Abstract
Artificial cells capable of both sensing and sending chemical messages to bacteria have yet to be built. Here we show that artificial cells that are able to sense and synthesize quorum signaling molecules can chemically communicate with V. fischeri, V. harveyi, E. coli, and P. aeruginosa. Activity was assessed by fluorescence, luminescence, RT-qPCR, and RNA-seq. Two potential applications for this technology were demonstrated. First, the extent to which artificial cells could imitate natural cells was quantified by a type of cellular Turing test. Artificial cells capable of sensing and in response synthesizing and releasing N-3-(oxohexanoyl)homoserine lactone showed a high degree of likeness to natural V. fischeri under specific test conditions. Second, artificial cells that sensed V. fischeri and in response degraded a quorum signaling molecule of P. aeruginosa (N-(3-oxododecanoyl)homoserine lactone) were constructed, laying the foundation for future technologies that control complex networks of natural cells.
Short abstract
We show that artificial cells that are able to sense and synthesize quorum signaling molecules can chemically communicate with V. fischeri, V. harveyi, E. coli, and P. aeruginosa.
Introduction
Artificial cells are encapsulated chemical systems that mimic cellular life. Most attempts at making artificial cells have focused on building some type of self-replicating system.1,2 Although self-replication is an important feature of life as we know it, self-replication alone is an insufficient criterion for assessing how lifelike a chemical system is.3 For example, cross-catalytic ribozyme ligases are capable of self-replication4 but do not alone constitute a living system. What is lacking is some sort of metric by which progress can be measured. One solution may be to describe chemical systems on a continuum where the typical binary categorization of alive and not alive is replaced by states that are increasingly lifelike. In this way, each iteration of constructing an artificial cell could be objectively and quantifiably evaluated in terms of likeness to a target natural cell. Such an approach is intuitive, because the emergence of life on Earth did not occur in a single event, but likely encompassed a series of steps, each bringing the chemical system closer to what is recognized as living today.5,6
It was previously suggested that a type of imitation game could be used to guide the construction of artificial cells in a way that bypasses the problems associated with a lack of a definition of life.7 In the original imitation game (or Turing test), the ability of a machine to deceive a judge (or interrogator) through textual communication into believing that the machine is a person was used to circumvent the problem of defining intelligence.8 In the cellular version, the ability of an artificial cell to deceive a natural cell is used to evaluate the artificial cell. Such a cellular Turing test is possible, because all cells communicate, from quorum sensing pathways in bacteria to pheromone responses in higher organisms.9 Further, artificial cells containing DNA and/or transcription–translation machinery can express genes,10,11 send chemical messages to bacteria,12,13 and interact with each other.14 Additionally, genetic constructs in water-in-oil emulsion droplets are able to either sense or send quorum molecules.15 Therefore, it should be possible to build genetically encoded artificial cells that can chemically communicate with bacteria. Since chemical communication leads to measurable changes in gene expression, next generation sequencing technologies can be used to quantifiably evaluate the extent of mimicry in a manner that is neither subjective nor binary. In other words, the cellular Turing test allows for the quantification of how lifelike the artificial cells are in comparison to a target living cell in a stratified manner.
Results and Discussion
Artificial Cells Can Sense Bacteria
To build artificial cells that mimic the ability of natural cells to chemically communicate, we attempted to reconstitute the well characterized quorum sensing pathways of Vibrio fischeri, Pseudomonas aeruginosa, and Escherichia coliin vitro. Genetic constructs were assembled with genes coding for the quorum responsive transcriptional activator or repressor plus additional accessory factors, as needed, and a transcriptional regulator binding site upstream of a gene encoding a fluorescent protein. In this way, the activity of each pathway could be assessed by the fluorescence arising from in vitro transcription–translation reactions. The N-3-(oxohexanoyl)homoserine lactone (3OC6 HSL) responsive system from V. fischeri was functional in vitro (Figure S1a,b). GFP expression in the presence of 10 μM 3OC6 HSL was 4-fold greater than in the absence of this quorum signal. Since the same transcriptional activator can sense another quorum molecule (N-octanoyl-l-homoserine lactone or C8 HSL) secreted from V. fischeri,(16) responsiveness to C8 HSL was assessed. Although the affinity of the transcriptional regulator LuxR for C8 HSL was low, a higher affinity mutant version of the protein (T33A S116A S135I LuxR or LuxR*)17 activated cell-free expression 7-fold in the presence of C8 HSL and 6-fold in the presence of 3OC6 HSL (Figure S1a,b). The ability to sense 3OC6 HSL could be removed by introducing an additional M65R substitution, as previously reported.17 Next, two P. aeruginosa quorum pathways were tested, including the N-(3-oxododecanoyl)homoserine lactone (3OC12 HSL) responsive LuxR and the N-butanoylhomoserine lactone (C4 HSL) responsive RhlR pathways. As previously observed,18 the genetic construct containing lasR was responsive to the quorum signal 3OC12 HSL in vitro, showing a 2-fold increase in protein expression (Figure S1c). However, the RhlR dependent system showed indistinguishable activity in the presence and absence of C4 HSL (Figure S1d). Finally, the autoinducer-2 (AI-2) system from E. coli was tested. While the expression of the transcriptional repressor LsrR fully inhibited protein expression, none of the tested constructs were derepressed by AI-2 (Figure S1e). The inclusion of the cAMP receptor protein (CRP) did not sufficiently improve derepression (Figure S1f). In summary, 3OC6 HSL, C8 HSL, and 3OC12 HSL were successfully detected by in vitro transcription–translation reactions.
Each functioning quorum sensing pathway was then encapsulated within cholesterol containing 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) phospholipid vesicles to determine whether quorum molecules could diffuse across the phospholipid membrane and activate gene expression of living cells. Here, activation resulted in the expression of firefly luciferase instead of GFP. Vesicles were incubated at 37 °C for 5 h in the presence and absence of the quorum molecule. The vesicles were then broken with Triton X-100 in the presence of luciferin and immediately measured for luminescence. Only in the presence of 3OC6 and C8 HSL was luminescence observed, indicating that the signaling molecules crossed the phospholipid membrane and activated gene expression (Figure S2a,b). 3OC6 HSL was previously shown to diffuse through the oil phase of water-in-oil emulsion droplets.15 Together, these results suggested that artificial cells should be able to sense quorum molecules that are naturally secreted from bacteria. To demonstrate that the sensing mechanism of artificial cells was capable of responding to V. fischeri, the supernatant of a V. fischeri culture was added to the suspension of vesicles. After 4 h of incubation, 69-, 19-, and 8-fold more luminescence was observed for artificial cells expressing LuxR, LuxR*, and M65R LuxR*, respectively, in response to the supernatant of V. fischeri than in the absence of the supernatant (Figure 1). The use of the supernatant of a V. fischeri culture removed the confounding effects of the natural luminescent properties of the bacterium itself. The data supported the ability of artificial cells made of phospholipid vesicles and transcription–translation machinery to sense molecules secreted from natural cells.
Figure 1.
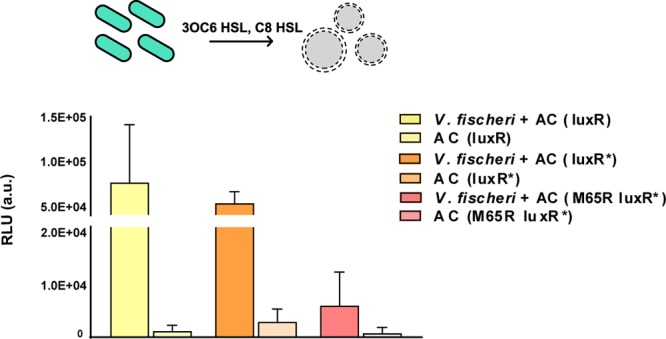
Artificial cells can sense quorum molecules released by natural cells. Artificial cells (AC) encoding either LuxR or LuxR* were able to sense the presence of V. fischeri. Negative control reactions were the artificial cells in the absence of the supernatant from V. fischeri (n = 3 biological replicates, mean ± SD). The schematic shows V. fischeri (teal, oblong) releasing quorum molecules that are sensed by artificial cells (gray, circle). RLU (relative luminescence units).
Artificial Cells Can Synthesize and Send Quorum Molecules to Natural Cells
Since communication requires the ability to both receive and send messages, we next probed whether it was possible to build artificial cells that could send chemical messages to bacteria in the form of quorum molecules. Genetic constructs encoding the synthesis machinery necessary to send chemical messages to V. fischeri, P. aeruginosa, and E. coli were assembled. V. fischeri synthesizes the N-acylhomoserine lactone 3OC6 HSL through the activity of LuxI, which uses S-adenosylmethionine and acyl chains donated from acyl carrier proteins as reactants.19 Similarly, P. aeruginosa synthesizes 3OC12 HSL through the activity of the LuxI homologue LasI. Additionally, P. aeruginosa synthesizes C4 HSL through a similar pathway that uses RhlI in place of LasI.20 The functionality of each genetic construct was assessed with reporter E. coli strains engineered to express GFP in response to a specific quorum molecule. After 6 h of transcription–translation at 37 °C of each genetic construct, an aliquot was added to the reporter strain and analyzed by flow cytometry. The 3OC6 HSL, 3OC12 HSL, and C4 HSL synthesis systems individually activated the expression of GFP of 90%, 50%, and 87% of the cells of the corresponding reporter bacterial strain, indicating that each genetically encoded quorum synthesis system was functional in vitro (Figure S3a). The AI-2 synthesis pathway used by E. coli is different and depends on the activity of three enzymes.21 The SAM-dependent methyltransferase converts S-adenosylmethionine to S-adenosylhomocysteine, which is in turn converted to S-ribosylhomocysteine by the enzyme Pfs. Lastly, LuxS produces AI-2 and homocysteine in a 1:1 ratio from S-ribosylhomocysteine. Pfs and LuxS can be fused together to form a larger polypeptide that efficiently synthesizes AI-2 in the presence of S-ribosylhomocysteine.22,23 We demonstrated that this fusion protein was active after in vitro transcription–translation by detecting synthesized AI-2 with the luminescent reporter Vibrio harveyi BB170 (Figure S3b).
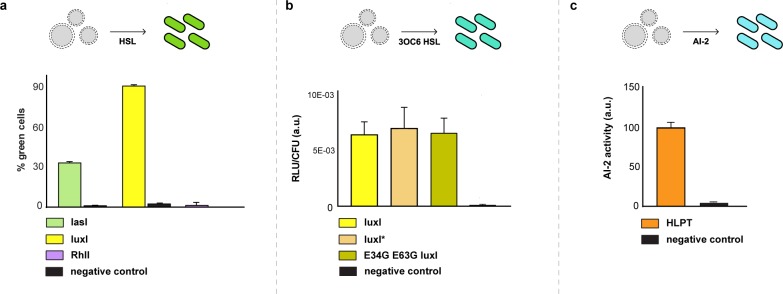
To ensure that each synthesized quorum molecule could escape lipid vesicles, the transcription–translation reactions were placed inside of vesicles. The loaded vesicles were mixed with reporter bacterial strains at 37 °C and analyzed by flow cytometry. Encapsulated genetic constructs for the synthesis of 3OC6 HSL and 3OC12 HSL resulted in approximately 90% and 35%, respectively, of fluorescent cells after 6 h of incubation, while the encapsulated C4 HSL synthesis system failed to induce detectable fluorescence of the reporter strain (Figure 2a). Two mutated versions of LuxI were also evaluated in an attempt to identify more active versions of this 3OC6 HSL synthesizing enzyme.24 Vesicles containing DNA encoding wild type LuxI, E34G E63G LuxI, and E34G E40G E63G LuxI (hereafter referred to as LuxI*) were incubated with a dilute culture of V. fischeri, and the induced luminescence of V. fischeri was evaluated. All three of the tested versions of LuxI induced similar levels of luminescence from V. fischeri (Figure 2b). The encapsulation of the genetically encoded AI-2 synthesis system resulted in the induction of luminescence of the AI-2 reporter strain of V. harveyi (Figure 2c). Therefore, the data indicate that artificial cells can be built to synthesize and release 3OC6 HSL, 3OC12 HSL, and AI-2. To ensure that the vesicles used to build the artificial cells could withstand the presence of bacteria, the release of encapsulated fluorophore from vesicles incubated with different bacteria was monitored. V. fischeri, V. harveyi, and E. coli did not degrade the vesicles under the conditions used for chemical communication within 6 h, whereas the presence of the opportunistic pathogen P. aeruginosa resulted in the degradation of the vesicles (Figure S4a,b).
Figure 2.
Artificial cells can synthesize and release quorum molecules to natural cells. (a) Artificial cells (AC) carrying genetic constructs for the synthesis of 3OC12 HSL, 3OC6 HSL, and C4 HSL were incubated with E. coli sensor strains and quantified by flow cytometry. (b) Artificial cells that expressed either LuxI, LuxI*, or E34G E63G LuxI for the synthesis of 3OC6 HSL successfully induced the production of luminescence in V. fischeri. (c) Artificial cells that expressed the AI-2 synthesizing fusion protein HLPT (His6-LuxS-PfS-Tyr5)22 were incubated with V. harveyi and monitored by luminescence. For all the experiments, n = 3 biological replicates, mean ± SD. RLU/CFU (relative luminescence units per colony forming unit per milliliter).
Artificial Cells Can Establish New Communication Networks between Natural Cells
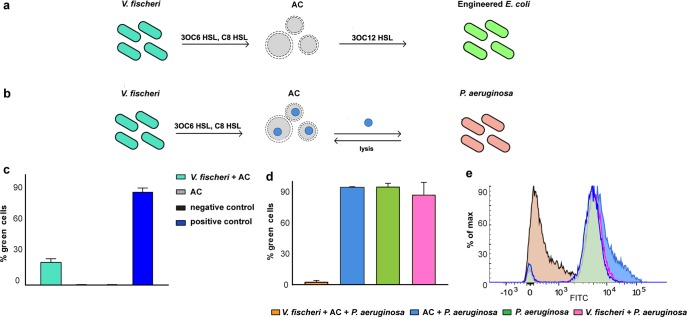
After demonstrating that the sensing and sending modules were functional inside of lipid vesicles, we next constructed artificial cells that were able to sense a quorum molecule and in response synthesize and release another quorum molecule. When properly engineered, such artificial cells would be able to mediate communication between two organisms that do not naturally communicate with each other. Further, the activity of the artificial cell would be easy to evaluate since the confounding influences of natural quorum pathways would be diminished. A genetic device that allowed for the synthesis of 3OC12 HSL in response to the presence of 3OC6 HSL was constructed. An engineered E. coli sensor strain for 3OC12 HSL was used as the receiver cell to avoid the cytotoxic effects of P. aeruginosa. The supernatant of V. fischeri was mixed with artificial cells and the E. coli reporter strain for 3OC12 HSL. 20% of the reporter strain expressed GFP, indicating that E. coli received a chemical message from the artificial cells in response to the 3OC6 HSL secreted by V. fischeri (Figure 3a,c). In the absence of the supernatant of V. fischeri, the artificial cells showed no activity. When the gene coding for the enzyme that synthesizes 3OC12 HSL was replaced by the fusion protein that produces AI-2, the resulting genetic circuit did not mediate communication with V. harveyi (Figure S5a,b).
Figure 3.
Artificial cells mediate communication between two different cell types. (a, b) A schematic of the experimental setup. (c) Communication between V. fischeri and engineered E. coli mediated by artificial cells was assessed by flow cytometry. (d, e) Artificial cells sense V. fischeri and in response degrade the 3OC12 HSL released by P. aeruginosa. Quantification was with an E. coli reporter strain by flow cytometry. For all the experiments, n = 3 biological replicates, mean ± SD. AC indicates artificial cells.
Artificial cells can be designed to disrupt the natural quorum pathways of P. aeruginosa. Acylhomoserine lactones are degraded by the Bacillus thuringiensis enzyme AiiA.25 After confirming that in vitro expressed AiiA was functional (Figure S6a), artificial cells were built to constitutively express AiiA so that the quorum molecules secreted by P. aeruginosa would be degraded. The LasR sensor for 3OC12 HSL was not encoded within the genetic content of the artificial cells since the membrane itself could serve as the sensor, that is, the membrane was disrupted by P. aeruginosa. When artificial cells expressing AiiA were incubated with P. aeruginosa, the extracellular levels of 3OC12 HSL were significantly reduced. In fact, in the absence of artificial cells, 90% of the E. coli reporter strain sensed 3OC12 HSL, whereas, in the presence of the artificial cells, only 18% of the reporter cells were activated (Figure S6b). Next, a 3OC6 HSL and C8 HSL responsive version of the artificial cells was prepared so that the signaling from one type of cell could result in the quenching of communication of another type of cell. A genetic construct expressing AiiA under the control of LuxR* allowed the artificial cells to decrease extracellular 3OC12 HSL by 95% in the presence of V. fischeri (Figure 3b,d,e). Although more work would be needed to convert such artificial cells into a useful technology, including the development of a membrane that can withstand P. aeruginosa, the data show that artificial cells could be built to interfere with biofilm formation in response to chemical signaling from another natural cell, since biofilm formation is strongly influenced by quorum signaling. However, more is possible. Engineered living cells have already been embedded in the gut microbiota26 and developed to treat inflammatory bowel disease27 and psoriasis,28 and to suppress appetite.29 Such technologies avoid flooding the organism with drug molecules, since therapeutic agents are only synthesized and released when and where needed. Artificial cells could do the same but within a more controllable chassis that does not replicate nor evolve.13
Artificial Cells Capable of Two-Way Communication Can Be Quantified by a Cellular Turing Test
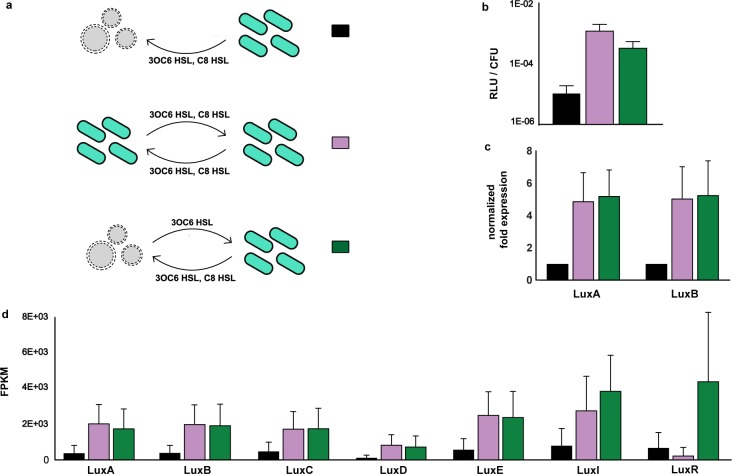
Having established that artificial cells can sense quorum molecules that are naturally secreted from bacteria, send chemical messages to natural bacteria, and mediate communication between two different bacterial species, we next sought to evaluate how lifelike such artificial cells are through a cellular Turing test. Therefore, artificial cells were constructed that could chemically communicate in a manner similar to V. fischeri. Four different genetic constructs that included the wild type or mutant versions of the receptor LuxR and the synthase LuxI were tested (Figure S7a). Artificial cells were added to a low density culture of V. fischeri exhibiting low luminescence and incubated for 3 h at 30 °C. The artificial cells containing DNA encoding LuxR* and LuxI* induced the greatest luminescent response per colony forming unit (CFU) and thus were best able to chemically communicate with V. fischeri (Figure S7b,c). Since the artificial cells could not replicate, the CFU solely reflected the number of viable natural cells. The extent of communication was influenced by the lipid composition of the membrane of the artificial cells, consistent with the diffusion of molecules across intact membranes (Figure S8). Further, identical reactions that were not encapsulated in vesicles were not able to engage in chemical communication with V. fischeri under the experimental conditions employed (Figure S8). The experiment was then repeated with the optimized genetic sequence so that the same samples could be evaluated by luminescence, RT-qPCR, and RNA sequencing. The luminescence data (Figure 4b) was confirmed by RT-qPCR (Figure 4c), which showed that the expression of luxA and luxB was similarly upregulated 5-fold both for communication mediated by artificial cells and for natural V. fischeri-V. fischeri communication. luxA and luxB were previously shown to be upregulated by 3OC6 HSL.30
Figure 4.
Two-way chemical communication for a cellular Turing test. (a) A schematic of the experimental setup showing chemical communication between V. fischeri and functional artificial cells (top, green), nonfunctional artificial cells (middle, black), and V. fischeri (bottom, magenta). Nonfunctional artificial cells could sense the presence of the quorum molecules released by V. fischeri and in response express T7 RNA polymerase, i.e., a response that had no bearing on V. fischeri. (b) Luminescence of V. fischeri in response to functional and nonfunctional artificial cells. (c) The activation of luxAB was assessed by RT-qPCR. Gene expression with respect to the negative control (V. fischeri in the presence of nonfunctional artificial cells) is shown. (d) RNA-seq analysis of the lux operon for communication between V. fischeri and nonfunctional artificial cells, V. fischeri, and functional artificial cells. For all the experiments, n = 6 biological replicates; mean ± SD. AC (artificial cells), FPKM (fragments per kilobase of transcript per million mapped reads), RLU/CFU (relative luminescence units per colony forming unit per milliliter).
RNA-seq can be used to quantify the extent to which artificial cells mimic natural cells. Although the luminescence and RT-qPCR data demonstrated that the artificial cells behaved at some level as natural cells, such data were clearly not sufficient to determine if the artificial cells were alive or not. To more quantitatively assess the performance of the artificial cells, the gene expression profile of natural cells in response to the activity of artificial cells was evaluated. Six replicates of the cellular Turing test were subjected to RNA-seq analysis. Incubation of V. fischeri with nonfunctional artificial cells resulted in 175 differently expressed coding sequences with respect to the undiluted, V. fischeri–V. fischeri communicating sample (Tables S1 and S2). Nonfunctional artificial cells contained transcription–translation machinery plus DNA encoding LuxR and T7 RNA polymerase under the control of a LuxR-responsive promoter. That is, nonfunctional artificial cells could sense quorum molecules but could not respond by synthesizing quorum molecules. The same experiment in the presence of functional artificial cells containing DNA encoding LuxR* and LuxI* showed 107 differently expressed coding sequences (Tables S1 and S3), meaning that the functional artificial cells better mimicked the influence of natural V. fischeri on V. fischeri than nonfunctional artificial cells. Although the RNA sequencing analysis, after false discovery rate (FDR) p value adjustment, did not identify statistically significant differences in the expression of the lux operon in response to functional and nonfunctional artificial cells, the increase in the number of reads from the six RNA-seq samples (Figure 4d) was similar to the activation measured by RT-qPCR (Figure 4c). In other words, although all of the comparisons had a FDR adjusted p value >0.05, the data were consistent with RT-qPCR data with p values of 0.0001 and 0.0006 for luxA and luxB, respectively. Further, the expression over the entire lux operon, with the exception of luxI and luxR, was more similar between natural V. fischeri and functional artificial cells than with nonfunctional artificial cells (Figure 4d). The luxI and luxR data were more difficult to interpret since these two genes were present in both V. fischeri and the functional artificial cells. A correlation between the gene expression profile of V. fischeri in response to nonfunctional and functional artificial cells showed that six of the seven genes of the lux operon fell off the correlation trend (Figure S9a), suggesting that the critical difference between the two types of artificial cells was their effect on quorum signaling, as expected. Additionally, the difference in the number of reads between V. fischeri–V. fischeri compared with V. fischeri–nonfunctional artificial cell and V. fischeri–functional artificial cell samples showed that the functional artificial cells better mimicked the effect on gene expression across the entire genome than the nonfunctional artificial cells (Figure S9b).
It is possible to calculate how lifelike the artificial cells are from the RNA-seq data. The nonfunctional artificial cells changed the expression of 175 coding sequences differently than V. fischeri. An artificial cell that functioned identically to V. fischeri would have induced zero differences in gene expression. If we consider the nonfunctional artificial cells as having 0% likeness to V. fischeri, then any reduction in the number of differences in gene expression would increase the degree of likeness of the artificial cell to V. fischeri. Such a calculation would indicate that the artificial cells here were 39% lifelike or V. fischeri-like ([(175 – 107)/175] × 100), but this value is clearly an overestimation because only two of the necessary components of the artificial cell were genetically encoded (LuxR* and LuxI*). The remaining components came from an extract of E. coli that was used to mediate transcription and translation. Engineered and naturally reduced bacterial genomes require over 100 genes to produce their transcription–translation machinery. In fact, the percentage of reduced genomes dedicated to gene expression is similar to the 39% lifelike value calculated here. For example, 41% of the synthetically produced, reduced Mycoplasma mycoides genome (i.e., JCVI-syn3.0) is necessary for gene expression.31 Similarly, one-third of the naturally reduced genomes of parasitic microorganisms, including Sulcia muelleri, Carsonella ruddii, and Buchnera aphidicola, are retained for gene expression.32−34 In other words, the data only make sense when put into the context of the entire genetic system required to support the synthesis of RNA, protein, and the products of protein enzymes, in this case quorum molecules. It thus follows that, even if it were possible to assemble an artificial cell containing a genome that can make its own transcription–translation machinery35 plus additional genes for quorum signaling, this artificial cell would still not pass the 50% mark with respect to V. fischeri. That is, it is more accurate to say that if the artificial cells used here were completely genetically encoded, then these artificial cells would be 39% V. fischeri-like, according to the described cellular Turing test.
As the complexity of artificial cells increases, more stringent versions of a cellular Turing test that better capture lifelike activity can be built. Here, the artificial cells were mixed with natural V. fischeri at an OD of 0.2–0.3 and incubated for 3 h at 30 °C before analysis. Under these conditions, replication was not required and the artificial cells did not need to survive for very long. A more stringent version of the cellular Turing would mix artificial cells with more dilute cultures of V. fischeri, or another target cell type, and would be assessed for activity after longer lengths of time. Such artificial cells would be capable of replication, which would also lead to daughter vesicles containing a greater fraction of machinery encoded within its own genome, as opposed to components purified from bacteria. It should be emphasized that such cellular Turing tests are not meant to function as a definition of life, but rather as a way to circumvent the problems associated with defining life. The choice of quorum signaling may appear arbitrary, particularly since not all organisms engage in quorum signaling, but all organisms do sense and respond to their chemical environment and interact with each other in some way that is processed on a chemical level. A version of the cellular Turing test described here may not be applicable to all organisms, but this test does provide an objective metric that does not emerge from qualitative lists of lifelike properties.
Conclusions
Our incomplete understanding of basic biochemical processes limits what can be built. Although we succeeded in assembling several different quorum pathways, the cycle of sensing and responding was only fully reconstituted for V. fischeri. One critical difficulty was the reconstitution of active sensing systems, even if the sensing mechanisms of the transcriptional activators and repressors were thought to be known.9 Conversely, every cell-free, quorum molecule synthesis pathway tested was functional. Although in vivo experiments are indispensable to the study of biology, in vivo experiments alone are often not sufficient to identify all of the molecular components needed for activity. Only by reconstituting a fully functional system in vitro can we begin to understand how the pieces fit together.36−39 Such an approach can extend beyond the characterization of individual biomolecules and pathways to our understanding of cellular life. In other words, we likely will not understand what is needed to make something alive until we can build a living cell from individual component parts. This requires an identification of the necessary genes and cytoplasmic components needed to synthesize a functioning cell from DNA.40 Impressive progress has been made in synthetic genomics,31,41 but the resulting living systems still depend on many genes with unknown function and many unidentified factors present in the living cell that receives the synthetic genome. The artificial cells described here suffer from similar complications; extract compositions are not fully known, and it is not currently possible to express in vitro functioning translation machinery.35 Removing these unknowns is necessary to build artificial cells that more fully break from the concept of vivum ex vivo. Building a fully defined artificial cell from scratch would lead to a much deeper understanding of life. A cellular Turing test can help guide progress toward such a goal.
Acknowledgments
We thank the Armenise-Harvard Foundation, the Simons Foundation (290358), COST action CM1304, and Synthetic Aesthetics (supported by EPSRC, EP/H01912X/1, and NSF, 0944139) for funding, and F. Chizzolini and D. Cecchi for help with preliminary experiments. This research was funded in part by the Autonomous Province of Trento, “Grandi Progetti 2012”, project “Characterizing and improving brain mechanisms of attention–ATTEND” and Ecomm. Aligned BAM files were submitted to the NCBI Sequence Read Archive (SRA) with the accession code SRP080795.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.6b00330.
Experimental details and supporting figures and tables (PDF)
Author Contributions
‡ R.L. and N.Y.M. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Noireaux V.; Maeda Y. T.; Libchaber A. Development of an artificial cell, from self-organization to computation and self-reproduction. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 3473–3480. 10.1073/pnas.1017075108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W.; Bartel D. P.; Luisi P. L. Synthesizing life. Nature 2001, 409, 387–390. 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- Luisi P. L.THE EMERGENCE OF LIFE: From Chemical Origins to Synthetic Biology; Cambridge University Press: Cambridge, U.K., 2006. [Google Scholar]
- Lincoln T. A.; Joyce G. F. Self-sustained Replication of an RNA Enzyme. Science 2009, 323, 1229–1232. 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal R.; Pross A.; Sutherland J. D. Towards an evolutionary theory of the origin of life based on kinetics and thermodynamics. Open Biol. 2013, 3, 130156. 10.1098/rsob.130156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protocells: bridging nonliving and living matter; Rasmussen S., Bedau M. A., Chen L., Deamer D., Krakauer D. C., Packard N. H., Stadler P. F., Eds.; MIT Press: Cambridge, MA, 2009. [Google Scholar]
- Cronin L.; Krasnogor N.; Davis B. G.; Alexander C.; Robertson N.; Steinke J. H. G.; Schroeder S. L. M.; Khlobystov A. N.; Cooper G.; Gardner P. M.; et al. The imitation game--a computational chemical approach to recognizing life. Nat. Biotechnol. 2006, 24, 1203–1206. 10.1038/nbt1006-1203. [DOI] [PubMed] [Google Scholar]
- Turing A. M. Computing Machinery and Intelligence. Mind 1950, 59, 433–460. 10.1093/mind/LIX.236.433. [DOI] [Google Scholar]
- Lentini R.; Yeh Martín N.; Mansy S. S. Communicating artificial cells. Curr. Opin. Chem. Biol. 2016, 34, 53–61. 10.1016/j.cbpa.2016.06.013. [DOI] [PubMed] [Google Scholar]
- Noireaux V.; Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 17669–17674. 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W.; Sato K.; Wakabayashi M.; Nakaishi T.; Ko-Mitamura E. P.; Shima Y.; Urabe I.; Yomo T. Synthesis of functional protein in liposome. J. Biosci. Bioeng. 2001, 92, 590–593. 10.1016/S1389-1723(01)80322-4. [DOI] [PubMed] [Google Scholar]
- Gardner P. M.; Winzer K.; Davis B. G. Sugar synthesis in a protocellular model leads to a cell signalling response in bacteria. Nat. Chem. 2009, 1, 377–383. 10.1038/nchem.296. [DOI] [PubMed] [Google Scholar]
- Lentini R.; Santero S. P.; Chizzolini F.; Cecchi D.; Fontana J.; Marchioretto M.; Del Bianco C.; Terrell J. L.; Spencer A. C.; Martini L.; et al. Integrating artificial with natural cells to translate chemical messages that direct E. coli behaviour. Nat. Commun. 2014, 5, 4012. 10.1038/ncomms5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y.; Li M.; Booth R.; Mann S. Predatory behaviour in synthetic protocell communities. Nat. Chem. 2016, 10.1038/nchem.2617. [DOI] [PubMed] [Google Scholar]
- Schwarz-Schilling M.; Aufinger L.; Mückl A.; Simmel F. C. Chemical communication between bacteria and cell-free gene expression systems within linear chains of emulsion droplets. Integr. Biol. (Camb) 2016, 8, 564–570. 10.1039/C5IB00301F. [DOI] [PubMed] [Google Scholar]
- Lupp C.; Ruby E. G. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 2004, 186, 3873–3881. 10.1128/JB.186.12.3873-3881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C. H.; Arnold F. H.; Leadbetter J. R. Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones. Mol. Microbiol. 2005, 55, 712–723. 10.1111/j.1365-2958.2004.04437.x. [DOI] [PubMed] [Google Scholar]
- Gray K. M.; Passador L.; Iglewski B. H.; Greenberg E. P. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J. Bacteriol. 1994, 176, 3076–3080. 10.1128/jb.176.10.3076-3080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A. L.; Hanzelka B. L.; Parsek M. R.; Greenberg E. P. Detection, purification, and structural elucidation of the acylhomoserine lactone inducer of Vibrio fischeri luminescence and other related molecules. Methods Enzymol. 2000, 305, 288–301. 10.1016/S0076-6879(00)05495-1. [DOI] [PubMed] [Google Scholar]
- Parsek M. R.; Val D. L.; Hanzelka B. L.; Cronan J. E.; Greenberg E. P. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 4360–4365. 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder S.; Shokat K.; Surette M. G.; Bassler B. L. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 2001, 41, 463–476. 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Fernandes R.; Bentley W. E. AI-2 biosynthesis module in a magnetic nanofactory alters bacterial response via localized synthesis and delivery. Biotechnol. Bioeng. 2009, 102, 390–399. 10.1002/bit.22078. [DOI] [PubMed] [Google Scholar]
- Gordonov T.; Kim E.; Cheng Y.; Ben-Yoav H.; Ghodssi R.; Rubloff G.; Yin J.-J.; Payne G. F.; Bentley W. E. Electronic modulation of biochemical signal generation. Nat. Nanotechnol. 2014, 9, 605–610. 10.1038/nnano.2014.151. [DOI] [PubMed] [Google Scholar]
- Kambam P. K. R.; Sayut D. J.; Niu Y.; Eriksen D. T.; Sun L. Directed evolution of LuxI for enhanced OHHL production. Biotechnol. Bioeng. 2008, 101, 263–272. 10.1002/bit.21901. [DOI] [PubMed] [Google Scholar]
- Pan J.; Huang T.; Yao F.; Huang Z.; Powell C. A.; Qiu S.; Guan X. Expression and characterization of aiiA gene from Bacillus subtilis BS-1. Microbiol. Res. 2008, 163, 711–716. 10.1016/j.micres.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Kotula J. W.; Kerns S. J.; Shaket L. A.; Siraj L.; Collins J. J.; Way J. C.; Silver P. A. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 4838–4843. 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidler L.; Hans W.; Schotte L.; Neirynck S.; Obermeier F.; Falk W.; Fiers W.; Remaut E. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 2000, 289, 1352–1355. 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- Rössger K.; Charpin-El-Hamri G.; Fussenegger M. A closed-loop synthetic gene circuit for the treatment of diet-induced obesity in mice. Nat. Commun. 2013, 4, 2825. 10.1038/ncomms3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schukur L.; Geering B.; Charpin-El Hamri G.; Fussenegger M. Implantable synthetic cytokine converter cells with AND-gate logic treat experimental psoriasis. Sci. Transl. Med. 2015, 7, 318ra201–318ra201. 10.1126/scitranslmed.aac4964. [DOI] [PubMed] [Google Scholar]
- Miyashiro T.; Ruby E. G. Shedding light on bioluminescence regulation in Vibrio fischeri. Mol. Microbiol. 2012, 84, 795–806. 10.1111/j.1365-2958.2012.08065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C. A. 3rd; Chuang R.-Y.; Noskov V. N.; Assad-Garcia N.; Deerinck T. J.; Ellisman M. H.; Gill J.; Kannan K.; Karas B. J.; Ma L.; et al. Design and synthesis of a minimal bacterial genome. Science 2016, 351, aad6253. 10.1126/science.aad6253. [DOI] [PubMed] [Google Scholar]
- McCutcheon J. P.; Moran N. A. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 19392–19397. 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Brocal V.; Gil R.; Ramos S.; Lamelas A.; Postigo M.; Michelena J. M.; Silva F. J. A Small Microbial Genome: The End of a Long Symbiotic Relationship?. Science 2006, 314, 312–313. 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- Nakabachi A.; Yamashita A.; Toh H.; Ishikawa H.; Dunbar H. E.; Moran N. A.; Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 2006, 314, 267. 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Jewett M. C.; Fritz B. R.; Timmerman L. E.; Church G. M.; Blanchard S.; Kim H.; Gonzalez R.; Puglisi J.; Chu S.; Carlson E.; et al. In vitro integration of ribosomal RNA synthesis, ribosome assembly, and translation. Mol. Syst. Biol. 2013, 9, 678–678. 10.1038/msb.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwille P. Jump-starting life? Fundamental aspects of synthetic biology. J. Cell Biol. 2015, 210, 687–690. 10.1083/jcb.201506125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann S. Systems of Creation: The Emergence of Life from Nonliving Matter. Acc. Chem. Res. 2012, 45, 2131–2141. 10.1021/ar200281t. [DOI] [PubMed] [Google Scholar]
- Loakes D.; Holliger P. Darwinian chemistry: towards the synthesis of a simple cell. Mol. BioSyst. 2009, 5, 686–694. 10.1039/b904024b. [DOI] [PubMed] [Google Scholar]
- Joyce G. F. Bit by Bit: The Darwinian Basis of Life. PLoS Biol. 2012, 10, e1001323. 10.1371/journal.pbio.1001323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C.; Church G. M. Towards synthesis of a minimal cell. Mol. Syst. Biol. 2006, 2, 45. 10.1038/msb4100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G.; Glass J. I.; Lartigue C.; Noskov V. N.; Chuang R.-Y.; Algire M. A.; Benders G. A.; Montague M. G.; Ma L.; Moodie M. M.; et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010, 329, 52–56. 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.