Abstract
Biofilms are composed of bacterial cells embedded in an extracellular polysaccharide matrix. A major component of the Escherichia coli biofilm matrix is PGA, a linear polymer of N-acetyl-d-glucosamine residues in β(1,6) linkage. PGA mediates intercellular adhesion and attachment of cells to abiotic surfaces. In this report, we present genetic and biochemical evidence that PGA is also a major matrix component of biofilms produced by the human periodontopathogen Actinobacillus actinomycetemcomitans and the porcine respiratory pathogen Actinobacillus pleuropneumoniae. We also show that PGA is a substrate for dispersin B, a biofilm-releasing glycosyl hydrolase produced by A. actinomycetemcomitans, and that an orthologous dispersin B enzyme is produced by A. pleuropneumoniae. We further show that A. actinomycetemcomitans PGA cross-reacts with antiserum raised against polysaccharide intercellular adhesin, a staphylococcal biofilm matrix polysaccharide that is genetically and structurally related to PGA. Our findings confirm that PGA functions as a biofilm matrix polysaccharide in phylogenetically diverse bacterial species and suggest that PGA may play a role in intercellular adhesion and cellular detachment and dispersal in A. actinomycetemcomitans and A. pleuropneumoniae biofilms.
Bacterial cells in a biofilm are surrounded by a self-synthesized, three-dimensional matrix that holds the cells together in a mass and firmly attaches the bacterial mass to the underlying surface (3). This matrix, referred to as the slime layer, glycocalyx, or extracellular polymeric substance (EPS) matrix, can comprise up to 90% of the biofilm biomass (8). In addition to its structural role, the EPS matrix provides biofilm cells with a protected microenvironment containing dissolved nutrients and secreted enzymes, as well as other biological molecules and abiotic substances originating from outside the biofilm. The EPS matrix may also contribute to the increased resistance to antibiotics and host defenses exhibited by biofilm cells (27, 32).
Polysaccharide is a major component of the EPS matrix in most bacterial biofilms (34). One of the best-characterized matrix polysaccharides is a hexosamine-containing polymer produced by several staphylococcal species, including Staphylococcus epidermidis and Staphylococcus aureus, and by Escherichia coli. This polymer, called PIA (also PNAG, PS/A, or SAA) in Staphylococcus spp. (2, 24, 28-30) and PGA in E. coli (36), consists of a linear chain of N-acetyl-d-glucosamine (GlcNAc) residues in β(1,6) linkage. Various forms of this GlcNAc polymer appear to differ in molecular weight, in the degree of deacetylation of the GlcNAc residues, and in the presence of O-acetyl substituents (24, 28, 30). PIA and PGA have been shown to play roles in abiotic surface attachment and intercellular adhesion (13, 30, 37), and PIA has been shown to protect S. epidermidis biofilm cells from host innate defenses, including phagocytosis and antimicrobial peptides (36).
In S. epidermidis, the production of PIA is encoded by a cluster of four tightly linked genes named icaADBC (10, 13, 30). IcaA is a transmembrane glycosyltransferase that synthesizes the GlcNAc polymer backbone (10). Sole expression of icaA induces only low enzymatic activity, but coexpression of icaA with icaD, a small gene that is translationally coupled to icaA, leads to a significant increase in enzymatic activity and is related to full phenotypic expression of PIA (10). IcaB is a secreted protein that is homologous to several polysaccharide N-deacetylases, including rhizobial NodB chito-oligosaccharide deacetylase, peptidoglycan GlcNAc deacetylase, and chitin deacetylase, suggesting that IcaB may be a deacetylase. IcaC is a predicted transmembrane protein with unknown function. In E. coli, the production of PGA is encoded by a cluster of four tightly linked genes named pgaABCD (37). PgaC exhibits 40% amino acid identity to S. epidermidis IcaA, suggesting that PgaC is also a glycosyltransferase. Although PgaD exhibits no primary amino acid sequence similarity to IcaD, both proteins are small (137 and 101 amino acid residues, respectively), and both contain two predicted transmembrane segments, which suggests that PgaD and IcaD may be functionally related. PgaA and PgaB are predicted to be outer membrane or secreted proteins (37). The N-terminal 300 amino acid residues of PgaB exhibit 25% identity to IcaB, suggesting that PgaB may also be a deacetylase. PgaA and the C-terminal 370 amino acid residues of PgaB are not homologous to any proteins in the National Center for Biotechnology Information (NCBI) public database.
A glycosyl hydrolase that degrades S. epidermidis PIA was recently identified (21). This enzyme, named dispersin B (DspB), is produced by the gram-negative oral bacterium Actinobacillus actinomycetemcomitans, the causative agent of a severe form of periodontal disease that affects adolescents (38). Treatment of S. epidermidis biofilms with recombinant A. actinomycetemcomitans dispersin B protein causes dissolution of the EPS matrix and detachment of biofilm cells from the surface (21). Treatment of A. actinomycetemcomitans biofilms with dispersin B also causes the detachment of cells from the surface (20), suggesting that A. actinomycetemcomitans biofilms contain a matrix polysaccharide that is structurally related to PIA. In this report, we present evidence that a cluster of four A. actinomycetemcomitans genes that is homologous to E. coli pgaABCD (19, 38, 40, and 12% amino acid identity, respectively) encodes the production of a hexosamine-containing exopolysaccharide that is structurally related to E. coli PGA and that this polysaccharide is a substrate for dispersin B. We also show that the A. actinomycetemcomitans PGA-like polysaccharide is not required for the attachment of cells to surfaces but may play a role in the aggregation of cells inside the biofilm colony and the detachment of cells from the colony. We further show that a similar hexosamine-containing polysaccharide constitutes a biofilm matrix component in the closely related bacterium Actinobacillus pleuropneumoniae, the causative agent of porcine pleuropneumonia (11).
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains used in this study are listed in Table 1. A. actinomycetemcomitans and S. epidermidis strains were grown in Trypticase soy broth (Becton Dickinson) supplemented with 6 g of yeast extract and 8 g of glucose per liter. Media were supplemented with 3 μg of chloramphenicol/ml, 40 μg of kanamycin/ml, 20 μg of nalidixic acid/ml, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), 0.01% Congo red dye, or 1.5% agar when appropriate. A. pleuropneumoniae strains, which were isolated from the lungs of pigs with porcine pleuropneumonia, were cultured in Mueller-Hinton broth supplemented with 6 g of yeast extract, 8 g of glucose, and 5 mg of NAD per liter. E. coli strains were grown in Luria-Bertani broth supplemented with ampicillin or chloramphenicol (50 μg/ml each) or 1 mM IPTG when appropriate. E. coli cultures were incubated in tubes with agitation. All other bacteria were incubated statically. A. actinomycetemcomitans was incubated in the presence of 10% CO2. All cultures were incubated at 37°C.
TABLE 1.
. Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| A. actinomycetemcomitans IDH781 | Clinical isolate (serotype d) | 12 |
| A. actinomycetemcomitans IDH781N | Spontaneous Nalr variant of IDH781 | M. Bhattacharjee and D. Figurski |
| A. actinomycetemcomitans JK1048 | IDH781N pgaC::R6Kγori/KAN; Kmr | This study |
| A. actinomycetemcomitans JK1049 | IDH781N pgaC::R6Kγori/KAN; Kmr | This study |
| A. actinomycetemcomitans JK1047 | IDH781N flp-1::Tn903φkan; Kmr | J. B. Kaplan, unpublished data |
| A. actinomycetemcomitans CU1000 | Clinical isolate (serotype f) | 6 |
| A. pleuropneumoniae IA1 | Clinical isolate (serotype 1) | Iowa State University |
| A. pleuropneumoniae IA5 | Clinical isolate (serotype 5) | Iowa State University |
| S. epidermidis NJ9709 | Clinical isolate | 21 |
| S. epidermidis 1457 | Clinical isolate | 26 |
| S. epidermidis 1457-M11 | 1457 icaA::Tn917 | 25 |
| E. coli DH5α | Used for pgaCD and icaAD expression | New England Biolabs |
| E. coli TRXWEC | pgaC::Tn10Cam: Cmr; used for expression of E. coli pgaABCD | 37 |
| E. coli BL21(DE3) | Used for overexpression of dispersin B | Novagen |
| Plasmids | ||
| LITMUS28 | E. coli cloning vector; Apr | New England Biolabs |
| pVK80 | LITMUS28 containing A. actinomycetemcomitans pgaCD | This study |
| pVK81-1 | pVK80 pgaC::R6Kγori/KAN | This study |
| pVK81-5 | pVK80 pgaC::R6Kγori/KAN | This study |
| pJAK16 | Broad-host-range expression vector; Cmr | 35 |
| pVK82 | pJAK16 containing A. actinomycetemcomitans pgaCD (Fig. 1) | This study |
| pVK91 | pJAK16 containing S. epidermidis icaAD | This study |
| pVK93 | pJAK16 containing A. pleuropneumoniae pgaCD (Fig. 1) | This study |
| pRK21761 | Use to mobilize plasmids from E. coli to A. actinomycetemcomitans | 35 |
| pET-28a | Used for overexpression of dispersin B | Novagen |
| pRC2 | pET-28a containing A. pleuropneumoniae dspB | This study |
| pUC19 | E. coli cloning vector; Apr | New England Biolabs |
| pPGA372 | pUC19 containing E. coli pgaABCD | 37 |
Nalr, nalidixic acid resistant; Kmr, kanamycin resistant; Cmr, chloramphenicol resistant; Apr, ampicillin resistant.
Targeted mutagenesis of A. actinomycetemcomitans pgaC.
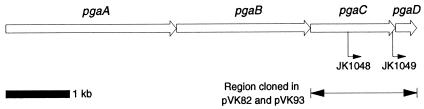
The pgaCD region of A. actinomycetemcomitans strain CU1000 was amplified by PCR using the forward primer 5′-TGACCGGATCCTCAAGCAGGTAAACCATAG-3′, which introduced a BamHI site (underlined) 27 bp upstream from the pgaC start codon, and the reverse primer 5′-TGACCGCTGCAGttaTTTTTTCTTTTTTCTCC-3′, which introduced a PstI site (underlined) immediately downstream from the pgaD stop codon (lowercase). PCRs were carried out as previously described (19). The PCR product was digested with BamHI and PstI and ligated into the BamHI/PstI sites of plasmid LITMUS28, resulting in plasmid pVK80 (Table 1). The DNA sequence of the insert from pVK80 (1,532 bp) was 99.3% identical (with six silent changes and five replacement changes) to that of the pgaCD region from strain HK1651 (Actinobacillus Genome Sequencing Project [http://www.genome.ou.edu/act.html]). Plasmid pVK80 was mutagenized by random in vitro insertion of the kanamycin resistance transposon R6Kγori/KAN by using an EZ-TN transposon insertion kit (Epicentre). Two mutant plasmids, pVK81-1 and pVK81-5, which contained transposon insertions in the middle and at the end of pgaC, respectively, were used to transform strain IDH781N to kanamycin resistance using a natural transformation protocol provided by Mrinal Bhattacharjee and David Figurski (Columbia University). Recombination of the transposons into the chromosomes of the transformants (designated JK1048 and JK1049) (Fig. 1) was confirmed by PCR using the forward primer 5′-GACGGTGATGCGGTATTGG-3′ and the reverse primer 5′-CCTCTATCCGGGCTGGTCCATAC-3′ (corresponding to bp 412 to 430 and 1207 to 1229 in the sequence with GenBank accession no. AY438014, respectively), which flanked the transposon insertion sites, and by DNA sequence analysis of the transposon junctions.
FIG. 1.
Genetic map of the pgaABCD locus of A. actinomycetemcomitans and A. pleuropneumoniae. The open arrows indicate open reading frames and directions of transcription. Gene names are indicated above. The arrows below pgaC indicate the locations and directions of transcription of transposon insertions in two A. actinomycetemcomitans mutant strains. The double arrow indicates the region cloned into plasmids pVK82 and pVK93. Scale bar = 1 kb.
Genetic complementation of A. actinomycetemcomitans pgaC.
The BamHI/PstI insert from pVK80 was ligated into the BamHI/PstI sites of the broad-host-range plasmid pJAK16, which placed pgaCD under the control of an IPTG-inducible tac promoter. The resulting plasmid (pVK82), or pJAK16 as a control, was mobilized into A. actinomycetemcomitans strain JK1048 by using the RK2 oriT-defective mutant plasmid pRK21761 as previously described (35). Plasmid-harboring strains were grown in media supplemented with chloramphenicol and IPTG.
Expression of A. actinomycetemcomitans pgaCD in E. coli.
Plasmid pVK82, or plasmid pJAK16 as a control, was transformed into E. coli strain DH5α. Transformants were incubated in broth for 16 h, diluted 1:50 in 3 ml of fresh broth, and incubated for an additional 5 h until the optical density of the culture (at 590 nm) reached 0.5. IPTG was added to a final concentration of 1 mM, and the cultures were incubated for an additional 2 h. Cells were harvested by centrifugation, washed three times with phosphate-buffered saline (PBS), and resuspended in 1 ml of PBS. The cells were treated with dispersin B as described below.
Expression of A. pleuropneumoniae pgaCD in E. coli.
Genomic DNA isolated from A. pleuropneumoniae strain IA1 was amplified by PCR using the forward primer 5′-TGACCGGATCCTCAAGCAGGTAAACCATAGGGATTGGTatgATTCTAGAAATATTCAG-3′, which inserts a BamHI restriction site (underlined) 27 bp upstream from the pgaC start codon (lowercase), and the reverse primer 5′-TGACCGCTGCAGttaAGGTTTTTTATGGCGAC-3′, which inserts a PstI restriction site (underlined) downstream from the pgaD stop codon (lowercase). The PCR product was digested with BamHI and PstI and ligated into the BamHI/PstI sites of pJAK16, resulting in plasmid pVK93. The DNA sequence of the insert from pVK93 was 99.6 to 100% identical to pgaCD homologues from four other A. pleuropneumoniae strains (representing serotypes 1, 5, and 7) in the NCBI public database. Plasmid pVK93 was transformed into E. coli strain DH5α, and transformants were cultured and induced with IPTG as described above.
Expression of S. epidermidis icaAD in E. coli.
Genomic DNA isolated from S. epidermidis strain NJ9709 was amplified by PCR using the forward primer5′-TGACCGGATCCTCAAGCAGGTAAACCATAGGGATTGGTatgCATATTTTTAACTTTTTAC-3′, which inserts a BamHI restriction site (underlined) 27 bp upstream from the icaA start codon (lowercase), and the reverse primer 5′-TGACCGCTGCAGtcaTATGTCACGACCTTTC-3′, which inserts a PstI restriction site (underlined) downstream from the icaD stop codon (lowercase). The PCR product was digested with BamHI and PstI and ligated into the BamHI/PstI sites of pJAK16, resulting in plasmid pVK91. Plasmid pVK91 was transformed into E. coli strain DH5α, and transformants were cultured and induced with IPTG as described above.
A. actinomycetemcomitans dispersin B purification and treatment.
A. actinomycetemcomitans dispersin B was purified as previously described (20). The protein concentration was determined by using a Bio-Rad Protein Assay kit. The specific activity of the enzyme was 970 U per mg of protein, where 1 U of enzyme activity was defined as the amount of enzyme needed to hydrolyze 1 μmol of 4-nitrophenyl-β-d-N-acetylglucosaminide to 4-nitrophenol and N-acetylglucosamine per min at 25°C in 50 mM sodium phosphate buffer (pH 4.5)-100 mM NaCl (20).
Bacterial cells were treated with 40 μg of dispersin B/ml in PBS for 30 min at 30°C. The cells were pelleted by centrifugation, and the supernatant was assayed for total hexosamine as described below. Crude polysaccharide samples were treated with 40 μg of dispersin B/ml in PBS for 30 min at room temperature. The samples were reprecipitated with sodium dodecyl sulfate (SDS) as described below, incubated at 100°C for 3 min, and then centrifuged. The supernatants were analyzed for total hexosamine as described below.
Expression and purification of A. pleuropneumoniae dispersin B.
Genomic DNA isolated from A. pleuropneumoniae strain IA1 was amplified by PCR using the forward primer 5′-GCTAGTCCATGGACTTACCTAAAAAAGAAAGCG-3′, which inserts an NcoI restriction site (underlined) at codon 26 of dspB, and the reverse primer 5′-GCAGGGATCCctaGTGGTGGTGGTGGTGGTGATGCGATTTCGGATCATTAG-3′, which inserts a BamHI restriction site (underlined) and six His codons (italics) flanking the dspB stop codon (lowercase). The PCR product was digested with NcoI and BamHI and ligated into the NcoI/BamHI sites of the T7 expression plasmid pET-28a, resulting in plasmid pRC2. The DNA sequence of the insert from pRC2 was 99.7 to 100% identical to sequences of dspB homologues from four other A. pleuropneumoniae strains in the NCBI public database. Plasmid pRC2, which encoded dispersin B (minus its leader peptide) fused to a hexahistidine metal-binding site at its C terminus, was transformed into E. coli strain BL21(DE3), and the fusion protein was purified by using Ni affinity chromatography as previously described (20).
Surface attachment and biofilm formation assays.
To quantitate surface attachment, 100-μl aliquots of serial twofold dilutions of inocula were transferred to the wells of a 96-well polystyrene microtiter plate (model 324662; Falcon) and incubated for 4 h. The wells were then washed with water and stained with crystal violet as previously described (16). The bound dye was solubilized in 100 μl of 100% ethanol, and the optical density of the ethanol-dye solution was measured using a Bio-Rad Benchmark microtiter plate reader set to 590 nm. To measure biofilm formation, cells were grown in 96-well microtiter plates for 16 h and then stained and quantitated as described above. Surface attachment and biofilm formation assays were performed three times with similar results.
Biofilm detachment assays.
Biofilms were grown for 16 h in the wells of a 96-well microtiter plate as described above. The biofilms were washed with water and then treated for 2 h at 37°C with 10 mM sodium metaperiodate in 50 mM sodium acetate buffer (pH 4.5) or with 100 μg of proteinase K (Sigma)/ml in 20 mM Tris (pH 7.5)-100 mM NaCl. After treatment, the biofilms were washed with water, stained with crystal violet, and quantitated as described above. Biofilm detachment assays were performed three times with similar results.
Autoaggregation assay.
One well of a six-well microtiter plate (model 353046; Falcon) was filled with 4 ml of fresh broth containing 107 CFU of bacteria/ml, and the plate was incubated for 16 h. The biofilm that formed on the surface of the well was rinsed with PBS and then scraped from the surface into 1 ml of PBS by using a cell scraper. A total of 200 μl of the cell suspension was transferred to a 0.5-ml polypropylene tube (model 6530; Corning), and the tube was vortexed at high speed for 10 s, incubated statically for 5 min, and photographed.
Immunofluorescence microscopy.
Aliquots of bacterial-cell suspensions (15 μl each) were heat fixed onto printed glass microscope slides, soaked in ice-cold methanol for 2 min, and then air dried. The fixed cells were incubated with 15 μl of anti-S. epidermidis PIA antiserum or the corresponding preimmune serum (diluted 1:50 in PBS) for 30 min at 37°C in a wet chamber (23). The slides were washed three times for 2 min each time with PBS and then air dried. Bound antibodies were detected using a fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin G antibody (Sigma) diluted 1:80 in PBS. After incubation for 30 min at 37°C in a wet chamber, the slides were washed twice with PBS and once with double-distilled water for 2 min. The slides were then air dried and viewed under a Zeiss fluorescence microscope at a magnification of ×1,000. PIA-producing S. epidermidis strain 1457 and its PIA-negative icaA insertion mutant, 1457-M11, were used as positive and negative controls, respectively (23).
Isolation of crude polysaccharide.
Confluent biofilms were grown in 100-mm-diameter tissue-culture-treated polystyrene petri dishes (model 430167; Corning) as previously described (17). The biofilms were washed with PBS, scraped from the surface into 5 ml of PBS with a cell scraper, and then transferred to a 15-ml conical centrifuge tube. The cells were pelleted by centrifugation for 10 min at 4,000 rpm in a Sorvall model RT7 centrifuge. The cell pellet was resuspended in 0.5 ml of 50 mM sodium acetate buffer (pH 5.8)-100 mM NaCl and then transferred to a 1.5-ml microcentrifuge tube. An equal volume of 10 mM Tris-HCl (pH 8.0)-5 mM EDTA-0.5% SDS was added, and the tube was vortexed briefly and incubated at 100°C for 3 min. The tube was centrifuged for 10 min at 15,000 rpm in a Sorvall MC12V microcentrifuge, and the supernatant was decanted. The pellet contained the crude polysaccharide. The yield was ∼300 μg of crude polysaccharide per mg (wet weight) of cells.
SDS-polyacrylamide gel electrophoresis analysis.
Polyacrylamide gels consisted of a 10-mm-long stacking gel (5% acrylamide) and a 35-mm-long running gel (15% acrylamide). The buffer system of Laemmli was employed (22). Prior to electrophoresis, crude polysaccharide samples (10 μg each) were resuspended in 25 mM Tris-HCl (pH 8.0)-5 mM β-mercaptoethanol-0.5% SDS and incubated at 100°C for 10 min. Samples were loaded directly onto the gel without centrifugation. After electrophoresis, the polysaccharides were stained with silver using a Bio-Rad Silver Stain kit.
Total hexosamine assay.
After centrifugation, total hexosamine was measured in the supernatant by using the Morgan-Elson colorimetric assay (33). All assays were performed at least three times with similar results.
Nucleotide sequence accession numbers.
The DNA sequence of pgaCD from A. actinomycetemcomitans strain CU1000 was deposited in GenBank under accession no. AY438014. The DNA sequences of pgaCD and dspB from A. pleuropneumoniae strain IA1 were deposited under accession no. AY618480 and AY618481, respectively.
RESULTS
A. actinomycetemcomitans pgaC mutants are defective in exopolysaccharide production.
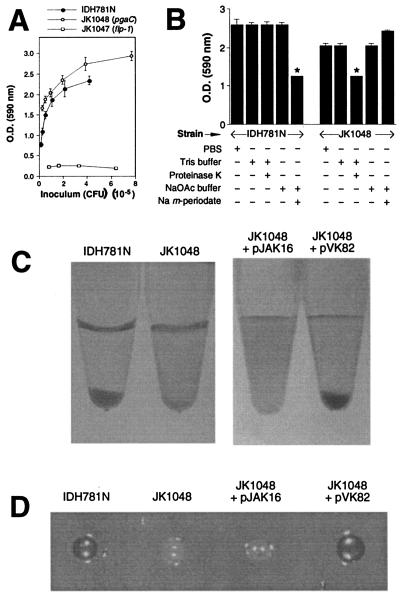
We constructed two isogenic mutants of A. actinomycetemcomitans strain IDH781N that contained transposon insertions in pgaC by using in vitro transposon mutagenesis, natural transformation, and homologous recombination (Fig. 1). The mutant strain JK1048, which contained a transposon insertion in the middle of pgaC, bound to polystyrene with the same avidity as the wild-type strain (Fig. 2A). Strain JK1047, which contains a transposon insertion in a gene (flp-1) required for type IV pilus production and biofilm formation (16), failed to bind to polystyrene. Unlike biofilms produced by the wild-type strain IDH781N, JK1048 biofilms were resistant to removal by the carbohydrate-modifying agent sodium metaperiodate and sensitive to detachment by proteinase K (Fig. 2B). These results are consistent with the hypotheses that JK1048 was deficient in exopolysaccharide production and that expression of pgaC was not required for surface attachment. Mutant JK1049, which contained a transposon insertion near the 3′ end of pgaC (Fig. 1), displayed the same adherence and autoaggregation phenotypes displayed by strain JK1048 (data not shown).
FIG. 2.
Phenotypes of wild-type A. actinomycetemcomitans strain IDH781N and pgaC mutant JK1048. (A) Surface attachment was measured in 96-well microtiter plates by staining with crystal violet. The optical density (O.D.) at 590 nm is proportional to the number of attached cells in the well. Strain JK1047 contains a transposon insertion in flp-1 which results in a complete loss of pilus production and surface attachment (16). The values indicate means (± standard deviations) for triplicate wells. (B) Detachment of preformed biofilms by proteinase K and sodium metaperiodate as measured by crystal violet staining. The values indicate means (plus standard deviations) for triplicate wells. The asterisks indicate values that were significantly less than the those of corresponding mock-treated control (P < 0.05; unpaired two-tailed Student t test). +, present; −, absent. (C) Autoaggregation of IDH781N and JK1048 (left) and of JK1048 carrying the vector plasmid pJAK16 or the complementary plasmid pVK82 (right). (D) Colonies of strains IDH781N and JK1048 and of strain JK1048 carrying the vector plasmid pJAK16 or the complementary plasmid pVK82 on Congo red agar. The dark colonies were red, and the light colonies were white. The colonies were ∼3 mm in diameter.
To test whether pgaC was involved in intercellular adhesion, we grew biofilms in polystyrene petri dishes and then scraped the biofilm cells from the polystyrene surface and transferred them to a tube (Fig. 2C, left). Five minutes after being vortexed, cells of the wild-type strain had settled to the bottom of the tube, whereas JK1048 cells remained in suspension. After 15 min, JK1048 cells had begun to settle, whereas cells of the flp-1 mutant JK1047 remained in suspension for long periods of time (data not shown). A plasmid carrying the wild-type A. actinomycetemcomitans pgaCD genes (pVK82) restored the ability of JK1048 cells to rapidly autoaggregate (Fig. 2C, right). These findings indicate that strain JK1048 exhibited a reduced autoaggregation phenotype.
To demonstrate that the pgaC mutant was deficient in exopolysaccharide synthesis, we grew wild-type strain IDH781N and mutant strain JK1048 on nutrient agar containing Congo red dye, which has an affinity for bacterial exopolysaccharides (1, 9, 39). Strain IDH781N produced red colonies on Congo red agar, whereas strain JK1048 produced white colonies (Fig. 2D). Plasmid pVK82 restored the ability of JK1048 colonies to bind Congo red dye.
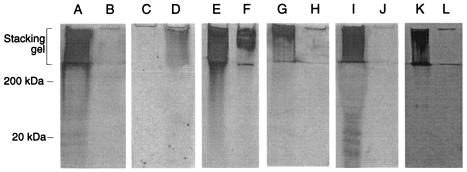
To confirm that the pgaC mutant was deficient in exopolysaccharide production, we analyzed crude polysaccharide isolated from the wild-type strain IDH781N and the mutant JK1048 by polyacrylamide gel electrophoresis (Fig. 3). Strain IDH781N produced an abundant, high-molecular-weight material that was retained in the stacking gel during electrophoresis (lane A). This material was absent from cells of strain JK1048 (lane B), and its production was restored by transformation of JK1048 with the complementary plasmid pVK82 (lanes C and D). This material was resistant to extraction by phenol (lanes E and F) and to digestion by proteinase K (data not shown), consistent with the hypothesis that it contained primarily polysaccharide. The material was degraded by dispersin B (lanes G and H) and was absent from IDH781N cells that were pretreated with dispersin B (lanes I and J). Cells of a different serotype of A. actinomycetemcomitans (strain CU1000) also produced the high-molecular-weight material (lane K), which was degraded by dispersin B (lane L). These findings indicate that A. actinomycetemcomitans pgaC encodes the production of a high-molecular-weight extracellular polysaccharide that is a substrate for dispersin B.
FIG. 3.
SDS-polyacrylamide gel electrophoresis analysis of A. actinomycetemcomitans crude polysaccharides. The location of the stacking gel and the approximate positions of molecular mass standards electrophoresed in an adjacent lane are indicated on the left. Polysaccharides were stained with silver. (A and B) Polysaccharide isolated from strain IDH781N (lane A) and pgaC mutant JK1048 (lane B). (C and D) Polysaccharide isolated from strain JK1048 harboring plasmid vector pJAK16 (lane C) or the complementary plasmid pVK82 (lane D). (E and F) Untreated polysaccharide isolated from strain IDH781N (lane E) and the same sample extracted with phenol (lane F). (G and H) Polysaccharide isolated from strain IDH781N was mock treated (lane G) or treated with dispersin B (lane H) prior to electrophoresis. (I and J) Polysaccharide isolated from IDH781N cells that were mock treated (lane I) or pretreated with dispersin B (lane J) prior to polysaccharide purification. (K and L) Polysaccharide isolated from CU1000 cells that were mock treated (lane K) or pretreated with dispersin B (lane L) prior to polysaccharide purification.
Enzymatic treatment of exopolysaccharide suggests the presence of a hexosamine-containing component.
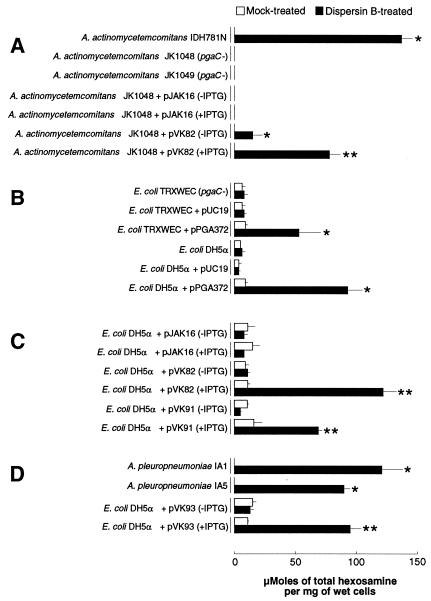
We treated A. actinomycetemcomitans cells with dispersin B and analyzed the cell supernatants for the presence of total hexosamine (Fig. 4A). Supernatants from mock-treated IDH781N cells contained no detectable hexosamine (<10 nmol per mg [wet weight] of cells), whereas supernatants from dispersin B-treated IDH781N contained high levels. Treatment of crude polysaccharide isolated from strain IDH781N resulted in the release of a similar amount of total hexosamine (126 ± 12 μmol per mg of polysaccharide). Cells of the pgaC mutant strain JK1048 released no detectable levels of total hexosamine, even after treatment with dispersin B (Fig. 4A). The complementary plasmid pVK82 restored the ability of JK1048 cells to release hexosamine after treatment with dispersin B. These data indicate that the pgaC-dependent exopolysaccharide produced by A. actinomycetemcomitans contains a hexosamine component.
FIG. 4.
Total hexosamine contents of supernatants from mock-treated and dispersin B-treated bacterial cells. (A) Wild-type A. actinomycetemcomitans strain IDH781N and isogenic pgaC mutants JK1048 and JK1049. Plasmid pVK82 contains the wild-type A. actinomycetemcomitans pgaCD genes. (B) Expression of E. coli pgaABCD(pPGA372) in two strains of E. coli. (C) Expression of A. actinomycetemcomitans pgaCD(pVK82) and S. epidermidis icaAD(pVK91) in E. coli DH5α. Values are means and ranges for duplicate samples. (D) Two wild-type strains of A. pleuropneumoniae and E. coli DH5α expressing the A. pleuropneumoniae pgaCD genes on a plasmid (pVK93). The asterisks indicate values that were significantly greater than the corresponding mock-treated control (P < 0.05; unpaired two-tailed Student t test). The double asterisks indicate values that were significantly greater than both the corresponding mock-treated control and the corresponding non-IPTG-induced control (P < 0.05).
Evidence that the A. actinomycetemcomitans pgaC-dependent polysaccharide is structurally related to E. coli PGA and S. epidermidis PIA.
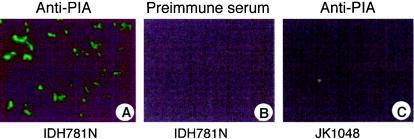
E. coli strains DH5α and TRXWEC (pgaC mutant) were transformed with plasmid pPGA372, which contains the E. coli pgaABCD locus and promoter region (37). The transformants released high levels of hexosamine when treated with dispersin B (Fig. 4B). The levels of hexosamine released by strains DH5α and TRXWEC harboring the vector plasmid pUC19 were similar, suggesting that pgaC is absent or not expressed in strain DH5α. The level of hexosamine released by DH5α harboring pPGA372 was similar to the level released by IPTG-induced DH5α cells harboring pVK82 (containing A. actinomycetemcomitans pgaCD) or pVK91 (containing S. epidermidis icaAD) (Fig. 4C). These findings suggest that E. coli pgaC, A. actinomycetemcomitans pgaC, and S. epidermidis icaA encode the production of similar hexosamine-containing polysaccharide substrates for dispersin B. In addition, wild-type A. actinomycetemcomitans IDH781N cells, but not cells of the pgaC mutant JK1048, cross-reacted with antiserum raised against S. epidermidis PIA (Fig. 5). Taken together, these data suggest that the pgaC-dependent polysaccharide produced by A. actinomycetemcomitans is structurally related to E. coli PGA and S. epidermidis PIA.
FIG. 5.
Detection of exopolysaccharide by immunofluorescence microscopy. (A) Wild-type strain IDH781N with anti-PIA antiserum. (B) Strain IDH781N with preimmune serum. (C) pgaC mutant strain JK1048 with anti-PIA antiserum.
Evidence that A. pleuropneumoniae biofilms produce a hexosamine-containing extracellular polysaccharide.
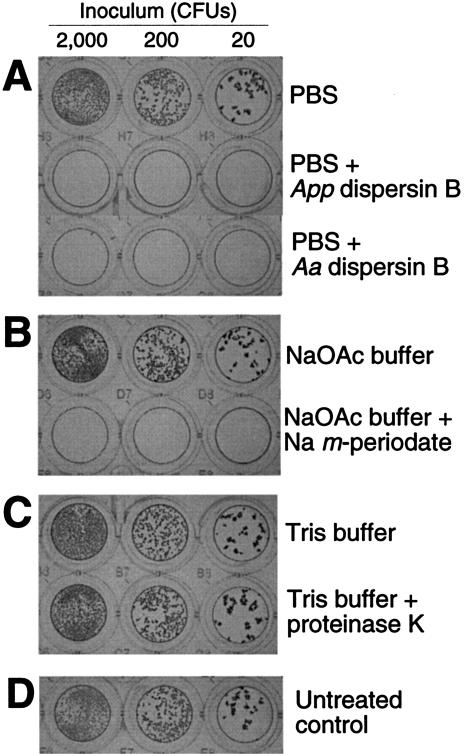
Two different wild-type strains of A. pleuropneumoniae released high levels of total hexosamine when treated with A. actinomycetemcomitans dispersin B (Fig. 4D), suggesting that A. pleuropneumoniae cells produce a hexosamine-containing extracellular polysaccharide similar to that produced by A. actinomycetemcomitans. The genome of A. pleuropneumoniae contains a locus that is homologous to A. actinomycetemcomitans pgaABCD (46, 46, 68, and 31% amino acid identity, respectively). When E. coli DH5α cells carrying the A. pleuropneumoniae pgaCD genes on a plasmid (pVK93) were treated with dispersin B, high levels of total hexosamine were released (Fig. 4D). These data suggest that the A. pleuropneumoniae pgaCD genes are orthologous to E. coli and A. actinomycetemcomitans pgaCD. The genome of A. pleuropneumoniae also contains a homologue of A. actinomycetemcomitans dspB (59% amino acid identity). Purified recombinant A. pleuropneumoniae DspB protein caused the detachment of A. pleuropneumoniae biofilms from surfaces (Fig. 6A). A. actinomycetemcomitans dispersin B also caused the detachment of A. pleuropneumoniae biofilms from surfaces (Fig. 6A), and A. pleuropneumoniae DspB caused the detachment of A. actinomycetemcomitans and S. epidermidis biofilms from surfaces (data not shown). These data suggest that A. pleuropneumoniae DspB exhibits the same substrate specificity as A. actinomycetemcomitans dispersin B. Like A. actinomycetemcomitans biofilms, biofilms produced by wild-type strains of A. pleuropneumoniae were sensitive to detachment by sodium metaperiodate and resistant to removal by proteinase K (Fig. 6B and C). Taken together, these data suggest that A. pleuropneumoniae biofilm cells produce a hexosamine-containing polysaccharide intercellular adhesin that is structurally and functionally related to the pgaC-dependent polysaccharide produced by A. actinomycetemcomitans.
FIG. 6.
Detachment of 18-h-old biofilm colonies of A. pleuropneumoniae strain IA5 grown in a 96-well microtiter plate. Biofilm colonies were treated with PBS with or without A. pleuropneumoniae (App) or A. actinomycetemcomitans (Aa) dispersin B (A); 50 mM sodium acetate (NaOAc) buffer, pH 4.8, with or without 10 mM sodium metaperiodate (B); 10 mM Tris-HCl (pH 7.6)-100 mM NaCl with or without 10 μg of proteinase K/ml (C). (D) Untreated controls. Biofilms were stained with crystal violet as previously described (16).
DISCUSSION
Our findings demonstrate that the pgaABCD locus of A. actinomycetemcomitans encodes the production of a high-molecular-weight, hexosamine-containing extracellular polysaccharide adhesin that is a substrate for dispersin B. This polysaccharide is structurally and functionally related to E. coli PGA and S. epidermidis PIA, suggesting that it is composed of a linear polymer of GlcNAc residues in β(1,6) linkage and that this linkage is the natural substrate for dispersin B. Because of the genetic similarity between the pgaABCD loci of A. actinomycetemcomitans and those of E. coli, this polysaccharide is referred to here as A. actinomycetemcomitans PGA.
Our data indicate that PGA is a component of A. actinomycetemcomitans biofilm colonies. Our findings suggest that PGA may play a role in intercellular adhesion within the biofilm colony and detachment of cells from the colony (17, 18, 20). PGA does not appear to play a major role in A. actinomycetemcomitans surface attachment (Fig. 2A) or biofilm colony formation (J. B. Kaplan, unpublished data), phenotypes that have been shown to be mediated by proteinaceous, adhesive type IV pili (5, 6, 14-16). The fact that pgaC mutants were sensitive to detachment by proteinase K (Fig. 2B) suggests that PGA may also function as a physical and/or chemical barrier which prevents the access of external agents to the bacterial cells and adhesive pili within the biofilm (3). PGA may account for the increased resistance to antimicrobial agents exhibited by A. actinomycetemcomitans biofilm cells compared to that exhibited by cells grown in planktonic form (4). A similar protective function was ascribed to S. epidermidis PIA (36).
Our findings constitute the first report of biofilm formation in A. pleuropneumoniae. Our data suggest that the A. pleuropneumoniae pgaCD and dspB genes are orthologues of A. actinomycetemcomitans pgaCD and dspB, respectively. The similar detachment phenotypes exhibited by biofilms produced by wild-type A. pleuropneumoniae and A. actinomycetemcomitans strains (Fig. 2B and 6B and C) further suggest that A. pleuropneumoniae forms biofilms by mechanisms analogous to those employed by A. actinomycetemcomitans. Biofilm formation has been shown to play an important role in the colonization and pathogenesis of A. actinomycetemcomitans (7, 31). Biofilm formation may also have relevance to the colonization, pathogenesis, and transmission of A. pleuropneumoniae.
Homologues of the pgaABCD locus are present in the genomes of several other beta and gamma Proteobacteria, including Bordetella pertussis, Bordetella parapertussis, Bordetella bronchiseptica, Burkholderia cepacia, Chromobacterium violaceum, Photobacterium profundum, Pseudomonas fluorescens, Ralstonia solanacearum, Xanthomonas axonopodis, Yersinia pestis, Yersinia enterocolitica, and Yersinia pseudotuberculosis. Our findings support the hypothesis that these homologous loci encode the production of hexosamine-containing exopolysaccharides that stabilize biofilms in these species (37).
Acknowledgments
We thank Mrinal Bhattacharjee and David Figurski (Columbia University) for providing the protocols and accompanying bacterial strains and plasmids used for natural transformation of A. actinomycetemcomitans; Xin Wang and Tony Romeo (Emory University) for providing E. coli strains and plasmids; Lorraine Hoffman and Tim Klinefelder (Iowa State University) for providing A. pleuropneumoniae strains; David Furgang for technical assistance; Bruce Roe, Fares Najar, Allison Gillaspy, Sandra Clifton, Tom Ducey, Lisa Lewis, and Dave Dyer of the Actinobacillus Genome Sequencing Project for providing sequence data prior to publication; and Daniel Fine for helpful assistance throughout the course of the project.
This work was supported in part by Public Health Service awards DE15124 (to J.B.K.) and DE12585 (to N.R.) and a grant from the Deutsche Forschungsgemeinschaft (SFB 470 Teilprojekt C10) to D.M.
REFERENCES
- 1.Arnold, J. W., and L. J. Shimkets. 1988. Inhibition of cell-cell interactions in Myxococcus xanthus by Congo red. J. Bacteriol. 170:5765-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldassarri, L., G. Donelli, A. Gelosia, M. C. Voglino, A. W. Simpson, and G. D. Christensen. 1996. Purification and characterization of the staphylococcal slime-associated antigen and its occurrence among Staphylococcus epidermidis clinical isolates. Infect. Immun. 64:3410-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 4.Fine, D. H., D. Furgang, and M. L. Barnett. 2001. Comparative antimicrobial activities of antiseptic mouthrinses against isogenic planktonic and biofilm forms of Actinobacillus actinomycetemcomitans. J. Clin. Periodontol. 28:697-700. [DOI] [PubMed] [Google Scholar]
- 5.Fine, D. H., D. Furgang, J. Kaplan, J. Charlesworth, and D. H. Figurski. 1999. Tenacious adhesion of Actinobacillus actinomycetemcomitans strain CU1000 to salivary-coated hydroxyapatite. Arch. Oral Biol. 44:1063-1076. [DOI] [PubMed] [Google Scholar]
- 6.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145:1335-1347. [DOI] [PubMed] [Google Scholar]
- 7.Fine, D. H., P. Goncharoff, H. Schreiner, K. M. Chang, D. Furgang, and D. H. Figurski. 2001. Colonization and persistence of rough and smooth colony variants of Actinobacillus actinomycetemcomitans in the mouths of rats. Arch. Oral Biol. 46:1065-1078. [DOI] [PubMed] [Google Scholar]
- 8.Flemming, H.-C., J. Wingender, T. Griegbe, and C. Mayer. 2000. Physico-chemical properties of biofilms, p. 19-34. In L. V. Evans (ed.), Biofilms: recent advances in their study and control. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 9.Freeman, D. J., F. R. Falkiner, and C. T. Keane. 1989. New method for detecting slime production by coagulase-negative staphylococci. J. Clin. Pathol. 42:872-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerke, C., A. Kraft, R. Sussmuth, O. Schweitzer, and F. Götz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide adhesin. J. Biol. Chem. 273:18586-18593. [DOI] [PubMed] [Google Scholar]
- 11.Haesbrouck, F., K. Chiers, I. Van Overbeke, and R. Ducatelle. 1997. Actinobacillus pleuropneumoniae infections in pigs: the role of virulence factors in pathogenesis and protection. Vet. Microbiol. 58:239-249. [DOI] [PubMed] [Google Scholar]
- 12.Haubek, D., K. Poulsen, S. Asikainen, and M. Kilian. 1995. Evidence for absence in northern Europe of especially virulent clonal types of Actinobacillus actinomycetemcomitans. J. Clin. Microbiol. 33:395-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Götz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 14.Kachlany, S. C., P. J. Planet, M. K. Bhattacharjee, E. Kollia, R. DeSalle, D. H. Fine, and D. H. Figurski. 2000. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in Bacteria and Archea. J. Bacteriol. 182:6169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, and D. H. Figurski. 2001. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 9:429-437. [DOI] [PubMed] [Google Scholar]
- 16.Kachlany, S. C., P. J. Planet, R. DeSalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40:542-554. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan, J. B., and D. H. Fine. 2002. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Appl. Environ. Microbiol. 68:4943-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan, J. B., M. F. Meyenhofer, and D. H. Fine. 2003. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J. Bacteriol. 185:1399-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan, J. B., M. B. Perry, L. L. MacLean, D. Furgang, M. E. Wilson, and D. H. Fine. 2001. Structural and genetic analyses of O polysaccharide from Actinobacillus actinomycetemcomitans serotype f. Infect. Immun. 69:5375-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan, J. B., C. Ragunath, K. Velliyagounder, D. H. Fine, and N. Ramasubbu. 2004. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 48:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Mack, D., K. Bartscht, C. Fischer, H. Rohde, C. de Grahl, S. Dobinsky, M. A. Horstkotte, K. Keil, and J. K. M. Knobloch. 2001. Genetic and biochemical analysis of Staphylococcus epidermidis biofilm accumulation. Methods Enzymol. 336:215-239. [DOI] [PubMed] [Google Scholar]
- 24.Mack, D., W. Fisher, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mack, D., M. Nedelmann, A. Krokotsch, A. Schwartzkopf, J. Heesemann, and R. Laufs. 1994. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 62:3244-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mah, T.-F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 28.Maira-Litrán, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark, D. A. Goldman, and G. B. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maira-Litrán, T., A. Kropec, D. Goldman, and G. B. Pier. 2004. Biologic properties and vaccine potential of the staphylococcal poly-N-acetyl glucosamine surface polysaccharide. Vaccine 22:872-879. [DOI] [PubMed] [Google Scholar]
- 30.McKenney, D., J. Hübner, E. Muller, Y. Wang, D. A. Goldman, and G. B. Pier. 1998. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66:4711-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreiner, H. C., K. Sinatra, J. B. Kaplan, D. Furgang, S. C. Kachlany, P. J. Planet, B. A. Perez, D. H. Figurski, and D. H. Fine. 2003. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc. Natl. Acad. Sci. USA 100:7295-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 33.Strominger, J. L., J. T. Park, and R. E. Thompson. 1959. Composition of the cell wall of Staphylococcus aureus: its relation to the mechanism of action of penicillin. J. Biol. Chem. 234:3263-3268. [PubMed] [Google Scholar]
- 34.Sutherland, I. W. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3-9. [DOI] [PubMed] [Google Scholar]
- 35.Thomson, V. J., M. K. Bhattacharjee, D. H. Fine, K. M. Derbyshire, and D. H. Figurski. 1999. Direct selection of IS903 transposon insertions by use of a broad-host-range vector: isolation of catalase-deficient mutants of Actinobacillus actinomycetemcomitans. J. Bacteriol. 181:7298-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vuong, C., J. M. Voyich, E. R. Fischer, K. R. Braughton, A. R. Whitney, F. R. DeLeo, and M. Otto. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6:269-275. [DOI] [PubMed] [Google Scholar]
- 37.Wang, X., J. F. Preston, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]
- 39.Zogaj, X., W. Bokranz, M. Nimtz, and U. Römling. 2003. Production of cellulose and curli fimbriae by members of the family Enterobactericeae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]