Abstract
About 130 kb of sequence information was obtained from the coliphage JS98 isolated from the stool of a pediatric diarrhea patient in Bangladesh. The DNA shared up to 81% base pair identity with phage T4. The most conserved regions between JS98 and T4 were the structural genes, but their degree of conservation was not uniform. The head genes showed the highest sequence conservation, followed by the tail, baseplate, and tail fiber genes. Many tail fiber genes shared only protein sequence identity. Except for the insertion of endonuclease genes in T4 and gene 24 duplication in JS98, the structural gene maps of the two phages were colinear. The receptor-recognizing tail fiber proteins gp37 and gp38 were only distantly related to T4, but shared up to 83% amino acid identity to other T6-like phages, suggesting lateral gene transfer. A greater degree of variability was seen between JS98 and T4 over DNA replication and DNA transaction genes. While most of these genes came in the same order and shared up to 76% protein sequence identity, a few rearrangements, insertions, and replacements of genes were observed. Many putative gene insertions in the DNA replication module of T4 were flanked by intron-related endonuclease genes, suggesting mobile DNA elements. A hotspot of genome diversification was located downstream of the DNA polymerase gene 43 and the DNA binding gene 32. Comparative genomics of 100-kb genome sequence revealed that T4-like phages diversify more by the accumulation of point mutations and occasional gene duplication events than by modular exchanges.
Phage T4 is among the best-characterized biological systems (20); for example, a recent review on the T4 phage genome alone quoted 1,240 references (27). In the era of sequence-driven microbiology and comparative genomics, it is therefore surprising that only a single further T4-type phage genome sequence has been analyzed, that of the Vibrio phage KVP40 (26). This phage differed substantially from T4 in genome size (244 versus 169 kb for T4), making direct comparisons difficult. In fact, 65% of the 386 open reading frames predicted for KVP40 had no known functions and lacked any database matches; 26% of the KVP40 open reading frames shared up to 60% amino acid identity with T4 proteins, defining it clearly as a distant relative of phage T4. However, beyond the conserved structural and DNA replication modules, T4 and KVP40 have diverged substantially from each other by the acquisition of large blocks of unrelated genes.
To better investigate the mode of evolution in this phage group, it will be necessary to compare the genomes from more closely related T4-type phages. Early work with heteroduplex mapping of Escherichia coli phages T2, T4, and T6 DNA revealed that their genomes differed at about 30 loci, showing both substitution and deletion loops (21). However, this analysis also suggested a close matching of the phage DNA sequence over about 85% of the genome length. The correlation of the physical and genetic maps allowed the location of the regions of diversity. In the last decade several of these regions were sequenced for different T-even phages, and the results correlated well with biological differences. For example, the distinct host ranges of T2, T4, and T6 were associated with distinct tail fiber gene constellations (the major difference being the role of gp37 and gp38 as phage adhesins) (28, 32).
The distinct glucosylation pattern of phage DNA was linked to unrelated forms of α-glucosyltransferases in T2 and T6 and T4 (34). Sequence analysis identified gene replacements in the DNA replication genes (31). When six variable loci in a set of 52 natural isolates of T-even phages that all grew on enterobacteria were investigated by PCR sequencing (30), four loci showed a dimorphic pattern (only two gene constellations). These were the regions between the DNA replication genes 59 and 32, the immunity locus upstream of the b-gt (β-glucosyltransferase) gene, the topoisomerase, and the tail locus. In contrast, two regions were polymorphic, with up to nine distinct gene constellations (ip3/2 and ip1 loci encoding internal head proteins).
On the other hand, some efforts were focused on the sequencing of particularly well conserved structural genes like the major head gene g23 in T4-like phages infecting an even wider range of host bacteria. Phylogenetic tree analysis of these single gene sequence data led to the distinction of T-even and pseudo-, schizo- and exo-T-even phages: these phages showed increasingly distant relationships with the reference T4 phage, covering first DNA sequence and then decreasing protein sequence identity (33). Graded relatedness that varied with the phylogenetic distance of the bacterial hosts was also reported in temperate dairy phages (5). In contrast, the standard theory of phage evolution developed in the 1970s for lambdoid coliphages (3) is based on abundant horizontal exchange of functional gene clusters (modules) (7) across wide phylogenetic distances of host bacteria (17).
This model therefore does not postulate evolutionary lineages of phages, but at best only an evolutionary history for individual phage modules. However, it is premature to postulate a distinct mode of evolution in lambdoid and T4-like coliphages as long as these inferences rely on the sequences of single phage genes. Despite the prominence of phage T4 for the history of molecular biology, only limited comparative whole genome analysis was published for T4-like E. coli phages. The currently most comprehensive study compared random clones representing 20% of the genome from the pseudo-T-even phage RB49 with T4. RB49 belongs to the three of the 52 T-even phages from the Toulouse collection growing on enterobacteria that do not react with the diagnostic g32 (major head gene) PCR test. RB49 shared only weak DNA but clear protein sequence identity with T4 (10).
In the present report we analyzed 100 kb of the genome sequence from phage JS98, a typical T4-like coliphage stool isolate from Bangladeshi children described in the accompanying paper (8), to provide a broader basis for the comparative genomics of T4-like coliphages.
MATERIALS AND METHODS
Phage selection and sequencing strategy.
Phage JS98, whose isolation is described in the accompanying manuscript (8), was chosen for detailed sequence analysis. Its preliminary characterization suggested a moderately divergent T4-like coliphage (8). JS98 phage DNA was sheared to about 1 kb before cloning into the plasmid vector pUC18; the plasmid was propagated in E. coli. The DNA was sequenced to an eightfold coverage. When the sequences were assembled, the JS98 genome was still represented by 20 contigs, suggesting a large number of host-lethal genes impeding cloning in E. coli. However, most contigs could be projected on the T4 map due to DNA sequence identity with T4 (Fig. 1). This projection predicted that several contigs were nearly adjacent. Indeed, a number of missing JS98 DNA sequences could be obtained by the sequencing of PCR amplicons obtained with primers placed at the ends of the 20 contigs. This approach reduced the number of contigs to 13.
FIG. 1.
Dot plot alignment of 133-kb DNA sequence information from phages JS98 and T4. Thirteen contigs from JS98 were projected on the T4 map, and the ordered contigs were artificially concatenated. The comparison window for the DNA sequence dot plot was 50 bp, and the stringency was 30 bp. Note that the complete sequence from T4 was plotted against the partial sequence from JS98. The horizontal lines cutting the JS98 sequence thus indicate gaps of nonsequenced JS98 DNA. The largest gaps are anticipated between contigs where the diagonal of the alignment showed a substantial parallel shift (e.g., between contigs 5 and 12). Note the DNA repeat over the two g24 copies. To facilitate the orientation, a scale in base pairs is provided for the T4 genome on the top x axis, and the location of the major T4 modules as classified before (16) is given at the bottom x axis.
Several JS98 contigs lacked DNA sequence relationship with phage T4 (Fig. 1, contigs 8, 20, 19, and 40). Sequence matches of the predicted proteins with T4 or its closely related phage RB69 (http://phage.bioc.tulane.edu/) (contig 8: 22% and 40% amino acid identity with nrdC.5, 6 from T4, log E value = −8 and −27, respectively; contig 20: 78% amino acid identity with e.6 homologue from RB69, log E = −25; contig 19: 56 and 76% amino acid identity with mlA and denA homologue from RB69, log E = −61 and −24, respectively; contig 40: 35% amino acid identity with arn.2-4 homologues from RB69) allowed a tentative ordering of all available contigs (Fig. 1). The remaining gaps correspond in T4 to regions containing genes of mainly unknown functions or functions that are likely to interfere with their cloning.
Sequence analysis.
Open reading frames have been predicted with the FrameD program (Toulouse Bioinfo INRA) with ATG, GTG, and TTG as possible start codons and a minimum size of 30 amino acids. Nucleotide and predicted amino acid sequences were compared in the databases of NCBI/GenBank; EMBL; PIR-Protein; SWISS-PROT; and PROPOSITE. The NCBI database contains the complete but as yet unpublished genome sequences from two E. coli T4-like phages (RB69 and RB49) and two Aeromonas T4-like phages (Aeh1 and 44RR 28.t). Additional database searches have been conducted with BLAST (1) and PSI-BLAST at the NCBI and FASTA (24). A search for tRNA genes was done with the tRNAscan-SE program (25). The motif search was performed with Pfam.
Nucleotide sequence accession number.
The sequence data were deposited at GenBank under accession numbers AY746495, AY746496, and AY746497.
RESULTS AND DISCUSSION
Overall, 133 kb (80%) of the genome from JS98 was sequenced. A projection of the sequences on the T4 map is shown in Fig. 1. This tentative assembly should be interpreted with caution because we lack experimental evidence (except for the PCR bridging of supposed adjacent contigs, see above) that the order of the contigs in JS98 is the same as the parallel sequences in T4. Three large contigs representing 100 kb of genome information were analyzed in detail: contig 9, a 48.7-kb segment covering the morphogenesis module; contig 38, a 20.4-kb region containing tail fiber and DNA transaction genes; and contig 12, a 25.4-kb section with DNA replication genes.
Structural gene cluster of phage JS98.
The structural genes in JS98 are found on two different contigs (contigs 9 and 38, Fig. 1). According to the tentative projection of JS98 on the T4 sequence, both structural gene clusters are separated by a segment of DNA transaction genes (Fig. 1). This separation of tail fiber genes from the rest of the structural genes is probably an ancient character because it was also observed in phage KVP40 (26).
Endonucleases.
Figure 2 shows an alignment of the structural genes in JS98 contig 9 and T4 DNA. In addition to the homing endonuclease genes (segB, -C, -D, and −E), this region of T4 has only three small genes of unknown function that lack counterparts in JS98 (genes 5.3, 24.2, and usvY.1). The homing endonuclease genes are not genuine phage DNA, but belong to selfish DNA elements that are frequently intron-associated (2, 11) These elements have efficiently invaded the T4 genome and are also found in the DNA transaction module opposite the tail fibre cluster (I-TevI and segG) (Fig. 3) and the DNA replication module (segA, mobB, mobC, and I-TevII) (Fig. 4). They consistently lack a counterpart in the corresponding JS98 DNA. Inconsistent distribution of these elements has been described for T2, T4 and T6 phages (29). No homing endonuclease has as yet been detected in the JS98 genome. Apparently there are ill-defined barriers to intron promiscuity in bacteriophages (12, 13).
FIG. 2.
Comparison of the morphogenesis modules from coliphages JS98 and T4. Alignment of the baseplate, tail, and head genes from T4 (upper line) and contig 9 from JS98 (lower line). Genes sharing protein sequence identity are linked by blue to red shading according to the color key provided at the bottom right. The likely function of the genes is indicated by the color of the arrows according to the color key at the bottom left. All T4 genes are annotated with their gene numbers, and selected genes are annotated with their functions. The top line provides a base pair scale and the positions of the first and last depicted T4 genes on the T4 genome.
FIG. 3.
Comparison of the tail fiber and the adjacent DNA transaction modules from phages JS98 and T4. Alignment of the tail fiber genes (blue), DNA transaction and transcription genes (pink), inserted endonuclease (orange), and lysis (green) genes of contig 38 from JS98 (top) and T4 (bottom). The colors of the connecting shadings indicate the percentage of amino acid identity (see Fig. 2 for a key to the color code). The bottom line provides a scale in base pairs. T4 genes are annotated with the T4 gene terminology. JS98 open reading frames lacking a T4 complement are annotated with the closest match to another T4-like phage (strain in capital letters, percentage amino acid identity in parentheses) or a minus sign if they are lacking any database match.
FIG. 4.
Comparison of the DNA replication module from phage JS98, T4, and RB69. Alignment of the DNA replication genes from phage RB69 (top), contig 12 from JS98 (center), and T4 (bottom). DNA replication genes are colored in yellow, DNA-modifying genes in violet, nucleotide metabolism genes in orange, endonuclease genes frequently found associated with mobile DNA are in light brown, and the immunity gene is in blue. The T4 open reading frames are annotated with the conventional gene name, JS98 open reading frames are numbered, and RB69 genes are quoted with the annotations given to them in the GenBank entry (accession number NC_004928). Genes that share sequence identity are connected by shading, and the degree of amino acid identity is color coded (for the key, see Fig. 2).
JS98 differs from T4 by a longer wac gene (encoding the phage neck-associated whiskers, which facilitate long tail fiber attachment; references for this and all other T4 genes mentioned are provided in reference 27), a shorter hoc gene (coding for an outer capsid protein in T4), and a possible duplication of g24 (encoding a head vertex protein). At the left side of the JS98 morphogenesis module lie genes for deoxynucleoside monophosphate kinase, gp57A and -B chaperons, and several auxiliary genes related to T4, but JS98 diverges from T4 3 kb further downstream (data not shown).
Head and tail.
The structural proteins from JS98 shared amino acid sequence identity with T4 ranging from a minimum of 32% for gene 29 (the tail length determinant in T4) to a maximum of 90% for gene 23 (encoding the major head subunit precursor). Overall, the head gene module showed the highest degree of sequence conservation, but the extent of conservation differed substantially from gene to gene (50 to 90% amino acid identity) (Fig. 2). The next most conserved genes were the four tail genes (51 to 72% amino acid identity), which are mixed with head genes. This intermingling of head and tail genes is probably an ancestral character because it was also observed in vibriophage KPV40 (26).
Baseplate.
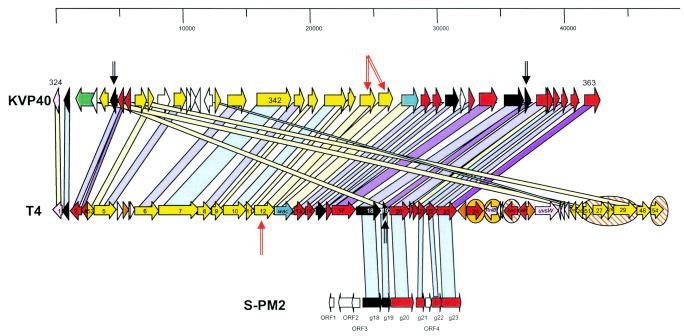
The protein sequence identity between JS98 and T4 was 35 to 72% amino acid identity for the baseplate “wedge” module and 32 to 61% for the baseplate “hub” module (Fig. 2). The separation of baseplate genes into two clusters interrupted by tail and head genes is not a conserved trait because in vibriophage KPV40 all baseplate genes come in a single gene cluster (Fig. 4). We need sequence data from a third group of distantly related T4-like phages like cyanophage S-PM2 (15) to decide what constellation is the ancient configuration.
At the DNA sequence level, the JS98 head module revealed 72 to 81% base pair identity with T4 DNA, showing only very few small gaps. Individual baseplate wedge genes reached up to 75% base pair identity over the entire gene, while baseplate hub genes shared DNA sequence identity only over segments of the individual genes, resulting in many gaps in the alignment (Fig. 1). Possibly, the less conserved baseplate proteins are less bound by tight protein-protein interactions and have more free surface area. The three-dimensional structure of the T4 baseplate was recently determined by cryoelectron microscopy (22), allowing us to test the correlation between the degree of sequence conservation and location in the baseplate structure.
In the wedge module gp9, gp11, and gp12 showed the lowest degree of sequence conservation, and these were also the most outside-oriented structures of the baseplate. gp12 is the short tail fiber, making the external and downside hexagonal garland of the baseplate. gp11 is the even more exposed linker protein connecting the gp12 fibers. gp9 is the most outside oriented protein of the upper, tail-oriented part of the baseplate. The central part of the baseplate hub is composed of the well-conserved gp5 and gp27, making the cell-puncturing device (19), and gp54 and gp48, making the contact to the tail sheath (22). The less well conserved gp26, gp51 (assembly catalyst?), gp28, and gp29 (tail length determinant) were not located in the baseplate cryostructure, precluding an analysis of the structure-conservation correlation.
Fibers.
The whisker fiber gene wac, although bracketed by the relatively well-conserved baseplate wedge and the head modules, shared only 32% amino acid identity with its T4 homologue. The JS98 tail fiber genes are found in a different genome segment, contig 38, bracketed, as in phage T4 by the RNase-encoding gene rnh and the lysis protein t (Fig. 3). Also, the tail fiber proteins were less well conserved than the rest of the phage structural proteins. In addition, a clear gradient of sequence conservation between T4 and JS98 was seen along the tail fiber structure, decreasing from the highest values for the proximal (gp34) over the hinge protein (gp35) to the small (gp36) and the large distal tail fiber subunit (gp37) showing the lowest degree of conservation. The JS98 gene opposite the T4 gene 38 lacks sequence relatedness with T4.
The tail fiber adhesin domains are located in different genes in T-even phages. In phage T4, the adhesin is located in the C-terminal domain of gp37, while in other T-even phages the adhesin is a separate protein, gp38, which comes in at least three different alleles represented by T2, Ac3, and T6/Ox2 (32). The corresponding predicted JS98 proteins showed clear sequence identity with RB69 (75 and 55% amino acid identity with gp37 and gp38, respectively), T6 (47 and 79% amino acid identity for gp37 and gp38, respectively), and phage Ox2 (83% amino acid identity with gp38), but only low or no amino acid identity with the corresponding T4 proteins. Notably, the host ranges of JS98 and RB69 overlap on the pathogenic E. coli strains; this was not the case between JS98 and T4 (data not shown). The adhesin JS98 is thus a variant of the T6 adhesin allele. This region is a hotspot of lateral gene transfer not only between T4-like phages, but also between a much wider group of morphologically distinct coliphages (14).
Evolutionary analysis of structural genes: point mutations and graded relatedness.
The accumulation of point mutations is apparently a major driving force in T4-type phage evolution. However, the degree of sequence diversification was not uniform. The head proteins were the most conserved structural proteins, probably due to substantial protein-protein interactions in the phage capsid. Table 1 shows the statistics of point mutations for the major structural components of the head and the tail sheath. It is apparent that the JS98 and T4 tail gene 18 homologues are separated by more base pair changes than the head gene 23 homologues. Two processes could account for this difference. One possibility is that the differences are the result of phages that diversified entirely separately and then recombined parts of their genomes. The nonhomogeneous sequence conservation over the structural genes could be cited as an argument for frequent recombination among this group of phages in nature. Alternatively, the different degrees of sequence diversification could reflect a differential selection pressure.
TABLE 1.
Base pair exchanges in the major head and tail genes between phages JS98 and T4a
| Region (bp) | No. of base pair differences
|
|||||
|---|---|---|---|---|---|---|
| 1st | 2nd | 3rd | Total | 3rd/syn | 3rd/nonsyn | |
| g23 | 68 | 49 | 185 | 302 | 128 | 1 |
| 1-250 | 18 | 11 | 32 | 61 | 18 | 0 |
| 250-500 | 6 | 4 | 28 | 38 | 25 | 0 |
| 500-750 | 17 | 15 | 32 | 64 | 15 | 0 |
| 750-1000 | 6 | 3 | 25 | 34 | 21 | 0 |
| 1000-1250 | 10 | 9 | 27 | 46 | 21 | 0 |
| 1250-1560 | 11 | 7 | 41 | 59 | 28 | 1 |
| g18 | 157 | 106 | 336 | 599 | 196 | 7 |
| 1-250 | 16 | 12 | 39 | 67 | 23 | 1 |
| 250-500 | 32 | 22 | 45 | 99 | 17 | 2 |
| 500-750 | 16 | 10 | 37 | 63 | 24 | 2 |
| 750-1000 | 22 | 17 | 48 | 87 | 31 | 0 |
| 1000-1250 | 24 | 17 | 43 | 84 | 21 | 2 |
| 1250-1500 | 15 | 4 | 42 | 61 | 30 | 0 |
| 1500-1750 | 17 | 12 | 42 | 71 | 25 | 0 |
| 1750-1980 | 15 | 12 | 40 | 67 | 25 | 0 |
The first column describes the investigated region, which was either the entire gene or successive DNA segments of 250 bp; the second to fourth columns give the number of differences at the first, second, and third codon positions, respectively, of the two phages. The fifth column lists the total number of changes observed over the indicated gene regions. The sixth and seventh columns give the number of synonymous and nonsynonymous changes, respectively, in the codons that differ only at the third base position.
Support for this interpretation can be derived from the analysis of individual genes. For example, in gene 23, the second and fourth 250-bp segments showed significantly fewer overall base pair changes and a significant preference for the observed base pair changes to occur at the third base pair position, indicative of selection pressure to maintain the amino acid sequence. In addition, from the 128 codons that differed between g23 from T4 and JS98 exclusively at the third base position, all but one were synonymous changes (Table 1). A similar, although less impressive, avoidance of nonsynonymous base pair changes was also observed for the codons that differed only at the third base position of the g18 sequences (Table 1). In contrast, the first segment of gene 23, which encodes a segment that is cleaved off during maturation of the prohead (20) and thus under less structural constraint, showed a greater number of base pair changes and the bias towards changes at the third codon position was less marked. This second interpretation is also backed by the observation of a similar gradient of sequence identity, although at a lower percentage level, in the T4-KVP40 comparison (head > tail > base plate > tail fiber) (Fig. 5). Even the head genes from a T4-like phage isolated from a very distant bacterial host, cyanobacteria (15), still shared some amino acid sequence identity with T4 (Fig. 5). Comparable series of graded relatedness were seen between structural genes from members of the phage family Siphoviridae and were interpreted as evidence for vertical elements in the evolution of phages (5).
FIG. 5.
Comparison of the structural gene modules in more distantly related T4-like phages. The T4 line is a copy from Fig. 2; it was aligned with the structural gene clusters from vibriophage KVP40 (top) and cyanophage S-PM2 (bottom). The putative gene functions and the amino acid sequence identity are color coded as specified in Fig. 3 (pale yellow here indicates 20 to 30% amino acid identity). Duplicated KVP40 genes are marked with two black or red arrows; the corresponding T4 gene was also marked. T4 genes located in a different genome position in KVP40 are marked with a yellow oval, and T4 genes lacking a corresponding KVP40 gene are indicated by a striped oval. Selected KVP40 and S-PM2 genes are annotated with their gene numbers used in the original publication (15, 27). The top scale is in base pairs.
Evolutionary analysis of structural genes: gene duplications and rearrangements.
The structural-gene clusters from T4-type phages were apparently also shaped by gene duplications and genome rearrangements. For example, T4 gene 24, encoding in T4 a head vertex subunit, was probably duplicated in JS98 (Fig. 2). However, the “duplicated” gp24 homologues in JS98 differ substantially in amino acid sequence and both show in addition sequence links to gp23. T4 gp23 and gp24 share 29% amino acid identity, suggesting that g23/g24 were already the result of an even earlier duplication event (16). Thus, instead of duplication of an ancestral JS98 g24 gene, the genes could be the consequence of sequential duplication and divergence of gene 23; the branching order is not certain.
One g24 copy was in addition split into two open reading frames and shared 70% amino acid identity with T4 gp24 and 28% amino acid identity with T4 gp23. The other g24 copy was integral and shared 55 and 28% amino acid identity with T4 gp24 and gp23, respectively. Both JS98 g24 copies shared DNA sequence identity (Fig. 1), while no DNA sequence identity was observed between them and JS98 g23, arguing for a more recent g24 duplication event. Duplication of structural genes seems to be a frequent observation in T4-type phage evolution, as demonstrated by the KVP40-T4 comparison (15) (Fig. 5). The KVP40 baseplate gene 12 is tandemly duplicated, while the tail gene 19 in KVP40 was duplicated with one copy in the baseplate and another in the head gene cluster (26).
gp24 plays an interesting role in T4 head maturation. The T4 prohead consists of an outer shell (gp23) and an inner scaffold (gp22). When gp24 is added to the prohead, maturation cleavage occurs: gp23 loses its N-terminal part, while gp22 is degraded completely (8, 20). Further functional data point to a link between gp24 and gp23: g24 bypass mutations map in g23. Specifically, one class (trb) maps in those regions of gp23 that share sequence similarity with gp24 (16). It therefore seems plausible that g24 evolved from g23 by developing a new accessory function. In fact, duplications liberate one copy of the duplicated genes from the constraints imposed by selection, since a reserve gene is still in place. Genes are therefore free to evolve new functions.
DNA replication module.
The 25-kb contig 12 contains many of the DNA replication and recombination genes known from T4 (Fig. 4). Specifically, JS98 encodes close homologues of the T4 replicative helicase (gp41), the RecA-like recombination protein UvsX, the DNA polymerase (gp43), its two clamp-loader subunits (gp62 and gp44), and the sliding clamp (gp45), as well as the recombination-nuclease proteins gp46 and gp47 and, at some map distance, the endonuclease gp49, involved in resolution of the recombination junctions. Amino acid identity ranges from 54% (gp45) to 76% (gp49). Furthermore, the relative gene order of the essential replication and recombination genes was conserved between JS98 and T4 (Fig. 4).
Major differences between the corresponding JS98 and T4 regions were observed at the level of nonessential genes not involved in DNA replication and recombination functions, which were found in the same genome region. In fact, three groups of T4 genes lacked counterparts in JS98. Notably, the genes β-gt and α-gt, involved in glucosylation of the hydroxymethyl cytosine residues in T4, are accompanied by the segA and mobB homing endonuclease genes, respectively, which suggests a mobile character for these regions (Fig. 4). Like RB69, JS98 contains more open reading frames in this region. Significant matches were observed with genes annotated as thymidylate synthase and methylthioadenine synthetase. Only open reading frame 10 showed a nonsignificant match with a sugar nucleotidyltransferase over part of its sequence (data not shown) Also, the auxiliary T4 genes nrdG and nrdD, which encode subunits of an anaerobic nucleotide reductase, lack complements in the corresponding genome region from JS98 (Fig. 4). In T4, nrdD was associated with the endonuclease gene mobC, again suggesting a mobile DNA element.
For the region located between uvsX and the DNA polymerase gene 43, all but one JS98 gene matched RB69 genes from the corresponding region (Fig. 4). However, RB69 differs in this region from JS98 by rearrangement, insertion, and replacement of genes (Fig. 4), defining this region as a likely hotspot for gene insertions in T4-like coliphages. There are other but much smaller regions where JS98 differed from both RB69 and T4, e.g., near the T4 pin gene, encoding a protease inhibitor.
As in T4, JS98 has genes in this genome segment that are not involved in DNA replication; the superinfection immunity gene imm, the translational repressor gene regA, rpbA (encoding a RNA polymerase binding protein), and the late sigma factor gene 55, and all were sequence related to T4 and RB69. Interestingly, regA- and g55-like genes were also seen at identical position in KVP40, suggesting ancient insertion events in the DNA replication module.
Near the tail fiber gene cluster, the T4 genome contains a number of genes that block or reduce DNA synthesis when mutated (27). To the former group belong the essential genes g32 (the single-stranded-DNA-binding protein which builds the scaffold of DNA replication) and g59 (the loader of the gp41 helicase). Closely related JS98 genes were identified on contig 38 with closely related map order and sequence relatedness (61 to 76% amino acid identity) (Fig. 3). To the latter group belong genes encoding the α subunit of the ribonucleotide reductase (NrdA), thymidylate synthetase, dihydrofolate reductase (Frd), and RNase H, involved in the processing of Okazaki fragments (Rnh). JS98 showed again T4-related proteins (50 to 86% amino acid identity). Differences between JS98 and T4 were two endonuclease genes in T4 (I-TevI and segG), gene replacements (JS98 frd.1 and -2, flanked by genes lacking any database match), single gene indels (e.g., T4 nrdA.2), and a duplication of frd.3 in JS98 (Fig. 3).
Diversification of T4-like phage genomes: an outlook.
The fact that three independent isolates of T4-like phages (T4, RB69 [http://phage.bioc.tulane.edu/], and JS98) isolated 40 years apart from two different geographical areas (New York and Dhaka) (23) can be aligned at the DNA level essentially over their entire genome suggests that modular exchanges, which are such a dominant feature in the evolution of the lambda-like supergroup of E. coli, are a less prominent process in T4-like phages. However, this observation should be interpreted with caution. Our current analysis of the JS98 sequence is biased towards the most conserved genome segments (structural and DNA replication genes). When extending the analysis to the remainder of JS98, we will probably see more substantial diversity, as already indicated by the worse DNA sequence alignment for the smaller contigs not analyzed in the present report (Fig. 1). In fact, the comparison of the T4-like phage genome maps on the Tulane University website (http://phage.bioc.tulane.edu/) revealed that the diversity of the sequenced phages was highest over those regions of the T4 genome lacking attributed gene functions.
Nevertheless, our current JS98 sequence analysis identified a number of regions where modular exchanges most likely had occurred. One such region encodes the receptor-recognizing fibers (Fig. 3). This region was previously identified as a recombination hotspot (14, 28, 32). Another likely modular exchange was seen at the level of the JS98 frd.1 and -2 genes (Fig. 3) Interestingly, in an independent study involving nineteen T4-like phages, three very different genome patterns have been reported for the frd.1 to frd.3 region, a nonessential T4 region (23). As in our comparison, most of frd.3 was conserved, while half of frd.2 and the frd2/frd.1 region showed strong polymorphism. Similarly polymorphic loci on the genomes from T4-like coliphages were previously identified by heteroduplex mapping (21) or the sequencing of PCR products in six different genome regions (30). However, the size of the modularly exchanged DNA segments seems to be smaller in T4-like phages than in lambdoid coliphages. Also, the number of distinct alleles seems to be lower; one larger survey identified predominantly two allelic forms. With respect to the head, tail, and baseplate genes or the major DNA replication genes, even a single allelic form was identified. This single structural gene cluster maintained protein sequence identity over substantial evolutionary distances separating the bacterial hosts (proteobacteria and cyanobacteria) (Fig. 5), suggesting a substantial element of vertical evolution for this genome region in T4-like phages.
In the lambda supergroup of Siphoviridae a much lower degree of structural protein sequence conservation was seen when phages were analyzed over a comparable phylogenetic distance of their bacterial hosts. In fact, lambda-like Siphoviridae from gram-positive and gram-negative bacteria shared essentially only a comparable organization of the head and tail gene map (5). In addition, a number of sequence-unrelated structural gene modules were identified even within a single host species like E. coli (18). Diversification by the accumulation of point mutations is clearly the dominant mode of evolution over diversification by modular exchanges for the T4-like structural genes.
We identified two likely candidates for gene duplications in the T4-JS98 comparison (g24 and frd.3) and the T4-KVP40 comparison revealed that this is in fact a common observation (26) (Fig. 5). We are not aware of similar observations in lambda-like Siphoviridae. This difference might be of functional importance. Duplicated genes allow the accumulation of many mutations in one copy of the gene that is liberated from the constraints to maintain a potentially essential function. This process might be necessary for phages that evolve new functions predominantly from genes already existing in the phage genome. Lambda-like phages, in contrast, seem to have opted for the import of new functions by lateral gene transfer from sources outside of the gene pool of lambda-like phages.
In contrast to the classical modular exchange reactions, where blocks of functionally homologous gene are laterally transferred, the term moron (more DNA) was coined in lambda-like phages to identify additional open reading frames endowed with their own promoter and terminator that can integrate in various positions of the phage genome (18). Candidates for extra DNA were also identified in T4-like phages. For example, the region between the uvsX and g43 replication genes, encoding the β-glucosyltransferase and deoxycytosine monophosphate hydroxymethylase involved in the T4-specific chemical modification of cytosine bases, looks like an insertion into T4 DNA. The same applies to the α-glucosyltransferase gene. In contrast to the morons from lambdoid phages, several of these extra DNA segments defined by the T4-JS98 comparison are flanked (α-gt and β-gt) or even bracketed (nrdD) by homing endonucleases. Insertion of these selfish DNA elements is a prominent process in T4 (see above). T4 might be quite permissive for this type of DNA because it occasionally introduces extra genes that developed useful functions for the phage, like the DNA-modifying enzymes that protect the phage DNA against destruction by restriction enzymes.
It is still premature or even wrong to conclude that T4- and lambda-like phages evolve with different modes. Such conclusions must be based on the comparison of more than three phage genomes. Completion of the sequencing of four further T4-like phage genomes (NCBI database, http://phage.bioc.tulane.edu/) will soon allow comparative genomics approaches with this workhorse of molecular biology.
Such data are not only of academic interest. T4-like phages are candidates for phage therapy of various E. coli infections (9). It will be important to understand how extra DNA can be introduced into the T4 phage genome and whether these extra genes are only to the benefit of the phage, as expected for an obligately virulent phage. Temperate phages, in contrast, can coexist with their lysogenic bacterial hosts. Some phage morons are therefore also of survival benefit for the bacterial host. In fact, temperate phages from bacterial pathogens frequently encode bacterial virulence factors (4, 6), which make such phages clearly unsuitable for phage therapy.
Acknowledgments
We thank the Swiss National Foundation for financial support of Sandra Chibani-Chennoufi and Carlos Canchaya (grant 5002-057832).
We thank Elizabeth Kutter for critical reading of the manuscript and Caroline Barretto for preparation of the GenBank file.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell-Pedersen, D., S. Quirk, J. Clyman, and M. Belfort. 1990. Intron mobility in phage T4 is dependent upon a distinctive class of endonucleases and independent of DNA sequences encoding the intron core: mechanistic and evolutionary implications. Nucleic Acids Res. 18:3763-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botstein, D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484-490. [DOI] [PubMed] [Google Scholar]
- 4.Brüssow, H., C. Canchaya, and W-D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brüssow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 6.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brüssow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casjens, S., G. F. Hatfull, and R. W. Hendrix. 1992. Evolution of dsDNA tailed-bacteriophage genomes. Semin. Virol. 3:383-397. [Google Scholar]
- 8.Chibani-Chennoufi, S., J. Sidoti, A. Bruttin, M.-L. Dillmann, E. Kutter, F. Qadri, S. A. Sarker, and H. Brüssow. 2004. Isolation of Escherichia coli bacteriophages from the stool of pediatric patients in Bangladesh. J. Bacteriol. 186:8287-8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chibani-Chennoufi, S., J. Sidoti, A. Bruttin, E. Kutter, S. Sarker, and H. Brüssow. 2004. In vitro and in vivo bacteriolytic activities of Escherichia coli phages: implications for phage therapy. Antimicrob. Agents Chemother. 48:2558-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desplats, C., C. Dez, F. Tétart, H. Eleaume, and H. M. Krisch. 2002. Snapshot of the genome of the pseudo-T-even bacteriophage RB49. J. Bacteriol. 184:2789-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eddy, S. R., and L. Gold. 1991. The phage T4 nrdB intron: a deletion mutant of a version found in the wild. Genes Dev. 5:1032-1041. [DOI] [PubMed] [Google Scholar]
- 12.Edgell, D. R., M. Belfort, and D. A. Shub. 2000. Barriers to intron promiscuity in bacteria. J. Bacteriol. 182:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foley, S., A. Bruttin, and H. Brüssow. 2000. Widespread distribution of a group I intron and its three deletion derivatives in the lysin gene of Streptococcus thermophilus bacteriophages. J. Virol. 74:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haggard-Ljungquist, E., C. Halling, and R. Calendar. 1992. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J. Bacteriol. 174:1462-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hambly, E., F. Tétart, C. Desplats, W. H. Wilson, H. M. Krisch, and N. H. Mann. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. USA 98:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haynes, J. A., and F. A. Eiserling. 1996. Modulation of bacteriophage T4 capsid size. Virology 221:67-77. [DOI] [PubMed] [Google Scholar]
- 17.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 19.Kanamaru, S., P. G. Leiman, V. A. Kostyuchenko, P. R. Chipman, V. V. Mesyanzhinov, F. Arisaka, and M. G. Rossmann. 2002. Structure of the cell-puncturing device of bacteriophage T4. Nature 415:553-557. [DOI] [PubMed] [Google Scholar]
- 20.Karam, J. D. 1994. Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 21.Kim, J. S., and N. Davidson. 1974. Electron microscope heteroduplex study of sequence relations of T2, T4, and T6 bacteriophage DNAs. Virology 57:93-111. [DOI] [PubMed] [Google Scholar]
- 22.Kostyuchenko, V. A., P. G. Leiman, P. R. Chipman, S. Kanamaru, M. J. van Ramino acidij, F. Arisaka, V. V. Mesyanzhinov, and M. G. Rossmann. 2003. Three-dimensional structure of bacteriophage T4 baseplate. Nat. Struct. Biol. 10:688-693. [DOI] [PubMed] [Google Scholar]
- 23.Kutter, E., K. Gachechiladze, A. Poglazov, E. Marusich, M. Shneider, P. Aronsson, A. Napuli, D. Porter, and V. Mesyanzhinov. 1995. Evolution of T4-related phages. Virus Genes 11:285-297. [DOI] [PubMed] [Google Scholar]
- 24.Lipman, D. J., and W. R. Pearson. 1985. Rapid and sensitive protein similarity searches. Science 227:1435-1441. [DOI] [PubMed] [Google Scholar]
- 25.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, E. S., J. F. Heidelberg, J. A. Eisen, W. C. Nelson, A. S. Durkin, A. Ciecko, T. V. Feldblyum, O. White, I. T. Paulsen, W. C. Nierman, J. Lee, B. Szczypinski, and C. M. Fraser. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Ruger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montag, D., I. Riede, M. L. Eschbach, M. Degen, and U. Henning. 1987. Receptor-recognizing proteins of T-even type bacteriophages. Constant and hypervariable regions and an unusual case of evolution. J. Mol. Biol. 196:165-174. [DOI] [PubMed] [Google Scholar]
- 29.Quirk, S. M., D. Bell-Pedersen, J. Tomaschewski, W. Ruger, and M. Belfort. 1989. The inconsistent distribution of introns in the T-even phages indicates recent genetic exchanges. Nucleic Acids Res. 17:301-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Repoila, F., F. Tétart, J. Y. Bouet, and H. M. Krisch. 1994. Genomic polymorphism in the T-even bacteriophages. EMBO J. 13:4181-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selick, H. E., G. D. Stormo, R. L. Dyson, and B. M. Alberts. 1993. Analysis of five presumptive protein-coding sequences clustered between the primosome genes, 41 and 61, of bacteriophages T4, T2, and T6. J. Virol. 67:2305-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tétart, F., C. Desplats, and H. M. Krisch. 1998. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: recombination between conserved motifs swaps adhesin specificity. J. Mol. Biol. 282:543-556. [DOI] [PubMed] [Google Scholar]
- 33.Tétart, F., C. Desplats, M. Kutateladze, C. Monod, H. W. Ackermann, and H. M. Krisch. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183:358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkler, M., and W. Ruger. 1993. Cloning and sequencing of the genes of beta-glucosyl-HMC-alpha-glucosyl-transferases of bacteriophages T2 and T6. Nucleic Acids Res. 21:1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeh, L. S., T. Hsu, and J. D. Karam. 1998. Divergence of a DNA replication gene cluster in the T4-related bacteriophage RB69. J. Bacteriol. 180:2005-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]