Abstract
Bacteria utilize specialized multi-protein machineries to synthesize the essential peptidoglycan (PG) cell wall during growth and division. The divisome controls septal PG synthesis and separation of daughter cells. In E. coli, the lipid II transporter candidate FtsW is thought to work in concert with the PG synthases penicillin-binding proteins PBP3 and PBP1b. Yet, the exact molecular mechanisms of their function in complexes are largely unknown. We show that FtsW interacts with PBP1b and lipid II and that PBP1b, FtsW and PBP3 co-purify suggesting that they form a trimeric complex. We also show that the large loop between transmembrane helices 7 and 8 of FtsW is important for the interaction with PBP3. Moreover, we found that FtsW, but not the other flippase candidate MurJ, impairs lipid II polymerization and peptide cross-linking activities of PBP1b, and that PBP3 relieves these inhibitory effects. All together the results suggest that FtsW interacts with lipid II preventing its polymerization by PBP1b unless PBP3 is also present, indicating that PBP3 facilitates lipid II release and/or its transfer to PBP1b after transport across the cytoplasmic membrane. This tight regulatory mechanism is consistent with the cell’s need to ensure appropriate use of the limited pool of lipid II.
Most bacteria surround their cytoplasmic membrane with a peptidoglycan (PG) sacculus which protects the cell from bursting due to the turgor and maintains cell shape. In order to propagate, bacteria have to enlarge and divide their cell envelope including their PG sacculus1. These processes are performed by specialized multiprotein complexes, the elongasome and divisome; each of them contains all the enzymatic activities required for the synthesis of new PG material and its insertion into the cell wall2. The substrate for PG synthesis is the precursor lipid II, which is synthesized on the inner face of the cytoplasmic membrane and subsequently translocated through the membrane by a flippase (FtsW and/or MurJ)3,4,5. Once on the periplasmic side of the membrane, lipid II is polymerized by the glycosyltransferase (GT) and the transpeptidase (TP, the target of penicillin) activities of the penicillin-binding proteins (PBPs)6,7 in coordination with interacting proteins of their respective networks, allowing growth and division of the bacterial cell2.
The divisome controls septal PG synthesis at mid-cell and its differentiation into the new poles of daughter cells. The components of the divisome span the cytoplasm, the cytoplasmic membrane, the periplasm and the outer membrane (in Gram-negative bacteria) and communicate by dynamic protein-protein interactions through these cell compartments. This organization allows the coordination between the cytoskeleton and the synthesis of precursors in the cytoplasm, their transport across the cytoplasmic membrane and septal peptidoglycan synthesis, and, in Gram-negative bacteria, the coordination with outer membrane invagination8,9. In Escherichia coli, the divisome includes over 20 proteins which assemble in an ordered and interdependent manner in two steps10; first, the tubulin-like FtsZ, ZipA, FtsA, ZapA-E and FtsEX localize at mid-cell underneath the cytoplasmic membrane. Later, the downstream components FtsK, FtsQ-FtsL-FtsB, FtsW-FtsI (PBP3) and FtsN join sequentially, some as preformed subcomplexes11,12, to form the mature divisome13. The bifunctional PG GTase/TPase PBP1b and its regulators (LpoB, CpoB, TolA) also associate with the divisome. PBP1b requires the PG TPase PBP3 which itself requires FtsW for septal localization14,15. PBP3 interacts in vitro with FtsW12, PBP1b15, and FtsN16 and with other proteins of the divisome2. Bacterial two-hybrid (BATH) analysis also revealed an interaction between PBP1b and FtsW12. FtsW is a polytopic membrane protein that belongs to the SEDS (shape, elongation, division, and sporulation) family and plays an essential role in cell division. It was shown to transport lipid II across membranes in vitro3. The inability of FtsW to transport bulky lipid II variants suggests the presence of a size-restricting pore that accommodates lipid II during transport17. Moreover, mutagenesis studies of FtsW showed that residues R145 and K153 of the transmembrane segment (TM) 4 are essential for flippase activity and the mutated proteins were incapable to rescue a conditional FtsW mutant strain17. The periplasmic loop (E240-A249) between transmembrane helices (TMs) 7 and 8 is essential for septal PG assembly and the loop between TMs 9 and 10 is required for the localization of PBP3 at the septum18.
A second candidate protein proposed to flip lipid II, MurJ, belongs to the multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily4,19. MurJ is essential for viability and peptidoglycan synthesis in E. coli. The heterologous expression of the O-antigen flippase Wzk from Helicobacter pylori in E. coli was able to complement the loss of MurJ20. A recently proposed third flippase candidate, Amj of Bacillus subtilis, is the founding member of new membrane protein family21. It was recently reported that the SEDS protein RodA is essential for viability of a B. subtilis mutant lacking all class A PBPs, and has a GTase activity22. Hence, different roles for these different families of integral membrane proteins involved with PG growth have been proposed, showing the need to clarify their precise biochemical activities.
Several PG enzymes are known to engage in functional interactions with divisome proteins. PBP1b is stimulated by FtsN and the outer membrane (OM) lipoprotein LpoB, which binds to its UB2H domain16,23,24,25, and the dynamic interactions between LpoB, CpoB and TolA with PBP1b facilitate the coordination between PG synthesis and OM invagination8. FtsQ, FtsL and FtsB associate in a stable ternary complex11 but their exact roles are unknown. FtsN is the last protein to localize to the division site and interacts with PBP1b, PBP3, FtsQ and FtsW2,16. It is believed to play an important role in the stability of the divisome and the initiation of the cell envelope constriction by activating septal PG synthesis through its interactions with FtsA and FtsQLB26,27. Large complexes containing more than two divisome proteins have been isolated in a few cases, the largest being a 1 MDa complex containing FtsZ, ZipA, FtsK, FtsQLB, and FtsN from E. coli28. In another study the proteins DivIB, DivIC, FtsL, PBP2x and FtsW from Streptococcus pneumoniae were co-expressed in E. coli and complexes composed of up to five proteins were isolated29.
While it is now evident that the PG synthase activities are regulated within the divisome and coordinated with the progression of the cell cycle through protein-protein interactions, the molecular mechanisms of these processes remain largely unknown. The E. coli septum PG synthesis components of the divisome includes FtsQ-FtsL-FtsB, FtsW-PBP3, PBP1b, FtsN, LpoB, CpoB and TolA (Fig. 1). In this work, we have investigated the divisome enzymes directly involved in septum synthesis, the synthases PBP1b and PBP3 and the lipid II flippase candidate FtsW, with the aim to understand how the interactions between them regulate the PG synthesis activities.
Figure 1. Schematic representation of the septal peptidoglycan synthesis core of the divisome.
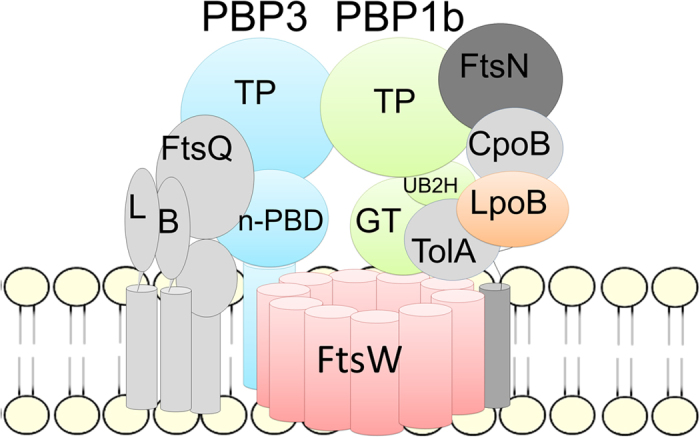
The proteins studied in this paper are shown in color, the others are in grey. TP, transpeptidase domain; GT, glycosyltransferase domain; L, FtsL; B, FtsB; n-PBD, non-penicillin binding domain.
Results
FtsW, PBP3 and PBP1b form a ternary complex in vitro
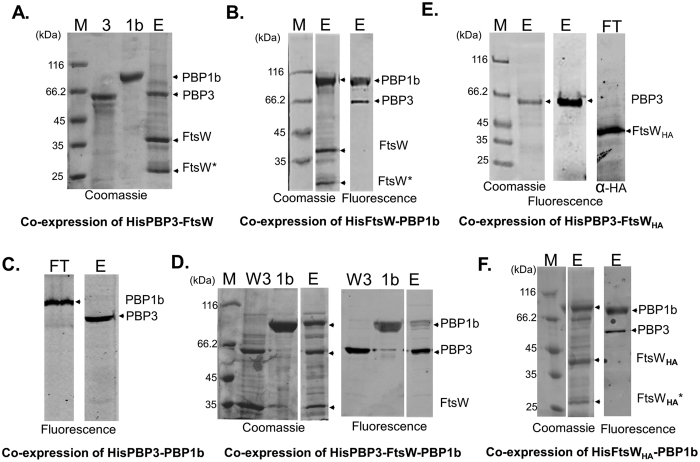
As a transporter candidate, FtsW is probably the last protein to interact with lipid II before its polymerization by the glycosyltransferase activity of a bifunctional PBP. PBP1b localizes to the septum and interacts with several divisome proteins and thus, it is likely to be the first enzyme to process lipid II after its translocation during cell division. PBP3 forms a complex with FtsW and its specific transpeptidase activity is essential for cell division. Binary interactions between FtsW, PBP3 and PBP1b have been shown by the bacterial two hybrids (BATH) system15,30,31 and confirmed in vitro12,15 except for the interaction between PBP1b and FtsW that was only shown by BATH12. We sought to complete the characterization of this protein triplet by analyzing the interaction between PBP1b and FtsW in vitro and to isolate the ternary complex. The proteins were co-expressed in E. coli followed by affinity co-purification with one of the two proteins containing an N-terminal His-tag (bait) and the second one left untagged (prey). It is noteworthy that the latter method was found to be more efficient in the detection of complexes than the purification of the separate proteins followed by reconstitution in vitro. This might be due to the fact that once a sufficiently stable complex is formed it can be solubilized without dissociating while, when the proteins are solubilized separately, the detergent might interfere with some of the interactions. The co-expression system was first tested using PBP3 and FtsW that were previously shown to form a sub-complex12. After co-expression and membrane solubilization with detergent (DDM) the proteins were loaded on a Nickel affinity column followed by washing steps and imidazole elution. The fractions were then analyzed using SDS-PAGE and fluorescent-ampicillin to label the PBPs. As expected, the results show that PBP3 and FtsW co-eluted (Fig. 2A), confirming their interaction. The same strategy was then applied to the HisFtsW-PBP1b and HisPBP3-PBP1b pairs. We found that HisFtsW and PBP1b co-eluted (Fig. 2B). However, in the case of HisPBP3-PBP1b only tagged PBP3 was recovered after elution, and PBP1b was found in the flow through (Fig. 2C). These results show, firstly, that untagged PBP1b does not bind nonspecifically to the column in the absence of interacting partner and therefore, that FtsW and PBP1b interact specifically. Secondly, the absence of detectable interaction between PBP1b and PBP3 by this method is probably due to weak and/or dynamic interaction between the two proteins. This conclusion is consistent with previous results showing that co-immunoprecipitation of PBP1b and PBP3 occurs after cross-linking15. Consequently, we conclude that the FtsW-PBP3 and FtsW-PBP1b interactions are stronger or less dynamic than the PBP1b-PBP3 interaction. Interestingly, labelling of the PBPs with fluorescent ampicillin after elution revealed that the native E. coli PBP3 co-purified with HisFtsW-PBP1b (Fig. 2B). This result was confirmed by the co-expression and co-purification of HisPBP3, FtsW and PBP1b, which confirmed the co-elution of the three proteins (Fig. 2D). Since PBP1b was not retained with HisPBP3 but only when FtsW was co-expressed, this suggests that PBP1b is bound to the ternary complex via FtsW. The construct of PBP1b also contained FtsN but this protein did not co-elute with the three proteins, suggesting that its interactions with the ternary complex is weak.
Figure 2. Interactions between FtsW, PBP3 and PBP1b.
The proteins were co-expressed in E. coli cells; one contains a His-Tag and was used as bait while the other proteins are untagged. The co-expressed proteins are indicated below each panel. The expressed protein cannot be detected in the total extract but only after affinity purification. The cytoplasmic membranes from cells expressing the indicated proteins were solubilized with detergent followed by a purification step over a Ni-affinity column. The eluted fractions or flow-through fractions were labeled by fluorescent ampicillin and analyzed by SDS-PAGE. The gels were first subjected to fluorescence imaging followed by Coomassie blue staining. 3, 1b and W3 indicate lanes with purified PBP3, PBP1b and FtsW-PBP3, respectively (size controls). Lanes E depict elution fractions from the Ni-affinity columns and FT the flow-through. M, protein standards. The bands of PBP1b, PBP3 and FtsW are indicated on the right side of the gels. FtsW* indicates an FtsW degradation product. In panel E, the flow-through fraction (FT) was analyzed by immunoblotting using antibodies against the epitope HA to detect FtsWHA.
The loop between TM 7 and 8 of FtsW is essential for the formation of the FtsW-PBP3 complex
The first 56 N-terminal amino acids of PBP3, containing the transmembrane (TM) segment, are essential for its localization and for the interaction with the TMs of FtsW32,33. A model of the interaction between the TMs of FtsW and PBP3 was proposed using a co-evolutionary approach34. In addition, the loop between TM9 and 10 of FtsW is involved in the interaction with PBP3 and PBP1b12,18. Mutagenesis studies in the large loop between TM7 and 8 implied its importance for the function of FtsW, but the exact role was not determined18,35. The insertion of the 9 amino acid hemagglutinin (HA) peptide in the TM7/8 loop between residues 293–294 of FtsW yields a functional protein (FtsWHA) as shown by in vivo complementation18. However, when FtsWHA was co-expressed with HisPBP3, only tagged PBP3 was recovered during the elution step (Fig. 2E) (under the same conditions, wild-type FtsW forms a stable complex with PBP3 (Fig. 2A)). This indicates that the binding between the two proteins was affected by the insertion of the HA peptide and that the TM7/8 loop is crucial for a strong interaction with PBP3. Since the PBP3-PBP1b and PBP3-FtsWHA interactions are too weak to observe pairwise complexes using the co-expression/co-purification method, we hypothesized that the simultaneous interaction of PBP3 with PBP1b and FtsWHA might stabilize a ternary complex. Indeed, when HisFtsWHA and PBP1b were co-expressed and co-purified, the two proteins co-eluted with PBP3 (Fig. 2F), supporting our hypothesis and confirming the presence of a ternary complex and not of two subpopulations of FtsW-PBP1b and FtsW-PBP3 complexes. To our knowledge, this is the first time that such a complex was isolated, suggesting that the transport of the lipid II precursor is coupled with its polymerization and insertion into the cell wall.
FtsW inhibits lipid II polymerization by PBP1b if not in complex with PBP3
In E. coli, the complex formed by PBP1b, PBP3 and FtsW constitutes the core-enzyme of the divisome that interacts with the lipid II substrate. In order to understand the effect of the interactions between these proteins on lipid II polymerization we analyzed the activity of PBP1b alone and in the presence of either FtsW or PBP3, or of the complex FtsW-PBP3. The glycosyltransferase activity of E. coli PBP1b was monitored by the continuous fluorescence method using dansyl lipid II as substrate36,37. The polymerase reaction is accompanied by a decrease in the fluorescence signal as a function of time. However, when FtsW from E. coli, Klebsiella pneumoniae or Salmonella enterica were added to the reaction mixture, the GT activity of PBP1b was inhibited in a concentration-dependent manner (Fig. 3A). Under the same conditions, PBP3 from E. coli slightly lowered the activity of PBP1b (Fig. 3B). Remarkably, the inhibitory effect of FtsW was suppressed in the presence of the co-purified E. coli FtsW-PBP3 complex and the activity of PBP1b was restored to a level close to that of PBP1b alone (Fig. 3B). FtsW inhibition could be the result of direct interaction with PBP1b or lipid II as FtsW potentially interacts with both.
Figure 3. Effect of FtsW, PBP3 and the FtsW-PBP3 complex on the activities of PBP1b.
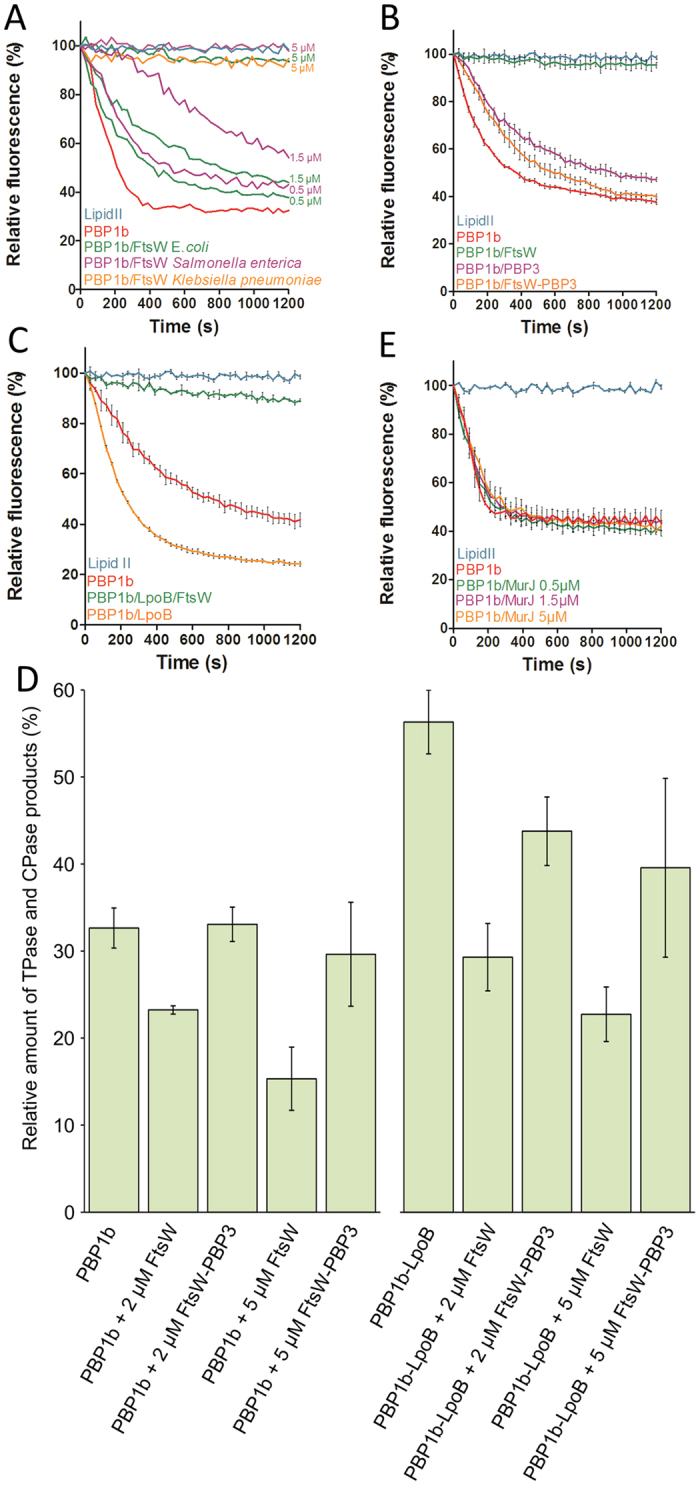
(A–C) Continuous fluorescence assay to measure the polymerization (GT activity) with dansyl-lipid II. The fluorescence decreases over time upon polymerization of dansyl-lipid II by PBP1b. A. inhibition of lipid II polymerization by FtsW from E. coli, S. enterica and K. pneumoniae. (B) In contrast to FtsW (5 μM), the polymerase activity of PBP1b is not inhibited by the FtsW-PBP3 complex (5 μM) and slightly decreases in the presence of PBP3 (5 μM). (C) PBP1b activation by LpoB (200 nM) does not suppress the inhibitory effect of FtsW (5 μM). (D) Effect of FtsW and the FtsW-PBP3 complex on TPase activity of PBP1b. The percentage of muropeptide products detected by HPLC analysis of PG produced in vitro by PBP1b in the presence of the proteins indicated beneath each bar (endpoint assay). Total TPase activity is the sum of activities derived from the TPase domain of PBP1b, including peptide cross-linking and carboxypeptidase activities. Values are the mean ± SD of three experiments. Examples of the corresponding HPLC chromatograms are depicted in Supplemental Figure S1. The effects of FtsW and FtsW-PBP3 on PBP1b’s PBP domain parallel the effects on GTase in A, consistent with the coupling of both domains. E. In contrast to FtsW, MurJ (0.5–5 μM) has no effect on the polymerase activity of PBP1b.
The lipoprotein LpoB interacts with PBP1b and stimulates its activity23,24,25. In order to test whether PBP1b activation by LpoB could reverse the inhibitory effect of FtsW on lipid II polymerization by PBP1b, the experiment was performed in the presence of LpoB. As previously published, LpoB activated the GT activity of PBP1b. FtsW was still able to inhibit PBP1b in the presence of LpoB (Fig. 3C).
The GT and TPase activity of class A PBPs is coupled38,39, and under certain conditions the stimulated PBP domain can perform carboxypeptidase (CPase) reactions40, thus we sought to test whether the inhibition of GTase activity by FtsW leads to a concomitant decrease in TPase/CPase activity by analyzing the composition of the PG produced. Indeed, after a 2 h incubation period with radioactive lipid II the total TPase/CPase activity of PBP1b was reduced by FtsW in a concentration dependent manner. Consistent with the GTase data this inhibitory effect was relieved by the presence of PBP3 (Fig. 3D). The same trend was observed in PG produced by PBP1b in the presence of LpoB, although in this experiment the presence of PBP3 did not fully restore peptide cross-linking to the uninhibited levels.
Interaction of FtsW with lipid II and its modulation by PBP3
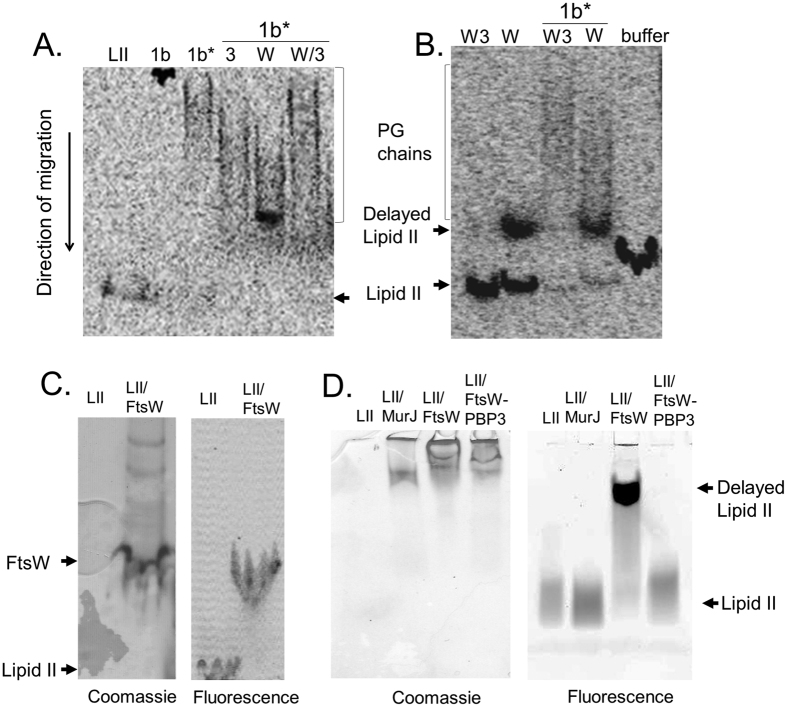
A second polymerase assay based on SDS-PAGE and radioactive lipid II was used to further analyze the effect of FtsW and PBP3 on lipid II polymerization by PBP1b. This assay allows the visualization of unused lipid II and the evaluation of the polymer glycan chain length when the TPase active site is inactivated (by the S510A mutation in PBP1bTP* or by penicillin). When both the GTase and TPase are active, the PG does not penetrate the gel, which indicates that it has been cross-linked (Fig. 4A). The PBP1bTP* activity was partially inhibited by addition of FtsW; the glycan chains formed were shorter when compared to the products of PBP1bTP* alone (Fig. 4A). Addition of PBP3 also resulted in a shortening of glycan chains but to a lesser extent than with FtsW (Fig. 4A). Moreover, we observed that FtsW modifies the mobility of lipid II in the gel while the FtsW-PBP3 complex had no effect on the activity of PBP1b or the mobility of lipid II (Fig. 4A and B). Similarly, the addition of FtsW to lipid II in the absence of PBP1b reduced the electrophoretic motility of lipid II in the gel, compared to its migration near the gel front in the sample with the FtsW-PBP3 complex (Fig. 4B). Coomassie staining of the gel shows the co-migration of FtsW and the fluorescent lipid II (Fig. 4C) suggesting a direct interaction between lipid II and FtsW. To further confirm this interaction, we used native PAGE. Interestingly, we found that the mobility of lipid II pre-incubated with FtsW was also affected under native conditions, and that the FtsW-PBP3 complex or MurJ had no effect on the mobility of lipid II (Fig. 4D) showing that the interaction between FtsW and lipid II is specific. These results are consistent with the observation that overproduction of FtsW arrests cell growth41. We also observed that high induction of FtsW arrests cell growth at an OD of ~1.5 but when FtsW-PBP3 was overexpressed, cells continue to grow (Fig. S2).
Figure 4. Effect of FtsW, PBP3 and the FtsW-PBP3 complex on the polymerization of lipid II by PBP1b and the mobility of lipid II (LII) analyzed by (SDS- or native) PAGE.
(A and B) radioactive lipid II revealed by storage phosphor imaging using Typhoon trio+ (GE Health Care). (A) SDS-PAGE analysis of PBP1b (1b) or PBP1bTP* mutant (1b*) (both at 100 nM) reaction products in the presence of PBP3 (3) (3 μM), FtsW (W) (3 μM), and the FtsW-PBP3 (W3) complex (3 μM). (B) effect of FtsW (3 μM) and the FtsW-PBP3 complex (3 μM) in the absence of PBP1b on the mobility of lipid II; buffer with excess detergent used as control. (C) Co-migration of FtsW and NBD-lipid II in SDS-PAGE. (D) analysis of NBD-lipid II (10 μM) migration by native-PAGE in the absence and the presence of FtsW, the FtsW-PBP3 complex or MurJ (all used at 5 μM). (C and D) fluorescent lipid II was first revealed by fluorescence imaging using Typhoon trio+, then the gels were stained with Coomassie blue.
All together our results provide evidence for a direct interaction between lipid II and FtsW and suggest that this interaction is responsible for the inhibition of PBP1b-catalyzed lipid II polymerization by FtsW. Moreover, the results also indicate that the interaction of FtsW with PBP3 modulates the interaction of the transporter candidate with its lipid II substrate.
While we have observed an interaction of E. coli FtsW with lipid II, this protein and two orthologues (K. pneumoniae FtsW or Salmonella enterica FtsW) were unable to catalyze the GTase reaction in vitro (Fig. 5). No polymerase activity was observed with FtsW-PBP3 complex either (Fig. 5A and Fig. S1). This contrasts the recent findings of a weak GT activity of a partially purified RodA protein from B. subtilis22 and the proposed in vivo GTase activity of E. coli RodA42. Our data suggest that in E. coli the GTase activity might be provided by the bi-functional class A PBPs and not by the two SEDS proteins, FtsW and RodA.
Figure 5. Analysis of the reactivity of FtsW with lipid II.
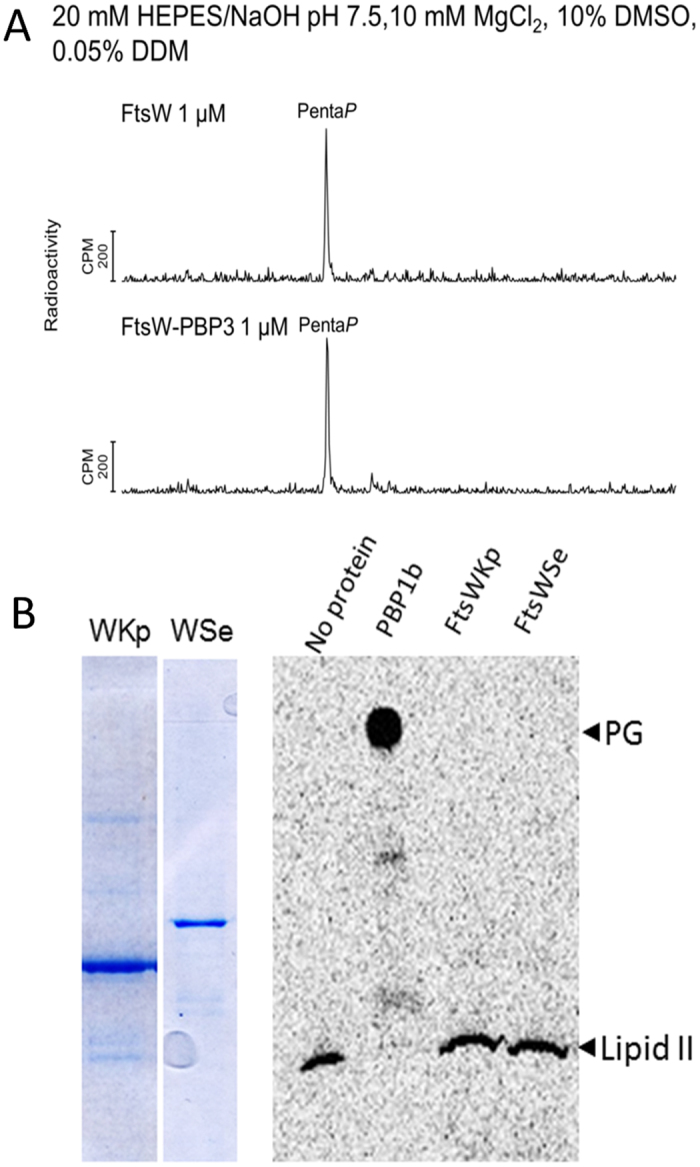
(A) HPLC chromatograms of the reaction products of radioactive lipid II incubated with E. coli FtsW or FtsW-PBP3. (B) right, TLC analysis of reaction products from radioactive lipid II incubated with FtsWKp or FtsWSe. Lipid II without protein and peptidoglycan (PG) synthesis by PBP1b is shown as control. Left, SDS-PAGE showing the purified K. pneumoniae FtsW (WKp) and S. enterica FtsW (WSe).
MurJ does not affect lipid II polymerization by PBP1b
The identity of the lipid II flippase is currently a matter of debate5. In E. coli, the SEDS proteins (FtsW and RodA) and MurJ were proposed as lipid II flippases3,4. The previous studies focused on the transport function of FtsW and MurJ using different in vitro and in vivo experiments, respectively. However, the different approaches did not produce consistent results to end the controversies about the identity of the lipid II transporter, which remains an open question. In this study, we have investigated the effect of MurJ on lipid II polymerization by PBP1b as described above for FtsW. We assumed that if MurJ is a lipid II transporter, it should interact with its substrate and thus could affect the activity of PBP1b as was observed with FtsW. When MurJ was added to the fluorescence assay containing PBP1b and dansyl lipid II, we found that, in contrast to the three different FtsW orthologues tested, MurJ has no effect on the polymerization of lipid II by PBP1b (Fig. 3E). Moreover, when MurJ was incubated with lipid II it did not exhibit any effect on the lipid II mobility in a native gel (Fig. 4D). These results suggest that MurJ may not interact directly with lipid II and may require unknown factors should it be a lipid II flippase. As FtsW and MurJ are both integral membrane proteins and were solubilized with the same detergent, our data highlight the specificity of the effect of FtsW on lipid II polymerization by PBP1b.
Discussion
New insights from FtsW-PBP3/PBP1b interactions suggest a functional coordination
In this work we have investigated the activities and interactions of core peptidoglycan synthesis proteins of the E. coli divisome. Our results show that PBP1b, PBP3 and FtsW form a ternary complex in vitro. Such a complex contains the necessary enzymatic activities to build the septum peptidoglycan, namely the GTase and TPase activities for glycan chain elongation and peptide cross-linking, respectively, provided by the bifunctional PBP1b and the division specific TPase activity of PBP3, in addition to the potential flippase activity of FtsW. This reinforces the idea that the three proteins work together during division in coordination with other divisome proteins. Our data is consistent with previous protein-protein interaction and localization data2,8 and support the hypothesis that class A and class B PBPs (here PBP1b and PBP3) participate in multi-enzyme complexes43,44. E. coli and other bacteria, require at least one class A PBP (PBP1a or PBP1b) for viability45,46. PBP1b can be replaced by PBP1a and therefore the latter is likely to be also recruited at the division site under specific control of the divisome.
We infer that our method of co-expression/-purification would detect only strong and stable interactions between proteins. Comparing the data from different overexpression pairs provided new insights into the stability of the interactions observed. The FtsW-PBP3 and FtsW-PBP1b complexes were readily isolated suggesting that they are maintained by considerably stronger interactions than those between PBP1b and PBP3. Consequently, PBP1b is likely recruited to the division site by the FtsW-PBP3 complex by multiple and simultaneous interactions, consistent with its PBP3-dependent mid-cell localization15. The loop between TM9 and 10 of FtsW is necessary for interaction with PBP318 and may also be involved in the interaction with PBP1b12. The largest periplasmic loop between TM7 and 8 (~66 residues) of FtsW is crucial for the function of the protein18. We showed here that the modification of this loop decreased the binding of FtsWHA to PBP3 to a point where the complex could no longer be detected, but the formation of the FtsWHA-PBP1b complex was not affected. These observations indicate that the loop between TM7 and 8 contributes to the interaction of FtsW with PBP3 but does not seem to play a role in the interaction with PBP1b. This is consistent with interaction studies between FtsW and PBP3 from Mycobacterium tuberculosis showing that the last two extracellular loops between TM 7/8 and TM 9/10 of FtsW also play important role in the binding of PBP347.
PBP3 modulates the interaction between FtsW and lipid II
The finding of the inhibitory effect of FtsW on the polymerization of lipid II by E. coli PBP1b, the suppression of the inhibitory effect of FtsW by PBP3, together with the interaction of FtsW with lipid II, strongly suggest a regulation mechanism of FtsW involving PBP3. Based on these results, we propose: (i) that FtsW alone interacts with high affinity with lipid II and prevents its polymerization by PBP1b, (ii) that the interaction of PBP3 with FtsW modulates the interaction between FtsW and lipid II so that the latter becomes accessible to PBP1b. The interaction of PBP3 with FtsW may induce a conformational change in FtsW that facilitates the release of lipid II and/or its transfer to PBP1b. FtsW and PBP3 are expressed at similar levels in cells and the two proteins form a complex prior to their localization at the division site, hence, only the active FtsW-PBP3 complex might be present in vivo. This is consistent with the observation that overproduction of FtsW inhibits cell growth41, in contrast, the overexpression of FtsW-PBP3 had no effect on the cell growth. The effect of overexpressed FtsW on cell growth may be the consequence of the sequestration of lipid II by the overexpressed protein. Thus, sequestration of lipid II when FtsW is overexpressed and its recovered availability when both FtsW and PBP3 are overexpressed seem to also happen in vivo.
In our study we have observed an interaction of E. coli FtsW with lipid II but this protein, and two analogs (FtsWKp and FtsWSe), were devoid of any GT activity. This contrasts the observed activity of RodA from B. subtilis22. These marked differences indicate that in E. coli, where at least one class A PBP is essential for viability, the GTase activity might be provided by the bi-functional class A PBPs and not by the two SEDS proteins, FtsW and RodA. Moreover, the activity data, together with the interaction detected between PBP1b and several divisome proteins, argue against the recent suggestion42 that the class A PBPs work ‘semi-autonomously’ outside the divisome/elongasome complexes and that the GT activity is provided by the SEDS proteins. In our view, other SEDS proteins need to be purified and systematically tested for GTase activity, lipid II binding and flippase activity to clarify these contradicting results and help better understand the function of these proteins in PG synthesis and cell morphogenesis. More effort should be put into the determination of the structure of SEDS protein, which could provide an insight on their function.
Based on our results and published data we propose a model where FtsW is a flippase interacting with its substrate, lipid II and that this interaction is regulated by PBP3 (see below).
Model of FtsW function and its regulation by PBP3
The potential transport function of FtsW was proposed to be mediated by a pore-like structure17. The large loop between TM7 and 8 could thus play a regulatory role by an open/close mechanism of the pore’s gate from the periplasmic side. As this loop interacts with PBP3, possibly via the N-terminal non-penicillin binding domain (n-PBD), this may induce a conformational change of the TM7/8 loop and opening of the pore, allowing the release of lipid II and/or its direct transfer to PBP1b. This mechanism would explain the observed effect of PBP3 on the interaction of FtsW with lipid II (Fig. 6). Alternatively, PBP3 binding could induce a structural rearrangement within the TM domain of FtsW leading to the lipid II release.
Figure 6. Model of modulation of the FtsW function by PBP3.
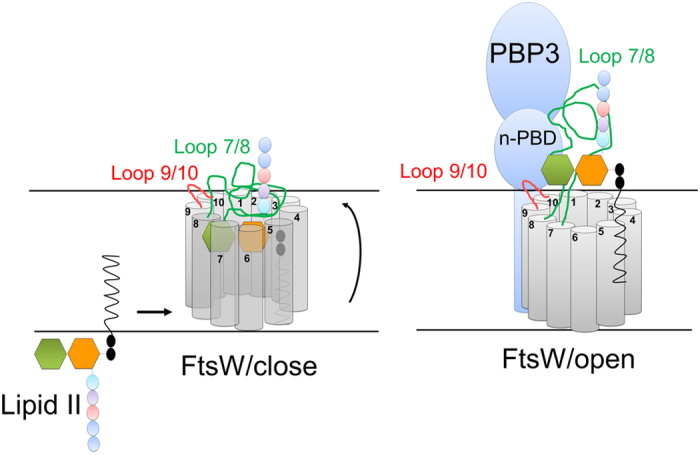
Left, in the absence of PBP3, lipid II is loaded into the pore of FtsW and translocated but cannot be released. Right, Interaction of PBP3 with FtsW induces conformational changes in the loop between TM 7 and 8, opening the pore gate and allowing the release of lipid II. The loop between TMs 9 and 10 is required for the localisation of PBP3 at the septum.
A similar mechanism is found in functionally unrelated transferases that use a polyprenyl–sugar carrier as substrate48,49. The archaeal AglB and the eubactertial PglB, two oligosaccharyltransferase (OST) belonging to family STT3 (stauroporine and temperature sensitive 3), catalyze protein N-glycosylation using polyprenyl-phospho-sugar as a donor. Modeling showed that their folds were related to that of FtsW34. They contain a characteristic long and dynamic loop between TM9 and 10 that oscillates between structured and unstructured states, facilitating the release of the glycosylated product from the active site48. ArnT is an OST with a fold similar to that of PglB and AglB but its function is different. ArnT is involved in the attachment of the undecaprenyl-linked L-Ara4N to lipid A. ArnT also exhibits a flexible periplasmic loop between TM7 and 8 that plays an important function by controlling the lipid A cavity by a structural change49.
FtsW and MurJ are both proposed to transport lipid II across the membrane that implicates that they should interact with their substrate. In consequence, this interaction should have an effect on the lipid II polymerization by the glycosyltransferase activity of PBP1b. This hypothesis was verified with FtsW but, in contrast, MurJ had no effect on the activity of PBP1b nor did it bind lipid II. This indicates that the two potential flippases work by different mechanisms. The modulation of the interaction between FtsW and lipid II by PBP3 could stimulate the flippase activity of FtsW. The lack of an interaction of MurJ with lipid II suggests that MurJ may have an indirect function in the process or that an unknown factor may be required for binding and probably the flippase activity, which would explain why MurJ alone was not active in the in vitro assay3. Alternatively, the turnover of lipid II by MurJ is fast and the release of lipid II does not require assistance from another protein.
In conclusion, we showed that FtsW, PBP3 and PBP1b form a ternary complex in vitro, FtsW interacts with lipid II and that this interaction is modulated by PBP3. We also showed that the loop between TM 7 and 8 is important for the interaction of FtsW with PBP3 and that this interaction may play role in the regulation of FtsW-lipid II interaction. Based on these results we propose a model where lipid II remains attached to FtsW after its transport across the cytoplasmic membrane and that its release and/or transfer to PBP1b is facilitated by PBP3 via allosteric conformational changes probably involving the loop between TM 7 and 8 of FtsW and/or the TM domain. This regulatory mechanism may ensure that the limited and precious lipid II precursor is not spuriously flipped to a non-productive end.
Experimental Procedures
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids are described in Table 1. Plasmids constructions are described in the supporting information. Oligonucleotides (Table S1) were purchased from Eurogentec (Angleur, Belgium). Bacteria were grown in Luria-Bertani (LB) medium supplemented with ampicillin (50 μg/ml) (from MP Biomedicals) or chloramphenicol (30 μg/ml) (from Sigma) or Kanamycin (50 μg/ml) (from MP Biomedicals).
Table 1. Strains and plasmids constructs.
| Strain/plasmids | Relevant genetic marker (s) or features | Sources/reference |
|---|---|---|
| E. coli Strains | ||
| Mach-I | ΔrecA1398 endA1 tonA Φ80ΔlacM15 ΔlacX74 hsdR (rK− K+) | Invitrogen |
| C43 (DE3) | F− ompT gal dcm hsdSB (rB− mB−) (DE3) | 56 |
| Lemo21 (DE3) | fhuA2 [lon] ompT gal (λ DE3) [dcm] ∆hsdS/pLemo (CamR) | New England Biolabs |
| Plasmids | ||
| pDML924 | pET28a, His-ponB | 50 |
| pDML924 (S510A) | pET28a, His-ponB (S510A) | This work |
| pDML2032 | pET22b, ftsN-His | This work |
| pDML2422 | pMCL210-ftsW-293HA | 18 |
| pDML2400 | pET28a, His-ftsW | 18 |
| pDML2040 | pETDuet, His-ftsI, ftsW | This work |
| pDML2041 | pETDuet, His-ftsW, ftsI | This work |
| pDML2043 | pETDuet, His-ftsI, ftsW (293HA) | This work |
| pDML2045 | pACYCDuet, ponB, ftsN-S. tag | This work |
| pCIP1000 | pETDuet, His-ftsW, ponB | This work |
| pCIP1083 | pETDuet, His-ftsWHA, ponB | This work |
| pET28MHL-WSe | pET28MHL, His-ftsWSe | This work |
| pET28MHL-WKp | pET28MHL, His-ftsWKp | This work |
| pCIP1051 | pETDuet, His-murJ | This work |
Reagents
Dansyl-lipid II, NBD-lipid II and [14C]-lipid II (0.155 μCi/nmol) were prepared as previously described50,51, Flavomycin was a gift from Aventis (Romainville, France). Fluorescein labelled ampicillin was prepared as previously described52.
Expression and purification of the proteins
E. coli PBP1b and PBP1b-S510A were expressed and purified as previously described50. LpoB (sol) was purified as described25.
E. coli FtsW, PBP3 or the PBP3-FtsW complex were expressed in E. coli strain C43 (DE3) harboring plasmids pDML2400, pDML2040, pDML2041 or pDML2043. Bacteria were grown at 37 °C, in LB medium supplemented with the appropriate antibiotic to an A600nm of 0.6. Then expression was induced for 4–5 hours by addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Cells were collected by centrifugation at 8000 × g for 20 minutes at 15 °C and resuspended in a buffer containing 50 mM Tris–HCl, pH 8.0, 50 mM NaCl, 2 mM MgCl2, 1.5 U ml−1 benzonase (EMD Millipore) and EDTA-free protease inhibitor Cocktail (Roche). The bacterial cells were lysed by three passages through a cell homogenizer (Emulsiflex C3 Avestin®). The cell lysate was spun down at 150,000× g for 1 hour at 4 °C and then the membranes were resuspended and washed in 25 mM Tris–HCl, pH 8.0, 500 mM NaCl, 10% glycerol and complete EDTA-free protease inhibitors. The membranes were collected by centrifugation at 150,000× g for 1 hour at 4 °C and solubilized in 50 mM Hepes pH 7.5, 500 mM NaCl, 10% glycerol, 40 mM n-dodecyl-β-D-maltopyranoside (DDM; Inalco®) and complete EDTA-free protease inhibitors. The mixture was incubated for 1 hour at room temperature followed by centrifugation at 150,000× g for 1 hour at 4 °C, the supernatant containing the solubilized membrane proteins or complexes were purified by Ni-affinity chromatography.
The samples were loaded onto a HisTrap column (GE HealthCare) conditioned in buffer A (50 mM Hepes, pH 7.5, 300 mM NaCl, 10% glycerol, 50 mM imidazole and 1 mM DDM). The proteins or complexes of proteins were eluted by an imidazole gradient (0.05–1 M). After SDS-PAGE analysis, pure fractions were pooled and desalted on a G25 Sephadex column and concentrated using an Amicon apparatus (EMD Millipore) with a 50 kDa cutoff membrane (for PBP3 and FtsW) or a 100 kDa cutoff (for the complex) and stored at −20 °C. The concentrations of the proteins or complexes were determined with the help of the BCA reagent kit (ThermoFisher Scientific).
The expression and purification procedures of the His-tagged FtsW-PBP1b and the ternary HisPBP3-FtsW-PBP1b complexes were similar to those described above, except that the membranes were solubilized with 40 mM of Lauryl Maltose Neopentyl Glycol (LMNG, Anatrace) instead of DDM and that the purification was carried out in buffer B (50 mM Hepes, pH 7.5, 200 mM NaCl, 10% glycerol and 0.2 mM LMNG).
Before the analysis of the fractions containing PBPs by SDS-PAGE, the samples were labelled with fluorescent ampicillin at 10 μM for 30 minutes at 37 °C.
FtsWs from Klebsiella pneumoniae and Salmonella enterica were expressed in E. coli Lemo21 (DE3) strain (New England Biolabs, Ipswich, MA). Cells were grown at 37 °C in LB medium until the cell density reached an A600nm of 0.4. The temperature was then decreased to 22 °C before induction with 0.8 mM IPTG and 100 μM rhamnose. After growth at 22 °C for 16 h, cells were harvested by centrifugation at 8000 × g. Cells were resuspended in cold lysis buffer (50 mM Tris-HCl pH 7.5, 500 mM NaCl, 5% glycerol, 5 mM MgCl2 and 2 mM β-mercaptoethanol) and disrupted by a homogeneizer (Emulsiflex C3 Avestin®). The cell lysate was centrifuged at 15,000× g for 15 min at 4 °C, and the supernatant was then ultracentrifuged at 150,000× g for 1 hour. The membrane fractions were resuspended in buffer C (25 mM Sodium Phosphate pH 7, 500 mM NaCl, 5% (vol/vol) glycerol and 2 mM β-mercaptoethanol) supplemented with 40 mM DDM. After another ultracentrifugation at 150,000 × g for 30 min, the supernatant was collected and purified by affinity chromatography on a HisTrap column (GE Healthcare). Fractions containing the purified protein were pooled, desalted on a Sephadex G25 column equilibrated with buffer C, concentrated using the Amicon apparatus with 50 kDa cutoff membrane (EDM Millipore) and stored at −20 °C.
E. coli MurJ was expressed in E. coli C43 (DE3) and purified on the Ni-NTA column as described above except that the expression was induced with 0.2 mM ITPG and that the protein purified in 25 mM Tris-HCl pH 7.5, 100 mM NaCl, 5% glycerol and 2 mM β-mercaptoethanol.
Continuous fluorescence assay
The PBP1b activity assays with dansyl lipid II as substrate were performed as described36,37 in a medium binding black 96-well microplate (Greiner Bio One). The samples contained 10 μM dansyl-lipid II, 50 mM Hepes pH 7.5, 200 mM NaCl, 10 mM CaCl2, 0.02% of decyl-PEG, 20% of dimethylsulfoxide (DMSO), 1000 unit ml−1 of penicillin G, when required, and 1 unit of N-acetylmuramidase of Streptococcus globisporus (Calbiochem). The proteins FtsW, PBP3, the FtsW-PBP3 complex or MurJ, were used at 0.5–5 μM concentrations and LpoB at 200 nM. The reactions were initiated by the addition of 50 nM PBP1b or PBP1b (S510A). The reaction was monitored by following the fluorescence decrease over 20–30 min at 30 °C using an Infinite 200 PRO Microplate reader (Tecan, Männedorf, Switzerland) with excitation at 340 nm and emission at 520 nm.
Analysis of glycan chains by SDS-PAGE
In vitro PG polymerization from radioactive lipid II was performed in 50 mM Hepes pH 7.5, 10 mM CaCl2, 200 mM NaCl, 0.02% decyl-PEG, 10% DMSO, 4 μM [14C]-lipid II (0.155 μCi. nmol−1) and 100 nM PBP1b or PBP1b (S510A). FtsW, PBP3, and the FtsW-PBP3 complex or MurJ were used at 3 μM. The samples were incubated at 37 °C for 1 hour and the reactions were stopped by the addition of Flavomycin (5 mM). The reaction products were analyzed by 9% SDS-PAGE as described53. The gels were dried, exposed to a storage phosphor screen (GE Healthcare) and imaged with a Typhoon Trio+ (GE Healthcare).
The interactions of lipid II with FtsW complex was also analyzed using the same procedure with modification: SDS was omitted in the gel and the loading buffer and was only added in the running buffer.
Analysis of the reactivity of FtsW with lipid II by TLC
4 μM [14C]-lipid II was incubated with 2 μM of purified E. coli, K. penumoniae or S. enterica FtsW proteins in 50 mM Hepes/NaOH pH7.0, 20 mM MgCl2, 20 mM CaCl2, 20% DMSO and 0.05% DDM for 2 h at 30 °C. PBP1b (100 nM) was incubated in the same condition as control for PG synthesis. The reaction products were separated by thin-layer chromatography (TLC) on silica gel plates (Fluka), using 2-propanol–ammonium hydroxide (25%)–water (6:3:1, vol/vol/vol) as the mobile phase. The TLC plates were exposed to a storage phosphor screen (GE Healthcare) for 16 h, and images were revealed using a Typhoon Trio imager. The images were analyzed using Image Quant TL software (GE Healthcare).
In vitro peptidoglycan synthesis assay with HPLC analysis of product
Performed as described previously54 with changes to the buffer conditions. Reactions were performed either in a solution of 10 mM Hepes/NaOH, 3 mM MgCl2, 200 mM NaCl, 0.04% DDM (w/v), pH 7.5 or in 20 mM Hepes/NaOH, 10 mM MgCl2, 25 mM NaCl, 10% DMSO, 0.05% DDM (w/v), pH 7.5. Lipid II substrate was dried and re- dissolved in 5 μL of 0.1% DDM prior to mixing with the appropriate enzyme/protein mixtures to give 15 μM final concentration. Combinations of 0.5 μM PBP1b with 2 or 5 μM of FtsW or FtsW-PBP3 complex in the presence or absence of 2 μM LpoB (sol). Reaction proceeded for 2 h at 37 °C. Processing of PG product for HPLC analysis was as described54.
Analysis of lipid II-proteins interaction by Native-PAGE
Native-PAGE was performed essentially as described55 using a 4–15% gel gradient in glycine buffer pH 10.6. The samples: NBD-lipid II (10 μM), or lipid II-protein (10 μM/5 μM) mixtures containing FtsW, the FtsW-PBP3 complex or MurJ were supplemented with 5% sucrose before electrophoresis. The gels were imaged with a Typhoon Trio+ (GE Healthcare) followed by Coomassie blue staining.
Western blotting
After SDS-PAGE, the proteins were electro-transferred to a PVDF membrane and probed by incubation with monoclonal anti-HA antibodies (Abcam). Immuno detection was done using peroxidase conjugated goat anti-mouse IgG secondary antibodies (Millipore). The proteins were visualized by enhanced chemiluminescence (ECL kit, GE Healthcare).
Additional Information
How to cite this article: Leclercq, S. et al. Interplay between Penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci. Rep. 7, 43306; doi: 10.1038/srep43306 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Badrish Soni, Sebastien De Boel, and Marie Schlosser for their help during this work. We are grateful to Martine Nguyen-Distèche and Jean-Marie Frère for comments on the manuscript. This work was supported by the Fonds de la Recherche Scientifique, FRFC n° 2.4543.12, the Belgian Science Policy program IAP 7/44 iPros and a Wellcome Trust Senior Investigator grant to WV (101824/Z/13/Z). MT is a research associate of the FRS_FNRS (Brussels, Belgium), SL is supported by FRIA (Fonds pour la formation à la Recherche dans l’Industrie et dans l’Agriculture) fellowship FRS_FNRS.
Footnotes
The authors declare no competing financial interests.
Author Contributions S.L., A.D., S.O., C.F. performed research. E.B. prepared dansyl and NBD labeled lipid II. W.V. and A.J.F.E., prepared LpoB, performed the TP experiments and edited the MS. M.T. designed the experiments, analyzed the data and wrote the paper.
References
- Vollmer W., Blanot D. & de Pedro M. A. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32, 149–167 (2008). [DOI] [PubMed] [Google Scholar]
- Egan A. J. F. & Vollmer W. The physiology of bacterial cell division. Ann N Y Acad Sci 1277, 8–28 (2013). [DOI] [PubMed] [Google Scholar]
- Mohammadi T. et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J 30, 1425–32 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham L.-T. et al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345, 220–2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N. Lipid Flippases for Bacterial Peptidoglycan Biosynthesis. Lipid Insights 8, 21–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage E. & Terrak M. Glycosyltransferases and Transpeptidases/Penicillin-Binding Proteins: Valuable Targets for New Antibacterials. Antibitics 5, 12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage E., Kerff F. F., Terrak M., Ayala J. A. & Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32, 234–258 (2008). [DOI] [PubMed] [Google Scholar]
- Gray A. N. et al. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. Elife 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding M. A., Ogata Y., Pecora N. D., Niki H. & de Boer P. A. J. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol 63, 1008–25 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarsman M. E. et al. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol 55, 1631–1645 (2005). [DOI] [PubMed] [Google Scholar]
- Buddelmeijer N. & Beckwith J. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol Microbiol 52, 1315–1327 (2004). [DOI] [PubMed] [Google Scholar]
- Fraipont C. et al. The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology 157, 251–9 (2011). [DOI] [PubMed] [Google Scholar]
- Goehring N. W., Gonzalez M. D. & Beckwith J. Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol Microbiol 61, 33–45 (2006). [DOI] [PubMed] [Google Scholar]
- Mercer K. L. N. & Weiss D. S. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J Bacteriol 184, 904–12 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsche U. et al. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol 61, 675–690 (2006). [DOI] [PubMed] [Google Scholar]
- Müller P. et al. The Essential Cell Division Protein FtsN Interacts with the Murein (Peptidoglycan) Synthase PBP1B in Escherichia coli. J Biol Chem 282, 36394–36402 (2007). [DOI] [PubMed] [Google Scholar]
- Mohammadi T. et al. Specificity of the transport of lipid II by FtsW in Escherichia coli. J Biol Chem 289, 14707–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastoret S. et al. Functional analysis of the cell division protein FtsW of Escherichia coli. J Bacteriol 186, 8370–8379 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvorup R. N. et al. The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur J Biochem 270, 799–813 (2003). [DOI] [PubMed] [Google Scholar]
- Elhenawy W. et al. The O-Antigen Flippase Wzk Can Substitute for MurJ in Peptidoglycan Synthesis in Helicobacter pylori and Escherichia coli. PLoS One 11, e0161587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske A. J. et al. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci USA 112, 6437–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske A. J. et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis-Bleau C. et al. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143, 1110–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A. et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143, 1097–109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan A. J. F. et al. Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B. Proc Natl Acad Sci USA 111, 8197–202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Persons L., Lee L. & de Boer P. A. J. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol Microbiol 95, 945–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang M.-J. & Bernhardt T. G. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol Microbiol 95, 925–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trip E. N. & Scheffers D.-J. A 1 MDa protein complex containing critical components of the Escherichia coli divisome. Sci Rep 5, 18190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirclerc-Savoye M. et al. Reconstitution of membrane protein complexes involved in pneumococcal septal cell wall assembly. PLoS One 8, e75522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lallo G., Fagioli M., Barionovi D., Ghelardini P. & Paolozzi L. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149, 3353–9 (2003). [DOI] [PubMed] [Google Scholar]
- Karimova G., Dautin N. & Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187, 2233–43 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette A. et al. Structural determinants required to target penicillin-binding protein 3 to the septum of Escherichia coli. J Bacteriol 186, 6110–7 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissel M. C., Wendt J. L., Mitchell C. J. & Weiss D. S. The transmembrane helix of the Escherichia coli division protein FtsI localizes to the septal ring. J Bacteriol 187, 320–8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov S. et al. Large scale determination of previously unsolved protein structures using evolutionary information. Elife 4, e09248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattar M. M. et al. Two polypeptide products of the Escherichia coli cell division gene ftsW and a possible role for FtsW in FtsZ function. J Bacteriol 179, 784–93 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz B., Markwalder J. A., Seitz S. P., Wang Y. & Stein R. L. A kinetic characterization of the glycosyltransferase activity of Eschericia coli PBP1b and development of a continuous fluorescence assay. Biochemistry 41, 12552–12561 (2002). [DOI] [PubMed] [Google Scholar]
- Egan A. J. F. & Vollmer W. Continuous Fluorescence Assay for Peptidoglycan Glycosyltransferases. Methods Mol Biol 1440, 171–84 (2016). [DOI] [PubMed] [Google Scholar]
- Bertsche U., Breukink E., Kast T. & Vollmer W. In vitro murein peptidoglycan synthesis by dimers of the bifunctional transglycosylase-transpeptidase PBP1B from Escherichia coli. J Biol Chem 280, 38096–38101 (2005). [DOI] [PubMed] [Google Scholar]
- Born P., Breukink E. & Vollmer W. In vitro synthesis of cross-linked murein and its attachment to sacculi by PBP1A from Escherichia coli. J Biol Chem 281, 26985–26993 (2006). [DOI] [PubMed] [Google Scholar]
- Egan A. J. F., Biboy J., van’t Veer I., Breukink E. & Vollmer W. Activities and regulation of peptidoglycan synthases. Philos Trans R Soc B Biol Sci 370, 20150031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattar M. M., Begg K. J. & Donachie W. D. Identification of FtsW and characterization of a new ftsW division mutant of Escherichia coli. J Bacteriol 176, 7140–7 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H. et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol 1, 16172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje J. V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev 62, 181–203 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzhaf M. et al. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol 85, 179–194 (2012). [DOI] [PubMed] [Google Scholar]
- Yousif S. Y., Broome-Smith J. K. & Spratt B. G. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J Gen Microbiol 131, 2839–45 (1985). [DOI] [PubMed] [Google Scholar]
- Reed P., Veiga H., Jorge A. M., Terrak M. & Pinho M. G. Monofunctional transglycosylases are not essential for Staphylococcus aureus cell wall synthesis. J Bacteriol 193, 2549–2556 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta P. et al. Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation and mediates the formation of a trimeric complex involving FtsZ, FtsW and PBP3 in mycobacteria. Mol Microbiol 62, 1655–73 (2006). [DOI] [PubMed] [Google Scholar]
- Matsumoto S. et al. Crystal structures of an archaeal oligosaccharyltransferase provide insights into the catalytic cycle of N-linked protein glycosylation. Proc Natl Acad Sci USA 110, 17868–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrou V. I. et al. Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation. Science (80-) 351, 608–612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrak M. et al. The catalytic, glycosyl transferase and acyl transferase modules of the cell wall peptidoglycan-polymerizing penicillin-binding protein 1b of Escherichia coli. Mol Microbiol 34, 350–364 (1999). [DOI] [PubMed] [Google Scholar]
- Breukink E. et al. Lipid II is an intrinsic component of the pore induced by nisin in bacterial membranes. J Biol Chem 278, 19898–19903 (2003). [DOI] [PubMed] [Google Scholar]
- Lakaye B. et al. Synthesis, purification and kinetic properties of fluorescein-labelled penicillins. Biochem J 300 Pt 1, 141–145 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett D. et al. Analysis of glycan polymers produced by peptidoglycan glycosyltransferases. J Biol Chem 282, 31964–31971 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biboy J., Bui N. K. & Vollmer W. In vitro peptidoglycan synthesis assay with lipid II substrate. Methods Mol Biol 966, 273–88 (2013). [DOI] [PubMed] [Google Scholar]
- Gallagher S. R. One-dimensional electrophoresis using nondenaturing conditions. Curr Protoc Mol Biol Chapter 10, Unit 10.2B (2001). [DOI] [PubMed] [Google Scholar]
- Miroux B. & Walker J. E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260, 289–298 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.