Abstract
Expression of toxT, the transcription activator of cholera toxin and pilus production in Vibrio cholerae, is the consequence of a complex cascade of regulatory events that culminates in activation of the toxT promoter by TcpP and ToxR, two membrane-localized transcription factors. Both are encoded in operons with genes whose products, TcpH and ToxS, which are also membrane localized, are hypothesized to control their activity. In this study we analyzed the role of TcpH in controlling TcpP function. We show that a mutant of V. cholerae lacking TcpH expressed virtually undetectable levels of TcpP, although tcpP mRNA levels remain unaffected. A time course experiment showed that levels of TcpP, expressed from a plasmid, are dramatically reduced over time without co-overexpression of TcpH. By contrast, deletion of toxS did not affect ToxR protein levels. A fusion protein in which the TcpP periplasmic domain is replaced with that of ToxR remains stable, suggesting that the periplasmic domain of TcpP is the target for degradation of the protein. Placement of the periplasmic domain of TcpP on ToxR, an otherwise stable protein, results in instability, providing further evidence for the hypothesis that the periplasmic domain of TcpP is a target for degradation. Consistent with this interpretation is our finding that derivatives of TcpP lacking a periplasmic domain are more stable in V. cholerae than are derivatives in which the periplasmic domain has been truncated. This work identifies at least one role for the periplasmic domain of TcpP, i.e., to act as a target for a protein degradation pathway that regulates TcpP levels. It also provides a rationale for why the V. cholerae tcpH mutant strain is avirulent. We hypothesize that regulator degradation may be an important mechanism for regulating virulence gene expression in V. cholerae.
Vibrio cholerae, which causes severe dehydrating diarrhea in humans, expresses important virulence determinants under the control of unusual transcription activator proteins, ToxR and TcpP, which reside in the inner membrane of the cell (16, 38). Membrane-localized ToxR and TcpP activate transcription of the toxT gene (11, 16), which encodes a direct activator of cholera toxin and toxin-coregulated pilus expression (11).
The model for the cooperative action between ToxR and TcpP in controlling toxT transcription is that TcpP binds to the toxT promoter just upstream of the basal promoter element and from this position interacts with and stimulates RNA polymerase. ToxR binds upstream of TcpP, and in the model, ToxR does not directly activate transcription but instead enables activation by TcpP, perhaps through protein-protein interaction (26,27). ToxR, TcpP, and other membrane-localized transcription activators are bitopic membrane proteins, with an amino-terminal cytoplasmic domain, called a winged helix-turn-helix domain, that is similar to a DNA-binding and transcription activation domain found in response regulator proteins such as OmpR and PhoB (32, 38). The amino-terminal domain is followed by a transmembrane domain, which, in turn, is followed by a periplasmic domain of indeterminate function. Attempts to identify a function for the periplasmic domain have been inconclusive, although some evidence suggests that it may be a signal-sensing domain. For example, replacing the periplasmic domain of ToxR with alkaline phosphatase leads to a constitutive phenotype in which toxin expression is no longer affected by some signals that influence expression in cells with wild-type ToxR (38). A fusion of the periplasmic domain of TcpP to β-lactamase is also active under conditions that are not normally permissive for expression of virulence genes in V. cholerae (16). Additionally, in the topologically similar CadC protein of Escherichia coli, which activates gene expression in response to low pH in the presence of lysine, mutations resulting in constitutive, signal-independent activity map to the periplasmic domain (10).
A feature of the periplasmic domain of ToxR-like proteins is their association with other membrane proteins that appear to regulate their function. For ToxR, this effector is ToxS, encoded downstream of toxR on the V. cholerae chromosome and required for wild-type ToxR activity (36). The TcpP effector is TcpH, the gene for which is cotranscribed with tcpP (4, 16). The precise mechanism of action of these membrane effector proteins is unclear. Based on evidence from experiments done with E. coli and Salmonella enterica serovar Typhimurium, ToxS is hypothesized to play a role in stabilizing ToxR (11, 13, 44). In E. coli, ToxS expression enables low levels of ToxR to activate transcription (11, 36) and this ability is associated with an increase in ToxR stability. More specifically, expression of ToxS has been shown to increase the stability of a ToxR-PhoA fusion and a λcI-ToxR fusion in E. coli (11, 13). Residues within the cytoplasmic amino-terminal domain of ToxR were identified as being required for this ToxS-dependent stability in E. coli (11). In addition, a Leu-to-Ser mutation at residue 33 of ToxS (ToxSL33S) conferred instability on ToxR in S. enterica serovar Typhimurium, again suggesting that interaction between ToxR and ToxS somehow regulates stability of the former by the latter (44).
Another hypothesis of ToxS function is that it enhances the dimerization of ToxR, thought to be a critical feature of ToxR activity (11, 13, 38). This hypothesis is based on the behavior of ToxR fusion proteins in E. coli, most notably one in which the periplasmic and transmembrane domains of ToxR were fused to the operator-binding site of the lambda repressor cI (13). This domain requires dimerization to bind DNA and repress transcription. ToxR-cI showed enhanced operator site binding in the presence of ToxS, suggesting that the presence of ToxS leads to a greater amount of dimerization of ToxR through its periplasmic or membrane domain. Later work examining ToxR conformation calls into question the necessity of dimerization for ToxR function in V. cholerae (12, 41, 42). Since virtually all of the work examining ToxS function has been performed with heterologous systems, the role of ToxS in V. cholerae remains unclear.
The relationship of TcpH to TcpP function has been less well characterized than that of ToxS to ToxR. Strains lacking tcpP are deficient for activation of toxT transcription (16), and strains with mutations in tcpH demonstrate irregular toxT transcription (4, 57) and are deficient for activating virulence factors downstream of toxT, including cholera toxin and toxin-coregulated pilus (4). tcpH mutants have also been shown to colonize infant mice at levels significantly below those of the wild type (4). These data demonstrate the importance of both tcpP and tcpH for toxT transcription activation. The TcpP requirement in virulence gene expression is better understood than is the TcpH requirement, although TcpH is likely needed because of its effect on TcpP function, as overexpression of tcpH enhances TcpP activity (16).
In this study we analyzed the role of TcpH in TcpP function and here report its role in a posttranslational mechanism that regulates TcpP levels in the cell. We show that a tcpH deletion mutant produces nearly undetectable steady-state levels of TcpP, in contrast to those of ToxR in V. cholerae in the absence of ToxS, and that this effect on TcpP concentration is not at the transcriptional level but rather at the posttranslational level. We present evidence that levels of TcpP are governed by a proteolytic mechanism that is antagonized by TcpH and that the target of TcpP degradation is its periplasmic, carboxy-terminal domain.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The V. cholerae classical strain O395 was used throughout this study. The E. coli strains DH5α and DH5αλpir were used for cloning, and SM10λpir was used for conjugation with V. cholerae. All strains were grown in Luria-Bertani (LB) medium at 30 or 37°C. To activate virulence gene expression, V. cholerae was cultured at 30°C in LB medium at pH 6.5. Mid-log cultures were used for all experiments. Plasmids used in this study include the suicide vector pKAS32 (51), the constitutive expression vector pACYC184, and the arabinose-inducible expression vector pBAD18Kan (15). Unless otherwise noted, 0.1% arabinose was used to induce expression of PBAD. Plasmid DNA was transferred into V. cholerae by electroporation or by filter conjugation with the E. coli strain SM10λpir. E. coli strains were transformed by standard methods (47). When appropriate, the following antibiotics were used at the concentrations indicated: streptomycin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 20 μg/ml; ampicillin, 100 μg/ml; and tetracycline, 10 μg/ml.
Construction of deletion mutants.
The ΔtcpH strain was constructed by cloning a fragment of DNA containing an internal deletion of tcpH generated by SOEing PCR (17), with the use of Expand Hi-Fidelity polymerase (Boehringer Mannheim), into the suicide vector pKAS32 (51). Construction of this mutant is described in detail elsewhere (57). Briefly, SOE (splicing by overlap extension) PCR was used to amplify a region of DNA spanning 689 bp upstream of the tcpH start codon to 572 bp downstream of the stop codon, containing an internal deletion of the tcpH gene. To facilitate cloning into the suicide vector pKAS32, the fragment was engineered with unique restriction sites at the 5′ and 3′ ends. The fragment was then cloned into pKAS32, and the resultant plasmid was then electroporated into the E. coli strain SM10λpir. This new strain was mated into V. cholerae strain O395 or O395 toxT::lacZ (16) by filter conjugation. Selection for integration was done on thiosulfate-citrate-bile salts-sucrose (Difco) plates containing ampicillin (50 μg/ml). Resolution of the cointegrate was selected for on LB plates containing streptomycin (1 mg/ml). Recombination and loss of the wild-type allele were confirmed by PCR analysis with primers flanking the deletion. The ΔtcpP (57) and ΔtoxR (26) strains used for this study have been previously described.
The ΔtoxS derivative was constructed as follows. A BamHI fragment from pVM16 (38), lacking the toxS coding sequence but carrying V. cholerae DNA from upstream and downstream of toxS, was cloned into the BglII site of the suicide plasmid pKAS32 (51). The resulting plasmid was introduced by electroporation into V. cholerae strain O395, with selection for ampicillin resistance encoded by the plasmid. Subsequent recombinants in which the wild-type allele was replaced with the ΔtoxS allele were identified as described previously (51). The presence of the mutant allele was then confirmed by PCR.
Construction of tcpP and toxR expression vectors.
Full-length tcpP and toxR genes were amplified from O395 chromosomal DNA with Expand Hi-Fidelity polymerase (Boehringer Mannheim). After amplification, the PCR products were digested and ligated into the inducible expression vector pBAD18Kan. The truncated forms of tcpP were constructed in the same manner by using the identical 5′ oligonucleotide and appropriate 3′ oligonucleotides. The truncations were made at amino acid positions 138, 169, 190, and 222. The tcpP-toxR chimeric gene was constructed by SOEing PCR to fuse the tcpP gene through the predicted transmembrane domain with the predicted periplasmic domain of toxR. The same strategy was employed to create the toxR-tcpP chimeric gene. The resulting chimeric DNA fragments were cloned into pBAD18Kan. The tcpH gene was cloned into the constitutive expression vector pACYC184.
Western blots.
Overnight cultures grown at 30°C in LB medium were subcultured at a dilution of 1:100 in LB (pH 6.5) and grown at 30°C for 4 to 5 h. For strains containing the pBAD18Kan vector, arabinose was added to the culture medium at the time of subculture. The mid-log culture (1 ml) was then pelleted and resuspended in 50 to 100 μl of 1× sample buffer (47). Whole-cell pellets were boiled for 5 min and loaded onto a 15% polyacrylamide gel. Loading volumes were adjusted to normalize for culture optical density at 600 nm (OD600). Proteins were transferred to Immobilon-P membranes (Millipore) and probed with rabbit anti-TcpP antibody (generated by Zymed) at 1:500, followed by goat anti-rabbit HRP-conjugated secondary antibody (Pierce) at 1:10,000. Proteins were detected with use of an ECL detection kit (Amersham).
RNA isolation and analysis.
Overnight cultures grown at 30°C in LB medium were subcultured at 1:100 in LB (pH 6.5) and grown at 30°C for 4 to 5 h. Mid-log-phase culture (15 ml) was then pelleted, and RNA was extracted by using TRIzol reagent (Gibco-BRL) according to the manufacturer's protocol. RNA samples were then treated with DNase I and quantified by measurements of OD260. Reverse transcriptase (RT)-PCR was performed with Superscript II reverse transcriptase (Gibco-BRL) according to the manufacturer's instructions. Each cDNA reaction was performed with 5 μg of RNA and 250 nmol of random oligonucleotides (Gibco-BRL). Control reactions lacking reverse transcriptase were performed in parallel. Equal amounts of the cDNA generated were then used in PCRs with primers to amplify either tcpP or, as a control, the α subunit of RNA polymerase.
Pulse expression experiments with arabinose and rifampin.
Overnight cultures grown at 30°C in LB medium were subcultured at 1:100 in LB (pH 6.5) and grown at 30°C for 3.5 h. l-(+)-Arabinose (Sigma) was added to a concentration of 0.1%, and cultures were held at 30°C for an additional hour. Next, rifampin (Sigma) was added to 200 μg/ml to stop transcription while cells remained under the given culture conditions. Samples were collected at various time points following the addition of rifampin. Culture sample (1 ml) was collected for OD600 determination and 1 ml was collected for Western blot analysis. Processing for Western blot analysis was done as described above.
Measurement of toxT-lacZ activation.
Overnight cultures grown at 30°C in LB medium were subcultured at 1:100 in LB (pH 6.5) and grown at 30°C for 4 to 5 h. Where appropriate, expression of PBAD was induced by the addition of arabinose at the time of subculture. Culture samples (50 to 100 μl) were used to determine β-galactosidase activity as previously described (34).
RESULTS
TcpP derivatives with altered periplasmic domains are unstable in V. cholerae.
Our investigation of TcpH function grew out of studies of localization requirements for the membrane-localized transcription factors ToxR and TcpP. Both are bitopic proteins with winged helix-turn-helix cytoplasmic domains required for DNA binding and transcription activation. ToxR and TcpP together activate toxT transcription, while ToxR alone controls transcription of the porin genes ompU and ompT, by activating the former and repressing the latter (37). Membrane localization of ToxR is a requirement for activation of the toxT promoter but not for regulation of the omp promoters (8).
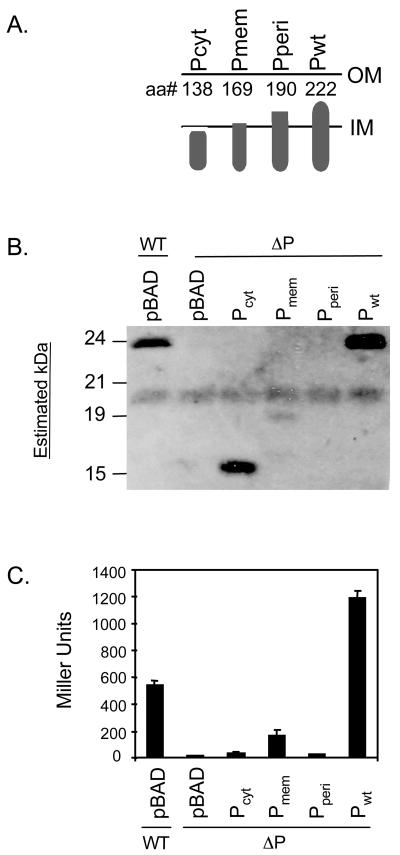
We sought to determine the localization requirements for TcpP-dependent transcription activation by constructing truncated forms of TcpP, shown in Fig. 1A. The cytoplasmic form of TcpP, TcpPcyt, was completely deficient for transcription activation, as determined in a V. cholerae toxT-lacZ fusion strain (Fig. 1C). The results for Tcpmem and TcpPperi, with deletion of all and half of the periplasmic domain, respectively, are shown in Fig. 1B and C. Based on their instability, no conclusions can be drawn about their ability to activate transcription, but the instability may reveal something about the overall structure or integrity of TcpP. These data suggest that the stability of TcpP is fragile and that the protein cannot tolerate gross structural changes to its periplasmic domain and, as a result, transcription activity may require the full-length version of the protein (Fig. 1C). Our observation that the TcpP cytoplasmic domain alone lacks the ability to activate transcription, along with prior work showing constitutive activity resulting from fusion of TcpP periplasmic domain with that of β-lactamase (16), led us to hypothesize that the periplasmic domain plays an important regulatory role in TcpP function. The observation that overexpression of tcpH, a primarily periplasmic protein, enhances TcpP activity provides evidence that TcpH may have a hand in TcpP stability and activity (16), as has been proposed for the ToxR effector protein ToxS (11, 13, 36, 44).
FIG. 1.
Effect of membrane localization on stability and activity of TcpP. (A) Schematic diagram of the TcpP truncates constructed. The truncates were expressed from the inducible plasmid pBAD18. (B) Western blot analysis of the ΔtcpP strain transformed with the truncates illustrated above. (C) β-Galactosidase assay of ΔtcpP strains containing a chromosomal toxT-lacZ reporter fusion transformed with the truncates described above.
Steady-state levels of full-length TcpP are dependent on TcpH in V. cholerae.
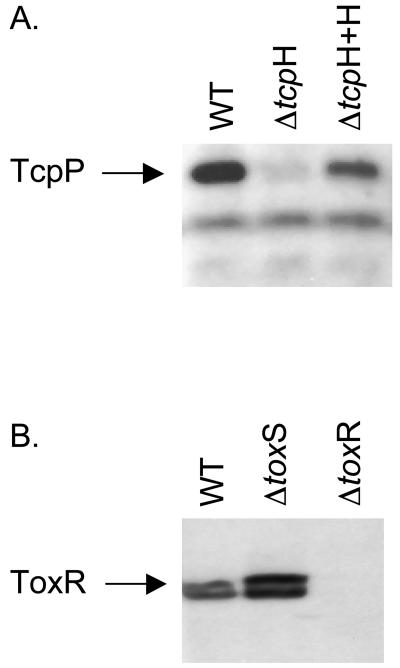
To begin assessing the role of TcpH in TcpP function, we examined the phenotype of a V. cholerae tcpH mutant. This mutant was previously shown to have an altered pattern of toxT transcription and, consequently, to express significantly reduced amounts of cholera toxin (4, 57). We determined the steady-state levels of TcpP in the wild type and the tcpH mutant by immunoblotting (Fig. 2A). This experiment showed that steady-state levels of TcpP are dependent on expression of TcpH, as the tcpH mutant strain expressed virtually undetectable levels of TcpP after growth to the mid-logarithmic phase in LB at pH 6.5 and 30°C, conditions that stimulate toxin and pilus production in wild-type V. cholerae (Fig. 2A). Restoration of TcpH production by plasmid complementation resulted in concomitant restoration of TcpP to near wild-type levels.
FIG. 2.
Effect of tcpH and toxS deletion on TcpP and ToxR, respectively. (A) Western blot analysis of TcpP levels in wild-type (WT), ΔtcpH and ΔtcpH + tcpH(pACYC184) cultures. (B) Western blot analysis of ToxR levels in the wild type (WT), ΔtoxS, and ΔtoxR. For both analyses, cells were grown to mid-log phase in LB medium at pH 6.5 at 30° C.
To determine if providing stability to their cognate activators is a common feature of the effector proteins ToxS and TcpH, we tested the levels of ToxR in a cell with a deletion of toxS. Unlike that which we observed for TcpP in the absence of TcpH, loss of ToxS had no measurable negative effect on steady-state levels of the ToxR protein (Fig. 2B), which is in agreement with observations from studying ToxR and ToxS function in serovar Typhimurium (44) but contrary to reports of experiments of the effects of ToxS on ToxR function in E. coli (11, 13). We conclude that TcpH ultimately regulates TcpP protein levels and that this type of control, which may be at the transcriptional or posttranscriptional level, is not a general characteristic of membrane-localized transcription factors in V. cholerae.
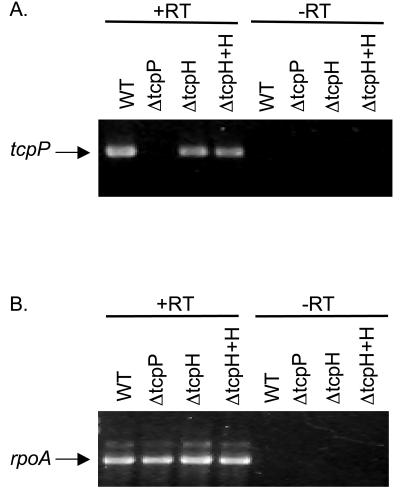
One explanation to account for diminished levels of TcpP in the tcpH null strain is that tcpH deletion affects tcpP transcription. To test this possibility, we performed RT-PCR with tcpP-specific primers on RNA from mid-logarthmic-phase cultures grown under inducing conditions. No difference in tcpP transcript levels between wild-type and tcpH mutant cultures was observed (Fig. 3). These findings rule out any significant effect of TcpH on tcpP transcription and suggest that TcpH contributes either to translation of the tcpP message or to stability of the protein.
FIG. 3.
Effect of tcpH deletion on the level of tcpP transcript. RT-PCR analysis of RNA transcript from wild-type (WT), ΔtcpP, ΔtcpH, and ΔtcpH + tcpH (pACYC184) cultures RNA was harvested from cultures grown to mid-log phase in LB medium at pH 6.5 and 30°C. (A) The TcpP-specific product is shown. (B) The RNA polymerase α-subunit (rpoA)-specific product is shown to demonstrate that levels of RNA have been normalized. +RT, with reverse transcriptase; −RT, without reverse transcriptase.
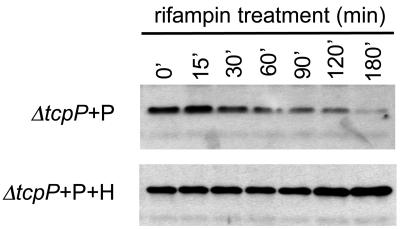
Based on prior observations regarding the relationship between ToxR and ToxS, we chose to investigate the effects of TcpH on TcpP protein stability. We performed a pulse expression experiment in which cloned tcpP was induced from the arabinose-inducible PBAD promoter in either the presence or absence of cloned, overexpressed TcpH (the chromosomal tcpH gene was intact for this experiment). After a pulse of tcpP induction by the addition of arabinose, rifampin was added to the cultures to inhibit further transcription initiation. Samples were taken at different times after rifampin addition and processed for immunoblot analysis (Fig. 4).
FIG. 4.
Co-overexpression of TcpH increases the stability of TcpP. Western blot analysis of the ΔtcpP tcpH+ strain transformed with tcpP alone (ΔtcpP + P) expressed from the inducible plasmid tcpP (pBAD18) or cotransformed with tcpP(pBAD18) and tcpH (ΔtcpP + P + H) expressed from pACYC184. Rifampin was added to stop transcription of mid-log cultures. Cultures remained at 30°C after the addition of rifampin.
In the absence of overexpressed TcpH, the level of TcpP steadily diminished over time after cessation of new transcription, so that by 180 min after the addition of rifampin, TcpP was nearly undetectable in culture extracts (Fig. 4, top). In contrast, when TcpP was induced in the presence of overexpressed TcpH, its level was unaffected by the addition of rifampin for the entirety of the experiment (Fig. 4, bottom), supporting the hypothesis that TcpH is important for the stability of TcpP in V. cholerae.
A TcpP-ToxR fusion protein is stable and active.
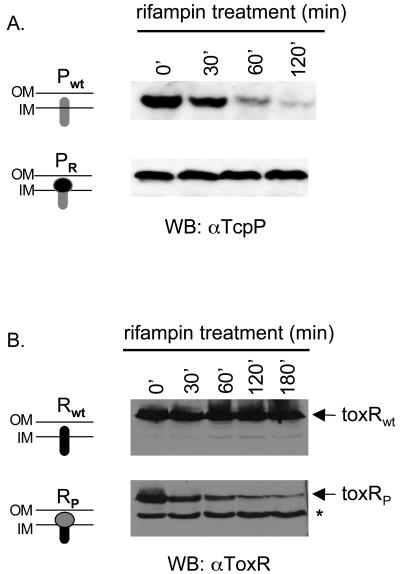
Our data to this point show that TcpH protects TcpP from turnover in V. cholerae and that ToxR is an inherently more stable protein in the absence of its cognate effector protein ToxS. Because TcpH is predicted to localize in the periplasm, we hypothesized that the periplasmic C terminus of TcpP is unstable in the absence of TcpH. We exchanged the C-terminal domain of TcpP with that of ToxR by splicing the two genes together, as described in Materials and Methods. The chimeric protein, TcpPR, was assayed for its stability compared to wild-type TcpP by the same pulse expression experimental protocol as described above for Fig. 4. The gene encoding TcpP or TcpPR was expressed from the PBAD promoter by arabinose induction, followed by the addition of rifampin to stop further transcription. Protein levels were determined over time by Western blotting of anti-TcpP sera. TcpP was again unstable in this experiment, which did not include co-overexpressed TcpH. By comparison, TcpPR was much more stable, supporting our hypothesis that the periplasmic domain of ToxR is more stable than that of TcpP (Fig. 5A).
FIG. 5.
Effect of the periplasmic domain on TcpP stability and activity. (A) The ΔtcpP mutant strain was transformed with either tcpPwt (pBAD18) containing the full-length wild-type version of tcpP or tcpPR (pBAD18) containing a chimeric version of tcpP in which the periplasmic domain has been replaced with that of ToxR. Western blot analysis of mid-log cultures was performed after the addition of rifampin to stop transcription. Cultures remained at 30°C after the addition of rifampin through the end of the time course. (B) The same experiment as described for panel A was performed with the ΔtoxR mutant strain transformed with either toxRwt-pBAD18 or toxRP (pBAD18).
To further demonstrate that the periplasmic domain of TcpP is a determinant of instability, the periplasmic domain of ToxR was replaced with that of TcpP and another pulse expression experiment was performed. Whereas ToxR was stable over the time period of the experiment, there was a marked decrease in ToxRP levels during the same period (Fig. 5B). Instability of the fusion protein may be reflected by the presence of the lower band, a possible degradation product (Fig. 5B). We do not believe this instability is attributable simply to alteration of the ToxR periplasmic domain or to fusion of a foreign domain, because other periplasmic chimeras of ToxR are not unstable; periplasmic fusions of ToxR with portions of β-lactamase, the activator protein GCN4, and alkaline phosphatase result in stable and active proteins in V. cholerae (8). Furthermore, when TcpH was coexpressed with ToxRP, this lower-molecular-weight species was undetectable (data not shown). We conclude from these experiments that the natural C terminus of TcpP confers instability on the remainder of the protein, that TcpH enhances stability of TcpP, and that the C terminus of ToxR is naturally more stable.
Protease Do, a V. cholerae homolog of DegP, does not act on TcpP.
Our experiments demonstrate that the instability of TcpP is due to its periplasmic domain, leading us to hypothesize that a periplasmic protease is responsible for TcpP turnover. In an effort to identify additional factors affecting TcpP stability, we focused on the periplasmic chaperone-protease DegP. Although the primary function of DegP in gram-negative prokaryotes is thought to be maintenance of healthy proteins in the periplasm (6), DegP has also been implicated in the virulence of numerous pathogenic organisms (3, 7, 21, 31, 43, 45). Examination of the V. cholerae degP homolog, protease do (ptd), indicated that ptd is not responsible for degradation of TcpP (data not shown).
DISCUSSION
Regulation of virulence gene expression in V. cholerae involves a multilevel regulatory cascade leading to toxT transcription by the membrane-localized regulatory proteins ToxR and TcpP. Of the two proteins, TcpP is more extensively regulated. Expression of toxR appears to be constitutive, although activity of the protein is regulated by ToxS (11, 13, 36, 44), while tcpP gene expression is controlled through the environmentally sensitive AphA and AphB proteins (25, 50) and by several other factors whose precise roles in this process are not as fully characterized as are those of AphA and AphB (1, 23, 24, 58). In this report we present evidence that levels of TcpP are also controlled by proteolysis and that TcpH can protect TcpP against this degradation. Prior work had identified tcpH as the site of a spontaneous mutation that rendered V. cholerae deficient for expression of ctxAB and tcpA and avirulent in the infant mouse model (4) and as being required for transcription of toxT (4, 57). Our findings provide the first mechanistic data to account for the link between TcpH and expression of the virulence genes ctxAB and tcpA in V. cholerae.
One conclusion that we draw from our results is that the functions of small effector proteins, such as ToxS and TcpH, in the service of their cognate membrane-localized activators are not equivalent. Lack of ToxS in the V. cholerae cell has no demonstrable effect on ToxR levels, whereas lack of TcpH leads to severely diminished levels of TcpP. One hypothesis to account for the finding regarding TcpH is that, in its absence, TcpP is folded poorly and is rapidly degraded as a consequence. Protecting TcpP from being degraded due to inefficient folding would suggest that TcpH may serve as a type of molecular chaperone for TcpP. A chaperone role has been proposed for ToxS, based on the observation that a mutant ToxS protein, ToxSL33S, led to instability of ToxR when the two proteins were coexpressed in serovar Typhimurium (44). In that study, the investigators observed no effect on ToxR levels in the absence of ToxS in serovar Typhimurium, similar to our finding for V. cholerae reported herein, but they did report that expression of ToxSL33S allowed for degradation of ToxR when the proteins were coexpressed. They postulated that the mutant protein may act to unfold the periplasmic domain of ToxR, thereby causing it to become susceptible to proteolysis. A mechanism for how this proposed unfolding may occur was not suggested; but taking all of their data together, Pfau and Taylor postulated that the role of ToxS is to enable the proper folding of ToxR needed for transcription activation but not for DNA binding (44).
TcpH may serve as a type of molecular chaperone for TcpP, but it appears to differ greatly in function from that hypothesized for ToxS by Pfau and Taylor (44). In that hypothesis, ToxR requires ToxS to facilitate the conformation necessary for transcription activation, not to fold into a stable structure, whereas the requirement for TcpH is critical for producing stable TcpP. The function of TcpH appears to be more similar to that of CooB or ExbB than that of ToxS. CooB is an apparent periplasmic chaperone required for CS1 pilus assembly in enterotoxigenic E. coli. CooB associates with pilus subunits, a necessary step in protecting them from periplasmic degradation (54). ExbB aids in membrane localization (22), stability (49), and activity of TonB (28), a protein required for transduction of energy from the inner to the outer membrane in E. coli. A proper ratio of ExbB:TonB is evidently necessary to protect protease-sensitive sites in TonB (49), and the interaction between these proteins appears to be long-lived, with the proteins remaining complexed throughout the energy transduction cycle.
Periplasmic chaperones associated with processes such as pilus biogenesis (18), outer membrane biogenesis (2, 5, 9, 39, 48), and secretion (19), have been identified but an overall understanding of the periplasmic folding process and inner membrane protein biogenesis is still emerging. None of the classical cytoplasmic chaperones, such as members of the hsp 60, hsp 70, or hsp 100 family, is found in the periplasm, consistent with the fact that this compartment lacks the ATP required for their activity. The periplasm does contain two other classes of folding catalysts, protein disulfide isomerases and peptidyl-prolyl isomerases, which act to catalyze thiol-disulfide exchanges and cis-trans isomerization, respectively. Apart from their roles in disulfide bond formation and peptidyl-prolyl isomerase activity, a few of these proteins also act as chaperones to promote folding and to prevent aggregation of unstable proteins (40, 46, 56). TcpH shares no homology or predicted functional domains with these folding factors or any other known proteins, offering no clues as to how TcpH functions.
Although we do not yet know the specific mechanism by which TcpH regulates the levels of TcpP in the cell, we do know that TcpP is indeed subject to proteolysis and we hypothesize that regulated degradation of TcpP may be a specific mechanism by which the cell maintains appropriate levels of downstream gene expression. It is clear from the extensive regulation upstream of tcpP in the virulence cascade of V. cholerae that a great deal of precision is required for proper activation of virulence genes. One can therefore imagine that there also exists deliberate ways of limiting expression of these genes. A study of the transcription profile of V. cholerae within human stools led the investigators to suggest that virulence gene expression is down-regulated as the microbes prepare for survival outside of the host (33). One recently defined mechanism for down-regulating virulence gene expression in V. cholerae is negative regulation by the LuxO-HapR quorum-sensing system. At high cell density, quorum-sensing regulators act to inhibit expression of the ToxR virulence cascade (24, 35, 58). We suggest that another mechanism could be regulated degradation of TcpP. That down-regulation of virulence may occur via modulation of TcpPH has been suggested previously, based on the observation that tcpH acquires frameshift mutations at a high frequency when cells are cultured overnight under conditions that induce expression of the ToxR regulon (4). From our data regarding the relationship between TcpP and TcpH, it follows that these mutations interfere with ability of TcpH to provide stability to TcpP.
We hypothesize that TcpP undergoes a regulated process of proteolysis whereby environmental signals control degradation of TcpP. TcpP turnover in this manner would allow the organism to make a quick transition in response to changes in environmental conditions. In lieu of a mechanism for regulating TcpP function by phosphorylation-de-phosphorylation as in classic two component regulators, TcpH-dependent stabilization may be a mechanism to control TcpP levels and, thereby, its activity. Several schemes might be imagined to address how steady-state TcpP levels are coordinated with favorable or unfavorable conditions for virulence gene expression. Protection of TcpP could be mediated by differential affinities between TcpP and TcpH under different environmental conditions, and there may even be competition between TcpH and the protease that degrades TcpP. The frameshift mutations that accumulate in TcpH under certain culture conditions (4) could be another way of modulating the TcpH-TcpP interaction according to the environment. Alternatively, regulation of degradation may lie on the side of the protease, rather than in TcpP or TcpH. If expression and/or activity of the protease is regulated, then fluctuations in its levels and activity corresponding to changes in conditions could dictate degradation of TcpP.
Our findings emphasize the role of posttranscriptional regulation in controlling global gene expression patterns. Specifically, we have uncovered a proteolytic mechanism with potential impact on virulence gene expression. There are many examples of proteolysis-regulating stress response, the cell cycle, and development in prokaryotes in response to temporal and spatial cues (20). One example of protein degradation mechanisms specifically associated with virulence control in a bacterial pathogen is provided by Pseudomonas aeruginosa. The algW and mucD genes encode proteins with homology to the periplasmic serine protease/chaperone DegP (29, 30, 52, 53). Inactivation of these genes causes derepression of alginate biosynthesis, leading to mucoidy, which is associated with chronic respiratory infections in cystic fibrosis (3).
The susceptibility of TcpP to proteolysis is likely attributable to being in a misfolded state long enough to be engaged by degradative proteases (14, 55). Identification of the protease(s) responsible for its degradation is of interest, and in this study, we focused on the chaperone-protease DegP, based on its location in the periplasm and involvement in a variety of processes, including pathogenesis (3, 7, 21, 31, 43, 45). Our results indicate that the V. cholerae degP homolog, protease Do (ptd), does not contribute to TcpP turnover, and analysis of other candidates is underway. Identifying the chaperones and protease(s) governing TcpP stability will provide insight into the dynamics of TcpP folding and stabilization by TcpH. More knowledge of these processes is critical for understanding the potential mechanism of proteolytic control of virulence gene regulation in V. cholerae.
Acknowledgments
We heartily thank Rutilio Fratti for innumerable discussions of this work and for helpful review of the manuscript and Brian Hammer for construction of the ΔtoxS strain. We also thank Ambrose Cheung for graciously providing laboratory space to carry out some of this work.
This study was supported by NIH grants AI31645 and AI45125 to V.J.D. N.A.B. is a trainee supported by the University of Michigan Genetics Training Grant (T32 GM07544) and Molecular Mechanisms of Microbial Pathogenesis Training Grant (T32 AI07528).
REFERENCES
- 1.Behari, J., L. Stagon, and S. B. Calderwood. 2001. pepA, a gene mediating pH regulation of virulence genes in Vibrio cholerae. J. Bacteriol. 183:178-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bothmann, H., and A. Pluckthun. 1998. Selection for a periplasmic factor improving phage display and functional periplasmic expression. Nat. Biotechnol. 16:376-380. [DOI] [PubMed] [Google Scholar]
- 3.Boucher, J. C., J. Martinez-Salazar, M. J. Schurr, M. H. Mudd, H. Yu, and V. Deretic. 1996. Two distinct loci affecting conversion to mucoidy in Pseudomonas aeruginosa in cystic fibrosis encode homologs of the serine protease HtrA. J. Bacteriol. 178:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll, P. A., K. T. Tashima, M. B. Rogers, V. J. DiRita, and S. B. Calderwood. 1997. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol. Microbiol. 25:1099-1111. [DOI] [PubMed] [Google Scholar]
- 5.Chen, R., and U. Henning. 1996. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol. Microbiol. 19:1287-1294. [DOI] [PubMed] [Google Scholar]
- 6.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10:443-455. [DOI] [PubMed] [Google Scholar]
- 7.Cortes, G., B. de Astorza, V. J. Benedi, and S. Alberti. 2002. Role of the htrA gene in Klebsiella pneumoniae virulence. Infect. Immun. 70:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawford, J. A., E. S. Krukonis, and V. J. DiRita. 2003. Membrane localization of the ToxR winged-helix domain is required for TcpP-mediated virulence gene activation in Vibrio cholerae. Mol. Microbiol. 47:1459-1473. [DOI] [PubMed] [Google Scholar]
- 9.De Cock, H., U. Schafer, M. Potgeter, R. Demel, M. Muller, and J. Tommassen. 1999. Affinity of the periplasmic chaperone Skp of Escherichia coli for phospholipids, lipopolysaccharides and non-native outer membrane proteins. Role of Skp in the biogenesis of outer membrane protein. Eur. J. Biochem. 259:96-103. [DOI] [PubMed] [Google Scholar]
- 10.Dell, C. L., M. N. Neely, and E. R. Olson. 1994. Altered pH and lysine signalling mutants of cadC, a gene encoding a membrane-bound transcriptional activator of the Escherichia coli cadBA operon. Mol. Microbiol. 14:7-16. [DOI] [PubMed] [Google Scholar]
- 11.DiRita, V. J., and J. J. Mekalanos. 1991. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell 64:29-37. [DOI] [PubMed] [Google Scholar]
- 12.Dziejman, M., H. Kolmar, H. J. Fritz, and J. J. Mekalanos. 1999. ToxR cooperative interactions are not modulated by environmental conditions or periplasmic domain conformation. Mol. Microbiol. 31:305-317. [DOI] [PubMed] [Google Scholar]
- 13.Dziejman, M., and J. J. Mekalanos. 1994. Analysis of membrane protein interaction: ToxR can dimerize the amino terminus of phage lambda repressor. Mol. Microbiol. 13:485-494. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman, S. 1999. Regulation by proteolysis: developmental switches. Curr. Opin. Microbiol. 2:142-147. [DOI] [PubMed] [Google Scholar]
- 15.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hase, C. C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In D. H. G. M.A. Innis, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, Calif.
- 18.Hung, D. L., J. S. Pinkner, S. D. Knight, and S. J. Hultgren. 1999. Structural basis of chaperone self-capping in P pilus biogenesis. Proc. Natl. Acad. Sci. USA 96:8178-8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs, M., J. B. Andersen, V. Kontinen, and M. Sarvas. 1993. Bacillus subtilis PrsA is required in vivo as an extracytoplasmic chaperone for secretion of active enzymes synthesized either with or without pro-sequences. Mol. Microbiol. 8:957-966. [DOI] [PubMed] [Google Scholar]
- 20.Jenal, U., and R. Hengge-Aronis. 2003. Regulation by proteolysis in bacterial cells. Curr. Opin. Microbiol. 6:163-172. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, K., I. Charles, G. Dougan, D. Pickard, P. O'Gaora, G. Costa, T. Ali, I. Miller, and C. Hormaeche. 1991. The role of a stress-response protein in Salmonella typhimurium virulence. Mol. Microbiol. 5:401-407. [DOI] [PubMed] [Google Scholar]
- 22.Karlsson, M., K. Hannavy, and C. F. Higgins. 1993. ExbB acts as a chaperone-like protein to stabilize TonB in the cytoplasm. Mol. Microbiol. 8:389-396. [DOI] [PubMed] [Google Scholar]
- 23.Kovacikova, G., and K. Skorupski. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol. Microbiol. 41:393-407. [DOI] [PubMed] [Google Scholar]
- 24.Kovacikova, G., and K. Skorupski. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46:1135-1147. [DOI] [PubMed] [Google Scholar]
- 25.Kovacikova, G., and K. Skorupski. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J. Bacteriol. 181:4250-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krukonis, E. S., R. R. Yu, and V. J. DiRita. 2000. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol. Microbiol. 38:67-84. [DOI] [PubMed] [Google Scholar]
- 27.Krukonis, E. S., and V. J. DiRita. 2003. DNA binding and ToxR responsiveness by the wing domain of TcpP, an activator of virulence gene expression in Vibrio cholerae. Mol. Cell 12:157-165. [DOI] [PubMed] [Google Scholar]
- 28.Larsen, R. A., M. G. Thomas, and K. Postle. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809-1824. [DOI] [PubMed] [Google Scholar]
- 29.Lipinska, B., O. Fayet, L. Baird, and C. Georgopoulos. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 171:1574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipinska, B., M. Zylicz, and C. Georgopoulos. 1990. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J. Bacteriol. 172:1791-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyon, W. R., and M. G. Caparon. 2004. Role for serine protease HtrA (DegP) of Streptococcus pyogenes in the biogenesis of virulence factors SpeB and the hemolysin streptolysin S. Infect. Immun. 72:1618-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Hackert, E., and A. M. Stock. 1997. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure 5:109-124. [DOI] [PubMed] [Google Scholar]
- 33.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schooluik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Miller, M. S., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 36.Miller, V. L., V. J. DiRita, and J. J. Mekalanos. 1989. Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J. Bacteriol. 171:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, V. L., R. K. Taylor, and J. J. Mekalanos. 1987. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell 48:271-279. [DOI] [PubMed] [Google Scholar]
- 39.Missiakas, D., J. M. Betton, and S. Raina. 1996. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol. Microbiol. 21:871-884. [DOI] [PubMed] [Google Scholar]
- 40.Missiakas, D., and S. Raina. 1997. Protein folding in the bacterial periplasm. J. Bacteriol. 179:2465-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ottemann, K. M., and J. J. Mekalanos. 1995. Analysis of Vibrio cholerae ToxR function by construction of novel fusion proteins. Mol. Microbiol. 15:719-731. [DOI] [PubMed] [Google Scholar]
- 42.Ottemann, K. M., and J. J. Mekalanos. 1996. The ToxR protein of Vibrio cholerae forms homodimers and heterodimers. J. Bacteriol. 178:156-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedersen, L. L., M. Radulic, M. Doric, and Y. Abu Kwaik. 2001. HtrA homologue of Legionella pneumophila: an indispensable element for intracellular infection of mammalian but not protozoan cells. Infect. Immun. 69:2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfau, J. D., and R. K. Taylor. 1998. Mutations in toxR and toxS that separate transcriptional activation from DNA binding at the cholera toxin gene promoter. J. Bacteriol. 180:4724-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purdy, G. E., M. Hong, and S. M. Payne. 2002. Shigella flexneri DegP facilitates IcsA surface expression and is required for efficient intercellular spread. Infect. Immun. 70:6355-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Schafer, U., K. Beck, and M. Muller. 1999. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 274:24567-24574. [DOI] [PubMed] [Google Scholar]
- 49.Skare, J. T., and K. Postle. 1991. Evidence for a TonB-dependent energy transduction complex in Escherichia coli. Mol. Microbiol. 5:2883-2890. [DOI] [PubMed] [Google Scholar]
- 50.Skorupski, K., and R. K. Taylor. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 31:763-771. [DOI] [PubMed] [Google Scholar]
- 51.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 52.Strauch, K. L., and J. Beckwith. 1988. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc. Natl. Acad. Sci. USA 85:1576-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strauch, K. L., K. Johnson, and J. Beckwith. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 171:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voegele, K., H. Sakellaris, and J. R. Scott. 1997. CooB plays a chaperone-like role for the proteins involved in formation of CS1 pili of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. USA 94:13257-13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wickner, S., M. R. Maurizi, and S. Gottesman. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 286:1888-1893. [DOI] [PubMed] [Google Scholar]
- 56.Wulfing, C., and A. Pluckthun. 1994. Protein folding in the periplasm of Escherichia coli. Mol. Microbiol. 12:685-692. [DOI] [PubMed] [Google Scholar]
- 57.Yu, R. R., and V. J. DiRita. 1999. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J. Bacteriol. 181:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]