Abstract
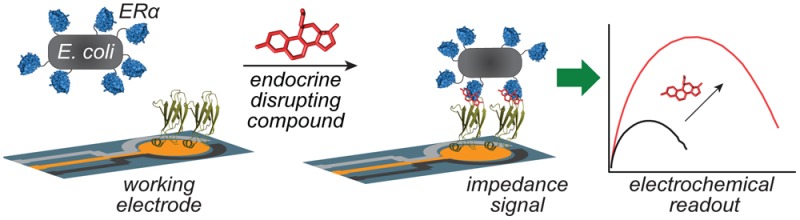
Endocrine disrupting compounds are found in increasing amounts in our environment, originating from pesticides, plasticizers, and pharmaceuticals, among other sources. Although the full impact of these compounds is still under study, they have already been implicated in diseases such as obesity, diabetes, and cancer. The list of chemicals that disrupt normal hormone function is growing at an alarming rate, making it crucially important to find sources of contamination and identify new compounds that display this ability. However, there is currently no broad-spectrum, rapid test for these compounds, as they are difficult to monitor because of their high potency and chemical dissimilarity. To address this, we have developed a new detection strategy for endocrine disrupting compounds that is both fast and portable, and it requires no specialized skills to perform. This system is based on a native estrogen receptor construct expressed on the surface of Escherichia coli, which enables both the detection of many detrimental compounds and signal amplification from impedance measurements due to the binding of bacteria to a modified electrode. With this approach, sub-ppb levels of estradiol and ppm levels of bisphenol A are detected in complex solutions. Rather than responding to individual components, this system reports the total estrogenic activity of a sample using the most relevant biological receptor. As an applied example, estrogenic chemicals released from a plastic baby bottle following microwave heating were detectable with this technique. This approach should be broadly applicable to the detection of chemically diverse classes of compounds that bind to a single receptor.
Short abstract
Endocrine disrupting compounds are rapidly detected electrochemically. We have developed a system using a native estrogen receptor construct expressed on the surface of E. coli to detect detrimental compounds at environmentally relevant levels.
Introduction
Endocrine disrupting chemicals (EDCs) are increasingly identified as potent and pervasive risks to human health. They enter the environment through numerous human activities, including pesticide use, agriculture, and fracking, and they are found in consumer products such as plastic kitchen products and food can linings.1−3 EDCs are especially dangerous because they are harmful at very low concentrations (picomolar to nanomolar), particularly to fetuses and newborns,4−8 and they are implicated in increased occurrences of obesity, diabetes, infertility, and cancer.9−11 The rapid and sensitive detection of these chemicals is therefore vital, ideally using equipment that is portable and inexpensive. Unfortunately, these compounds are particularly difficult to measure because they are not defined by a common chemical structure, but instead by their activity.12,13 To address this obstacle, we have developed a new detection paradigm for the sensitive, broad-spectrum detection of EDCs based on a native estrogen receptor alpha (ERα) construct expressed on the surface of Escherichia coli. These engineered bacterial sensors enable the detection of many detrimental compounds as well as signal amplification from impedance measurements as they bind to modified electrodes. Rather than responding to individual components, this approach reports the total estrogenic activity of a sample using the biological receptor itself. Additional features of this sensing strategy include sample volumes of only 10 μL, rapid response rates, and the use of low-cost, disposable electrodes. As such, it is the first broad-spectrum EDC assay that is appropriate for field use.
The current standards for EDC detection are cell-based assays (originally the E-SCREEN assay14 and, more recently, transactivation assays15,16 and yeast-based assays17,18) and radioactive19 and fluorescent competition assays.20,21 The cell-based transactivation involves the transcription of a reporter gene, such as a luciferase gene, following the addition of the compound in question. While effective, this analytical method is problematic for rapid, point-of-care application, as it can require multiple days of cell culture, specialized equipment, and trained laboratory personnel. Similar problems arise with fluorescent polarization assays, in which fluorescently labeled 17β-estradiol is displaced from specific antibodies by estrogenic compounds. This method requires several conjugation reactions and optimization steps, and a specialized fluorometer is necessary for measurement. As alternatives, efforts have been made to develop rapid EDC detectors, including both fluorescent and electrochemical sensors.2,22 While these platforms have had success in detecting specific compounds or chemical families, most are based on the binding of a single type of small molecule to a particular antibody or DNA aptamer, precluding broad detection of estrogenic activity (EA). Furthermore, antibodies can introduce cost and storage difficulties, and many platforms require analyte labeling with an electrochemical probe or fluorophore for detection.
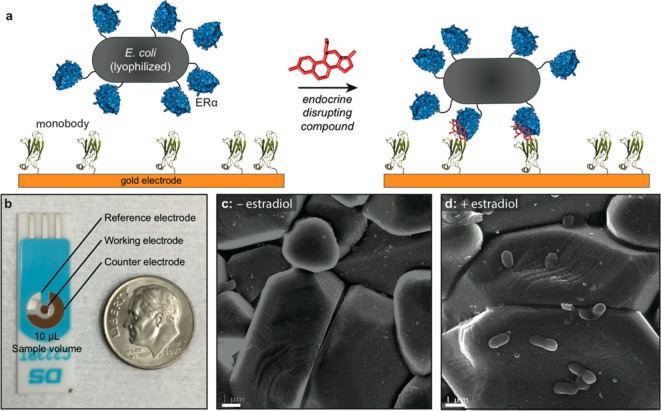
The approach described herein is based on a novel electrochemical sandwich assay (Figure 1a) and involves the use of lyophilized E. coli to cause changes in the surface impedance upon binding. Several unique aspects of this strategy enable the detection of a range of estrogenic compounds at exceptionally low concentrations. The E. coli surfaces are engineered to display the ERα capture agent, which facilitates detection of any compounds that associate with its binding pocket.23 The use of lyophilized E. coli limits their viability and increases storage life. The second component of the sandwich assay is an electrochemical working electrode modified with a previously reported protein that binds to ERα only when a ligand is present.24,25 This protein is attached through the interactions of a cysteine thiol with a disposable gold electrode surface (Figure 1b). The specificity of the monobody is observable by scanning electron microscopy of the working electrode surfaces. In the presence of estradiol (E2), E. coli was observed on the surface, while in the absence of E2, no E. coli bound the surface (Figure 1c,d).
Figure 1.
Overview of the electrochemical sandwich assay. (a) Monobodies assembled on a gold electrode surface capture EDCs bound to estrogen receptor α (ERα) that is surface expressed on E. coli. The binding of the large bacterial cells is easily detected by impedance spectroscopy. (b) The device is constructed on disposable electrodes and requires a 10 μL sample volume. Scanning electron microscopy images are shown for the monobody-modified gold electrode surface treated with lyophilized E. coli (c) in the absence of estradiol and (d) in the presence of 10 μM estradiol. Scale bars represent 1 μm.
Results and Discussion
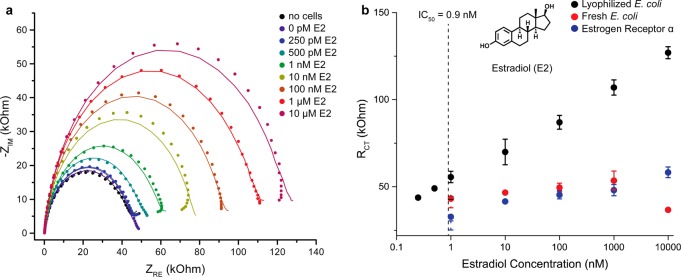
The use of lyophilized E. coli as a scaffold for the ERα protein resulted in significantly more sensitive measurements of E2 compared to the binding of ERα alone (Figure 2). The enhanced sensitivity is due to a substantially increased impedance response from the recruitment of the large E. coli cells to the gold surface, as compared to the significantly smaller free protein (Figure 2b). Additionally, no signal change is observed in the presence of E2 but with no E. coli or ERα added (Figure S3). Both fresh and lyophilized E. coli were tested, and a dependence on the number of cells used for detection was observed (Figure S1). For lyophilized cells, the optimal number of cells was found to be 104/mL. The number of ERα proteins surface expressed on fresh E. coli was determined to be approximately 70,000 using a fluorescent coumarin–E2 conjugate,26 while on the lyophilized E. coli it was slightly lower (50,000/cell) (Figure S2). This level of surface expression is expected, as the maximum number of ice nucleation proteins that were fused to ERα is on the order of 100,000.27,28 Fresh E. coli resulted in a small impedance response as compared to the lyophilized cells (Figure 2b) likely due to their motility, which reduces their binding to the electrode surface. This hypothesis was supported by comparing detection with E. coli killed with sodium azide to E. coli rendered nonviable, but alive and motile, by a low dose of chloramphenicol. The chloramphenicol-treated E. coli behaved as the untreated, live E. coli, and the sodium azide treated cells behaved similarly to the lyophilized cells (Figure S3). Consistent with this behavior, no E. coli from a live sample were observed on electrodes by electron microscopy.
Figure 2.
Electrochemical sandwich assay for endocrine disrupting compounds (EDCs). (a) Nyquist plots of estradiol detection with the platform at concentrations ranging from 0 pM to 10 uM, along with the CPE fits used to determine the charge transfer radius (RCT). (b) Estradiol concentration dependent RCT for ERα (blue), ERα on live E. coli (red), and ERα on lyophilized E. coli (black). Error bars represent the SD for n = 3 replicates.
Detection of the binding event was accomplished with electrochemical impedance spectroscopy (EIS) in ferricyanide/ferrocyanide solution. This technique is rapid (providing readout in minutes), sensitive, and label-free.29,30 Nyquist plots were generated from each EIS scan performed, and the data were fit to a constant phase element (CPE) circuit model (Figure 2a). The charge transfer resistance (RCT) was derived from the CPE fits and was found to be proportional to the amount of ERα bound to the electrode and, therefore, the amount of substrate present. Using RCT as a proxy for the concentration of substrate, we were able to detect 500 pM E2 with a large linear range of detection up to 10 μM (Figure 2b). As the required sample volume is especially low (10 μL), we were able to detect femtomoles of estradiol at the detection limit.
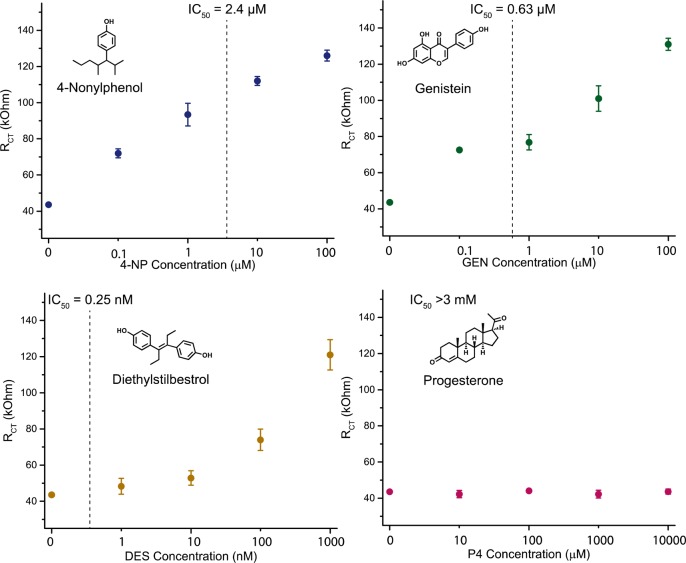
The system was found to be especially versatile, with detection of chemicals that have disparate chemical structures but similar bioactivity. The EDCs tested that bind ERα are 4-nonylphenol (4-NP), genistein (GEN), diethylstilbestrol (DES), and bisphenol A (BPA). Progesterone (P4) was used as a negative control, as P4 is not a substrate for ERα binding. In Figures 3 and 4a (turquoise), each EDC was tested over a range of concentrations selected on the basis of their respective IC50 values (shown as vertical lines). All agonists tested (4-NP, GEN, DES, and BPA) produced linear responses over an extended concentration range, with increasing RCT as EDC concentration increased. Some nonlinearity was observed at low concentrations of GEN, which could be due to complexities in the ternary complex formation. For DES, reduced linearity was observed as the detection limit was approached. Each of these compounds was detectable at exceptionally low concentrations, and most could be quantified below their IC50 values. DES was detectable to concentrations ten times its IC50 value. Unlike the EDCs that bind ERα, this platform shows no response to progesterone, indicating its specificity for estrogenic compounds. Similarly, this platform showed no response to the antagonist tamoxifen (TAM), indicating that the conformation of the ERα–antagonist complex does not bind the monobody on the electrode surface (Figure S5).
Figure 3.
Endocrine disrupting compound concentration dependent RCT for ERα on lyophilized E. coli with compounds that bind ERα: 4-nonylphenol (4-NP, top left), genistein (GEN, top right), and diethylstilbestrol (DES, bottom left). Progesterone, which does not bind ERα, shows no RCT response (bottom right). Error bars represent the SD for n = 3 replicates.
Figure 4.
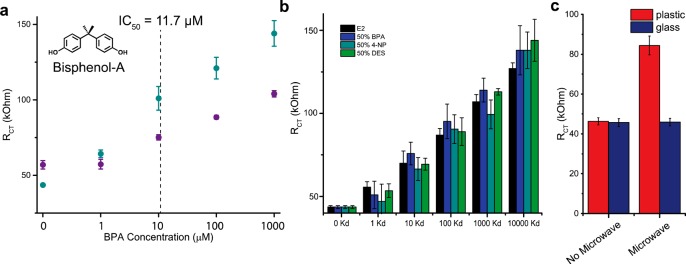
EDC detection from complex solutions. (a) Endocrine disrupting compound concentration dependent RCT for ERα on lyophilized E. coli with BPA in buffer (turquoise) and in infant formula (purple). (b) Combinations of BPA, 4-NP, DES, and GEN with comparable estradiol concentrations (black). Each solution contains 50% of one EDC, with 16.67% of each of the other three EDCs (where 100% would represent the known IC50 concentration) . Samples with 50% BPA (dark blue), 50% 4-NP (turquoise), and 50% DES (green) are shown. (c) Estrogenicity of plastic (red) and glass (blue) baby bottles before and after microwaving. Error bars represent the SD for n = 3 replicates.
In contaminated systems, EDCs rarely occur as a single compound. Rather, they are often mixed, providing an aggregate effect. The combined interaction of all the EDCs present with the ERα protein yields a response that can be benchmarked as a concentration of the native substrate, E2, that would produce similar activity. This equivalent response is termed the “estrogenic activity” (EA) of the solution. The sensor was therefore evaluated for its ability to determine EA of complex mixtures. The EDCs previously measured (BPA, 4-NP, DES, and GEN) were combined and compared with comparable estradiol concentrations. Each solution contained 50% of one EDC (relative to its IC50 value), with 16.67% of each of the other three EDCs. The absolute concentrations of the components appear in Table S1. The RCT values for the combined EDCs were compared to the equivalent concentration of E2 as a measure of the EA of the solution. Independent of the ratio of EDCs in the solution, the RCT was found to be comparable to the equivalent concentration of E2 (Figure 4b). This platform therefore shows the distinct advantage of providing a readout of the total EA from a complex mixture of components even when their specific identities are unknown.
Importantly, this approach enables detection of target compounds present in complex mixtures of proteins and small molecules. As EDCs are especially deleterious for proper development, their presence has been especially problematic in infant products. As one relevant example, the detection of BPA was evaluated in infant formula (Figure 4a). BPA was added to reconstituted formula from a commercial source in varying concentrations. The ability of the system to detect BPA was linear above the IC50 value, despite the addition of protein, lipid, and small molecule components. Below this concentration, the signal was indistinguishable from background, likely due to surface passivation from proteins in the formula.
As a final experiment, we evaluated the ability of the system to detect EA from an everyday source without prior knowledge of the contaminants. In the literature, E-SCREEN assays have shown that certain “BPA-free” plastic baby bottles release EDCs upon microwave heating.31 We sought to replicate this experiment using the faster and lower volume electrochemical assay described herein. Prior to microwave heating the plastic bottle, the buffer had no observable EA. However, after microwaving for ten 2 min periods, the buffer in the plastic bottle had significant EA, comparable to 100 nM E2. In contrast, the buffer in a glass bottle contained no EA before or after microwaving (Figure 4c).
Through this work, we have developed a new approach for determining the estrogenic activity of endocrine disrupting compounds. By combining impedance spectroscopy based detection with the signal amplification provided by a lyophilized E. coli scaffold, large responses in the charge transfer resistance of the electrode are observed, even in the presence of sub-ppb estradiol. The system provides the first reported sensor that responds broadly to all EDCs, and since it is based on inexpensive disposable electrode technology, it can be used in the field. The 10 μL sample size is far smaller than that needed for cell-based growth assays, and the readout is available in minutes, not days. Furthermore, the application of lyophilized E. coli as a scaffold for our protein provides a new method of signal amplification, and is crucially important for reaching the low detection limits that these compounds require. The system also shows promising compatibility with complex sample matrices, such as infant formula. This new sensing approach should be applicable to other diverse families of compounds that bind to a single receptor, such as PPARγ, and current efforts in our laboratory are exploring these possibilities.
Methods
Plasmid Preparation
Monobody Encoding pSKB3 Vector
The gene encoding for the ERα-estradiol selective monobody protein, a sequence adapted from Koide et al.,19,20 was synthesized by IDT Technologies with BamHI and XhoI restriction sites at the 5′- and 3′-ends and subcloned into a pSKB3 vector—a variation of Novagen’s pET-28a vector with the thrombin site exchanged for a TEV proteolysis site. The insert and vector backbone were double digested (BamHI/XhoI), heat inactivated at 80 °C for 5 min, ligated with QuickLigase (NEB) at a 5:1 molar ratio, and transformed into XL1Blue competent cells. Plating on kanamycin agar plates yielded individual colonies, which were cultured, DNA purified (NucleoSpin, MacheryNagel), and sequenced (Quintara BioSciences).
INPNC-ERα Encoding pSKB3 Vector
The synthetic gene (IDT Technologies) of ERα (organism, Homo sapiens; gene, ESR1, accession number P03372; residue number, 301–552) was subcloned with NheI and NotI restriction sites at the 5′- and 3′-ends into a pSKB3 vector containing an N-terminal maltose binding protein (MBP). The resulting vector furnished the following amino acid sequence: MASS-(His)6-TEV-MBP-Linker-ERα (where “Linker” = N10-LGASGSG).
The gene insert coding for the ice nucleation protein with the NC-terminal fusion (INPNC: fusion of the N-terminal membrane domain INPN and the C-terminal extracellular domain INPC) was synthesized by IDT Technologies with Nco1 and Nhe1 restriction sites at the 5′- and 3′-sites and was subcloned into the MBP-ERα pSKB3 vector above. The MBP gene was removed in the process. The resulting vector encodes for the following amino acid sequence: MAA-INPN-RS-INPC-SSN10LGASGSG-ERα. The INPNC insert and vector backbone were double digested (NcoI/NheI), heat inactivated at 65 °C for 15 min, ligated with QuickLigase (NEB) at a 5:1 molar ratio, and transformed into XL1Blue competent cells. Plating on kanamycin agar plates yielded individual colonies, which were cultured, DNA purified (NucleoSpin, MacheryNagel), and sequenced (Quintara BioSciences).
Protein Expression and Purification
Plasmids were transformed into E. coli BL21 (DE3) competent cells. Starter cultures (20 mL of LB, 50 mg/L kanamycin) were grown from single colonies overnight at 37 °C and used to inoculate 1 L of TB medium (50 mg/L kanamycin). Cultures were grown to an OD ∼ 0.5, cooled to 25 °C for 20 min, induced with 0.5 mM IPTG, and expressed overnight (18 h) at 25 °C. Cells were harvested by centrifugation for 15 min at 4000 rcf at 4 °C. The protein was purified directly without freezing.
Purification of Monobody Protein
The pellet was transferred to PBS buffer and centrifuged for 10 min at 4300 rcf. The resulting pellet was lysed in 30 mL of lysis buffer, referred to hereafter as buffer B (20 mM bicine, pH 8.5, 500 mM NaCl, 10 mM imidazole), supplemented with one tablet of EDTA-free SigmaFast Protease Inhibitor (Sigma-Aldrich), 5 mM PMSF, and 2 mg of lysozyme. Without incubation, the resuspension was lysed with an Avestin C3 homogenizer followed by a 20 min centrifugation at 24,000 rcf at 4 °C. The supernatant was filtered through a 40 μm Steriflip filter (Millipore) and loaded onto a 5 mL NiNTA column (Protino, Machery Nagel) connected to an Akta purifier preequilibrated with buffer B. The column was washed with 50 mL (10 column volumes) of 20 mM bicine (pH 8.5), 500 mM NaCl, 10 mM imidazole, 10 mM β-ME. The protein was eluted with 20 mM bicine (pH 8.5), 500 mM NaCl, 250 mM imidazole, 10 mM β-Me. Imidazole was removed by exchanging against 20 mM bicine (pH 8.5), 500 mM NaCl, with a 10DG desalting column (BioRad). For purposes of lyophilization, the protein was directly exchanged against 20 mM bicine (pH 8.5) and 100 mM trehalose, followed by flash freezing with liquid N2 and lyophilization (Labconco) overnight. Typical protein yields are 800 μM (3 mL total) from a 1 L culture, with a purity of ∼98% by SDS–PAGE and LC–MS (ESI-TOF) (6224 TOF and 1200 series HPLC, Agilent Technologies).
Expression of Cell Surface Displayed INPNC-ERα
INPNC-ERα was expressed from single BL21 colonies. The proteins were grown in 50 mL of TB in the presence of 50 mg/L kanamycin. At an OD600 ∼ 0.5 the culture was equilibrated to 25 °C for 20 min, induced with 0.5 mM IPTG, and expressed overnight (18 h) at 25 °C. The cells were centrifuged for 5 min at 4000 rcf and resuspended either in M9 minimal medium for direct use or in 20 mM HEPES (pH 7.5) with 100 mM trehalose for lyophilization. E. coli cells were lyophilized by flash freezing 100 μL aliquots in liquid nitrogen at an OD600 ∼ 0.1. The lyophilized samples were stored at either −20 °C or −80 °C. Sodium azide (NaN3) treated cells were incubated with 5 mg/mL NaN3 to induce toxicity. Chloramphenicol-treated cells were incubated with 10 μg/mL of chloramphenicol for 30 min, a concentration below toxicity but sufficient to inhibit protein synthesis.
Cell Viability of Lyophilized E. coli Cultures
The lyophilized sample produced from 100 μL at OD600 = 0.1 was dissolved in 600 μL of M9 minimal medium, and 125 μL volumes were streaked onto kanamycin agar plates. No colonies were observed after 24 h; colonies observed after 48 h were counted and compared against equivalently plated cells streaked from glycerol stocks.
Reconstitution of E. coli Cells
Aliquots of E. coli were reconstituted by dissolution in 100 μL of 20 mM HEPES (pH 7.5) to an OD of 0.1. Cells were incubated on ice for 20 min prior to further dilution.
Determination of Surface-Expressed ERα
An estradiol–coumarin conjugate (prepared as described in ref (21)) was added to a final concentration of 10 μM to either freshly harvested or lyophilized E. coli at an OD600 of 0.01 in M9 medium. Following a 20 min incubation, cells were purified from unbound E2–coumarin by spin filtration (10K; 5 min, 5000 rcf). The fluorescence of the E2–coumarin labeled cells was measured and compared to a standard curve of estradiol–coumarin fluorescence in M9 medium. Using the concentration of estradiol–coumarin, the number of receptors per E. coli cell was estimated.
Electrode Preparation
Disposable gold electrodes (1.3 mm diameter, cold annealed, DropSens) were preliminarily prepared in 0.5 M H2SO4 by scanning from 1.3 V to −0.2 V (vs internal reference, 100 mV scan rate, 9 scans). Electrodes were subsequently washed with Milli-Q water. Lyophilized monobody was diluted in 20 mM HEPES, 300 mM NaCl (pH 7.5) to a final concentration of 50 μM. A 10 μL portion was added to the electrode surface. Electrodes were placed in humidifier boxes and incubated overnight at 4 °C. Prior to detection using the monobody-modified electrodes, the electrodes were rinsed with 3 aliquots of 100 μL of 20 mM HEPES (pH 7.5).
Incubation of Analytes
Following reconstitution of lyophilized E. coli samples, the cells were diluted to the desired final concentration (104 cells/mL, although cells were tested at concentrations ranging from 103 to 107) in 20 mM HEPES (pH 7.5). For the detection of single endocrine disrupting chemicals (estradiol, DES, GEN, BPA, or 4-NP), the chemical was dissolved in DMSO to a 1000× dilution of the final concentration, such that the concentration of DMSO was constant at 0.1% v/v in solution with E. coli, including negative controls (in which either no cells or no EDC was added).
The EDC of interest was incubated with the E. coli for 20 min in solution prior to application to the electrode. A 10 μL portion of the E. coli solution containing the analyte compounds was then added to the electrode surface. E. coli solutions were incubated on the electrode surface for 20 min at ambient temperature in a humidifier box. The electrodes were subsequently rinsed with 3 aliquots of 100 μL of 20 mM HEPES (pH 7.5).
Electrochemical Impedance Spectroscopy
Electrochemical impedance spectroscopy (EIS) was performed with a Gamry Reference 600 potentiostat. The buffer consisted of 4 mM each of K3Fe(CN)6/K4Fe(CN)6 in 0.1 M KCl. Electrochemical measurements were acquired at the open circuit potential of the electrode and measured for 60 s prior to EIS. EIS measurements were made from 50,000 to 0.2 Hz with 10 points per decade and a 10 mV ac voltage. Electrochemical data analysis, including circuit modeling, was performed using the Gamry Echem Analyst software. Charge transfer resistance (RCT) was derived from a constant phase element (CPE) with diffusion circuit model fit.
Acknowledgments
This work was supported by Hound Labs and the NSF (CHE-1413666). A.L.F. was supported by the Arnold O. Beckman Foundation. A.C.H. was supported by Administral Anstalt Liechtenstein. Dr. Martin J. Mulvihill is also acknowledged for helpful discussions.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.6b00322.
Figures S1–S5, Table S1, and plasmid and insert sequences (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Melnick R.; Lucier G.; Wolfe M.; Hall R.; Stancel G.; Prins G.; Gallo M.; Reuhl K.; Ho S.-M.; Brown T.; Moore J.; Leakey J.; Haseman J.; Kohn M. Summary of the National Toxicology Program’s report of the endocrine disruptors lowdose peer review. Environ. Health Perspect. 2002, 110, 427–431. 10.1289/ehp.02110427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic M.; Eljarrat E.; Lopez de Alda M. J.; Barceló D. Endocrine disrupting compounds and other emerging contaminants in the environment: A survey on new monitoring strategies and occurrence data. Anal. Bioanal. Chem. 2004, 378, 549–562. 10.1007/s00216-003-2184-7. [DOI] [PubMed] [Google Scholar]
- Welshons W. V.; Nagel S. C.; vom Saal F. S. Large effects from small exposures. III. Endocrine mechanisms mediating effects of bisphenol A at levels of human exposure. Endocrinology 2006, 147, S56–S69. 10.1210/en.2005-1159. [DOI] [PubMed] [Google Scholar]
- Vandenberg L. N.; Colborn T.; Hayes T. B.; Heindel J. J.; Jacobs D. R. Jr.; Lee D. H.; Shioda T.; Soto A. M.; vom Saal F. S.; Welshons W. V.; Zoeller R. T.; Myers J. P. Hormones and endocrine-disrupting chemicals: low-dose effects and nonmonotonic dose responses. Endocr. Rev. 2012, 33, 378–455. 10.1210/er.2011-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabanič D.; Rupnik M.; Klemenčič A. K. Negative impact of endocrine-disrupting compounds on human reproductive health. Reprod., Fertil. Dev. 2011, 23, 403–416. 10.1071/RD09300. [DOI] [PubMed] [Google Scholar]
- Colborn T.; vom Saal F. S.; Soto A. M. Developmental effects of endocrine-disrupting chemicals in wildlife and humans. Environ. Health Perspect. 1993, 101, 378–384. 10.1289/ehp.93101378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan J. A. Environmental signaling: What embryos and evolution teach us about endocrine disrupting chemicals. Endocr. Rev. 2001, 22, 319–341. 10.1210/edrv.22.3.0432. [DOI] [PubMed] [Google Scholar]
- Schug T. T.; Johnson A. F.; Birnbaum L. S.; Colborn T.; Guillette L. J. Jr.; Crews D. P.; Collins T.; Soto A. M.; vom Saal F. S.; McLachlan J. A.; Sonnenschein C.; Heindel J. J. Minireview: endocrine disruptors: past lessons and future directions. Mol. Endocrinol. 2016, 30, 833–847. 10.1210/me.2016-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold R.; Padilla-Banks E.; Jefferson W.; Heindel J. Effects of endocrine disruptors on obesity. Int. J. Androl. 2008, 31, 201–208. 10.1111/j.1365-2605.2007.00858.x. [DOI] [PubMed] [Google Scholar]
- Roy J. R.; Chakraborty S.; Chakraborty T. R. Estrogen-like endocrine disrupting chemicals affecting puberty in humans--a review. Med. Sci. Monit. 2009, 15, RA137–145. [PubMed] [Google Scholar]
- Birnbaum L. S.; Fenton S. E. Cancer and developmental exposure to endocrine disruptors. Environ. Health Perspect. 2003, 111, 389–394. 10.1289/ehp.5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair R. M.; Fang H.; Branham W. S.; Hass B. S.; Dial S. L.; Moland C. L.; Tong W.; Shi L.; Perkins R.; Sheehan D. M. The Estrogen Receptor Relative Binding Affinities of 188 Natural and Xenochemicals: Structural Diversity of Ligands. Toxicol. Sci. 2000, 54, 138–153. 10.1093/toxsci/54.1.138. [DOI] [PubMed] [Google Scholar]
- Matthews J.; Celius T.; Halgren R.; Zacharewski T. Differential estrogen receptor binding of estrogenic substances: a species comparison. J. Steroid Biochem. Mol. Biol. 2000, 74, 223–234. 10.1016/S0960-0760(00)00126-6. [DOI] [PubMed] [Google Scholar]
- Soto A. M.; Sonnenschein C.; Chung K. L.; Fernandez M. F.; Olea N.; Serrano F. O. The E-SCREEN assay as a tool to identify estrogens: an update on estrogenic environmental pollutants. Environ. Health Perspect. 1995, 103, 113–122. 10.1289/ehp.95103s7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paech K.; Webb P.; Kuiper G. G. J. M.; Nilsson S.; Gustafsson J.-A.; Kushner P. J.; Scanlan T. S. Differential Ligand Activation of Estrogen Receptors ERα and ERβ at AP1 Sites. Science 1997, 277, 1508–1510. 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- Andersen H. R.; Vinggaard A. M.; Rasmussen T. H.; Gjermandsen I. M.; Bonefeld-Jørgensen E. C. Effects of Currently Used Pesticides in Assays for Estrogenicity, Androgenicity, and Aromatase Activity in Vitro. Toxicol. Appl. Pharmacol. 2002, 179, 1–12. 10.1006/taap.2001.9347. [DOI] [PubMed] [Google Scholar]
- Kolle S. N.; Kamp H. G.; Huener H. A.; Knickel J.; Verlohner A.; Woitkowiak C.; Landsiedel R.; van Ravenzwaay B. In house validation of recombinant yeast estrogen and androgen receptor agonist and antagonist screening assays. Toxicol. In Vitro 2010, 24, 2030–2040. 10.1016/j.tiv.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Beresford N.; Routledge E. J.; Harris C. A.; Sumpter J. P. Issues arising when interpreting results from an in vitro assay for estrogenic activity. Toxicol. Appl. Pharmacol. 2000, 162, 22–33. 10.1006/taap.1999.8817. [DOI] [PubMed] [Google Scholar]
- Kinnberg K.Evaluation of in vitro assays for determination of estrogenic activity in the environment; Working Report No. 43; 2003. http://www2.mst.dk/udgiv/publications/2003/87-7972-922-3/pdf/87-7972-923-1.pdf. Accessed 10/18/16.
- Bolger R.; Wiese T. E.; Ervin K.; Nestich S.; Checovich W. Rapid screening of environmental chemicals for estrogen receptor binding capacity. Envorin. Health Perspect. 1998, 106, 551–557. 10.1289/ehp.98106551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varriale A.; Pennacchio A.; Pinto G.; Oliviero G.; D’Errico S.; Majoli A.; Scala A.; Capo A.; Pennacchio A.; Di Giovanni S.; Staiano M.; D’Auria S. A Fluorescence Polarization Assay To Detect Steroid Hormone Traces in Milk. J. Agric. Food Chem. 2015, 63, 9159–9164. 10.1021/acs.jafc.5b03689. [DOI] [PubMed] [Google Scholar]
- Scognamiglio V.; Antonacci A.; Patrolecco L.; Lambreva M. D.; Litescu S. C.; Ghuge S. A.; Rea G. Analytical tools monitoring endocrine disrupting chemicals. TrAC, Trends Anal. Chem. 2016, 80, 555–567. 10.1016/j.trac.2016.04.014. [DOI] [Google Scholar]
- Pillon A.; Boussioux A.-M.; Escande A.; Aït-Aïssa S.; Gomez E.; Fenet H.; Ruff M.; Moras D.; Vignon F.; Duchesne M.-J.; Casellas C.; Nicolas J. C.; Balaguer P. Binding of estrogenic compounds to recombinant Estrogen Receptor-α: Application to environmental analysis. Environ. Health Perspect. 2005, 113, 278–284. 10.1289/ehp.7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide A.; Abbatiello S.; Rothgery L.; Koide S. Probing protein conformational changes in living cells by using designer binding proteins: Application to the estrogen receptor. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 1253–1258. 10.1073/pnas.032665299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.; Koide A.; Nettle K. W.; Greene G. L.; Koide S. Conformation-specific affinity purification of proteins using engineered binding proteins: application to the estrogen receptor. Protein Expression Purif. 2006, 47, 348–354. 10.1016/j.pep.2005.10.021. [DOI] [PubMed] [Google Scholar]
- MacDonald J. I.; Munch H. K.; Moore T.; Francis M. B. One-step site-specific modification of native proteins with 2-pyridinecarboxaldehydes. Nat. Chem. Biol. 2015, 11, 326–331. 10.1038/nchembio.1792. [DOI] [PubMed] [Google Scholar]
- van Bloois E.; Winter R. T.; Kolmar H.; Fraaije M. W. Decorating microbes: surface display of proteins on Escherichia coli. Trends Biotechnol. 2011, 29, 79–86. 10.1016/j.tibtech.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Li L.; Kang D. G.; Cha H. J. Functional display of foreign protein on surface of Escherichia coli using N-terminal domain of Ice Nucleation Protein. Biotechnol. Bioeng. 2004, 85, 214–221. 10.1002/bit.10892. [DOI] [PubMed] [Google Scholar]
- Lisdat F.; Schaefer D. The use of electrochemical impedance spectroscopy for biosensing. Anal. Bioanal. Chem. 2008, 391, 1555–1567. 10.1007/s00216-008-1970-7. [DOI] [PubMed] [Google Scholar]
- Guo X.; Kulkarni A.; Doepke A.; Halsall H. B.; Iyer S.; Heineman W. R. Carbohydrate-based label-free detection of Escherichia coli ORN 178 using Electrochemical Impedance Spectroscopy. Anal. Chem. 2012, 84, 241–246. 10.1021/ac202419u. [DOI] [PubMed] [Google Scholar]
- Yang C. Z.; Yaniger S. I.; Jordan V. C.; Klein D. J.; Bittner G. D. Most plastic products release estrogenic chemicals: a potential health problem that can be solved. Environ. Health Perspect. 2011, 119, 989–996. 10.1289/ehp.1003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.