Abstract
A 3-week coliphage survey was conducted in stool samples from 140 Bangladeshi children hospitalized with severe diarrhea. On the Escherichia coli indicator strain K803, all but one phage isolate had 170-kb genomes and the morphology of T4 phage. In spot tests, the individual T4-like phages infected up to 27 out of 40 diarrhea-associated E. coli, representing 22 O serotypes and various virulence factors; only five of them were not infected by any of these new phages. A combination of diagnostic PCR based on g32 (DNA binding) and g23 (major capsid protein) and Southern hybridization revealed that half were T-even phages sensu strictu, while the other half were pseudo-T-even or even more distantly related T4-like phages that failed to cross-hybridize with T4 or between each other. Nineteen percent of the acute stool samples yielded T4-like phages, and the prevalence was lower in convalescent stool samples. T4-like phages were also isolated from environmental and sewage water, but with low frequency and low titers. On the enteropathogenic E. coli strain O127:K63, 14% of the patients yielded phage, all of which were members of the phage family Siphoviridae with 50-kb genomes, showing the morphology of Jersey- and beta-4 like phages and narrow lytic patterns on E. coli O serotypes. Three siphovirus types could be differentiated by lack of cross-hybridization. Only a few stool samples were positive on both indicator strains. Phages with closely related restriction patterns and, in the case of T4-like phages, identical g23 gene sequences were isolated from different patients, suggesting epidemiological links between the patients.
Diarrhea continues to be one of the most common causes of morbidity and mortality among infants in developing countries (36). Many etiological agents have been implicated, but Escherichia coli has been consistently identified as the first or, after rotavirus, second most frequent cause of childhood diarrhea. For example, in the world's largest diarrhea clinic in Dhaka, Bangladesh, E. coli accounted for 29% of the pediatric diarrhea cases (3, 7). Among the E. coli strains, the major pathotypes were enteropathogenic (EPEC) and enterotoxigenic (ETEC) E. coli (4, 21). Treatment of E. coli diarrhea with oral rehydration solution has substantially reduced mortality from dehydration but has little or no effect on the pathogen itself (6). Antibiotics and antibodies are of limited use (12), and an E. coli diarrhea vaccine is still in the development phase (34).
Nearly 100 years ago, Félix d'Hérelle, the codiscoverer of bacterial viruses (bacteriophages), proposed phages for the treatment of bacterial diseases (17). In fact, the World Health Organization in the 1940s conducted a large field study with phages against cholera in rural India, and empirical phage therapy is still used against Shigella infections in Eastern Europe (37). Phages against E. coli diarrhea were explored in western veterinary applications (5, 35), but not yet in human medicine. One reason is the limited knowledge on phage-host interaction in the intestine (15). Even the most basic data on the genetic diversity of E. coli phages in the stool of diarrhea patients are lacking (19, 20). This lack of data is surprising because E. coli phages lambda and T4 are among the best-characterized biological systems (24). Furthermore, T4-like phages have often been isolated from human stools (25). Data on the genetic diversity of coliphages isolated from pediatric diarrhea patients are thus not only interesting for their potential medical use, but will also add to our understanding of the natural diversity of phages in the environment. Such data are especially interesting in the context of a phage-bacterium pair that has been intensively investigated under laboratory conditions (13). The current study reports on the isolation and basic genetic characterization of T4-like phages from pediatric diarrhea patients in Bangladesh.
MATERIALS AND METHODS
Phages.
The stool samples (10 g) were homogenized in TS (8.5 g of NaCl and 1 g tryptone per liter) to a final volume of 30 ml. Stool and water samples were centrifuged for 15 min at 14.500 × . and filtered through a Millex AP20 prefilter followed by a 0.45-μm Minisart filter. Subsequently, the samples were stored at 4°C. The phages were propagated and enumerated on indicator strains K803 and O127:K63 according to standard protocols (24) based on Hershey medium. The strains were designated JS (the initials of Josette Sidoti, who conducted the phage survey) followed by the patient number. In general, only a single phage plaque was amplified per patient unless the plaques differed clearly in appearance, in which case the isolates were differentiated by suffixes (e.g., JS114.1 to JS114.3). When the second isolate came from a different stool sample (from the convalescent phase), the code number was given a prime. The selection procedure thus targeted the isolation of distinct, independent phage types. Only in two test cases were several similar-appearing phage plaques deliberately propagated from the same stool sample (patients 110 and 122). For phages isolated from environmental water, the initials were followed by a D (Dhaka) or L (Lausanne).
Pulsed-field gel electrophoresis.
Blocks of 1% agarose containing 106 PFU of the indicated phage were incubated overnight in lysis buffer (0.05 M EDTA, 10 mM Tris, pH 8.0, 1% sodium dodecyl sulfate, and 50 μg of proteinase K per ml) and washed three times for 1 h in 10 ml of TE (10 mM Tris, pH 8.0, 1 mM EDTA). The blocks were then analyzed by electrophoresis in 1% agarose (0.5% TBE; 45 mM Tris-borate, 1 mM EDTA) for 20 h at 6 V/cm and 14°C with a pulse time of 1 to 20 s in a CHEF-DRII apparatus from Bio-Rad. The gel was stained for 30 min with ethidium bromide (0.5 μg/ml).
Electron microscopy.
Negative staining of phages was done with a mixture of solutions A and B (A: 2% ammonium molybdate at pH 7.0 or 2% phosphotungstic acid; B: 11% Bacitracine in distilled water) or a solution of 3% uranyl acetate. The Formvar-coated carbon grids were examined in a Philips CM12 transmission electron microscope at 80 kV. The dimensions of the phages were calibrated with T4 phage particles (24).
Southern blot.
PacI-digested phage DNA was separated on 0.8% (wt/vol) agarose gels. Transfer to Zeta-probe membrane was done according to the Bio-Rad protocol. Probes were labeled with α-32P with the random-primed DNA labeling system (Boehringer Mannheim) and purified with Nuc Trap probe purification columns (Stratagene). Prehybridization and hybridization conditions were 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5 mM EDTA (pH 8), 0.1% SDS, and 25% skim milk, overnight at 68°C. The membranes were then rinsed with 2× SSC and washed for 1 h at 65°C with 2x SSC and 0.1% SDS, then for 1 h in 0.5x SSC and 0.1% SDS, and finally for 15 min in 0.1x SSC and 0.1% SDS. The membranes were dried and then exposed to Biomax-MS autoradiography film (Kodak).
PCR and oligonucleotide primers.
PCRs were performed in a final volume of 50 μl with either 1 μl of T4-type phage lysate (107 PFU/ml) heated at 95°C for 20 min or 1 μl of phage DNA as a template. For the amplification of gene 32, we used the primers FR60 and FR61 described by Monod et al. (30). The conditions involved in this PCR were 35 cycles consisting of a 30-s denaturation period at 96°C, 2 min of annealing at 54°C, and 5 min of polymerization at 72°C. For the amplification of gene 23, we used the primers Mzia1 and cap8, binding to sites located in the best-conserved region of this gene, and the reaction conditions as described (39).
RESULTS AND DISCUSSION
Survey of coliphages in the stool samples of pediatric diarrhea patients.
Stool samples were obtained during a 3-week survey in February 2000 from 157 randomly chosen Bangladeshi children hospitalized with acute gastroenteritis in the general ward of the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B), diarrhea hospital in Dhaka. The age range (1 month to 4 years) and sex ratio (96 boys and 61 girls) of the patients corresponded to the usual epidemiological situation in this hospital (3). Stool samples from 17 cholera and shigella patients were excluded from the survey to increase the fraction of patients infected with EPEC and ETEC. According to the experience in this hospital (3, 4, 7, 21) this preselection ensures that about every third patient was likely to be infected with these two pathogens. For logistic reasons, no systematic diagnosis of the infectious agents was conducted with the enrolled patients.
After exclusion of the cholera and shigella patients, the stool samples from the remaining 140 patients were screened for coliphages in a spot assay (Table 1). We used the nonpathogenic laboratory E. coli K803, a derivative of K-12, as a primary indicator cell. K803 lacks any restriction-modification system, only has the core parts of the lipopolysaccharide, lacks O antigens, and is able to plate most coliphages isolated to date, including virtually all of the known T4-like phages, many of which were isolated on pathogenic E. coli (Kutter laboratory, unpublished studies that include over 100 such phages). All positive samples in the spot tests were confirmed by plaque assay. Twenty-six of the 140 patients (19%) yielded stool samples that were positive for phage plaques on strain K803, but titers were generally low (range, 30 to 5 × 104 PFU/g of stool). Ten patients showed phage titers exceeding 103 PFU/g of stool.
TABLE 1.
In vitro phage susceptibility of diarrhea-causing E. coli strains.
| Strain | No. of phages infecting | Sensitivity to phage
|
|||||
|---|---|---|---|---|---|---|---|
| 146.1 (T4) | 65.1 (T4) | 114.1 (T4) | 150 (T4) | 61.1 (β-4) | 114.2 (Jersey) | ||
| EPEC O18:K77 | 16 | X | X | X | |||
| EPEC O20:K84 | 4 | X | |||||
| EPEC O26:K60 | 10 | X | |||||
| EPEC O55:K59 | 7 | X | X | ||||
| EPEC O86:K61 | 13 | X | X | X | X | X | |
| EPEC O111:K58 | 10 | X | X | ||||
| EPEC O112:K66 | 6 | X | X | ||||
| EPEC O119:K69 | 5 | X | |||||
| EPEC O124:K72 | 0 | ||||||
| EPEC O125:K70 | 5 | X | |||||
| EPEC*:A/E+ | 16 | X | X | X | |||
| EPEC*:A/E+ | 6 | X | |||||
| EPEC*:A/E+ | 18 | X | X | X | X | X | |
| EPEC*:A/E+ | 1 | ||||||
| EPEC* | 13 | X | X | X | X | ||
| EPEC* | 13 | X | X | X | |||
| ETEC O6:H16 | 15 | X | X | X | |||
| ETEC O8:H9 | 0 | ||||||
| ETEC O15:H11 | 12 | X | X | X | |||
| ETEC O25:H42 | 12 | X | X | X | |||
| ETEC O78:H12 | 11 | X | |||||
| ETEC O115:H51 | 8 | X | |||||
| ETEC O20:H11 | 7 | X | X | ||||
| ETEC O27:H7 | 1 | ||||||
| ETEC O128:H18 | 0 | ||||||
| ETEC O63:H− | 8 | X | X | ||||
| ETEC O148:H28 | 14 | X | X | X | X | ||
| ETEC O153:H12 | 9 | X | |||||
| ETEC*LT+/ST+;CFA 1 | 5 | X | |||||
| ETEC*LT−/ST+;CS6 | 9 | ||||||
| ETEC*LT−/ST+;PCFO166 | 13 | X | X | X | |||
| ETEC*LT−/ST+;CFA1 | 13 | X | X | X | X | ||
| ETEC*LT/ST+;CS4:CS6 | 12 | X | |||||
| ETEC*LT+/ST+;CS1;CS3 | 13 | X | X | X | |||
| ETEC*LT+/ST+;CS5;CS6 | 0 | ||||||
| ETEC*LT+/ST+ | 17 | X | X | X | |||
| ETEC*LT+/ST+ | 1 | ||||||
| ETEC*LT+/ST− | 14 | X | X | X | |||
| ETEC*LT+/ST− | 0 | ||||||
| ETEC*LT+/ST− | 12 | X | |||||
The first column identifies the E. coli strain by O, K, or H serotype, pathotype, toxins or other virulence factors (see Materials and Methods for details). Strains from the ICDDR,B are labeled with an asterisk. The second column indicates the number of phages from 32 tested stool phage isolates that infected the indicated cells. The following columns mark cells that are sensitive to the indicated phage in the spot test. The individual phages are specified with their JS code number and their morphological diagnosis (in parentheses).
On the enteropathogenic E. coli strain O127:K63, a common isolate from childhood diarrhea (33), 18 out of 129 patients (14%, 11 samples were exhausted) showed stool specimens that tested positive for phage plaques. The titers on this strain (30 to 2 × 104 PFU/g of stool) were comparable to those on the laboratory E. coli strain. However, most patients yielded either phages on K803 (. = 22) or on the O127 strain (. = 14). Only four patients yielded phages both on O127 and on K803. This observation is not too surprising because T4, for example, infected K803 but not O127. When the results from both indicator strains were combined, 29% of the patients yielded coliphages. This percentage is close to the expected percentage of EPEC/ETEC infections in pediatric diarrhea patients in Dhaka. This suggests that the isolated phages were produced on the infecting E. coli strains and not on the gut E. coli commensal. We lack experimental evidence that proves this link, but previous (15) and ongoing (A. Bruttin, unpublished results) experiments in healthy adult mice and humans indicate that the normal coliphage detection rate on the K803 indicator is well below 5%.
For 52 patients, an additional stool sample was obtained at release from the hospital. On K803, the phage detection rate was lower in the stool sample from the convalescent than from the acute phase of the diarrhea episode (10 versus 25% prevalence, no titer difference). This low number seems inconsistent with coliphages being responsible for ending the suspected E. coli diarrhea episode; however, since the fecal coliphages were not titered on the specific infecting E. coli strains, there are other possible explanations for the results.
We identified in the literature only a single stool survey of phages in diarrhea patients (19). This study was conducted with adult patients suffering from traveler's diarrhea and showed substantially higher phage prevalence and phage titers in the stool than our survey. Their use of 10 indicator strains instead of two as in our study probably explains much of the difference.
Survey of coliphages in environmental water samples.
To complement the stool survey, a total of 104 environmental water samples were collected from the neighborhood of the patients. The samples included drinking (. = 10), well (. = 10), drain (. = 10), refuse (. = 10), canal (. = 11), pond (. = 7), sewage (. = 10), market (. = 10), street (. = 5), river (. = 6), and ditch (. = 5) water. Despite an expected high fecal pollution of environmental water in Dhaka (because people defecate in trenches along the streets), only a low level of coliphages was detected in the environmental water samples investigated. On the K803 indicator strain, only 4% gave a positive spot test (three drain, one ditch, and one sewage sample). Enrichment techniques did not increase the detection rate substantially; only one further phage was isolated. The detection rate was no higher than in a country with a high hygiene standard; in Lausanne, Switzerland, two out of four sewage samples yielded phages at low titers. This observation probably reflects the limited survival capacity of E. coli outside of the intestine (40) and the relatively rapid decay of phages in aqueous environments (41, 42). One might therefore question whether phages isolated from the patients were transmitted via contaminated environmental water.
Isolation of coliphages.
We treated multiple phage isolates from the same patient as epidemiologically linked samples even when they were derived from different stool samples (acute and convalescent) or when isolated on different indicators, giving 40 independent phage isolates: 22 from patients positive only on K803, 14 only on O127, and the four that were positive on K803 and O127. However, while all 22 K803 phage plaques could be amplified, only 8 of the 14 O127 phage plaques resulted in an amplifiable phage isolate after two isolation attempts. We were only able to isolate phage from two out of the four patients yielding plaques on both indicator strains. In both patients (114 and 76), the phages isolated on the two different indicator strains differed clearly in genome size (50 versus 170 kb; see below), increasing the number of independent phage isolates to 34 (22 + 8 + 2 × 2).
With the exception of patient 122, stool samples plated on K803 showed clear plaques and the isolated phages showed a 170-kb DNA genome size in pulsed-field gel analysis (see Fig. 1B, lanes a to d, for examples). From six patients (65, 94, 102, 110, 140, and 146), we isolated two or three phages from plaques that clearly differed in size. Since these isolates also showed distinct PCR reactivity with T4-specific primers (data not shown), it seems that a few patients harbored more than one T4-type phage in the same stool sample. In a control experiment, identical-looking plaques from the same stool sample were isolated for two patients. The phages isolated showed an identical restriction pattern (e.g., isolates JS110.1 and JS110.2; data not shown) and identical PCR reactivity (see Fig. 4C).
FIG. 1.
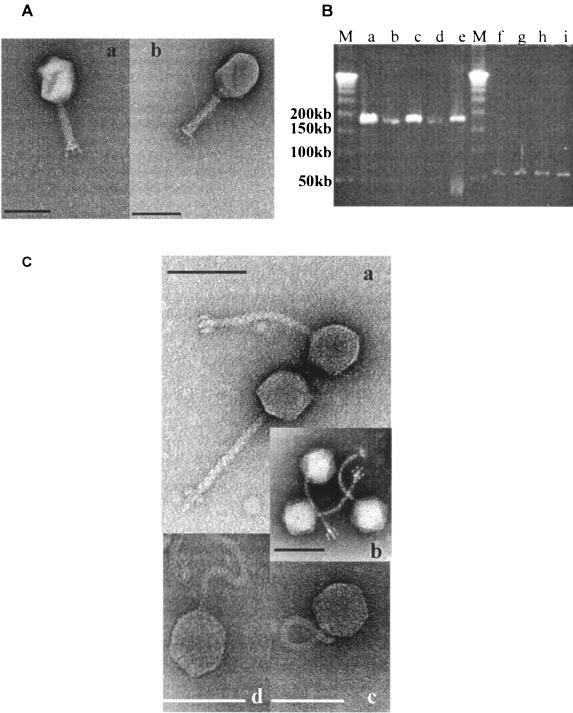
Genome size and morphology of the phages isolated from the stools of pediatric diarrhea patients. (A) Transmission electron microscopy picture of CsCl density gradient-purified myophages JS98 (a) and JSL.3 (b). Negative staining was with uranyl acetate. The size bar correspond to 100 nm. (B) Pulsed-field gel electrophoresis of myophages (lanes a, c, d, and e; lane b, T4 phage control) and of siphophages (lanes f to i) isolated from the stools of different patients. All isolates are from different patients lane a, JS4; lane 6, JS94.1; lane d, JSD.1; lane e, JS94.3; lane f, JS83.1; lane g, JS20; lane h, JS77.2; lane i, JS122.1. M, size markers (phage lambda concatemers, 50-kb DNA ladder; Promega). (C) Transmission electron microscopy picture of CsCl density gradient-purified bacteriophage JS77.1 after negative staining with uranyl acetate (a), phage JS83.2 after negative staining with phosphotungstic acid (b), and phage JS61.2 (c) and phage JS122.1 (d) after negative staining with ammonium molybdate coupled with bacitracin. The bar corresponds to 100 nm.
FIG. 4.
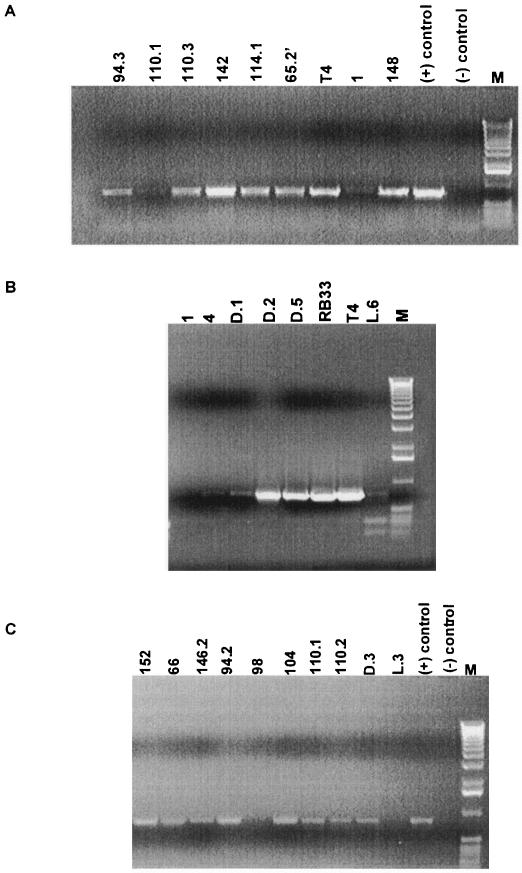
PCR analysis of the phages with a 170-kb genome. A. PCR with primers FR60 and FR61 amplifying gene 32 from stool samples. Gel analysis of the PCR fragments obtained with heat-treated phage particles from eight different stool phage isolates derived from seven different patients identified by their patient number at the top of the lanes. The lane marked with + is the positive control with T4 phage DNA and − is the negative control containing the master mix without DNA. M, 1-kb lambda DNA ladder. B. PCR with primers FR60 and FR61 amplifying gene 32 from four phages isolated from environmental water samples identified by their geographical origin at the top of the lanes. RB33 and T4 are the positive controls. 1 and 4 are negative controls with stool phages JS1 and JS4. C. PCR with primers Mzia1 and CAP8 amplifying gene 23 with heat-treated phage particles from eight stool isolates of seven different patients identified by their code number, two water phages identified by their geographical origin, T4 phage as a positive control, and no phage particles as the negative control (−). M, 1-kb lambda DNA ladder.
Patient 122 yielded turbid plaques on the K803 lawn of two sizes. Phages from both plaque sizes showed 50-kb genomes (Fig. 1B, lane i) and comparable morphology (data not shown). These phages did not cross-hybridize with phage lambda DNA.
In contrast, all phages isolated on O127:K63 showed 50-kb-long genomes (Fig. 1B, lanes f to h). They all made clear plaques, suggesting virulent phages, whereas half of the stool phage isolates from the patients with traveler's diarrhea were reported to be temperate phages (19). The lack of isolation of lambdoid coliphages in our study was not due to immunity functions in the indicator cell, since we used a K803 derivative that lacks the lambda prophage and that plates such phages well. This difference could therefore reflect biological differences between adult and childhood diarrhea, but the significance of this finding should not be overinterpreted because each study only gives a snapshot of the phages that are circulating in a given environment at the moment of sampling that plate on the specific hosts used. It is also likely that specific phage titers ebb and flow in an ecosystem.
Electron microscopy.
Electron microscopy showed that the phages with 170-kb genomes were Myoviridae (phages with long contractile tails) closely resembling coliphage T4 (Fig. 1A). All phages with a 50-kb genome were Siphoviridae (phages with long, noncontractile tails). Eight Siphoviridae isolates resembled the virulent Salmonella enterica serovar Typhimurium phages Jersey (1) and MB78 (23) (Fig. 1Ca). The characteristic trait is a fork-like baseplate that resolves in frontal view into six structures surrounding the tail's end (Fig. 1Cb). No such phage has previously been reported for E. coli (H. Ackermann, personal communication), demonstrating that even in intensively investigated research areas like coliphages, a moderately large ecological survey can still lead to “new” phage types. JS61.2 resembled coliphages from the beta-4 group, for which the extremely flexible tail is diagnostic (2) (Fig. 1Cc). The only phage forming turbid plaques (JS122) has a 61-nm head and a 128-nm twisted tail (Fig. 1Cd).
Host range.
Selected stool phage isolates were tested in the spot test for their host ranges on 40 diarrhea-causing E. coli strains, including 12 enteropathogenic E. coli (EPEC) strains representing the major serotypes isolated worldwide from pediatric diarrhea patients (33). This set of strains covered 10 different somatic O antigens and 10 different capsular K antigens (Table 1). In addition, the test collection contained 12 major enterotoxigenic E. coli (ETEC) serotypes isolated from either pediatric gastroenteritis patients or adults suffering from traveler's diarrhea (Table 1). The ETEC strains represented 11 further O antigens, 10 distinct H antigens, and various combinations of heat-stable (ST) and heat labile (LT) toxin producers (collected by B. Rowe, Central Public Health Laboratory, London, United Kingdom). Twelve further strains represent the predominant E. coli isolates from the Dhaka hospital, which were typed by DNA probes for the presence of heat-labile and heat-stable enterotoxins, colonization factor antigen, coli surface antigens, and the attaching-effacing genes according to published methods (18, 22) (Table 1).
In the spot test, 50% of the strains were infected by more than 10 phage isolates, while only 12% were not infected by any of the isolated phages (Table 1). Many but not all (e.g., JS150) of the T4-like Myoviridae infected numerous pathogenic E. coli strains. However, none of the isolated phages infected Salmonella enterica serovar Typhimurium or Enterobacter sakazaki strains. In contrast, the different fecal Siphoviridae infected very few pathogenic E. coli strains (Table 1).
The capacity of T4-like phages to infect E. coli strains covering a wide range of O serotypes is important for the prospect of phage therapy. Success with this approach hinges on the broad lytic capacity of obligate virulent phages because temperate phages are carriers of too many virulence factors (8). This broad lytic activity of strains within a species might not be linked to the isolation history of the phages from diarrhea patients. It possibly reflects a generally broader host range of Myoviridae than of Siphoviridae, as was previously observed in Listeria monocytogenes (27), Staphylococcus (31), Vibrio cholerae (28), and cyanobacteria (38).
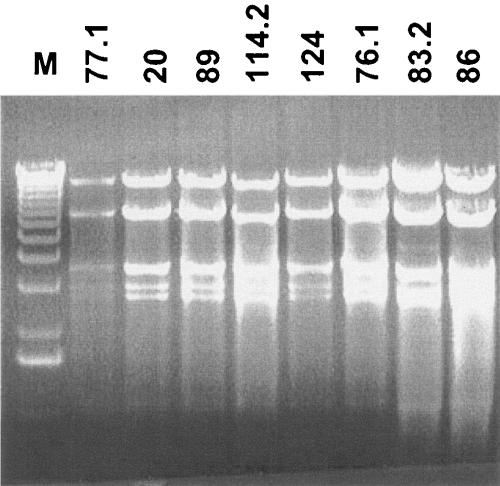
Restriction analysis.
Isolates of phages with 50-kb genomes derived from eight different patients (prototype JS77.1) on the indicator cell O127 showed identical restriction patterns (Fig. 2). Given the host specificity of known O antigen-specific phages, it may not be surprising that many phages that infect one O serotype are closely related. These phages do not represent a prophage from O127 because the majority of the isolated plaques did not yield a phage isolate when their propagation was tried on O127. In addition, patient 61 yielded on O127 indicator a siphophage with a 50-kb genome, but it showed a clearly distinct EcoRV restriction pattern (data not shown). Also, the 50-kb siphophage isolated from the stool of patient 122 on the K803 indicator strain showed a distinct restriction pattern (data not shown). Southern blot hybridizations confirmed that isolates JS122.1, JS61.2, and JS77.1 belong to three different DNA homology groups (data not shown).
FIG. 2.
Restriction analysis of siphophages with 50-kb genomes. EcoRV restriction digests of phages isolated from stool samples of eight different patients identified with their JS code number at the top of the lanes. M, size markers, 1-kb ladder.
T4 contains glucosylated hydroxymethyl cytosine instead of cytosine (11). This modification makes T4 DNA resistant to digestion with most restriction enzymes. Only a few enzymes are known to cut T4 DNA (EcoRV, NdeI, PacI, SwaI, SspI, SphI, and DraI) (26) (30). However, of nine T4-like phage isolates selected from different patients, none was digested by EcoRV, NdeI, SwaI, or SphI, and only two were digested by SspI, while all were at least partially cut by PacI (Fig. 3A). The best DNA digestion of the T4-like phages was obtained with the restriction enzyme DraI (data not shown). With this enzyme, several T4-like phages isolated from different patients showed closely related if not identical restriction patterns (e.g., JS9, JS12, JS94, JS104, and JS110, data not shown). The repetitive isolation of Myoviridae and Siphoviridae with similar if not identical restriction patterns from different patients suggests a possible epidemiological link between the patients. One could imagine that phages “travel” together with the diarrhea-causing E. coli strains by a person-to-person infection route via direct fecal-oral transmission. This process is not unlikely because the stomach acidity does not represent a substantial barrier for either phages or E. coli (15).
FIG. 3.
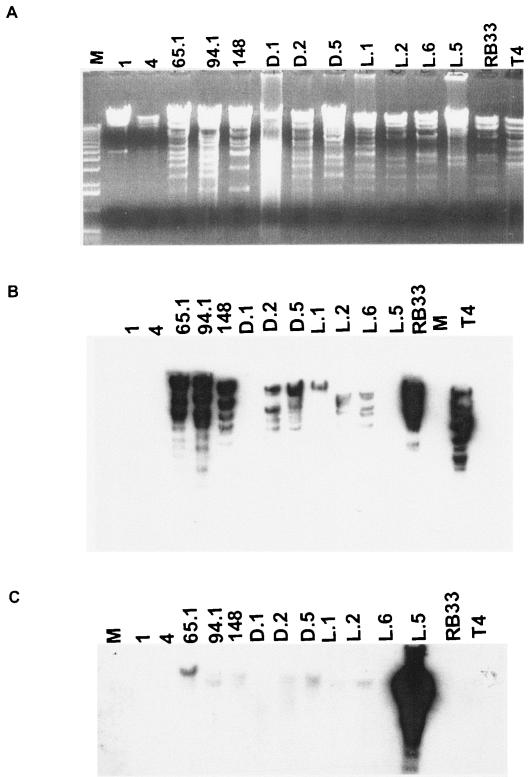
Restriction and Southern analysis of the stool and environmental water myophages with 170-kb genomes. A. PacI digests of five stool myophages from different children identified with their JS code number, seven environmental water myophages identified with their geographical origin (D, Dhaka; L, Lausanne) at the top of the lanes, and two reference phages (RB33 and T4). M, 1-kb lambda DNA ladder. B and C. Corresponding Southern blot hybridization with radiolabeled phage T4 DNA (B) or phage JSL.5 (C) as a probe.
Southern analysis.
We probed the PacI-digested phage DNA on Southern blots with labeled T4 DNA. Some isolates showed extensive cross-hybridization with the T4 probe, while others showed less extensive cross-hybridization or lacked DNA homology with T4 at the high stringency hybridization conditions (Fig. 3B). One of the phages that did not cross-hybridize with T4 DNA was the sewage phage isolate JSL.5. The genetic distance between JSL.5 and T4 and the other phages was confirmed when the Southern blot was probed with JSL.5 DNA. Only weak or no cross-hybridization was observed with the majority of the phages, including phages JS1, JS4, and JSD.1 (Fig. 3C) which also failed to hybridize with T4 DNA. There apparently exists a substantial genetic diversity among T4-like phages isolated on E. coli, as already suggested by the sequencing data published for the T4-like E. coli phage RB49 (16); for further information on this subject, see the website http://phage.bioc.tulane.edu/ and the accompanying report (14).
PCR.
Partial sequencing of T4-like phages identified graded genetic relatedness within T4-like phages and diagnostic PCR was developed from these sequence comparisons (32). PCR results with a primer pair (FR60 and FR61) from the T4 gene 32 (single-stranded binding protein) sequence, found to be diagnostic for the T-even subgroup of the T4-type phages, are shown in Fig. 4A and B. Phages that cross-hybridized with T4 DNA on the blots gave a clear positive PCR result in this assay, while phage isolates that did not cross-hybridize with T4 gave no or weak PCR bands (see for example phage JS1 in Fig. 3B and Fig. 4A). The phage isolates negative for a PCR product of gene 32 were then retested with a PCR based on a pair of primers (Mzia1 and cap8) in the T4 gene 23 (major head protein) sequence which allow the detection of the T-even, pseudo-T-even, and schizo-T-even subgroups of the T4-type phages (39).
Only the sewage phage isolate JSL.3 and the stool phage isolate JS98 were negative for this gene 23-specific PCR with heat-treated phages (Fig. 4C). When repeating the test with purified phage DNA as template, only JSL.3 was negative in the g23 PCR. Negative PCR results do not prove unequivocally that the PCR target is not present. On the other hand, a positive PCR result does show that the two phages are very likely related at the PCR target sequence. In both cases, it is advisable to confirm PCR results by Southern hybridization.
Overall, 12 of 31 (39%) T4-like stool phage isolates tested were classified by PCR into the T-even group sensu stricto (g23 and g32 PCR positive). The remaining Myoviridae represent more distantly related phages of the T4 phage group (g23-positive and g32-negative in PCR). The degree of genome diversification within the latter group of T4-like coliphages was investigated by an exploratory sequence analysis of the g23 PCR amplification products from fourteen g32 PCR-negative phages. The phages from eight different patients (JS4, JS9, JS32, JS76.2′, JS94.2, JS98, JS104, and JS110.1) yielded an identical 680-bp sequence that differed by 19% from the reference T4 g23 DNA sequence (accession number AY744074).
The isolation of eight such apparently closely related phages from eight independent patients is at first glance surprising, because many authors have commented in the past on the apparent high diversity of phage types in the natural environment. However, in DraI digestion several of these independent phage isolates shared a related if not an identical restriction pattern. These two observations suggest that we are dealing here with a stool phage that was relatively prevalent in Dhaka at the time of the survey. In fact, at some point on the time and space scales that diversity must break down, that is, the diverse population must be made of a collection of clones of essentially identical phages. This prediction seems to fit the observations, but it should not be overinterpreted because the use of only two indicator strains might have narrowed the isolation of more diverse phages. In addition, further regions of these eight apparently closely related phages should be sequenced to settle whether sequence identity extends to other regions of their genomes as well.
To explore the genetic nature of this apparently prevalent Dhaka phage isolate, we chose phage JS98 for genome sequencing. As JS98 differs from the reference T4 phage by 19% at the nucleotide level in the highly conserved major head gene, we reasoned that its sequence will allow us insights into the tempo and mode of evolution in T4-like phages. On the other hand, JS98 is, in contrast to vibriophage KVP40, sufficiently close to T4 to reveal the mechanisms that shape the short-term evolution of T4-like genomes (28). The accompanying report (14) describes the JS98-T4 genome comparison.
The g23 PCR product of four T4-like phage isolates from environmental water in Dhaka were sequenced. Isolates JSD.2 and JSD.5 were PCR g32+/g23+ and therefore as expected shared higher identity with the major head gene from T4 (94 to 96% nucleotide identity) than JS98 (accession numbers AY744075 and AY744076). Two other isolates, JSD.1 and JSD.3, were PCR g32/g23+. They closely resembled the g23 sequence from JS98; JSD.1 matched JS98 at 95% of nucleotides, while JSD.3 differed from the JS98 reference sequence at only 2 out of 730 sequenced bp of g23 (accession numbers AY744077 and AY744078). The most distant T4-like phages were two isolates from sewage water in Lausanne. Phage JSL.5 still yielded a g23 PCR product; it was as distant from T4 as from JS98 and differed from either phage T4 or JS98 at 138 of the 730 sequenced bp of g23 (>19% difference) (accession number AY744079). JSL.3 in contrast failed to amplify either PCR product.
Outlook.
For several reasons, T4-like E. coli phages are interesting candidates for phage therapy. They can be easily isolated from stool or sewage samples. Contrary to Siphoviridae isolated from the same source material, T4-like phages tend to infect many different serotypes of pathogenic E. coli even when propagated on a nonpathogenic E. coli strain like the K-12 derivative K803. This observation is a definitive asset for the safe production of phages for therapeutic purposes. In marked contrast to temperate E. coli phages (8-10), T4 phages have not been reported to encode any virulence genes important for bacterial pathogenicity (29), a crucial aspect for the safe use of phages. In addition, the T4-like phages described in the current report survived unprotected gastrointestinal passage in mice (15). Oral phages did not eliminate the endogenous E. coli gut flora, while an invading oral E. coli strain was controlled by T4-like phages in situ (15). Finally, a small phase I clinical trial demonstrated efficient gut passage and no adverse effects of oral T4 phage exposure in healthy adult human volunteers (Bruttin et al., unpublished data). After further safety studies in healthy and then diarrhea-stricken children, the stage is set to explore the potential of T4-based phage therapy in carefully controlled phase II clinical trials.
Acknowledgments
We thank Hans Ackermann for advice on morphological diagnosis of phages, H. M. Krisch for advice on the choice of the primers for PCR diagnostics of the T4-like phages, and Chelsea Thomas and Sarah Larson from the Evergreen lab for help in the initial phase of the work.
We thank the Swiss National Foundation for financial support of Sandra Chibani-Chennoufi (grant 5002-057832).
REFERENCES
- 1.Ackermann, H. W., L. Berthiaume, and S. S. Kasatiya. 1972. Morphology of lysotypic phages of Salmonella paratyphi B (Felix and Callow chart). Can. J. Microbiol. 18:77-81. [DOI] [PubMed] [Google Scholar]
- 2.Ackermann, H. W., and T. M. Nguyen. 1983. Sewage coliphages studied by electron microscopy. Appl. Environ. Microbiol. 45:1049-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert, M. J., A. S. Faruque, S. M. Faruque, R. B. Sack, and D. Mahalanabis. 1999. Case-control study of enteropathogens associated with childhood diarrhea in Dhaka, Bangladesh. J. Clin. Microbiol. 37:3458-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albert, M. J., S. M. Faruque, A. S. Faruque, P. K. Neogi, M. Ansaruzzaman, N. A. Bhuiyan, K. Alam, and M. S. Akbar. 1995. Controlled study of Escherichia coli diarrheal infections in Bangladeshi children. J. Clin. Microbiol. 33:973-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrow, P., M. Lovell, and A. Berchieri, Jr. 1998. Use of lytic bacteriophage for control of experimental Escherichia coli septicemia and meningitis in chickens and calves. Clin. Diagn. Lab Immunol. 5:294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhan, M. K., D. Mahalanabis, O. Fontaine, and N. F. Pierce. 1994. Clinical trials of improved oral rehydration salt formulations: a review. Bull. W.H.O. 72:945-955. [PMC free article] [PubMed] [Google Scholar]
- 7.Black, R. E., M. H. Merson, A. S. Rahman, M. Yunus, A. R. Alim, I. Huq, R. H. Yolken, and G. T. Curlin. 1980. A two-year study of bacterial, viral, and parasitic agents associated with diarrhea in rural Bangladesh. J. Infect. Dis. 142:660-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd, E. F., and H. Brüssow. 2002. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 10:521-529. [DOI] [PubMed] [Google Scholar]
- 9.Brüssow, H., C. Canchaya, and W.-D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brüssow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson, K., E. A. Raleigh, and S. Hattman. 1994. Restriction and modification, p. 369-381. In J. D. Karam (ed.), Bacteriophage T4. ASM Press, Washington, D.C.
- 12.Casswall, T. H., S. A. Sarker, S. M. Faruque, A. Weintraub, M. J. Albert, G. J. Fuchs, N. H. Alam, A. K. Dahlstrom, H. Link, H. Brussow, and L. Hammarström. 2000. Treatment of enterotoxigenic and enteropathogenic Escherichia coli-induced diarrhoea in children with bovine immunoglobulin milk concentrate from hyperimmunized cows: a double-blind, placebo-controlled, clinical trial. Scand. J. Gastroenterol. 35:711-718. [DOI] [PubMed] [Google Scholar]
- 13.Chibani-Chennoufi, S., A. Bruttin, M. L. Dillmann, and H. Brüssow. 2004. Phage-host interaction: an ecological perspective. J. Bacteriol. 186:3677-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chibani-Chennoufi, S., C. Canchaya, A. Bruttin, and H. Brüssow. 2004. Comparative genomics of the T4-like Escherichia coli phage JS98: implications for the evolution of T4 phages. J. Bacteriol. 186:8276-8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chibani-Chennoufi, S., J. Sidoti, A. Bruttin, E. Kutter, S. Sarker, and H. Brüssow. 2004. In vitro and in vivo bacteriolytic activities of Escherichia coli phages: implications for phage therapy. Antimicrob. Agents Chemother. 48:2558-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desplats, C., C. Dez, F. Tetart, H. Eleaume, and H. M. Krisch. 2002. Snapshot of the genome of the pseudo-T-even bacteriophage RB49. J. Bacteriol. 184:2789-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duckworth, D. H. 1999. History of virology: bacteriophages, p. 725-730. In A. Granoff and R. G. Webster (ed.), Encyclopedia of virology. Academic Press, Memphis, Tenn.
- 18.Faruque, S. M., K. Haider, M. J. Albert, Q. S. Ahmad, A. N. Alam, S. Nahar, and S. Tzipori. 1992. A comparative study of specific gene probes and standard bioassays to identify diarrhoeagenic Escherichia coli in paediatric patients with diarrhoea in Bangladesh. J. Med. Microbiol. 36:37-40. [DOI] [PubMed] [Google Scholar]
- 19.Furuse, K. 1987. Distribution of coliphages in the environment: general considerations, In S. M. Goyal, C. P. Gerba, and G. Bitton (ed.), Phage ecology. Wiley Interscience, New York, N.Y. p. 87-124.
- 20.Furuse, K., S. Osawa, J. Kawashiro, R. Tanaka, A. Ozawa, S. Sawamura, Y. Yanagawa, T. Nagao, and I. Watanabe. 1983. Bacteriophage distribution in human faeces: continuous survey of healthy subjects and patients with internal and leukaemic diseases. J. Gen. Virol. 64:2039-2043. [DOI] [PubMed]
- 21.Hoque, S. S., A. S. Faruque, D. Mahalanabis, and A. Hasnat. 1994. Infectious agents causing acute watery diarrhoea in infants and young children in Bangladesh and their public health implications. J. Trop. Pediatr. 40:351-354. [DOI] [PubMed] [Google Scholar]
- 22.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joshi, A., J. Z. Siddiqi, G. R. Rao, and M. Chakravorty. 1982. MB78, a virulent bacteriophage of Salmonella typhimurium. J. Virol. 41:1038-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karam, J. D. 1994. Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 25.Kutter, E., K. Gachechiladze, A. Poglazov, E. Marusich, M. Shneider, P. Aronsson, A. Napuli, D. Porter, and V. Mesyanzhinov. 1995. Evolution of T4-related phages. Virus Genes 11:285-297. [DOI] [PubMed] [Google Scholar]
- 26.Kutter, E., T. Stidham, B. Guttman, E. Kutter, D. Batts, S. Peterson, T. Djavakhishvili, F. Arisaka, V. Mesyanzhinov, W. Rüger, and G. Mosig. 1994. Genomic map of bacteriophage T4, In J. D. Karam (ed.), Bacteriophage T4. ASM Press, Washington, D.C. p. 491-519.
- 27.Loessner, M. J., M. Rudolf, and S. Scherer. 1997. Evaluation of luciferase reporter bacteriophage A511::luxAB for detection of Listeria monocytogenes in contaminated foods. Appl. Environ. Microbiol. 63:2961-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, E. S., J. F. Heidelberg, J. A. Eisen, W. C. Nelson, A. S. Durkin, A. Ciecko, T. V. Feldblyum, O. White, I. T. Paulsen, W. C. Nierman, J. Lee, B. Szczypinski, and C. M. Fraser. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Ruger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monod, C., F. Repoila, M. Kutateladze, F. Tétart, and H. M. Krisch. 1997. The genome of the pseudo T-even bacteriophages, a diverse group that resembles T4. J. Mol. Biol. 267:237-249. [DOI] [PubMed] [Google Scholar]
- 31.Pantucek, R., A. Rosypalova, J. Doskar, J. Kailerova, V. Ruzickova, P. Borecka, S. Snopkova, R. Horvath, F. Gotz, and S. Rosypal. 1998. The polyvalent Iphage phi 812: its host-range mutants and related phages. Virology 246:241-252. [DOI] [PubMed] [Google Scholar]
- 32.Repoila, F., F. Tétart, J. Y. Bouet, and H. M. Krisch. 1994. Genomic polymorphism in the T-even bacteriophages. EMBO J. 13:4181-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robins-Browne, R. M. 1987. Traditional enteropathogenic Escherichia coli of infantile diarrhea. Rev. Infect. Dis. 9:28-53. [DOI] [PubMed] [Google Scholar]
- 34.Savarino, S. J., E. R. Hall, S. Bassily, T. F. Wierzba, F. G. Youssef, L. F. Peruski, Jr., R. Abu-Elyazeed, M. Rao, W. M. Francis, H. El Mohamady, M. Safwat, A. B. Naficy, A. M. Svennerholm, M. Jertborn, Y. J. Lee, and J. D. Clemens. 2002. Introductory evaluation of an oral, killed whole cell enterotoxigenic Escherichia coli plus cholera toxin B subunit vaccine in Egyptian infants. Pediatr. Infect. Dis. J. 21:322-330. [DOI] [PubMed] [Google Scholar]
- 35.Smith, H. W., M. B. Huggins, and K. M. Shaw. 1987. Factors influencing the survival and multiplication of bacteriophages in calves and in their environment. J. Gen. Microbiol. 133:1127-1135. [DOI] [PubMed] [Google Scholar]
- 36.Snyder, J. D., and M. H. Merson. 1982. The magnitude of the global problem of acute diarrhoeal disease: a review of active surveillance data. Bull. W.H.O. 60:605-613. [PMC free article] [PubMed] [Google Scholar]
- 37.Sulakvelidze, A., Z. Alavidze, and J. G. Morris, Jr. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan, M. B., J. B. Waterbury, and S. W. Chisholm. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047-1051. [DOI] [PubMed] [Google Scholar]
- 39.Tétart, F., C. Desplats, M. Kutateladze, C. Monod, H. W. Ackermann, and H. M. Krisch. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183:358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wommack, K. E., R. T. Hill, T. A. Muller, and R. R. Colwell. 1996. Effects of sunlight on bacteriophage viability and structure. Appl. Environ. Microbiol. 62:1336-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]