. Soedarjo, P. M. Fox, and D. T. Webb. 2003. The mid genes of Rhizobium sp. strain TAL1145 are required for degradation of mimosine into 3-hydroxy-4-pyridine and are inducible by mimosine. Microbiology 149:537-546. [DOI] [PubMed] [Google Scholar]
- 12.Bowen, G. D., and A. D. Rovira. 1976. Microbial colonisation of plant roots. Annu. Rev. Phytopathol. 14:121-144. [Google Scholar]
- 13.Bringhurst, R. M., Z. G. Cardon, and D. Gage. 2001. Galactosides in the rhizosphere: utilization by Sinorhizobium meliloti and development of a biosensor. Proc. Natl. Acad. Sci. USA 98:4540-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark, S. R. D., I. J. Oresnik, and M. F. Hynes. 2001. RpoN of Rhizobium leguminosarum bv. viciae strain VF39SM plays a central role in FnrN-dependent microaerobic regulation of genes involved in nitrogen fixation. Mol. Gen. Genet. 264:623-633. [DOI] [PubMed] [Google Scholar]
- 15.di Guan, G. C., P. Li, P. D. Riggs, and H. Inouye. 1988. Vectors that facilitate the expression and purification of foreign peptides in E. coli by fusion to maltose-binding protein. Gene 67:21-30. [DOI] [PubMed] [Google Scholar]
- 16.Dowling, D. N., and W. J. Broughton. 1986. Competition for nodulation of legumes. Annu. Rev. Microbiol. 40:131-157. [DOI] [PubMed] [Google Scholar]
- 17.Finan, T. M., B. Kunkel, G.F. de Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fry, J., M. Wood, and P. S. Poole. 2001. Investigation of myo-inositol catabolism in Rhizobium leguminosarum bv. viciae and its effect on nodulation competitiveness. Mol. Plant-Microbe Interact. 14:1016-1025. [DOI] [PubMed] [Google Scholar]
- 19.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, P. Abola, F Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thébault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 20.Gardina, P., C. Conway, M. Kosman, and M. Manson. 1992. Aspartate and maltose-binding protein interact with adjacent sites in the Tar chemotactic signal transducer of Escherichia coli. J. Bacteriol. 174:1528-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geer, L. Y., M. Domrachev, D. J. Lipman, and S. H. Bryant. 2002. CDART: protein homology by domain architecture. Genome Res. 12:1619-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiménez-Zurdo, J. I., F. M. Garcia-Rodríguez, and N. Toro. 1997. The Rhizobium meliloti putA gene: its role in the establishment of the symbiotic interaction with alfalfa. Mol. Microbiol. 23:85-93. [DOI] [PubMed] [Google Scholar]
- 23.Jiménez-Zurdo, J. I., P. van Dillewijn, M. J. Soto, M.R. de Filipe, J. Olivares, and N. Toro. 1995. Characterization of a Rhizobium meliloti proline dehydrogenase mutant altered in nodulation efficiency and competitiveness on alfalfa roots. Mol. Plant-Microbe Interact. 8:492-498. [DOI] [PubMed] [Google Scholar]
- 24.Jones, J. D. G., and N. Gutterson. 1987. An efficient mobilizable cosmid vector and its use in rapid marker exchange in Pseudomonas fluorescens strain HV37a. Gene 61:299-306. [DOI] [PubMed] [Google Scholar]
- 25.Kemner, J. M., X. Liang, and E. W. Nester. 1997. The Agrobacterium tumefaciens virulence gene chvE is part of a putative ABC-type sugar transport operon. J. Bacteriol. 179:2452-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knee, E. M., F. Gong, M. Gao, M. Teplitski, A. R. Jones, A. Foxworthy, A. J. Mort, and W. D. Bauer. 2001. Root mucilage from pea and its utilization by rhizosphere bacteria as a sole carbon source. Mol. Plant-Microbe Interact. 14:775-784. [DOI] [PubMed] [Google Scholar]
- 27.Long, S. R. 1996. Rhizobium symbiosis: Nod factors in perspective. Plant Cell 8:1885-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNeil, M., A. G. Darvill, S. C. Fry, and P. Albersheim. 1984. Structure and function of the primary cell walls of plants. Annu. Rev. Biochem. 53:114-122. [DOI] [PubMed] [Google Scholar]
- 29.Meade, H. M., S. R. Long, G. B. Ruvkin, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Springs Harbor Laboratory, Cold Springs Harbor, N.Y.
- 31.Moënne-Locoz, Y., I. J. Baldani, and R. W. Weaver. 1995. Sequential heat curing of Tn5mob-sac labeled plasmids from Rhizobium to obtain derivatives with various combinations of plasmids and no plasmids. Lett. Appl. Microbiol. 20:175-179. [Google Scholar]
- 32.Mortensen, L., G. Dandanell, and K. Hammer. 1989. Purification and characterization of the DeoR repressor of Escherichia coli. EMBO J. 8:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mowbray, S. L., and D. E. Koshland. 1987. Additive and independent responses in a single receptor: aspartate and maltose stimuli on the Tar protein. Cell 50:171-180. [DOI] [PubMed] [Google Scholar]
- 34.Oresnik, I. J., T. C. Charles, and T. M. Finan. 1994. Second site mutations specifically suppress the Fix− phenotype of Rhizobium meliloti ndvF mutations on alfalfa: identification of a conditional ndvF-dependent mucoid colony phenotype. Genetics 136:1233-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oresnik, I. J., L. A. Pacarynuk, S.A.P. O'Brien, C. K. Yost, and M. F. Hynes. 1998. Plasmid encoded catabolic genes in Rhizobium leguminosaurm bv. trifolii: evidence for a plant-inducible rhamnose locus involved in competition for nodulation. Mol. Plant-Microbe Interact. 11:1175-1185. [Google Scholar]
- 36.Oresnik, I. J., S. Twelker, and M. F. Hynes. 1999. Cloning and characterization of a Rhizobium leguminosarum gene encoding a bacteriocin with similarities to RTX toxins. Appl. Environ. Microbiol. 65:2833-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng, W., Y. Lee, and E. W. Nester. 1998. The phenolic recognition profiles of Agrobacterium tumefaciens VirA protein are broadened by a high level of the sugar-binding protein ChvE. J. Bacteriol. 180:5632-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips, D. A., E. S. Sande, J. A. C. Vriezen, F. J. de Bruijn, D. Le Rudulier, and C. M. Joseph. 1998. A new genetic locus in Sinorhizobium meliloti is involved in stachydrine utilization. Appl. Environ. Microbiol. 64:3954-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poole, P., and D. Allaway. 2000. Carbon and nitrogen metabolism in Rhizobium. Adv. Microb. Physiol. 43:117-163. [DOI] [PubMed] [Google Scholar]
- 40.Power, J. 1967. The l-rhamnose genetic system of E. coli K-12. Genetics 55:557-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 42.Robleto, E. K., E. S. Kmiecik, E. S. Oplinger, J. Nienhuis, and E. W. Triplett. 1998. Trifoltoxin production increases nodulation competitiveness of Rhizobium etli CE3 under agricultural conditions. Appl. Environ. Microbiol. 64:2630-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenblueth, M., M. F. Hynes, and E. Martínez-Romero. 1998. Rhizobium tropici teu genes involved in specific uptake of Phaseolis vulgaris bean-exudate compounds. Mol. Gen. Genet. 258:587-598. [DOI] [PubMed] [Google Scholar]
- 44.Rutberg, B. 1997. Antitermination of transcription of catabolic operons. Mol. Microbiol. 23:413-421. [DOI] [PubMed] [Google Scholar]
- 45.Ruvkun, G. B., and F. M. Ausubel. 1981. A general method for site-directed mutagenesis in prokaryotes. Nature 289:85-88. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. A. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 47.Sawada, H., and Y. Takagi. 1964. The metabolism of l-rhamnose in E. coli. III. l-Rhamnulose-phosphate aldolase. Biochim. Biophys. Acta 64:26-32. [DOI] [PubMed] [Google Scholar]
- 48.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo engineering: transposon mutagenesis in gram-negative bacteria. Bio/Techniques 1:784-791. [Google Scholar]
- 49.Soedarjo, M., and D. Borthakur. 1998. Mimosine, a toxin produced by the tree-legume Leucaena provides a nodulation competition advantage to mimosine-degrading Rhizobium strains. Soil Biol. Biochem. 30:1605-1613. [Google Scholar]
- 50.Streit, W. R., C. M. Joseph, and D. A. Phillips. 1996. Biotin and other water-soluble vitamins are key growth factors for alfalfa colonization by Rhizobium meliloti 1021. Mol. Plant-Microbe Interact. 9:330-338. [DOI] [PubMed] [Google Scholar]
- 51.Sundararajan, T. A., A. M. C. Rapin, and H. M. Kalakar. 1962. Biochemical observations on E. coli mutants defective in uridine diphosphoglucose. Proc. Natl. Acad. Sci. USA 48:2187-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takagi, Y., and H. Sawada. 1964. The metabolism of l-rhamnose in E. coli. I. l-Rhamnose isomerase. Biochim. Biophys. Acta 92:10-17. [DOI] [PubMed] [Google Scholar]
- 53.Takagi, Y., and H. Sawada,. 1964. The metabolism of l-rhamnose in E. coli. II. l-Rhamnulose kinase. Biochim. Biophys. Acta 64:18-25. [DOI] [PubMed] [Google Scholar]
- 54.Triplett, E. W., and M. Sadowsky. 1992. Genetics of competition for nodulation of legumes. Annu. Rev. Microbiol. 46:399-428. [DOI] [PubMed] [Google Scholar]
- 55.Van Egeraat, A. W. S. M. 1975. The possible role of homoserine in the development of Rhizobium leguminosarum in the rhizosphere of pea seedlings. Plant Soil 42:381-386. [Google Scholar]
- 56.Van Rhijn, P., and J. Vanderleyden. 1995. The Rhizobium-plant symbiosis. Microbiol. Rev. 59:124-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venter, A. P., S. Twelker, I. J. Oresnik, and M. F. Hynes. 2001. Analysis of the genetic region encoding a novel rhizobiocin from Rhizobium leguminosarum bv. viciae strain 306. Can. J. Microbiol. 47:495-502. [DOI] [PubMed] [Google Scholar]
- 58.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific Publications, Oxford, England.
- 59.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 60.Yanisch-Perron, C., J. Viera, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of M13mp18 and pUC19. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 61.Yarosh, O. K., T. C. Charles, and T. M. Finan. 1989. Analysis of C4-dicarboxylate transport genes in Rhizobium meliloti. Mol. Microbiol. 3:813-823. [DOI] [PubMed] [Google Scholar]
- 62.Yost, C. K., P. Rochepeau, and M. F. Hynes. 1998. Rhizobium leguminosarum contains a group of genes that appear to code for methyl-accepting chemotaxis proteins. Microbiology 144:1945-1956. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, Y. C., C. Conway, M. Rosato, Y. Suh, and M. D. Manson. 1992. Maltose chemotaxis involves residues in the N-terminal and C-terminal domains on the same faces of maltose-binding protein. J. Biol. Chem. 267:22813-22820. [PubMed] [Google Scholar]