Abstract
The gab operon (gabDTPC) in Escherichia coli functions in the conversion of γ-aminobutyrate to succinate. One component of gab operon regulation involves the RpoS sigma factor, which mediates activation at high cell density. Transposon mutagenesis was used to identify new genes that regulate gab operon expression in rich media. A Tn5tmp insertion in the hldD (formerly rfaD) gene increased gabT::lacZ expression 12-fold. The hldD gene product, an ADP-l-glycerol-d-mannoheptose-6-epimerase, catalyzes the conversion of ADP-d-glycerol-d-mannoheptose to ADP-l-glycerol-d-mannoheptose, a precursor for the synthesis of inner-core lipopolysaccharide (LPS). Defined mutations in hldE, required for heptose synthesis, and waaF, required for the addition of the second heptose to the inner core, also resulted in high-level gabT::lacZ expression. The hldD, hldE, and waaF mutants exhibited a mucoid colony phenotype due to production of a colanic acid capsule. However, in the hldD::cat background, the high-level expression of gabT::lacZ was independent of the regulatory components for colanic acid synthesis (rcsA, rcsB, and rcsC) and also independent of manC (cpsB), a structural gene for colanic acid synthesis. Activation of gabT::lacZ in the hldD::cat background was dependent on the RpoS sigma factor. The hldD::cat mutation resulted in a sixfold increase in the levels of a translational RpoS-LacZ fusion and had a marginal effect on a transcriptional fusion. This study reveals a stress-induced pathway, mediated by loss of the LPS inner core, that increases RpoS translation and gab operon expression in E. coli.
The gabDTPC operon in Escherichia coli functions in the conversion of γ-aminobutyrate (GABA) to succinate (3, 17). The gabD gene encodes a succinate:semialdehyde dehydrogenase that generates succinate from succinate-semialdehyde. The gabT gene encodes a succinate:semialdehyde aminotransferase that catalyzes the formation of succinate semialdehyde from GABA. The gabP gene encodes a GABA permease (3). In addition to the catabolism of γ-aminobutyrate, the gab operon has been proposed to contribute to polyamine homeostasis during nitrogen-limited growth (25) and to maintain high internal glutamate concentrations under stress conditions (14).
Regulation of the gab operon occurs at multiple levels. At least three promoters that transcribe the gab operon have been identified. The RpoS-encoded sigma factor σS or σ38 transcribes the gab operon, and recent studies indicate that two RpoS-dependent promoters are present upstream of the gab operon (2, 14, 23). In addition, a σ70-dependent promoter, together with the Nac regulatory protein, contributes to expression under nitrogen-limiting conditions (25, 33). In nitrogen-rich environments, two accumulated extracellular signals, indole and a second unidentified signal, independently contribute to activation of the gab operon at high cell density in an RpoS-dependent manner (2, 31). Expression of the gab genes is also enhanced at high pH (28). Negative regulation of gab expression is mediated by the GabC repressor (23), also termed CsiR (14). It has been proposed that activation by extracellular signals and RpoS is dependent on loss of the GabC repressor (25).
In this study, a genetic approach was used to search for genes that regulate expression of a gabT::lacZ fusion under conditions of nitrogen excess. The parent strain for these studies was PR1, a strain unable to produce indole (Table 1). Therefore, PR1 relies on a second unknown signal for activation of gabT::lacZ by cell-cell signaling (31). We were originally interested in mutations that altered the production or response to the second signal. However, in this study, mutations were identified that altered gabT::lacZ expression by a previously unreported mechanism. Mutations in hldD or hldE, which truncate the inner core of lipopolysaccharide (LPS), resulted in high-level expression of the gabT::lacZ fusion along with the induction of mucoidy. However, the colanic acid capsule itself did not have a role in the high-level expression of gabT::lacZ. In addition, the overexpression of gabT::lacZ in the hldD::cat background was independent of RcsA and the RcsB/C two-component system. This pathway for activation of gabT::lacZ in the hldD::cat background was dependent on RpoS, and the loss of hldD increased RpoS translation.
TABLE 1.
Strains used in this study
| E. coli strain | Genotype | Source or reference |
|---|---|---|
| ZK126 | tna-2 ΔlacU169 | 31 |
| VS20320 | rcsB::mini-Tn10tet | V. Stout |
| PR1 | ZK126 gabT::mini-Tn5lacZ-tet/1 | This study |
| MJ2 | PR1 hldD::Tn5tmp | This study |
| MJ11 | PR1 hldD::cat | This study |
| MJ12 | MJ11 rcsB::mini-Tn10tet | This study |
| MJ13 | PR1 hldE::cat | This study |
| MJ18 | MJ11 cpsB(manC)::tet | This study |
| MJ19 | PR1 waaL::cat | This study |
| MJ20 | PR1 waaF::cat | This study |
| MJ27 | MJ11 rcsA::tet | This study |
| MJ31 | MJ11 rpoS::Tn10 | This study |
| MJ32 | MJ2 rcsC::cat | This study |
| DDS1626 | rpoS-lacZ | D. Sledjeski |
| PR1000 | DDS1626 hldD::cat | This study |
Isolation of a mutant with high-level expression from a gabT::lacZ fusion.
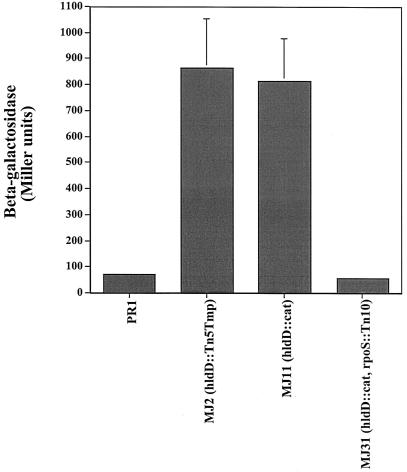
To search for genes that regulate a gabT::lacZ transcriptional fusion in E. coli strain PR1, a library of mini-Tn5tmp insertions was constructed using the EZ::TN DHFR-1 Tnp Kit (Epicentre Technologies). The Tn5tmp transposome was electroporated into E. coli PR1, and cells were plated on Luria-Bertani (LB) agar containing trimethoprim (50 mg/ml) and 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal). From approximately 14,600 trimethoprim-resistant colonies, a pale blue, mucoid colony was isolated and designated MJ2. To determine whether the pale blue phenotype was related to gabT::lacZ expression, PR1 and MJ2 cells were assayed for β-galactosidase in LB broth. Unexpectedly, despite the pale blue phenotype on X-Gal plates, the expression of gabT::lacZ was 12-fold higher in MJ2 (864 ± 188 U) than in PR1 (72 ± 4 U) (Fig. 1). The discrepancy between colony color and β-galactosidase expression was due to a masking of the colony color by the capsular material.
FIG. 1.
Loss of hldD leads to activation and gabT::lacZ. The indicated strains were grown to early log phase (A600 = 0.35) in 0.5× LB medium at pH 7.5 and analyzed for β-galactosidase by the method of Miller (15). Data shown are means of duplicate samples from three independent experiments. Error bars, standard deviations.
Loss of hldD leads to increased expression of gabT::lacZ.
MJ2 was found to be resistant to P1 lysis, and repeated attempts to use transduction to confirm linkage of trimethoprim resistance to the MJ2 phenotypes were unsuccessful. The Tn5tmp insertion was cloned, and sequence analysis of the transposon junction indicated an insertion at position 1286 in the 2,037-bp hldD (formerly rfaD) gene. HldD is an ADP-l-glycerol-d-mannoheptose-6-epimerase that catalyzes the conversion of ADP-d-glycerol-d-mannoheptose to ADP-l- glycerol-d-mannoheptose, a precursor for the synthesis of inner lipopolysaccharide (5). The phenotypes of MJ2, such as the formation of mucoid colonies, resistance to P1 lysis, and sensitivity to detergents, were consistent with hldD mutants described in the literature (5, 6, 18, 20, 24). Since P1 transduction could not be used to move the hldD::Tn5tmp mutation into a fresh background, a hldD::cat mutation was constructed in strain PR1 by the one-step inactivation procedure of Datsenko and Wanner (7). PCR products were generated from pKD3 using the primers listed in Table 2. The hldD::cat mutation in PR1 was verified by PCR, and this strain was designated MJ11. Like MJ2, strain MJ11 was mucoid, resistant to P1 lysis, and sensitive to detergents and exhibited constitutive expression of the gabT::lacZ fusion (Fig. 1). Therefore, we conclude that an insertion in hldD was responsible for the increased expression of gabT::lacZ. In addition, the above phenotypes of the hldD::cat allele, i.e., the mucoid phenotype, detergent sensitivity, and increased gabT::lacZ expression, were restored to the wild type by a plasmid containing the E. coli hldD gene (data not shown).
TABLE 2.
PCR primers used for one-step inactivation of genes
| Gene | Primera | Sequence (5′→3′)b |
|---|---|---|
| hldD | F | CCACGAAGGCGCGTGCTCTTCCACCACCGAGTGGGACGGCACATATGAATATCCTCCTTAG |
| R | AACGGAAGCCAACAATCTGCGAGTTCGCTTCAGGCAGGATTTGTGTAGGCTGGAGCTGCTTC | |
| hldE | F | ATGAAAGTAACGCTGCCAGAGTTTGAACGTGCAGGAGTGATGCATATGAATATCCTCCTTAG |
| R | CTTCTTGATGATGTTGGTCGTCGAGCAACCGTCTTCAAAGTTGTGTAGGCTGGAGCTGCTTC | |
| waaL | F | AATGACCGCATTTCTTTTGGTGTAGGAACGGCAACAGGAGCCATATGAATATCCTCCTTAG |
| R | AATTCCCATCAGACCTTTCAGTGACCCTGCTTCAATTATCTTGTGTAGGCTGGAGCTGCTTC | |
| rcsA | F | GTTGATGACCTTGCCATAGCTTGTGATTCACAGCGCCCTTCGGTTTTTTCGTTTTCAGAG |
| R | CGGTCTTGGCTTTGATATTCATTTGGTCAGAGATTTGAATCGCTGCCCGAGATGCGCCGC | |
| manC | F | AACCGAATCTGGAAACCGCTCAGGCCTATGTGGCAAGCGGCGGTTTTTTCGTTTTCAGAG |
| R | CTGTACCGCGTTACGGTCGGCAATCAGCACCGCATCTTTGCGCTGCCCGAGATGCGCCGC | |
| waaF | F | GCCGGAAGTTAACGAAGCTATTCCTATGCCTCTCGGTCACGCATATGAATATCCTCCTTAG |
| R | TCATGATCTTTCGCCGAGCCAAACAGAACCACCTGATAACCTGTGTAGGCTGGAGCTGCTTC | |
| rcsC | F | TGTTCAGAGCATTGGCGTTAGTGCTCTGGCTGTTGATTGCTTCATATGAATATCCTCCTTAG |
| R | CGTCAGCGTCTGTTTTATCACATCCAGCGTTACCGGCTTCGACAGTGTGTAGGCTGGAGCTGCTTC |
F and R represent the forward and reverse primers, respectively.
The underlined sequences are the primer sequences that amplify the antibiotic resistance cassette.
Expression of the gabT::lacZ fusion used in this study was previously shown to be dependent on the RpoS sigma factor (2). To determine whether the high-level gabT::lacZ expression in the hldD::cat background was RpoS dependent, an hldD::cat rpoS::Tn10 double mutant was constructed. The activation of gabT::lacZ was abolished in the double mutant, with levels of β-galactosidase (58 ± 2 Miller units) that were slightly lower than those in the wild type (72 ± 4 Miller units) (Fig. 1).
The loss of the inner-core domain of lipopolysaccharide results in constitutive gabT::lacZ expression.
hldD is the first gene in the hldDFCL operon, and the HldD enzyme (ADP-l-glycerol-d-mannoheptose-6-epimerase) catalyzes the last step in the biosynthetic pathway of the heptose LPS precursor (1, 11, 22). The increased gabT::lacZ expression in the hldD mutant could directly result from the loss of heptose. However, an alternative possibility, given the unexpected role in regulation, is that the HldD enzyme carries out a second undefined activity that accounts for the changes in gabT::lacZ expression. To address the possibility that a general loss of heptose can lead to this phenotype, a heptoseless mutant was created by inactivation of the hldE (rfaE) gene using the PCR-mediated one-step procedure (7). HldE is a bifunctional enzyme with d-β-d heptose phosphate-7-kinase and d-β-d heptose-1-phosphate adenylyltransferase activities. This enzyme is required for the second and fourth step in the biosynthesis of ADP-l-β-d-heptose (11). Strain MJ13 hldE::cat was mucoid and exhibited a 5.7-fold increase in expression of the gabT::lacZ fusion. Therefore, loss of heptose is sufficient to confer constitutive activation of gabT::lacZ.
The insertional mutations in hldD are likely to be polar on the downstream waaFCL genes that are involved in the assembly of the heptose precursor into LPS. WaaC transfers the first heptose to the inner-core backbone, and WaaF transfers the second heptose to the core (26, 32). WaaL is required for the addition of O antigen to the core. To address the role of the downstream genes in gabT::lacZ expression, the waaF and waaL genes were individually mutated by the insertion of a chloramphenicol resistance cassette via PCR-mediated mutagenesis (7). The waaF::cat mutation in MJ20 resulted in a 5.6-fold increase in gabT::lacZ expression relative to that in the wild type. In contrast, the waaL::cat mutation in MJ19, which prevents the addition of O antigen to the core, had no effect on gabT::lacZ expression. Since E. coli K-12 strains are already devoid of O antigen, this result was expected. However, the results with the waaF and hldD mutations are consistent with truncation of the LPS inner core leading to gabT::lacZ activation.
The colanic acid capsule and the Rcs regulatory proteins are not required for gabT activation in hldD mutants.
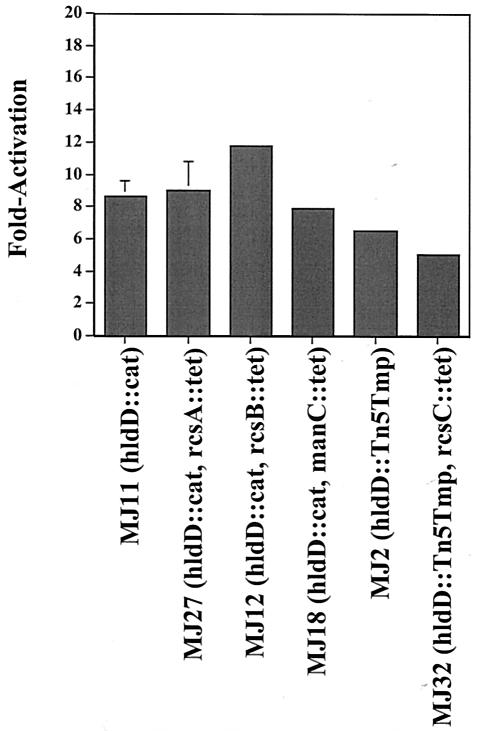
A prominent phenotype of the hldD mutant was the production of capsular polysaccharide. Synthesis of the colanic acid capsule is controlled by the regulators of colanic acid synthesis (rcs) genes composed of the activator RcsA, the sensor kinase RcsC, and the response regulator RcsB (4, 8, 9, 10, 19, 30). First, to determine whether the production of colanic acid in hldD::cat cells was responsible for the constitutive activity of gabT::lacZ, a double-mutant MJ18 strain (hldD::cat manC::tet) was constructed. The manC gene, formerly called cpsB, encodes a mannose-1-phosphate guanosyltransferase required for production of colanic acid (29). As expected, MJ18 exhibited a nonmucoid phenotype. The gabT::lacZ fusion in MJ18 hldD::cat manC::tet was activated 7.9-fold above the levels seen in PR1, indicating that colanic acid itself was not responsible for the high-level gabT::lacZ expression in the hldD background (Fig. 2). The role of the rcs genes in gabT::lacZ activation was also examined. The loss of rcsA or rcsB in the hldD::cat background did not reduce the high-level gabT::lacZ expression, and the loss of rcsB enhanced expression (Fig. 2). The loss of rcsC in the hldD::Tn5tmp background resulted in levels of gabT::lacZ expression that were slightly reduced, with 5.1-fold activation versus the 6.5-fold activation in MJ2 hldD::Tn5tmp (Fig. 2).
FIG. 2.
The colanic acid capsule and the Rcs regulatory proteins are not required for gabT::lacZ activation in the hldD::cat background. Cells were grown in 0.5× LB medium at pH 7.5, harvested for β-galactosidase analysis at early log phase (A600 = 0.35), and assayed by the method of Miller (15). The reported values represent the fold activation of the gabT::lacZ fusion in the various strains relative to the expression in the isogenic parent PR1. The baseline value of β-galactosidase in PR1 gabT::lacZ was 68 ± 1 U.
Loss of hldD increases RpoS translation.
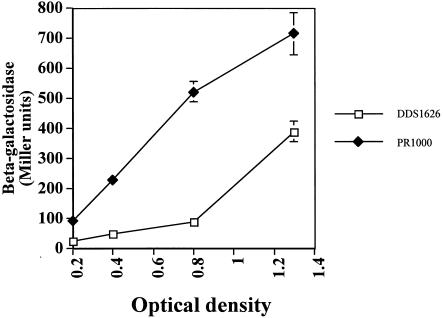
To further investigate the role of RpoS in the increased expression of gabT in the hldD background, we addressed whether RpoS itself was increased in hldD mutants. An RpoS-LacZ translational fusion was used to examine expression in wild-type and hldD::cat backgrounds. As shown in Fig. 3, the expression of RpoS-LacZ was markedly increased in the hldD::cat background relative to that in the wild type. The maximum difference was observed at an optical density at 600 nm of 0.8, where a sixfold increase was evident in the hldD mutant (Fig. 3). The effect of hldD::cat on rpoS transcription was also examined by reverse transcription-PCR analysis. The hldD:cat allele has less than a twofold effect on rpoS mRNA levels (data not shown).
FIG. 3.
Loss of hldD increases RpoS translation. Cultures of either DDS1626 (RpoS-LacZ) or PR1000 (RpoS-LacZ, hldD::cat) were grown in 0.5× LB medium, and β-galactosidase activities were assayed at the indicated optical densities. The reported values represent the average of quadruplicate samples from two independent experiments.
These data combined with those in Fig. 1 suggest that RpoS overexpression in the hldD::cat background is a primary mechanism for the activation of gabT::lacZ. Two additional experiments were conducted to verify that increased RpoS expression results in overexpression of the gabT::lacZ fusion. First, RpoS expression is increased at low temperature (27). Expression of the gabT::lacZ fusion was increased fourfold at 25°C compared to that at 37°C (51 ± 0.35 versus 13 ± 1 Miller units). Second, the response regulator RssB (SprE) targets RpoS for proteolysis, and rssB mutants accumulate increased levels of RpoS (16, 21). The expression of gabT::lacZ was elevated fivefold in an rssB/sprE mutant compared to that in the wild type (data not shown).
Concluding remarks.
In this study, a gabT::lacZ fusion was used as a biosensor to identify loci involved in its regulation. Mutations in hldD, hldE, and waaF, which result in loss of the heptose precursor for the inner core of lipopolysaccharide, resulted in activation of a gabT::lacZ fusion. This pathway for gabT::lacZ activation was independent of RcsA, RcsB, and RcsC. However, one possible caveat in the interpretation of a role for RcsA is the fact that the hldD rcsA double mutant formed small colonies that were genetically unstable and gave rise to larger colonies. Although great care was taken in assaying the hldD rcsA double mutant, including assays on primary transductants, we cannot rule out the possibility that second-site suppressor mutations restored some level of gabT::lacZ expression in this background.
In the hldD::cat background, the activation of gabT::lacZ required a functional RpoS (Fig. 1). Moreover, the expression of an RpoS-LacZ translational fusion was increased sixfold in the hldD::cat background (Fig. 3). These data indicate that the increased expression of gabT::lacZ was likely due to increased levels of RpoS. To our knowledge, the increase in RpoS translation in response to loss of the LPS inner core has not been previously described. Studies by Majdalani et al. have demonstrated that a small regulatory RNA, rprA, regulates RpoS translation and that cell envelope stress increased the expression of rprA in a manner that was dependent on the RcsC/YojN/RcsB phosphorelay (12, 13). Moreover, it has also been demonstrated that loss of the LPS inner core acts directly, or indirectly, to regulate activity of the RcsC/YojN/RcsB phosphorelay (19, 24). Therefore, the mechanism that increases RpoS translation in response to loss of the LPS inner core could involve the RcsC/YojN/RcsB phosphorelay. However, this phosphorelay is clearly not required for the increased expression of the gab operon in the hldD::cat background (Fig. 2). This information, together with the fact that the high-level gab expression in the hldD::cat background is strictly RpoS dependent (Fig. 1), suggests that a second pathway for increasing RpoS expression in the hldD::cat background exists. Osmotic shock, a condition that may mimic loss of the LPS inner core, has previously been shown to increase RpoS translation in a manner that required both rprA and a second small regulatory RNA, dsrA (12, 27). Our preliminary studies indicate that the individual loss of rprA or dsrA had only a minor effect on the increased RpoS translation in the hldD::cat background. However, both rprA and dsrA may be required for the increased RpoS translation in mutants defective in the LPS inner core. The identification of gene products that are required for the increased RpoS translation in the hldD::cat background will provide important information on how cells transmit alterations in LPS to regulation of RpoS and the gab operon.
Acknowledgments
This work was supported by NSF awards 9904766 and 0406047 (P.N.R.) and NIH award TW-000-11.
We thank Valerie Stout for the rcsB mutant.
REFERENCES
- 1.Austin, E. A., J. F. Graves, L. A. Hite, C. T. Parker, and C. A. Schnaitman. 1990. Genetic analysis of lipopolysaccharide core biosynthesis by Escherichia coli K-12: insertion mutagenesis of the rfa locus. J. Bacteriol. 172:5312-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baca-DeLancey, R. R., M. M. South, X. Ding, and P. N. Rather. 1999. Escherichia coli genes regulated by cell-to-cell signaling. Proc. Natl. Acad. Sci. USA 96:4610-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartsch, K., A. von Johnn-Marteville, and A. Schulz. 1990. Molecular analysis of two genes of the Escherichia coli gab cluster: nucleotide sequence of the glutamate:succinic semialdehyde transaminase gene (gabT) and characterization of the succinic semialdehyde dehydrogenase gene (gabD). J. Bacteriol. 172:7035-7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brill, J. A., C. Quinlan-Walshe, and S. Gottesman. 1988. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 170:2599-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman, W. G., Jr. 1983. The rfaD gene codes for ADP-l-glycero-d-mannoheptose-6-epimerase. An enzyme required for lipopolysaccharide core biosynthesis. J. Biol. Chem. 258:1985-1990. [PubMed] [Google Scholar]
- 6.Coleman, W. G., Jr., and L. Leive. 1979. Two mutations which affect the barrier function of the Escherichia coli K-12 outer membrane. J. Bacteriol. 139:899-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottesman, S., and V. Stout. 1991. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol. Microbiol. 5:1599-1606. [DOI] [PubMed] [Google Scholar]
- 9.Gottesman, S., P. Trisler, and A. Torres-Cabassa. 1985. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J. Bacteriol. 162:1111-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupte, G., C. Woodward, and V. Stout. 1997. Isolation and characterization of rcsB mutations that affect colanic acid capsule synthesis in Escherichia coli K-12. J. Bacteriol. 179:4328-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kneidinger, B., C. Marolda, M. Graninger, A. Zamyatina, F. McArthur, P. Kosma, M. A. Valvano, and P. Messner. 2002. Biosynthesis pathway of ADP-l-glycero-beta-d-manno-heptose in Escherichia coli. J. Bacteriol. 184:363-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majdalani, N., S. Chen, J. Murrow, K. St John, and S. Gottesman. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39:1382-1394. [DOI] [PubMed] [Google Scholar]
- 13.Majdalani, N., D. Hernandez, and S. Gottesman. 2002. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46:813-826. [DOI] [PubMed] [Google Scholar]
- 14.Metzner, M., J. Germer, and R. Hengge. 2004. Multiple stress signal integration in the regulation of the complex σS-dependent csiD-ygaF-gabDTP operon in Escherichia coli. Mol. Microbiol. 51:799-811. [DOI] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1977. Experiments in molecular genetics, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Muffler, A., D. Fischer, S. Altuvia, G. Storz, and R. Hengge-Aronis. 1996. The response regulator RssB controls stability of the sigma(S) subunit of RNA polymerase in Escherichia coli. EMBO J. 15:1333-1339. [PMC free article] [PubMed] [Google Scholar]
- 17.Niegemann, E., A. Schulz, and K. Bartsch. 1993. Molecular organization of the Escherichia coli gab cluster: nucleotide sequence of the structural genes gabD and gabP and expression of the GABA permease gene. Arch. Microbiol. 160:454-460. [DOI] [PubMed] [Google Scholar]
- 18.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pegues, J. C., L. S. Chen, A. W. Gordon, L. Ding, and W. G. Coleman, Jr. 1990. Cloning, expression, and characterization of the Escherichia coli K-12 rfaD gene. J. Bacteriol. 172:4652-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pratt, L. A., and T. J. Silhavy. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. USA 93:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roncero, C., and M. J. Casadaban. 1992. Genetic analysis of the genes involved in synthesis of the lipopolysaccharide core in Escherichia coli K-12: three operons in the rfa locus. J. Bacteriol. 174:3250-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schellhorn, H. E., J. P. Audia, L. I. Wei, and L. Chang. 1998. Identification of conserved, RpoS-dependent stationary-phase genes of Escherichia coli. J. Bacteriol. 180:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnaitman, C. A., and J. D. Klena. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider, B. L., S. Ruback, A. K. Kiupakis, H. Kasbarian, C. Pybus, and L. Reitzer. 2002. The Escherichia coli gabDTPC operon: specific gamma-aminobutyrate catabolism and nonspecific induction. J. Bacteriol. 184:6976-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sirisena, D. M., K. A. Brozek, P. R. MacLachlan, K. E. Sanderson, and C. R. Raetz. 1992. The rfaC gene of Salmonella typhimurium. Cloning, sequencing, and enzymatic function in heptose transfer to lipopolysaccharide. J. Biol. Chem. 267:18874-18884. [PubMed] [Google Scholar]
- 27.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 28.Stancik, L. M., D. M. Stancik, B. Schmidt, D. M. Barnhart, Y. N. Yoncheva, and J. L. Slonczewski. 2002. pH-dependent expression of periplasmic proteins and amino acid catabolism in Escherichia coli. J. Bacteriol. 184:4246-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres-Cabassa, A. S., and S. Gottesman. 1987. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J. Bacteriol. 169:981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, D., X. Ding, and P. N. Rather. 2001. Indole can act as an extracellular signal in Escherichia coli. J. Bacteriol. 183:4210-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkinson, R. G., P. Gemski, Jr., and B. A. Stocker. 1972. Non-smooth mutants of Salmonella typhimurium: differentiation by phage sensitivity and genetic mapping. J. Gen. Microbiol. 70:527-554. [DOI] [PubMed] [Google Scholar]
- 33.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]