Abstract
Vancomycin is used increasingly to treat invasive infections caused by multidrug-resistant Streptococcus pneumoniae. Although no vancomycin-resistant strains have been isolated to date, tolerant strains that fail to die rapidly and that cause relapsing disease have been described. The vex123-pep27-vncRS locus, consisting of an ABC transporter, a presumed signaling peptide, and a two-component system, respectively, has been implicated in vancomycin tolerance. Recent findings, however, challenged this model. The data presented here indicate that erythromycin in the growth medium induces a vancomycin-tolerant phenotype and that loss of function of Pep27 or VncRS does not alter autolysis. However, a role for the ABC transporter encoded by the vex123 genes in tolerance was confirmed. A vex3 mutant was considerably more tolerant to vancomycin treatment than the wild-type strain T4, and the strength of the phenotype depended on the orientation of the resistance cassette used to construct the mutant. Microarray results suggested a number of genes that might be involved in tolerance in the vex3 mutant. Although the exact function and regulation of the vex123-pep27-vncRS locus remains to be determined, several factors influence the autolysis behavior of S. pneumoniae, including the bacterial capsule, erythromycin, and the lytA and vex3 gene products.
Streptococcus pneumoniae, the causative agent of middle ear infections, pneumonia, bacteremia, and meningitis, has demonstrated increased resistance to previously effective antibiotics (1, 33). Vancomycin has been used more frequently for severe, invasive disease where immediate intervention is required, since no resistant strains have been reported to date. Exposure to vancomycin or β-lactams results in autolysis of sensitive cells (6). Strains which do not undergo autolysis after treatment with these antibiotics show increased survival and are termed tolerant (10, 20, 32). This phenotype is of potential medical significance since tolerance can lead to treatment failures and facilitate the development of antibiotic resistance (20). Recently, reports of vancomycin tolerance have been published, raising anxiety about treatment options, particularly for meningitis (2, 10, 11, 20).
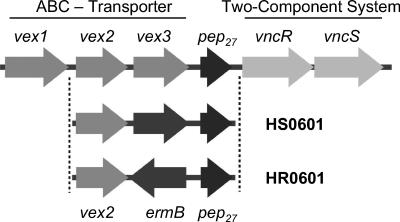
In a study seeking the signaling cascade that activates the autolytic pathway by analysis of loss-of-function mutants, a genetic locus consisting of six genes was identified (Fig. 1) (22, 23). Three of the genes, vex1, vex2, and vex3, have similarity to the transmembrane and ATP-binding proteins of ABC transporters, while the fourth gene, pep27, was predicted to encode a peptide of 27 amino acids. The last two genes in the locus, vncR and vncS, have similarity to response regulator and histidine kinase subunits of a two-component system. The proposed function of the proteins was to measure cell density via a quorum-sensing mechanism and to initiate autolysis. The model predicted that Pep27 was secreted from the cell by the Vex123 transporter system and that the peptide accumulated outside of the cell. At a threshold concentration in the environment, Pep27 was expected to activate the membrane-bound signal sensor VncS. The signal would then be relayed to the DNA-binding protein VncR, which was hypothesized to be a repressor of genes involved in autolysis. The model also predicted that antibiotics, such as penicillin or vancomycin, could short-circuit this activation process and trigger autolysis before stationary growth phase was reached (22, 23).
FIG. 1.
Organization of the vex-vnc locus in S. pneumoniae. The Vex1 and Vex3 proteins are predicted to function as transmembrane permeases, while the Vex2 protein contains motifs typical for an ABC cassette protein. The three proteins together represent an ABC transport system. Pep27 was reported to be a peptide involved in cell-cell signaling, and VncR and VncS are subunits of a two-component signal transduction system. The vex-vnc locus in the mutant strains HS0601 and HR0601 is depicted schematically to indicate the replacement of vex3 by ermB. The arrows indicate the directions of gene transcription.
Recent work has challenged this model. In this second study, Robertson et al. (26) were unable to detect a biological effect of Pep27 on a growing culture of S. pneumoniae, and deletions in the vncRS genes did not alter lysis with respect to vancomycin. As an alternative explanation, it was suggested that erythromycin in the growth medium, which is required to maintain the stability of the mutants created by insertion-duplication mutagenesis, caused a vancomycin-tolerant phenotype regardless of the specific gene that was mutated (26).
This inconsistency between the two studies prompted a detailed reanalysis of the vex-vnc locus and the behavior of individual mutants with respect to vancomycin treatment. Our data confirm that the presence of erythromycin increases the tolerance of a strain to vancomycin. Newly constructed mutations in the genes pep27, vncR, or vncS had no effect on vancomycin tolerance. The role of the VncRS two-component system remains to be determined, since transcriptional analyses of various mutants also did not demonstrate regulation of vex123 or other genes. However, the replacement of vex3 by an ermB resistance cassette did yield a strain that was vancomycin tolerant, even in the absence of erythromycin. Transcriptional analyses showed that several genes are differentially regulated in this tolerant vex3 mutant.
MATERIALS AND METHODS
Bacterial strains and PCR ligation mutagenesis.
The strains of S. pneumoniae used in this study are listed in Table 1. Null mutants in the genes for vex3, pep27, vncR, vncS, or lytA were generated in S. pneumoniae strain T4 by PCR ligation mutagenesis (18). Gene SP2155, coding for a truncated immunoglobulin A1 (IgA1) protease (29) (http://www.tigr.org), was mutated to serve as the negative control (strain HS2155). lytA, coding for the LytA autolysin, was mutated as the positive control (strains HS1937 and HR1937). DNA fragments of ∼1 kb representing sequences 5′ and 3′ of the gene to be deleted were amplified by PCR using TaKaRa LA Taq DNA polymerase (Takara, Shiga, Japan) and primers (available on request). PCR products were purified by using Wizard PCR Preps (Promega, Madison, Wis.) and digested with either NgoMIV or EagI (New England BioLabs, Beverly, Mass.). The ermB expression cassette from plasmid pTCV-lac (25) was PCR amplified and digested with XmaI and PspOMI. 5′ and 3′ PCR fragments were ligated to the ermB cassette to yield ligation products of approximately 3 kb. Alternatively, PCR products were fused together by gene splicing by overlap extension (SOEing) (12, 13). Mutagenic PCR products were transformed into S. pneumoniae T4 by using standard methods (3). Erythromycin-resistant clones were selected on tryptic soy agar plates containing 4% sheep blood and 1 μg of erythromycin (Sigma)/ml. MICs were determined by using the E-test. The parental strain T4 was sensitive (MIC, 0.125 μg/ml) to erythromycin, while all mutant strains, containing the ermB gene, were resistant (MICs, 48 mg/ml or higher) to erythromycin. Mutant clones were confirmed by DNA sequence analysis using primers that annealed outside the DNA region used for mutagenesis. This analysis was done to ensure the correct location and sequence of the ermB gene and surrounding DNA. All sequencing reactions were carried out at the Hartwell Center for Biotechnology at St. Jude Children's Research Hospital.
TABLE 1.
Streptococcus pneumoniae strains used in this study
| Strain | Characteristics | Reference |
|---|---|---|
| T4 | Clinical isolate from the blood of a 30-year-old male patient, capsular serotype 4, highly virulent in mouse model | 29 |
| T4R | Unencapsulated derivative of strain T4, hyperlytic compared to strain T4 | 9 |
| R6 | Unencapsulated derivative of strain D39, capsular serotype 2, avirulent laboratory strain | 14 |
| Tupelo | Clinical isolate from a 10-month-old girl with meningitis, capsular serotype 14, multidrug resistant, naturally vancomycin tolerant | 20 |
| HS2155 | T4 mutant, truncated immunoglobulin A1 protease gene (SP2155) replaced by ermB cassette | This study |
| HS1937 | T4 mutant, autolysin lytA gene (SP1937) replaced by ermB cassette | This study |
| HR1937 | T4 mutant, autolysin lytA gene (SP1937) replaced by ermB cassette, ermB in reverse orientation relative to lytA | This study |
| HS0601 | T4 mutant, vex3 gene (SP0601) replaced by ermB cassette | This study |
| HR0601 | T4 mutant, vex3 gene (SP0601) replaced by ermB cassette, ermB in reverse orientation relative to vex3 | This study |
| HS0602 | T4 mutant, pep27 gene (SP0602) replaced by ermB cassette | This study |
| HS0603 | T4 mutant, vncR gene (SP0603) replaced by ermB cassette | This study |
| HS0604 | T4 mutant, vncS gene (SP0604) replaced by ermB cassette | This study |
| HR0604 | T4 mutant, vncS gene (SP0604) replaced by ermB cassette, ermB in reverse orientation relative to vncS | This study |
Bacterial growth and kill curves.
Overnight cultures of S. pneumoniae were grown from −70°C glycerol stocks. Cultures were routinely grown overnight in 10 ml of C+Y medium in KIMAX culture tubes (18 by 150 mm) at 37°C and 5% CO2 without agitation. The following day, fresh cultures were started by diluting the overnight cultures 1:100 in prewarmed C+Y medium. The turbidity of the cultures was measured in 30-min intervals with a Turner model 340 spectrophotometer at 620 nm. Vancomycin (5 μg/ml; 10× MIC; Sigma) was added at an optical density at 620 nm (OD620) of 0.25 to 0.30, corresponding to mid-logarithmic growth phase and approximately 3 × 107 CFU/ml. Bacterial viability (CFU/ml) was determined at the time of the addition of vancomycin and 4 h thereafter by plating 10-μl samples on tryptic soy broth-blood agar plates (15) and counting the colonies at 18 to 24 h.
RNA isolation.
Bacterial cells were harvested for RNA isolation during logarithmic growth phase at an OD620 of 0.45 to 0.5, which corresponds to 7.5 × 107 CFU/ml. RNA isolation was performed by using the QIAGEN RNeasy Mini Kit (QIAGEN, Inc., Valencia, Calif.) with the following modifications. Bacteria were lysed by shaking for 5 min in the presence of 400 mg of 0.1-mm zirconia-silica beads (BioSpec Products) by using a Mini-Beadbeater 3110BX (BioSpec), followed by incubation at 70°C for 10 min. The lysate was centrifuged through a QIAshredder (QIAGEN) and processed according to the manufacturer's instructions, with an on-column DNase digestion step. Quantitation of RNA samples was performed by using a UV spectrophotometer (UV-1601; Shimadzu, Kyoto, Japan) at OD260.
Northern blot.
For Northern blot analysis, 5 μg of total RNA was separated on a 1.2% MOPS (morpholinepropanesulfonic acid)-formaldehyde gel and transferred to nylon membranes (Duralon-UV; Stratagene, La Jolla, Calif.) according to standard procedures (28). Hybridization reactions with ermB- or vncS-specific probes were carried out by using Church's buffer (5) for 16 h at 66°C. Blots were washed with 1× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])-0.1% sodium dodecyl sulfate at 66°C and exposed at −80°C to BioMax MR film (Eastman Kodak Company, Rochester, N.Y.) with intensifying screens.
Microarray analysis.
Whole-genome S. pneumoniae cDNA microarrays were received as a grant from the Pathogen Functional Genomics Resource Center (PFGRC; http://pfgrc.tigr.org/; The Institute for Genomic Research, Rockville, Md.). Microarray analyses were performed according to MIAME (minimum information about a microarray experiment) standards by using the protocols provided by PFGRC with minor modifications. cDNA was generated from bacterial RNA with random hexamers and SuperScript II (Invitrogen, Carlsbad, Calif.) in the presence of 5-(3-aminoallyl)-dUTP (Ambion, Inc.). Cy-3 and Cy-5 monoreactive fluorescent dyes were then coupled to aminoallyl group-containing cDNA products according to PFGRC standard operating procedure M007 (8). The fluorolabeled products were then quantified spectrally, and equimolar quantities of Cy-3- and Cy-5-labeled products were combined and used for hybridization according to standard operating procedure M008 (7). All microarray experiments were performed in triplicate with independent biological samples, including one dye flip experiment in which the Cy-3 and Cy-5 labels were reversed to account for dye bias. After hybridization, raw TIFF images from the scanner were loaded into GenePix Pro version 4.1 (Axon, Inc., Union City, Calif.) for analysis. Spot grids (.gal files) were manually fitted to the microarray images. Spots were automatically flagged in instances of high background or stray fluorescent signals. The resulting GenePix result files (.gpr files) were exported into a DecisionSite for Functional Genomics version 7.2 module (Spotfire, Inc., Somerville, Mass.). Further analyses were performed to normalize the data set in an intensity-dependent manner (34) and to perform statistical analysis by analysis of variance (ANOVA). Genes identified as statistically significant were further subjected to false discovery rate corrections. Genes with at least a twofold change in expression and an ANOVA P value of <0.05 were considered significantly differentially expressed.
Real-time PCR analysis.
RNA quantitation by real-time PCR analysis was done on an ABI Prism 7700 sequence detection system (Perkin-Elmer, Foster City, Calif.) by using TaqMan reverse transcriptase and SYBR green reagents according to the protocols provided by the supplier (Applied Biosystems, Foster City, Calif.). PCR primers were designed to yield amplification products of approximately 100 bp and were tested in preliminary experiments to control for primer-dimer formation and amplification efficiency. RNA was isolated as described above, quality tested on agarose gels, and DNase-treated by using a DNA-free kit from Ambion (Austin, Tex.). TaqMan reverse transcriptase was used to transcribe 2.5 ng of total RNA into cDNA; the enzyme was omitted in control reactions. PCRs were performed by adding 5 μl of cDNA to 25 μl of SYBR Green PCR Master Mix in a total reaction volume of 50 μl. Thermocycler conditions were set and data analysis was done according to the manufacturer's recommendations. All real-time PCR data were normalized to the gyrA gene as an internal control and are shown relative to the data for the wild-type strain T4 or the control strain HS2155, which were grown in the absence of antibiotics. All data are expressed on a log2 scale and are the means and standard deviations for three independent biological samples.
Cloning procedures.
Plasmid pWHM302 was designed to overexpress the vncR gene in S. pneumoniae. The −35 and −10 elements of the fcsK promoter (4) were PCR amplified, and in the process the sequence for the ribosome binding site changed to GGAGG. This minimal promoter lacks predicted operator sites and is active in the absence of added inducers. The vncR gene from strain T4 was amplified and fused to the minimal fcsK promoter by overlap-extension PCR. The resulting PCR product possessed SalI and BamHI restriction sites for cloning into the shuttle vector pDL278. The PCR amplifications did not introduce point mutations as determined by DNA sequencing. S. pneumoniae clones were selected on tryptic soy broth-blood agar plates containing 500 μg of spectinomycin/ml after transformation with either pDL278 (empty vector) or pWHM302.
Western blot analysis.
Cultures were grown in 5 ml of C+Y medium to an OD620 of 0.30. Cells were harvested by centrifugation and pellets were resuspended in 100 μl of 20 mM HEPES (pH 8.0) containing protease inhibitors (Complete Mini; Roche). The resuspension was then sonicated on ice in three 20-s bursts. The protein content of the supernatant was determined after centrifugation, and 5 μg of protein was run on a 10% PreCast gel (Bio-Rad). After completion of the run, proteins were blotted onto polyvinylidene difluoride membrane by using standard methods. Immunodetection was performed on the membrane by using anti-VncR antibodies at a 1:2000 dilution overnight at 4°C. Goat anti-rabbit horseradish peroxidase was used as the secondary antibody at a 1:10000 dilution for 30 min at room temperature. Membranes were exposed to film after the membrane was reacted with detection reagent (SuperSignal West Pico Chemiluminescent; Pierce) according to the manufacturer's instructions.
RESULTS
Erythromycin causes a vancomycin-tolerant phenotype.
The insertion-duplication mutagenesis method relies on plasmid integration into the chromosome via homologous recombination. Although the requirement for continuous antibiotic selection to prevent reversible loss of the integrated plasmid is a disadvantage of this method (17), it has received longstanding, widespread use, including for analysis of antibiotic action (22, 23). Recent work by Robertson and coworkers (26) suggested that the presence of erythromycin to maintain such mutations results in a tolerant phenotype independent of the gene interrupted by the plasmid. To specifically address this question for the vex-vnc locus, genetically stable mutants were generated by using the PCR ligation mutagenesis method described by Lau et al. (18), which uses a PCR product containing a selectable marker that is flanked by DNA fragments that are homologous to regions 5′ and 3′ of the target gene. Homologous recombination completely replaces the target gene with the selectable marker, creating a genetically stable mutant. The ermB cassette from plasmid pTCV-lac (25) was chosen as the selectable marker to allow for bacterial growth in the presence or absence of erythromycin. This cassette consists of a strong promoter, a leader peptide, and the ermB gene and allows for erythromycin-induced expression of the ermB gene. Using this method, we replaced the vex3, pep27, vncR, vncS, and lytA genes and the control gene SP2155, encoding a truncated IgA1 protease, with an ermB cassette. In contrast to previous experiments that used the unencapsulated laboratory strain R6 (23, 26), we created mutants with the virulent strain T4 to study tolerance in a clinically relevant isolate.
For all mutants, cultures grown in the presence of erythromycin had a prolonged doubling time and a decreased loss of OD620 and CFU/ml after vancomycin treatment compared to the cultures grown in the absence of erythromycin (Table 2). These results confirm previous observations that erythromycin in the growth medium causes a low level of tolerance to vancomycin (26).
TABLE 2.
Decrease in OD and CFU after vancomycin treatment for 4 h
| Strain | Growth conditiona | Doubling time (min) | Percent loss of OD620 | Log decrease in CFU/ml ± SDb |
|---|---|---|---|---|
| T4 | 50 | 70 | 1.9 ± 0.6 | |
| T4R | 35 | 96 | 6.2 ± 0.5 | |
| R6 | 46 | 95 | 2.7 ± 0.4 | |
| Tupelo | 49 | 4 | 0.4 ± 0.1 | |
| HS2155 | 44 | 86 | 1.1 ± 0.1 | |
| HS2155 | + Erm | 74 | 60 | 0.6 ± 0.2 |
| HS1937 | 50 | 18 | 0.4 ± 0.1 | |
| HS1937 | + Erm | 123 | −6 | −0.1 ± 0.1 |
| HR1937 | 51 | 7 | 0.4 ± 0.1 | |
| HS0601 | 46 | 67 | 0.8 ± 0.1 | |
| HS0601 | + Erm | 62 | 46 | 0.9 ± 0.2 |
| HR0601 | 51 | 37 | 0.4 ± 0.1 | |
| HS0602 | 44 | 89 | 2.6 ± 0.1 | |
| HS0602 | + Erm | 69 | 78 | 1.2 ± 0.1 |
| HS0603 | 45 | 78 | 2.0 ± 0.4 | |
| HS0603 | + Erm | 68 | 77 | 1.3 ± 0.2 |
| HS0604 | 44 | 84 | 2.6 ± 0.2 | |
| HS0604 | + Erm | 76 | 74 | 1.8 ± 0.1 |
| HR0604 | 43 | 79 | 2.0 ± 0.2 |
+ Erm, grown in the presence of erythromycin.
At 4 h. Mean values for three independent experiments are shown.
The effect of erythromycin was distinguishable and additive to loss of function of LytA, the major autolysin of S. pneumoniae. Mutants that lack a functional lytA gene are tolerant and have been used as the gold standard in tolerance experiments. The T4 mutant of lytA, strain HS1937, underwent lysis at a much slower rate than the wild-type strain T4, and the rate of lysis was further decreased in the presence of erythromycin (Table 2).
Response of vex3, pep27, vncR, or vncS mutants to vancomycin challenge in the absence of erythromycin selection.
Given the increased tolerance invoked by erythromycin, a valid test of vancomycin tolerance would require an assay of the survival of the mutants in the absence of selection by erythromycin. In the absence of erythromycin, interruption of pep27 in strain HS0602, vncR in strain HS0603, or vncS in strain HS0604 did not decrease autolysis. All three strains lysed at least as rapidly as the wild-type strain T4 after the addition of vancomycin (Table 2). These results confirm those of Robertson et al. (26) and indicate that the phenotype observed by Novak et al. (23) most likely arose due to the presence of erythromycin in the growth medium.
In contrast to the other mutants, the vex3 mutant HS0601 was the only strain that underwent less vancomycin-induced lysis than the wild-type strain even in the absence of erythromycin (Table 2). Although HS0601 reproducibly underwent less lysis than the parental strain T4, the differences in OD620 values were not statistically significant. The viability of the HS0601 mutant decreased by 0.8 log unit, compared to a decrease of 1.9 log units for the wild-type strain T4 and 0.4 log unit for the tolerant lytA mutant HS1937. These data show that although pep27, vncR, and vncS are not directly involved in autolysis as previously suggested (22, 23), vex3 might play a role in autolysis.
Overexpression of downstream genes due to a polar effect of the ermB cassette.
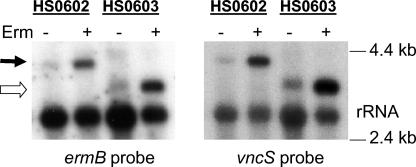
Transcripts emanating from the ermB promoter might extend into downstream genes, especially when ermB expression is induced in the presence of erythromycin. These polar effects may obscure the true phenotype of a gene deletion. To test if the ermB cassette had a polar effect, we isolated RNA from strains HS0602 (pep27 replaced by ermB) and HS0603 (vncR replaced by ermB) grown in the presence or absence of erythromycin and performed Northern blot analysis (Fig. 2). An ermB-specific probe detected a transcript of 4 kb in the HS0602 mutant and a 3-kb transcript in the HS0603 mutant, both of which were also detected with a vncS-specific probe. The large transcript in the HS0602 mutant corresponds to a polycistronic transcript that includes ermB, vncR, and vncS, while the transcript seen in HS0603 corresponds to an mRNA that contains the ermB and vncS genes. The intensities of the bands were relatively weak in RNA samples from cultures grown in the absence of erythromycin and drastically increased when cultures were grown in the presence of erythromycin. These results suggest that insertion of the ermB cassette leads to the overexpression of 3′ genes, even in the absence of erythromycin induction of the ermB promoter. This hypothesis was supported by real-time PCR analysis (see below). Therefore, further clarification of the tolerance phenotype of the vex3 mutant was undertaken to determine any possible contribution of overexpression of downstream genes.
FIG. 2.
Northern blot analysis of transcriptional read-through from the ermB promoter. RNA from the pep27 mutant HS0602 and the vncR mutant HS0603 was isolated after the cells were grown in the absence or presence of erythromycin, and samples were subjected to Northern blot analysis with probes specific for ermB or vncS. High-molecular-weight bands represent ermB-vncR-vncS (filled arrow) or ermB-vncS (open arrow) polycistronic mRNAs. rRNA is shown as the loading control.
Generation of mutants which carry an inverted ermB cassette.
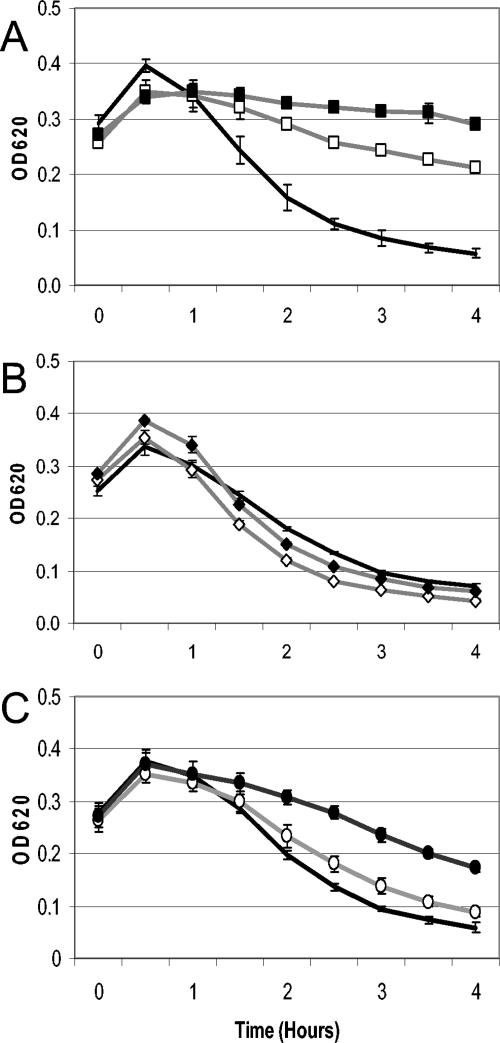
To distinguish the polar effect of the ermB cassette from the phenotype of the deletion mutants, an additional set of mutants was created with the ermB cassette cloned in the opposite orientation relative to the gene that was replaced (an example is shown in Fig. 1). Inversion of the ermB cassette did not change the slow rate of lysis of the lytA mutant HR1937 relative to HS1937 or of the rapidly lytic vncS mutant HR0604 relative to HS0604 (Fig. 3A and B, respectively), indicating that mutants bearing this inverse cassette demonstrated their true phenotype. Lysis and killing of the vex3 mutant HR0601 (ermB inverted with respect to vex-vnc) indicated that it was even more tolerant than the vex3 mutant HS0601 (ermB not inverted) (Fig. 3C). HR0601 lost only 37% of its initial optical density and showed a decrease of only 0.4 log CFU/ml, thereby fulfilling the standard definition for tolerance (11) (Table 2). The accentuation of tolerance of HR0601 over HS0601 demonstrated that the orientation of the resistance cassette was able to influence the phenotype of the mutant despite the fact that both strains carried a deletion mutation of the vex3 gene.
FIG. 3.
Effect of the orientation of the ermB cassette on autolysis. Wild-type strain T4 (black line) served as the control for all panels. (A) Autolysis of the lytA autolysin mutants HS1937 (open squares) and HR1937 (filled squares). (B) Autolysis of the vncS histidine kinase mutants HS0604 (open diamonds) and HR0604 (filled diamonds). (C) Autolysis of the vex3 transmembrane protein mutants HS0601 (open circles) and HR0601 (filled circles). All samples were treated with 10× MIC of vancomycin as the cultures reached an OD620 of 0.275 to 0.30. Means and standard deviations for three independent experiments are shown.
vex12 and vncRS are overexpressed in the vex3 mutant HS0601, but not in HR0601.
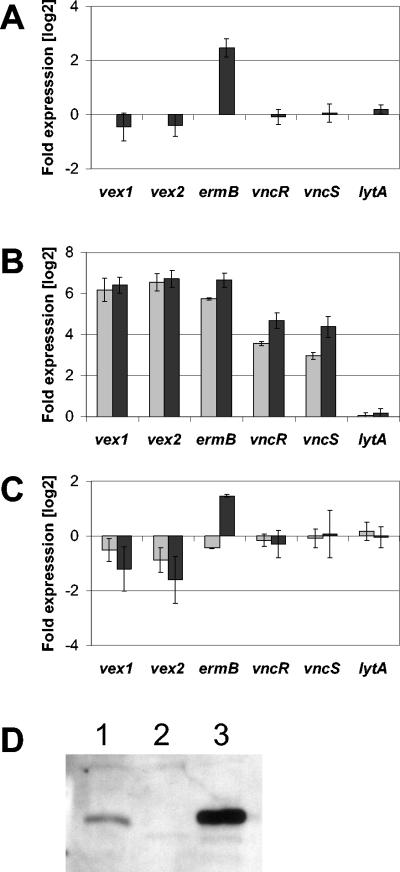
Overexpression of genes downstream of the ermB cassette might account for the difference in tolerance of HS0601 and HR0601. Expression of the vex and vnc genes in the control mutant HS2155 and the two vex3 mutants HS0601 and HR0601 was determined by reverse transcriptase PCR (RT-PCR) (Fig. 4A). Data were first normalized to the gyrA gene and then to strain HS2155, which was grown in the absence of antibiotics. While expression of the vex and vnc genes was not significantly changed in the control strain HS2155 with or without erythromycin, the amount of ermB transcript increased 5.6-fold in the presence of the drug, showing that erythromycin induces the ermB promoter and that erythromycin by itself does not alter the transcription of vex123-vncRS. Transcription of the lytA autolysin was indistinguishable for all three strains tested, regardless of the presence of erythromycin. This result demonstrates that the observed effects are specific to the vex-vnc locus and the ermB gene within it.
FIG. 4.
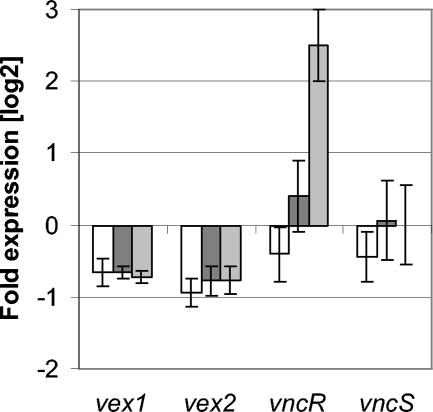
Expression of vex12 and vncRS in vex3 mutants. (A) The control mutant HS2155 was grown in the absence (baseline) and presence (dark gray bars) of erythromycin, and total RNA was analyzed by real-time PCR analysis using primers specific for vex1, vex2, ermB, vncR, vncS, or lytA. Gene expression in strains HS0601 (B) and HR0601 (C) is depicted relative to RNA from strain HS2155 grown in the absence of erythromycin (baseline). Cells were grown in the absence (light gray bars) or presence (dark gray bars) of 1 μg of erythromycin/ml prior to RNA isolation. All data were normalized to the gyrA transcript and are expression levels on a log2 scale; the means and standard deviations for three independent experiments are shown. (D) Western blot of cellular extracts from strains T4 (lane 1), vncR mutant HS0603 (lane 2), and vex3 mutant HS0601 (lane 3) probed with VncR-specific antibodies.
Expression of vncR and vncS was 12- and 8-fold higher in the HS0601 mutant than in the control strain, and these values more than doubled (were 26- and 22-fold higher) in the presence of the antibiotic, indicating that the erythromycin induced transcription of vncRS as the result of transcriptional read-through from ermB. Consistent with this result, transcription levels of vncRS in the HR0601 mutant remained identical to those of the control strain. This finding suggests that the deletion of vex3 per se does not change the expression levels of the vex-vnc locus. A Western blot analysis of cellular extracts from strains T4, HS0603, and HS0601, using a VncR-specific antibody, showed a correlation between gene transcription and protein expression levels (Fig. 4B). Thus, the more tolerant mutant HR0601 showed no change in vncRS transcription, while the less tolerant HS0601 did show such a change. This result suggests either that tolerance is not a result of change in expression of vncRS or that increased expression of vncRS counteracts tolerance.
Effect of VncR on the transcription of vex or vnc.
Expression of vex1 and vex2 in strain HS0601 in the absence of erythromycin was increased 76- to 96-fold over that in the control strain, and it increased a further 12-fold when erythromycin was present in the culture medium (expression of the deleted vex3 gene was not detectable).
No such increased expression of vex genes was detected for strain HR0601, raising the question of whether vex1 and vex2 were being upregulated by VncR. To determine if increased levels of VncR and VncS were linked to increased vex gene transcription and decreased tolerance of the vex3 mutant HS0601, a mutant was constructed to overexpress vncR from a nonintegrated plasmid. The −35 and −10 elements from the fcsK promoter and the vncR gene were cloned into the shuttle vector pDL278, yielding plasmid pWHM302. RNA from strains T4, HR0601, HR0601 (pDL278), and HR0601 (pWHM302) were used for real-time PCR analysis to determine if increased expression of vncR resulted in increased expression of vex1 or vex2 (Fig. 5). The vncR gene was 5.7-fold overexpressed in strain HR0601 (pWHM302) compared to strain T4, while vncR expression did not change significantly in strain HR0601 (pDL278). Expression of vex1, vex2, or vncS was not significantly altered in any of the three strains tested, demonstrating that the overexpression of vncR does not drive increased expression of these genes under the conditions tested. Vancomycin kill curves demonstrated that tolerance to vancomycin was not affected by the overexpression of vncR in HR0601 (pWHM302) (data not shown). These results suggest that fivefold overexpression of vncR alone is not sufficient to induce expression of the vex123-vncRS genes and that the expression level of VncR is independent of tolerance.
FIG. 5.
Effect of vncR overexpression on lytA, vex12 and vncRS mRNA levels. RNA from strains T4 (baseline), HR0601 (white), HR0601 (pDL278) (dark gray), and HR0601 (pWHM302) (light gray) was analyzed by real-time PCR analysis. Data were normalized to the gyrA gene and to RNA from strain T4. The data are expression levels on a log2 scale; the means and standard deviations for three independent experiments are shown.
Microarray analysis of vex3, vncR, and vncS mutants.
To examine genes whose expression is altered in the tolerant mutant HR0601, which might explain tolerance, microarray analysis was used to determine genes controlled by the vncRS two-component system (Table 3). Deletion of vncR in strain HS0603 upregulated four genes more than twofold: genes encoding ribosomal protein L31 and three cell wall surface anchor family proteins (SP0462 to SP0464) that appear to form one operon. Two genes, pbuX and purK, were downregulated in the vncS mutant HS0604. Although both genes are involved in purine metabolism, their connection to vncRS remains to be determined. These scant changes were in agreement with the absence of a phenotype with these deletions.
TABLE 3.
Microarray results showing genes that are differentially regulated in the vncR mutant HS0603, the vncS mutant HS0604, and the vex3 mutants HS0601 and HR0601 in the absence of erythromycina
| Microarray IDb | Annotation | Fold change for:
|
|||
|---|---|---|---|---|---|
| HS0603 | HS0604 | HS0601 | HR0601 | ||
| vex-vncRS locus | |||||
| SP0598 | Hypothetical protein | 62.0 | |||
| SP0599 | Transmembrane protein, vex1 | 114.7 | |||
| SP0600 | ABC transporter, ATP-binding protein, vex2 | 121.6 | |||
| SP0601 | Transmembrane protein, vex3 | −33.0 | −10.4 | ||
| SP0603 | DNA-binding response regulator, vncR | −150.6 | 14.8 | ||
| SP0604 | Sensor histidine kinase, vncS | −45.5 | 14.5 | ||
| Transport functions | |||||
| SP0729 | Cation-transporting ATPase, E1-E2 family | −2.5 | |||
| SP1062 | ABC transporter, ATP-binding protein | 2.1 | |||
| SP1215 | Transporter, FNT family, putative | −2.7 | |||
| SP1241 | Amino acid ABC transporter, permease protein | 2.9 | |||
| SP1242 | Amino acid ABC transporter, ATP-binding protein | 2.4 | |||
| SP1394 | Amino acid ABC transporter | −1.7 | 2.0 | ||
| SP1527 | ABC transporter, oligopeptide-binding protein, aliB | −2.0 | |||
| SP1572 | Nonheme iron-containing ferritin | 2.5 | |||
| SP1848 | Xanthine permease, pbuX | −2.0 | |||
| SP1871 | Iron-compound ABC transporter | −3.0 | |||
| SP2108 | Maltodextrin ABC transporter, malX | −1.7 | |||
| SP2109 | Maltodextrin ABC transporter, malC | −2.0 | |||
| SP2110 | Maltodextrin ABC transporter, malD | −1.8 | |||
| Ribosomal proteins | |||||
| SP0441 | Ribosomal protein L28, rpmB | −2.4 | |||
| SP1107 | Ribosomal protein L27, rpmA | 2.3 | |||
| SP1299 | Ribosomal protein L31, rpmE | 2.0 | |||
| Biosynthesis and degradation of polysaccharides | |||||
| SP0107 | LysM domain protein | 2.8 | |||
| SP0268 | Alkaline amylopullulanase, putative | −2.4 | |||
| SP2106 | Glycogen phosphorylase family protein | −2.2 | |||
| SP0240 | Phosphoglycerate mutase family protein | 2.1 | |||
| SP0285 | Alcohol dehydrogenase, zinc containing | −2.2 | |||
| SP0447 | Ketol acid reductoisomerase, ilvC | 2.1 | |||
| SP0502 | Glutamine synthetase, glnA | 3.4 | |||
| SP0730 | Pyruvate oxidase, spxB | −2.1 | |||
| SP0837 | DNA topology modulation protein FlaR, putative | 2.3 | |||
| SP0965 | Endo-beta-N-acetylglucosaminidase, lytB | 2.0 | |||
| SP1052 | Phosphoesterase, putative | 2.1 | |||
| SP1113 | DNA-binding protein HU, hup | 2.1 | |||
| SP1470 | Thiamine biosynthesis protein ApbE, putative | −2.6 | |||
| SP1471 | Oxidoreductase, putative | −3.1 | |||
| SP1472 | Oxidoreductase, putative | −2.8 | |||
| SP1963 | CBS domain protein | 2.2 | |||
| SP2026 | Alcohol dehydrogenase, iron containing | −4.0 | |||
| SP2056 | N-acetylglucosamine-6-P deacetylase, nagA | −2.1 | |||
| SP2157 | Alcohol dehydrogenase, iron containing | −2.6 | |||
| SP2216 | Secreted 45-kDa protein, usp45 | 2.4 | |||
| Hypothetical proteins | |||||
| SP0088 | Hypothetical protein | 2.0 | |||
| SP0097 | Conserved domain protein | −3.7 | |||
| SP2107 | 4-Alpha-glucanotransferase, malQ | −2.0 | |||
| Regulatory functions | |||||
| SP0501 | Transcriptional regulator, MerR family | 3.3 | |||
| SP0837 | DNA topology modulation protein FlaR, putative | 2.3 | |||
| SP1061 | Protein kinase, putative | 2.7 | |||
| SP1362 | Putative transcriptional regulator, mecA | 2.0 | |||
| SP1809 | Transcriptional regulator | 2.0 | |||
| SP1821 | Sugar-binding transcriptional regulator, LacI family | 2.1 | |||
| SP2234 | Transcriptional regulator, TetR family | −2.2 | |||
| SP2235 | Response regulator, comE | 2.9 | |||
| SP2236 | Putative sensor histidine kinase, comD | 2.5 | |||
| Cell surface proteins | |||||
| SP0462 | Cell wall surface anchor family protein | 2.7 | 4.6 | ||
| SP0463 | Cell wall surface anchor family protein | 3.1 | 4.2 | ||
| SP0464 | Cell wall surface anchor family protein | 2.6 | 4.3 | ||
| SP2190 | Choline binding protein A, cbpA | 3.3 | |||
| Various functions | |||||
| SP0054 | Phosphoribosylaminoimidazole carboxylase, purK | −2.5 | |||
| SP0098 | Hypothetical protein | −4.4 | |||
| SP0099 | Hypothetical protein | −4.2 | |||
| SP0100 | Conserved hypothetical protein | −3.2 | |||
| SP0430 | Hypothetical protein | −2.2 | |||
| SP0448 | Hypothetical protein | 2.4 | |||
| SP0728 | Hypothetical protein | −2.0 | |||
| SP0742 | Conserved hypothetical protein | 2.8 | |||
| SP0861 | Hypothetical protein | 2.2 | |||
| SP0958 | Hypothetical protein | −2.2 | |||
| SP1053 | Conserved domain protein | 2.1 | |||
| SP1059 | Hypothetical protein | 3.3 | |||
| SP1140 | Hypothetical protein | 2.0 | |||
| SP1141 | Hypothetical protein | 2.1 | |||
| SP1145 | Hypothetical protein | 2.0 | |||
| SP1793 | Hypothetical protein | 2.0 | |||
| SP1810 | Hypothetical protein | 2.0 | |||
| SP2191 | Conserved hypothetical protein | 2.3 | |||
| SP2232 | Conserved hypothetical protein, authentic frameshift | −4.3 | |||
| SP2233 | Hypothetical protein | −3.4 | −3.7 | ||
Mean changes (n-fold) from three independent biological samples are shown. Genes that are differentially regulated more than twofold are shown. Statistical significance was verified by ANOVA.
Microarray ID, identification number from microarray analysis.
Seventeen genes were differentially regulated in the vex3 mutant HS0601, including three hypothetical proteins, cell wall anchor family proteins, and the vex123-vncRS locus itself. The expression of vncRS was increased 14-fold and that of vex12 was increased more than 100-fold, values in good agreement with the RT-PCR results. The expression of the gene encoding hypothetical protein SP0598 was also increased (62-fold), although this result may be an artifact since 66 bp of the 162-bp reading frame for SP0598 overlap with the vex1 gene.
Microarray analysis of RNA from the mutant strain HR0601 showed that 58 genes were differentially regulated greater than twofold. Only the gene encoding hypothetical protein SP2233 was downregulated greater than twofold in both vex3 mutants HS0601 and HR0601. SP1394, annotated as the gene encoding amino acid transporter-amino acid binding protein, is the only gene that was downregulated in HS0601 and upregulated in HR0601 at statistically significant levels. Differential regulation of these 57 genes may represent part of the cellular response to the loss of vex3 gene expression that results in tolerance.
DISCUSSION
The main objective of this study was to reexamine the role of the vex-vnc locus in vancomycin tolerance in S. pneumoniae. Early reports had indicated that the vex123, pep27, and vncRS genes played a major role in autolysis and vancomycin tolerance (22, 23). Later studies by Robertson and coworkers (26) challenged this model, suggesting that erythromycin in the culture medium engendered the vancomycin-tolerant phenotype. Our results confirmed elements of both studies. In agreement with the work of Robertson et al., deletions in pep27, vncR, and vncS did not confer vancomycin tolerance. Consistent with the work of Novak et al., deletion of vex3 resulted in mutants that showed various levels of vancomycin tolerance. The basis for this reduced rate of vancomycin-induced lysis remains to be determined.
In studies of the 13 complete pneumococcal histidine kinase response regulator pairs (16, 31), deletion of the vncRS two-component system did not change viability, growth rate, competence (16), or virulence in the lung (31). The vncRS genes were deemed not essential (30) and not important for virulence in a mouse respiratory tract infection model (31). Our microarray analyses failed to suggest a function for VncRS, stressing that the role of this two-component system in pneumococcal biology remains to be determined. A point mutation at position 440 in VncS was thought to cause the vancomycin tolerance of three type 9V clinical isolates on the basis of the similar phenotype of the analogous laboratory mutant SPSJ01 (23). However, since the mutant was generated by insertion duplication mutagenesis by using plasmid pJDC9, which contains an erm resistance gene, this interpretation appears to be no longer valid and the underlying defect of these clinical isolates remains to be determined. The tolerance of the laboratory-constructed vncS deletion mutant (22) can be attributed to either of two artifacts introduced by the antibiotic selection. Ng and coworkers determined that a number of genes are differentially regulated in response to erythromycin (21). One or more of these genes could be involved in vancomycin tolerance. Alternatively, the reduced rate of autolysis could be the result of the decreased growth rate of cultures exposed to erythromycin. In the case of the vex3 mutants HS0601 and HR0601, however, tolerance is not the result of a decrease in growth rate since the strains' growth rates are comparable to those of the wild-type strain T4.
A recent publication by Ortega et al. (24) reported that the cell wall-inhibiting antibiotics penicillin or cefotaxime resulted in a great decrease in CFU when the antibiotics were given individually compared to results with the same antibiotics given jointly with erythromycin. This finding agrees with the present and published findings (19, 26) and highlights the need to better understand the effects that antibiotics have on bacteria.
Interpretation of the tolerant phenotype of the HS0601 mutant generated by gene replacement must consider the polar effects generated by the antibiotic resistance cassette. Overexpression of vncS, located 3′ to the resistance cassette, was demonstrated with the pep27 and vncR mutants HS0602 and HS0603, respectively. The ermB promoter used to generate the mutants in this study was shown to be constitutively active at low levels and inducible by erythromycin. Read-through transcription from this promoter may explain the high level of vncRS transcripts evident by RT-PCR and microarray analyses of the vex3 mutant HS0601. However, fivefold overexpression of vncR in strain HR0601 (pWHM302) did not increase the rate of transcription of the vex or vnc genes, suggesting that vncS needs to be overexpressed as well or that other factors are involved in the high level of expression of the vex12 genes in strain HS0601. It also remains possible that higher expression of vncR or vncRS may be required to change vex12 expression.
Robertson et al. (26) reported that vex123 expression is derepressed in the absence of vncS, a result that we were unable to confirm by microarray analysis. This inconsistency may be due to several factors, such as differences in pneumococcal strains (R6 versus T4) or growth media (CDM versus C+Y) or the method of generating mutants (partial versus complete gene replacement). Alternatively, the same factors that lead to overexpression of vex12 and vncRS in the vex3 mutant HS0601 might have played a role in Robertson's work as well.
The vex3 mutant HS0601 produced large amounts of vex12 mRNA, but was tolerant to vancomycin only to a small degree. In contrast, the vex3 mutant HR0601 produced less vex12 mRNA than the control or wild-type strain, but was more tolerant to vancomycin. One possible explanation for these results is that the Vex123 ABC transporter is involved in tolerance, a phenotype that is evident in strains lacking vex3. This phenotype is strongest in HR0601 and might be partially attenuated in HS0601 because overexpression of Vex1 might be able to compensate for the loss of Vex3. The two proteins have a similar domain organization, with an FtsX-like permease domain at the carboxy terminus, and share 28% sequence identity at the amino acid level (http://smart.embl-heidelberg.de/). Verification of this model and determination of the function of Vex123 in pneumococcal cell biology will require further investigation.
Further evidence for the role of the vex123 genes in tolerance comes from a study by Rodriguez et al. (27), in which the vex123-pep27 genes from tolerant and nontolerant strains were sequenced. The authors found two alleles each for the vex2 and pep27 genes, one allele combination exemplified by strain T4 and the other by strain R6. The mixed pairing of R6 pep27 and T4 vex2 alleles was frequently found with tolerant isolates, whereas nontolerant isolates mostly possessed pairs of alleles restricted to either T4 or R6 only. The T4 pep27 and R6 vex2 alleles were never found together in any strain, indicating a strong negative selection. These findings suggest that the sequence of the vex locus is an epidemiological marker for tolerance. The present study indicates that this marker may be related to the biological function of Vex123, the absence of which leads to tolerance.
The results presented in the present study indicate that the original model of a death signaling pathway consisting of the vex123-pep27-vncRS gene products must be corrected. Tolerance can arise by loss of function of Vex3. It is possible that high expression levels of Vex1 can functionally substitute for the lack of Vex3, thereby attenuating the tolerant phenotype. This phenotype is not related to increased expression of VncR, since the VncRS two-component system does not appear to regulate transcription of vex123. Vancomycin tolerance is a multifactorial phenomenon arising from the effects of the bacterial capsule (6), erythromycin treatment (23), the autolysin LytA (32), and the ABC transport system Vex123. The connection between these factors and stationary phase autolysis remains to be determined.
Acknowledgments
This work was supported by grant 5R01AI039482-07 from the National Institute of Allergy and Infectious Diseases, Cancer Center grant P30 CA21765, and the American Lebanese Syrian Associated Charities.
REFERENCES
- 1.Adam, D. 2002. Global antibiotic resistance in Streptococcus pneumoniae. J. Antimicrob. Chemother. 50(Suppl.):1-5. [DOI] [PubMed] [Google Scholar]
- 2.Boost, M. V., W. M. Ko, and M. M. O'Donoghue. 2003. Penicillin and vancomycin tolerance among clinical isolates of Streptococcus pneumoniae in Hong Kong. Hong Kong Med. J. 9:415-418. [PubMed] [Google Scholar]
- 3.Bricker, A. L., and A. Camilli. 1999. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172:131-135. [DOI] [PubMed] [Google Scholar]
- 4.Chan, P. F., K. M. O'Dwyer, L. M. Palmer, J. D. Ambrad, K. A. Ingraham, C. So, M. A. Lonetto, S. Biswas, M. Rosenberg, D. J. Holmes, and M. Zalacain. 2003. Characterization of a novel fucose-regulated promoter (PfcsK) suitable for gene essentiality and antibacterial mode-of-action studies in Streptococcus pneumoniae. J. Bacteriol. 185:2051-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernebro, J., I. Andersson, J. Sublett, E. Morfeldt, R. Novak, E. Tuomanen, S. Normark, and B. H. Normark. 2004. Capsular expression in Streptococcus pneumoniae negatively affects spontaneous and antibiotic-induced lysis and contributes to antibiotic tolerance. J. Infect. Dis. 189:328-338. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert, J., J. Hasseman, and R. Cline. 18 September 2002, posting date. Microbial DNA probe hybridization. [Online.] http://pfgrc.tigr.org/protocols/M008.pdf.
- 8.Gilbert, J., J. Hasseman, and R. Cline. 18 September 2002, posting date. Microbial RNA aminoallyl labeling for microarrays. [Online.] http://pfgrc.tigr.org/protocols/M007.pdf.
- 9.Gosink, K. K., E. R. Mann, C. Guglielmo, E. I. Tuomanen, and H. R. Masure. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 68:5690-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriques, N. B., R. Novak, A. Ortqvist, G. Kallenius, E. Tuomanen, and S. Normark. 2001. Clinical isolates of Streptococcus pneumoniae that exhibit tolerance of vancomycin. Clin. Infect. Dis. 32:552-558. [DOI] [PubMed] [Google Scholar]
- 11.Hidalgo, M., E. Castaneda, and C. A. Arias. 2003. Tolerance to vancomycin in a multiresistant, Colombian isolate of Streptococcus pneumoniae. J. Antimicrob. Chemother. 52:300-302. [DOI] [PubMed] [Google Scholar]
- 12.Horton, R. M. 1995. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3:93-99. [DOI] [PubMed] [Google Scholar]
- 13.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 14.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jett, B. D., K. L. Hatter, M. M. Huycke, and M. S. Gilmore. 1997. Simplified agar plate method for quantifying viable bacteria. BioTechniques 23:648-650. [DOI] [PubMed] [Google Scholar]
- 16.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 17.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 18.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 19.Lin, E., R. J. Stanek, and M. A. Mufson. 2003. Lack of synergy of erythromycin combined with penicillin or cefotaxime against Streptococcus pneumoniae in vitro. Antimicrob. Agents Chemother. 47:1151-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCullers, J. A., B. K. English, and R. Novak. 2000. Isolation and characterization of vancomycin-tolerant Streptococcus pneumoniae from the cerebrospinal fluid of a patient who developed recrudescent meningitis. J. Infect. Dis. 181:369-373. [DOI] [PubMed] [Google Scholar]
- 21.Ng, W. L., K. M. Kazmierczak, G. T. Robertson, R. Gilmour, and M. E. Winkler. 2003. Transcriptional regulation and signature patterns revealed by microarray analyses of Streptococcus pneumoniae R6 challenged with sublethal concentrations of translation inhibitors. J. Bacteriol. 185:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novak, R., E. Charpentier, J. S. Braun, and E. Tuomanen. 2000. Signal transduction by a death signal peptide: uncovering the mechanism of bacterial killing by penicillin. Mol. Cell 5:49-57. [DOI] [PubMed] [Google Scholar]
- 23.Novak, R., B. Henriques, E. Charpentier, S. Normark, and E. Tuomanen. 1999. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature 399:590-593. [DOI] [PubMed] [Google Scholar]
- 24.Ortega, M., F. Marco, A. Soriano, J. Gomez, M. Almela, and J. Mensa. 2004. In vitro antagonism between beta-lactam and macrolide in Streptococcus pneumoniae: how important is the antibiotic order? Int. J. Antimicrob. Agents 24:178-180. [DOI] [PubMed] [Google Scholar]
- 25.Poyart, C., and P. Trieu-Cuot. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156:193-198. [DOI] [PubMed] [Google Scholar]
- 26.Robertson, G. T., J. Zhao, B. V. Desai, W. H. Coleman, T. I. Nicas, R. Gilmour, L. Grinius, D. A. Morrison, and M. E. Winkler. 2002. Vancomycin tolerance induced by erythromycin but not by loss of vncRS, vex3, or pep27 function in Streptococcus pneumoniae. J. Bacteriol. 184:6987-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez, C. A., R. Atkinson, W. Bitar, C. G. Whitney, K. M. Edwards, L. S. Mitchell, J. Li, J. Sublett, C. S. Li, T. Liu, P. J. Chesney, and E. I. Tuomanen. 2004. Tolerance to vancomycin in pneumococci: detection with a molecular marker and assessment of clinical impact. J. Infect. Dis. 190:1481-1487. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 30.Thanassi, J. A., S. L. Hartman-Neumann, T. J. Dougherty, B. A. Dougherty, and M. J. Pucci. 2002. Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res. 30:3152-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 32.Tomasz, A., A. Albino, and E. Zanati. 1970. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227:138-140. [DOI] [PubMed] [Google Scholar]
- 33.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, C. Lexau, A. Reingold, L. Lefkowitz, P. R. Cieslak, M. Cetron, E. R. Zell, J. H. Jorgensen, and A. Schuchat. 2000. Increasing prevalence of multidrug-resistant Streptococcus pneumoniae in the United States. N. Engl. J. Med. 343:1917-1924. [DOI] [PubMed] [Google Scholar]
- 34.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]