Abstract
Comparative sequence analysis of IS50 transposase-related protein sequences in conjunction with known structural, biochemical, and genetic data was used to determine domains and residues that play key roles in IS50 transposase function. BLAST and ClustalW analyses have been used to find and analyze six complete protein sequences that are related to the IS50 transposase. The protein sequence identity of these six homologs ranged from 25 to 55% in comparison to the IS50 transposase. Homologous motifs were found associated with each of the three catalytic residues. Residues that play roles in transposase-DNA binding, protein autoregulation, and DNA hairpin formation were also found to be conserved in addition to other residues of unknown function. On the other hand, some homologous sequences did not appear to be competent to encode the inhibitor regulatory protein. The results were also used to compare the IS50 transposase with the more distantly related transposase encoded by IS10.
Transposable elements have played an important role in generating genetic diversity through their ability to cause insertions, deletions, inversions, and chromosome fusions. These elements are ubiquitous, and in some cases transposable elements and their inactive descendants can constitute the majority of a genome (12). The presence of a transposable element in a given genome is the product of lateral gene transfer. In bacteria, for instance, transposable elements may be introduced by means of DNA transformation, plasmid transfer, or phage infection. If an element was acquired recently by a given genome, it may well be absent from the genomes of close relatives.
One class of transposable element (“cut and paste” transposons) moves from one DNA molecule to another (or from site to site within a DNA molecule) by a process that involves excision of the transposon DNA from the original site and integration of the transposon DNA into the second site. These DNA excision and integration processes are catalyzed by a transposon-specific protein called a transposase (26). Understanding how transposases function is of general importance because many transposases are members of a superfamily of proteins that includes human immunodeficiency virus type 1 integrase and possibly the RAG-1 protein responsible for the DNA cleavage events in the immune system V(D)J joining process. That is, this family of proteins share overall architectural properties, detailed active-site structures, and catalytic mechanisms (13, 16, 26, 33).
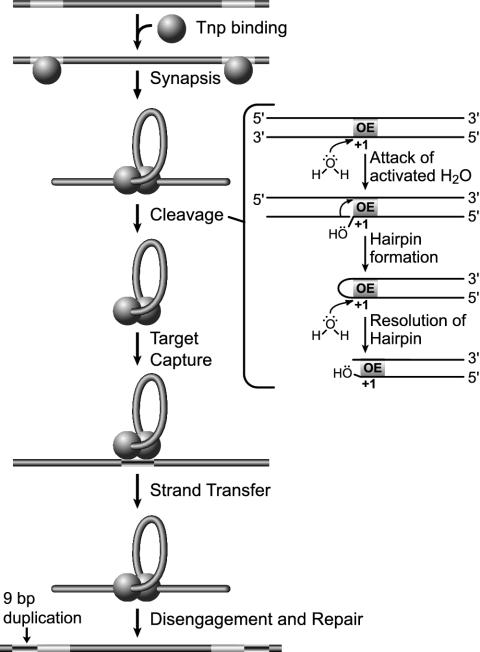
IS50 is a component of the composite transposon Tn5. IS50 transposase (also called Tn5 transposase) is an excellent model system because both biochemical and genetic data and detailed structural information are available (32, 33). As a result, the overall mechanism of IS50 transposition is well understood (see Fig. 1) and the roles of many protein residues in this process have been determined. However, structure-function studies of IS50 transposase are far from complete. For instance, the residues that participate in intramolecular regulatory contacts or transposase recognition contacts with donor DNA or target DNA are currently unknown.
FIG. 1.
IS50 transposition mechanism. IS50 transposition is a multistep process including binding of transposase monomers to the transposon end recognition sequences (so-called cis binding), dimerization of the monomeric transposase-DNA complexes to form the synaptic complex (at this stage trans transposase-DNA contacts are formed), catalysis of the three phosphoryl transfer reactions (3′-end nicking, hairpin formation, and hairpin cleavage), leading to release of the transposon DNA from the donor DNA backbone, target DNA capture, strand transfer of transposon DNA 3′ ends into the target, disengagement of the transposase molecules, and repair of the adjacent single-stranded DNA gaps. This figure is similar to one that was published before (33).
Bioinformatic analyses of IS50 transposase homologs offer powerful approaches for studying protein structure and biochemical function. For example, assuming that IS50 homologous sequences are derived from a common ancestry, the related protein sequences can be compared to infer conserved amino acid motifs that are likely of general importance. Conserved motifs can then be located on the three-dimensional structure with a particular interest in relating them to the known active site, DNA binding domains, protein-protein contact regions, etc., in order to better understand regions conserved during the evolution of IS50 transposase. These homologs could also help determine what residues are not conserved despite the fact that they appear to be important in IS50 transposase functionality. Finally, the genome databases offer some insight into the “natural” distribution and diversity of mobile elements such as IS50.
Previously, IS50 transposase had been classified as a member of the IS4 family of transposases (34). However, IS50 transposase has a limited degree of homology to other IS4 family members (such as IS10 transposase) primarily involving the so-called YREK motif around the third catalytic residue (note that the YREK motif is located within the C1 region [10, 24, 34]). A more detailed comparison between IS50 and IS10 transposases is presented later in this communication. A recent comparison of IS50 transposase to the other members of the IS4 transposase family incorrectly located the first catalytic residue for Tn5 transposase (10). Thus, this earlier work, given the low degree of similarity outside of the C1 region, would not provide the desired detailed information for elucidating structure-function characteristics.
MATERIALS AND METHODS
The IS50 transposase sequence used in these studies (accession no. AAB60064) is from Ahmed and Podemski (1). All other sequences were found through the NCBI database. The IS50 transposase homolog sequence accession numbers are listed in Table 1. The BLAST-P analysis (2) was used through the NCBI web site (http://www.ncbi.nlm.nih.gov/BLAST/) and the ClustalW analysis (37) was used through the EBI web site (http://www.ebi.ac.uk/sevices/index.html). Default parameters were applied in both cases.
TABLE 1.
Tn5 transposase ortholog sources
| Alignment | Source | Reference | Accession no. |
|---|---|---|---|
| Clustal W | Photorhabdus luminescens | 14 | NC_005126 |
| Gloeobacter violaceus | 26 | NC_005125 | |
| Wolbachia phage WO, wTai | 24 | BAA89624 | |
| Sargasso Sea isolate EAK | 38 | EAK27828 | |
| Sargasso Sea isolate EAI | 38 | EAI27958 | |
| Sargasso Sea isolate EAJ | 38 | EAJ39384 | |
| Pairwise | Vibrio vulnificus | 10 | NC_005139 |
| Legionella pneumophila | NCBI database | CUCGC_272624 | |
| Escherichia coli CFT073 | 39 | NC_004431 | |
| Escherichia coli O157:H7 | 29 | NC_002695 | |
| Chloroflexus aurantiacus | NCBI database | NZ_AAAH01000339 | |
| Synechococcus sp. strain PCC 7002 | NCBI database | jmarg_32049 | |
| Gemmata obscuriglobus | NCBI database | TIGR_214688 | |
| Nostoc punctiforme | NCBI database | NC_003272 | |
| Azotobacter vinelandii | NCBI database | AAAU02000001 | |
| Bacteroides thetaiotaomicron | 42 | NC_004663 |
The structural analyses were performed with the Chimera software (http://www.cgl.ucsf.edu/chimera [19]) and the coordinates for the IS50 transposase-DNA paired-end complex (PDB accession code 1MUH [14]).
RESULTS AND DISCUSSION
The IS50 transposase database comparison was performed in the following steps. First, IS50 transposase homologs were identified by using a BLAST-P similarity search (2) based upon the IS50 transposase amino acid sequence (1) against the 159 completed microbial and lower eukaryotic sequences that, as of 1 April 2004, had been completed and deposited in the NCBI database. The following criteria were imposed on all of the sequences that were identified in this similarity search. The cutoff score was e < 0.001, the sequences were full length (with homologous matches extending from the N terminus to the C terminus of IS50 transposase) with no nonsense codons, and no exact duplicate sequences were included. Partial sequence matches and matches from the incomplete genome database were saved and used later in the analysis.
The deliberately conservative approach of including only apparently full-length homologous sequences was used to avoid the inclusion of sequences in which the transposase may have been inactivated due to deletions. The concern was that inactivated deleted sequences could accumulate mutations altering functionally important homologies. N-terminal sequences starting at residue 26 are known to play a critical role in the required transposon DNA binding step (30), and C-terminal sequences, including residue 468 (out of 476), are known to be required for the essential transposase-transposase dimerization step (36).
With the above strategy, BLAST-P analysis revealed two full-length annotated IS50 transposase matches in the genomes of Photorhabdus luminescens (15), and Gloeobacter violaceus (27) (Table 1). The search was then expanded to include the newly released results from the random environmental Sargasso Sea sequencing project (39). From this analysis, three additional unique, full-length sequences similar to IS50 transposase were added to the database. Finally, the IS50 transposase-like sequence previously reported in the Wolbacchia prophage WO genome (25) was added to the collection.
The protein sequences of these six IS50 transposase matches were aligned with the ClustalW (37) program available through EBI. The calculated alignments are shown in Fig. 2. Several things are apparent from this analysis. First, all three of the catalytic residues are located within conserved motifs (see below and Fig. 2). These conserved motifs around the active-site residues will be discussed below. Other apparently conserved residues are also indicated in Fig. 2 and will be discussed in greater detail below.
FIG. 2.
A. Clustal W alignment of the IS50 transposase sequence versus six full-length IS50 transposase-like sequences. Active-site residues are white on a black background. Other sequence identities are boxed with * underneath. Residues with a shaded background (and either : or . underneath) denote chemical conservation. Conserved motifs that are discussed further are residues 20 to 31 (IS50 transposase numbering scheme), 38 and 39, 58 to 63, 92 to 103 (DTT), 160 to 166, 183 to 193 (DREAD), 207 to 211, 296 to 306, 319 to 335 (YREK), 342 to 351, 354 to 366, 417 to 425, 429 to 434, and 449 to 452. B. DTT, DREAD, and TYEK active-site motifs.
Finally, apparently truncated sequences were analyzed relative to IS50 transposase by pairwise comparisons. These truncated sequences from Vibrio vulnificus (11), Legionella pneumophila (NCBI database), Escherichia coli CFT073 (40), E. coli O157:H7 (30), Chloroflexus aurantiacus (NCBI database), Synechococcus sp. strain PCC 7002 (NCBI database), Gennata obscuriglobus (NCBI database), Nostoc punctiforme (NCBI database), Azotobacter vinelandii (NCBI database), and Bacteroides thetaiotaomicron (43) (Table 1) and several truncated sequences from the random environmental collection also had scores of e < 0.001. These additional sequences demonstrated the same homology matches as shown in Fig. 2 (data not shown).
It is of interest that only two protein sequences were found in our study that had a high degree (>55%) of identity to IS50 transposase. These two sequences were partial sequence fragments from the random environmental sample (39). One sequence was 147 residues long (IS50 transposase is 476 residues long) and was 98% identical to the IS50 transposase sequence. The second sequence was 100% identical to IS50 transposase but was only 65 residues long. The failure to find more close homologs to IS50 transposase is consistent with the fact that IS50 is seemingly rare, having only been isolated twice. The first isolate was from an R factor carried in a Klebsiella pneumoniae strain (5), and the second isolate was from a different R factor carried in an E. coli strain isolated from a pig (J. Davies, personal communication).
One concern about the above analysis is that the IS50 transposase homologs were not tested for functionality. However, the observation that the active-site residues and the motifs around these residues were completely conserved suggests that the homologs are functional transposases derived from shared ancestry. One exception to this probable transposase functionality involves the partial transposase-like sequences found in Wolbachia pipientis that were not used in our analysis (see below).
It is of note that the IS50 transposase homologs were found in the genomes of but a few widely divergent bacteria. This observation supports the hypothesis that the presence of transposase homologs in these genomes reflects lateral gene transfer. A particularly interesting bacterium whose genome contains fragments of an IS50 transposase-like gene is Wolbachia pipientis. Wolbachia pipientis is an obligate endosymbiont of arthropods (41) and nematodes (9) and thus would not a priori be expected to participate in the lateral transfer of genetic information. However, the Wolbachia pipientis genome is littered with mobile DNA (42), including prophages (7), that may have been the vehicle for introducing a transposable element. Consistent with this hypothesis is the observation of an IS50 transposase homolog in the WO prophage genome of Wolbachia wTai, but not in that of the related Wolbachia wKue prophage genome (25). The transposase gene contained on the prophage genome is full length and is imbedded in an IS50-like sequence (S. Bordenstein and W. Reznikoff, data not shown). In addition, there are IS50 transposase-like gene sequences contained on the Wolbachia pipientis chromosome. These are truncated and contain nonsense codons (42) and thus are likely inactive remnants of transposase genes.
Homologies found among IS50-like transposases.
In this section, the sequence homologies discovered in the Clustal W analysis and confirmed by the pairwise analyses will be specifically analyzed.
Motifs containing the three catalytic residues.
Transposase and retroviral integrase proteins are known to contain three key catalytic residues. These are the DDE (aspartate, aspartate, and glutamate) residues that chelate two Mg2+ ions necessary for the phosphoryl transfer reaction catalyses that occur in transposition (and retroviral DNA integration) (13, 18, 35, 44). In IS50 transposase these catalytic residues are known from genetic and structural studies to be D97, D188, and E326 (13, 14, 28, 31).
Comparison of the IS50 transposase sequence with the six homologs listed in Table 1 demonstrates that there are clearly conserved amino acid motifs around all three of the catalytic residues (Fig. 2). These motifs are (L/V/I) (L/F) (—) (I/L) (E/Q) (D97) (T) (T/S) (—) (L/I) (S/N/T) (F/Y) (residues 92 to 103; IS50 transposase numbering scheme) surrounding the first catalytic residue (subsequently called the DTT motif); (V/A/T) (I/V) (—) (V/I) (—) (D188) (R) (E) (A/S) (D) (I/M/L) (residues 183 to 193) surrounding the second catalytic residue (subsequently called the DREAD motif); and the (Y) (—) (—) (R) (W) (—) (I/V) (E326) (—) (—) (H) (—) (—) (—) (K) (T/S) (G) motif (residues 319 to 335) surrounding the third catalytic residue (called the YREK motif [34]). The YREK motif was previously described as an important signature within the C1 region of the IS4 family transposases (10, 24, 34).
The present communication is the first identification of the conserved DTT and DREAD motifs. The second T residue of the DTT motif is the only residue other than the catalytic residues within the DTT and DREAD motifs for which any biochemical data exists. This amino acid (T99) contacts the phosphate backbone of the recognition end sequence DNA in the IS50 transposase-end sequence DNA cocrystal structure (14). The presence of several charged residues within the DREAD motif suggests that they may interact with DNA as would occur during target and/or donor DNA binding. A role for this region in target DNA binding is supported by the recent isolation of a mutation at residue R189 that changes target sequence specificity (Goryshin and Reznikoff, unpublished results). Target DNA binding is a critical step in transposition about which little is known.
In Fig. 3, various residues mentioned above are highlighted as colored spheres in the IS50 transposase-end sequence DNA cocrystal structure. The DDE catalytic residues are presented in this and all subsequent structural figures as yellow spheres. The DTT motif is presented as orange spheres, and the DREAD motif is presented as magenta spheres. Other sequences highlighted in Fig. 2 will be discussed below, as will the YREK motif.
FIG. 3.
Molecular location of DTT and DREAD active-site motifs. The locations of the indicated residues are located on the molecular structure of the IS50 transposase bound to the precleaved recognition end DNA sequences. In this and all subsequent figures, the catalytic residues D97, D188, and E326 are shown in yellow space-filled spheres. Conserved motifs are presented in the indicated colors.
Residues 20 to 31 and 58 to 63.
The initial step in IS50 transposase-mediated transposition is the binding of monomeric transposase to the transposon end recognition sequence. These initial contacts between the transposase and the end recognition sequence DNA are primarily mediated by N-terminal amino acids and are defined as cis contacts. The X-ray cocrystal structure of the IS50 transposase-recognition end sequence paired end complex shows direct transposase-DNA contacts that include residues 26, 27, 30, 57, 58, and 62 (14). The importance of the N-terminal sequences in DNA binding is also supported by genetic studies (29, 38, 47). The Clustal W analysis of the IS50-like transposase homologs demonstrates homologies including residues 20 to 31 and 58 to 63; in other words, within the domain that makes cis contacts with the recognition end sequence DNA. Residues 20 to 31 (dark blue spheres) and 58 to 63 (medium blue spheres) are highlighted in Fig. 4. The cognate end DNA sequences that are presumably recognized by the IS50-like transposases have not been identified, but the conservation of residues within the DNA binding domain suggests a degree of homology for the DNA sequences as well.
FIG. 4.
Conserved N-terminal DNA binding motifs.
Residues 38, 39, and 449 to 452.
It has been hypothesized that a key mechanism for downregulating IS50 transposition involves intramolecular inhibitory contacts between the N terminus and the C terminus. Full-length monomeric wild-type transposase does not detectably bind transposon end DNA in vitro unless the C terminus is deleted (46) and does not detectably dimerize unless the N terminus is deleted (8). This intramolecular interaction is thought, from molecular modeling studies, to involve residue W450 with amino acids near residue 40 (32). Genetic studies support this model in that substitutions at residues 41, 42, 449, 450, 451, and 454 all manifest a hyperactive phenotype as if they decrease this inhibitory interaction (Gradman and Reznikoff, unpublished). Thus, it is of interest that residues 38, 39, and 449 to 452 are conserved among the IS50 transposase homologs, suggesting that the proposed intramolecular inhibition model is a general property of this group of transposases.
Residues 160 to 166 and 207 to 211.
Residues 160 to 166 and 207 to 211 are also conserved (Fig. 2), but there are no biochemical data to suggest a function for these amino acids. As demonstrated in Fig. 3, these residues (160 to 167, cyan spheres; 207 to 211, dark blue spheres) come in close contact with the active-site motifs DTT and DREAD. Two possible roles can be entertained to explain the conservation of these residues. They could provide important structural support for the active-site residues D97 and D188. Alternatively, or in addition, some of these residues are charged or polar and thus might interact with either donor backbone DNA during formation of the synaptic complex or with target DNA just prior to strand transfer. In fact, a mutation of residue K212 has been found that changes the target site specificity of the transposition reaction (Goryshin and Reznikoff, unpublished).
Residues 296 to 306.
Residue W298 (cyan sphere in Fig. 5) is known to play an important role in forming a stacking relationship with a flipped base at position 2 in the transposon end recognition sequence DNA (3). This is an important trans transposase-DNA contact. trans transposase contacts occur as a consequence of the second transposase monomer's binding the end recognition DNA near the cleavage site within the confines of the active site (14). The W-thymine stacking stabilizes the “flipped” base arrangement that likely destabilizes the DNA near where cleavage will occur and facilitates hairpin formation (see Fig. 1). Hairpin formation is a key step in the double-strand breakage that is characteristic of IS50 and IS10 transposon excision from donor DNA (6, 22).
FIG. 5.
Conserved C1 region. Residues 296 to 306 (except 298) are in dark blue. W298, which stabilizes the T2 flipped base, is in cyan. The YREK motif (except for the yellow 326) is in purple.
Residues 296 to 306 are also conserved among the IS50 homologs (note that W298, L300, T302, P305, V306, and E307 are also found in the IS10 transposase sequence [see Fig. 6]). It is not known what role residues 296 to 297 and 299 to 303 (blue spheres in Fig. 5) play, although they may participate in facilitating the base flipping discussed above.
FIG. 6.
Clustal W alignment of IS50 and IS10 C1 regions. The aligned residues are from 298 to 355 (Tn5 transposase numbering scheme). The conservation of the residues is indicated as described for Fig. 2A.
Residues 319 to 335.
The sequence 319 to 335 (purple spheres in Fig. 5) contains the YREK motif encompassing the catalytically critical aspartic acid. Many of the residues in this region form trans transposase-DNA contacts. trans contacts occur as the end recognition DNA sequence is placed in the active site during the formation of the dimeric synaptic complex (see Fig. 1) (14). The high conservation of this region extends to other members of the IS4 transposase family, as discussed below (34). In addition, some DNA contacts are formed by these residues that also exist in other retrovirus integrase superfamily proteins. For instance, the residue equivalent to K333 in human immunodeficiency virus type 1 integrase can be cross-linked to the tip of the human immunodeficiency virus type 1 DNA (20).
Residues 342 to 351.
Residues 342 to 351 (blue spheres in Fig. 6) from X-ray crystallographic structural analyses encompass three residues thought to form cis transposase end recognition sequence contacts (14). These residues are 342, 344, and 348. Mutations in 342 and 348 cause defects in transposition, presumably via defects in DNA binding. (Sterling and Reznikoff, unpublished data).
Residues 354 to 366, 417 to 425, and 429 to 434.
Residues 354 to 356, 417 to 425, and 429 to 434 appear to be conserved, but there exist no genetic, biochemical, or structural explanation for this conservation.
A homology that was not found in some IS50-like transposases: an AUG residue at or near residue 56
One of the key mechanisms that regulates IS50 transposase activity is the synthesis of the inhibitory regulatory protein (inhibitor or p2) (21). The inhibitor protein is thought to inactivate transposase through the formation of inactive inhibitor-transposase heterodimers (8). Inhibitor translation initiation occurs at residue 56 of the transposase sequence (23). Although the inhibitor is translated in the same reading frame as the transposase, the bulk of inhibitor production is encoded by a distinct mRNA species programmed by a promoter that overlaps and competes with the transposase mRNA promoter (23). The production of the inhibitor results in a reduction in the frequency of IS50 transposition and couples this downregulation to the Dam methylation state of the transposase promoter site and thus to DNA replication (45). The ability of IS50 transposase gene homologs to separately encode an inhibitor protein has not been experimentally studied; however, an analysis of the sequences indicates that this potential is not shared by all of the homologs, since three of the homologs (encoded in the Gloeobacter genome, the Wolbachia wTai phage WO genome, and one of the pooled environmental sample genomes) lack a methionine (or a valine) residue near position 56.
However, it should be noted that for IS50, the production of N-terminally truncated versions of transposase that potentially could inhibit transposition also occurs by means of proteolytic digestion of the transposase (38). It is quite possible that this type of proteolytic digestion of transposase also occurs for the transposase homologs that are studied in this communication.
Relationship to IS10 transposase.
The above BLAST-P analysis did not reveal IS10 transposase to be a close homolog of IS50 transposase. IS10 and IS50 transposases have been classified as belonging to distinct groups within the IS4 transposase family (10, 24). In order to investigate the relationship between IS50 and IS10 transposases, a BLAST pairwise comparison was performed and the previous IS50-IS10 transposase primary and secondary structure comparison (17) was reviewed. These two transposase proteins appear to have three different regions of interest. There is a central region (between IS50 W298 and I355) that demonstrates high homology between IS50 and IS10 transposases (30% identity) (Fig. 6). This central region is very similar to the C1 region described previously (10, 24, 34). This sequence includes the YREK motif that was used to define the IS4 transposase family and W298, which, as mentioned above, is believed to play an important role in the hairpin cleavage mechanism. N-terminal residues 1 to 297 of IS50 transposase and C-terminal residues 356 to 476 of IS50 transposase share limited homologies with IS10 transposase, although most proposed secondary structures and the DD active-site residues are shared (17). Of note is that IS10 transposase lacks the DTT and DREAD motifs as well as other regions of homology described above.
One model to explain the IS50-IS10 transposase relationship is to posit that they evolved from the same ancestral transposase sequence but that the requirements for hairpin formation as an intermediate resulted in greater conservation of the sequences around the YREK motif then for sequences in other portions of the proteins.
A second model posits that IS50 and IS10 transposases are related through gene fusion events. That is, the 298 to 355 sequence evolved from a common ancestral sequence that in one or the other transposase became fused to two sequences from a different transposase lineage. These observations also explain why the first critical catalytic D residue of IS50 transposase was misidentified in the IS4 transposase family analysis (10, 24). The significant divergence of the N-terminal and C-terminal sequences between IS50 and IS10 transposases suggests that the IS50 synaptic complex molecular structure may not be a good detailed model for the IS10 synaptic complex. Similarly, some of the detailed biochemistry deduced for IS10 transposase may not be precisely shared by IS50 transposase, with the exception that both IS50 and IS10 transposases are known to catalyze DNA strand breakage through a DNA hairpin mechanism (see Fig. 1). Notice that the sequence between W298 and I355 is thought to play a key role in the hairpin mechanism. It has recently been hypothesized that a similar sequence plays a critical role in hairpin binding by the tyrosine recombinase-like protein ResT from Borrelia burgdorferi (4).
Conclusion.
The bioinformatics analysis of IS50 transposase identified residues known from previous genetic, biochemical, and structural studies to be important for IS50 transposition. In addition the bioinformatics analysis laid the framework for future structure-function studies of this important protein. For instance, it identified the presence of motifs in which the first two catalytic residues are located and it suggested that an intramolecular regulatory N terminal-C-terminal interaction has been conserved. These and other homologies that were found will be useful in guiding site-specific mutagenesis studies. Finally, the analysis offered insights into the ecology and evolution of IS50 transposase homologs. For example, the IS50 transposase-like sequences, while rare, are distributed throughout the genomes of vastly divergent terrestrial and aquatic bacteria with both free-living and intracellular lifestyles. In addition, it identified a putative event of lateral IS50 transfer mediated by a bacteriophage in the genome of the endosymbiont Wolbachia.
Acknowledgments
The work described in this communication was performed while the senior author was a visiting scientist at the Marine Biological Laboratory. He owes special thanks to all members of the Josephine Bay Paul Center for Comparative Molecular Biology and Evolution, especially Mitchell Sogin for his hospitality and Matthew Beverly and Richard Fox for their help.
J.A. was supported by a grant from the National Science Foundation (MCB0084089) administered by W.S.R. S.R.B. held a National Research Council Research Associateship Award. W.S.R. is the Evelyn Mercer Professor of Biochemistry and Molecular Biology. Additional thanks are given to the NASA Astrobiology Institute (Cooperative Agreement NNA04CC04A to Mitchell L. Sogin) and the W. M. Keck Ecological and Evolutionary Genetics Facility within the Josephine Bay Paul Center for Comparative Molecular Biology and Evolution at the M.B.L. Molecular graphics images were produced by using the UCSF Chimera package from the Computer Graphics Laboratory, University of California, San Francisco (supported by NIH grant P41 RR-01081).
REFERENCES
- 1.Ahmed, A., and L. Podemski. 1995. The revised nucleotide sequence of Tn5. Gene 154:129-130. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped Blast and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ason, B., and W. S. Reznikoff. 2002. Mutational analysis of the base flipping event found in Tn5 transposition. J. Biol. Chem. 277:11284-11291. [DOI] [PubMed] [Google Scholar]
- 4.Bankhead, T., and G. Chaconas. 2004. Mixing active site components: A recipe for the unique enzymatic activity of a telomere resolvase. Proc. Natl. Acad. Sci. USA 101:13768-13773. [DOI] [PMC free article] [PubMed]
- 5.Berg, D. E., J. Davies, B. Allet, and J.-D. Rochaix. 1975. Transposition of R factor genes to bacteriophage λ. Proc. Natl. Acad. Sci. USA 72:3628-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhasin, A., I. Y. Goryshin, and W. S. Reznikoff. 1999. Hairpin formation in Tn5 transposition. J. Biol. Chem. 274:37021-37029. [DOI] [PubMed] [Google Scholar]
- 7.Bordenstein, S.R. and J. J. Wernegreen. 2004. Bacteriophage flux in endosymbionts (Wolbachia): lateral transfer, recombination rates, and the “intracellular arena” hypothesis. Mol. Biol. Evol. 21:1981-1991. [DOI] [PubMed]
- 8.Braam, L. A. M., I. Y. Goryshin, and W. S. Reznikoff. 1999. A mechanism for Tn5 inhibition. J. Biol. Chem. 274:86-92. [DOI] [PubMed] [Google Scholar]
- 9.Casiraghi, M., O. Bain, R. Guerrero, C. Martin, V. Pocacqua, S. L. Gardner, A. Franceschi, and C. Bandi. 2004. Mapping the presence of Wolbachia pipientis on the phylogeny of filarial nematodes: evidence for symbiont loss during evolution. Int. J. Parasitol. 34:191-203. [DOI] [PubMed] [Google Scholar]
- 10.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 11.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. T. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig, N. L., R. Craigie, M. Gellert, and A. M. Lambowitz (ed.). 2002. Mobile DNA II. ASM Press, Washington, D.C.
- 13.Davies, D. R., L. M. Braam, W. S. Reznikoff, and I. Rayment. 1999. The three-dimensional structure of a Tn5 transposase-related protein determined to 2.9 A resolution. J. Biol. Chem. 274:11904-11913. [DOI] [PubMed] [Google Scholar]
- 14.Davies, D. R., I. Y. Goryshin, W. S. Reznikoff, and I. Rayment. 2000. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289:77-85. [DOI] [PubMed] [Google Scholar]
- 15.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J. F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Médigue, A. Lanois. K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 11:1307-1313. [DOI] [PubMed] [Google Scholar]
- 16.Gellert, M. 2002. V(D)J recombination, p. 705-729.. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D. C..
- 17.Haniford, D.B. 2002. Transposon Tn10, p. 457-483.. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C..
- 18.Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: Transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. [DOI] [PubMed] [Google Scholar]
- 19.Huang, C. C., G. S. Couch, E. F. Petterson and T. E. Ferrin. 1996. Chimera: an extensible molecular modeling application constructed using standard components. p. 724. Pacific Symposium on Biocomputing. World Scientific Publishing, Singapore, Singapore.
- 20.Jenkins, T. M., D. Esposito, A. Engelman, and R. Craigie. 1997. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photocrosslinking. EMBO J. 16:6849-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, R. C., J.C.-P. Yin, and W. S. Reznikoff. 1982. The control of Tn5 transposition in Escherichia coli is mediated by protein from the right repeat. Cell 30:873-882. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy, A. K., A. Guhathakurta, N. Kleckner, and D. B. Haniford. 1998. Tn10 transposition via a DNA hairpin intermediate. Cell 95:125-134. [DOI] [PubMed] [Google Scholar]
- 23.Krebs, M. P., and W. S. Reznikoff. 1986. Transcriptional and translational sites of IS50. Control of transposase and inhibitor expression. J. Mol. Biol. 192:781-791. [DOI] [PubMed] [Google Scholar]
- 24.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masui, S., S. Kamoda, T. Sasaki, and H. Ishikawa. 2000. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J. Mol. Evol. 51:491-497. [DOI] [PubMed] [Google Scholar]
- 26.Mizuuchi, K., and T. A. Baker. 2002. Chemical mechanisms for mobilizing DNA, p. 12-23.. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C..
- 27.Nakamura, Y., T. Kaneko, S. Sato, M. Mimuro, H. Miyashita, T. Tsuchiya, S. Sasamoto, A. Watanabe, K. Kawashima, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, N. Nakazaki, S. Shimpo, C. Takeuchi, M. Yamada, and S. Tabata. 2003. Complete genome structure of Gloeobacter violaceus PCC7421, a cyanobacterium that lacks thylakoids. DNA Res. 10:137-145. [DOI] [PubMed] [Google Scholar]
- 28.Naumann, T. A., and W. S. Reznikoff. 2002. Tn5 transposase active site mutants. J. Biol. Chem. 277:17623-17629. [DOI] [PubMed] [Google Scholar]
- 29.Naumann, T. A., and W. S. Reznikoff. 2002. Tn5 transposase with an altered specificity to transposon ends. J. Bacteriol. 184:233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Pósfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 31.Peterson, G., and W. Reznikoff. 2003. Tn5 transposase active site mutations suggest position of donor backbone DNA in synaptic complex. J. Biol. Chem. 278:1904-1909. [DOI] [PubMed] [Google Scholar]
- 32.Reznikoff, W.S. 2002. Tn5 transposition, p. 403-422.. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D. C..
- 33.Reznikoff, W. S. 2003. Tn5 as a model for understanding DNA transposition. Mol. Microbiol. 47:1199-1206. [DOI] [PubMed] [Google Scholar]
- 34.Rezsohazy, R., B. Hallet, J. Delcour, and J. Mahillon. 1993. The IS4 family of insertion sequences: Evidence for a conserved transposase motif. Mol. Microbiol. 9:1283-1295. [DOI] [PubMed] [Google Scholar]
- 35.Rice, P., R. Craigie, and D. R. Davies. 1996. Retroviral integrases and their cousins. Curr. Opin. Struct. Biol. 6:76-83. [DOI] [PubMed] [Google Scholar]
- 36.Steiniger-White, M., and W. S. Reznikoff. 2000. The C-terminal alpha helix of Tn5 transposase is required for synaptic complex formation. J. Biol. Chem. 275:23127-23133. [DOI] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Twining, S. S., I. Y. Goryshin, A. Bhasin, and W. S. Reznikoff. 2001. Functional characterization of arginine 30, lysine 40, and arginine 62 in Tn5 transposase. J. Biol. Chem. 276:23135-23143. [DOI] [PubMed] [Google Scholar]
- 39.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 40.Welch, R. A., V. Burland, G. Plunkett, III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17021-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Werren, J. H., and D. M. Windsor. 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. R. Soc. Lond. B Biol. Sci. 267:1277-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, M., L. V. Sun, J. Vamathevan, M. Riegler, R. Deboy, J. C. Brownlie, E. A. McGraw, W. Martin, C. Esser, N. Ahmadinejad, C. Wiegand, R. Madupu, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, A. S. Durkin, J. F. Kolonay, W. C. Nelson, Y. Mahamoud, P. Lee, K. Berry, M.B Young, T. Utterback, J. Weidman, W. C. Nierman, I. T. Paulsen, K. E. Nelson, H. Tettelin, S.L. O'Neill, and J. A. Eisen. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLOS Biology 2:327-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 44.Yang, W., and T. A. Steitz. 1995. Recombining the structures of HIV integrase, RuvC and RnaseH. Structure 3:131-134. [DOI] [PubMed] [Google Scholar]
- 45.Yin, J.C.-P., M. P. Krebs, and W. S. Reznikoff. 1988. The effect of dam methylation on Tn5 transposition. J. Mol. Biol. 199:35-46. [DOI] [PubMed] [Google Scholar]
- 46.York, D., and W. S. Reznikoff. 1996. Purification and biochemical analysis of a monomeric form of Tn5 transposase. Nucleic Acids Res. 24:3790-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, M., and W. S. Reznikoff. 1997. Tn5 transposase mutants that alter DNA binding specificity. J. Mol. Biol. 271:362-373. [DOI] [PubMed] [Google Scholar]