Abstract
Small regulatory RNAs (sRNAs) have recently been shown to be the main controllers of several regulatory pathways. The function of sRNAs depends in many cases on the RNA-binding protein Hfq, especially for sRNAs with an antisense function. In this study, the genome of Borrelia burgdorferi was subjected to different searches for sRNAs, including direct homology and comparative genomics searches and ortholog- and annotation-based search strategies. Two new sRNAs were found, one of which showed complementarity to the rpoS region, which it possibly controls by an antisense mechanism. The role of the other sRNA is unknown, although observed complementarities against particular mRNA sequences suggest an antisense mechanism. We suggest that the low level of sRNAs observed in B. burgdorferi is at least partly due to the presumed lack of both functional Hfq protein and RNase E activity.
During the last few years, many new small regulatory RNAs (sRNAs) have been discovered in the model organism Escherichia coli. Their functions range from being antisense RNAs, affecting translation of or degrading target mRNA, to binding and sequestering proteins (21). The activities of the newly discovered antisense RNAs seem to be dependent on the function of Hfq, which belongs to the Sm family of RNA-binding proteins (16, 24). The Hfq protein stimulates duplex formation between the sRNA and its target mRNA, either by binding the sRNA, as in the cases of OxyS and Spot42 (17, 24), or by binding the target mRNA, as in the case of sodB (11). In many instances, the action of Hfq overcomes the low complementarity between the target mRNA sequence and the sRNA sequence, but binding of Hfq to the sRNA might also protect the sRNA against digestion by RNase E (9, 15). RNase E has been shown to be involved in the digestion of sRNA-mRNA duplexes (13). The mechanism of how RNase E accesses its target remains elusive, although it is suggested that Hfq and RNase E compete for the same binding site on the sRNA (9, 15). It is generally believed that most organisms contain sRNAs, although it has been suggested that some, if not all, sRNA molecules in Rickettsia conorii are nonfunctional (7).
Borrelia burgdorferi is a spirochete occasionally transmitted to humans by Ixodes ticks and may cause Lyme disease (3). An unusual feature of B. burgdorferi is its relatively small genome consisting of an approximately 1-Mb large chromosome and an additional 0.6 Mb dispersed on 21 plasmids (4, 10).
In this work, we conducted multiple analyses of the B. burgdorferi genome in order to identify new small RNAs. Although several thorough searches were carried out, we observed only two new sRNAs in addition to the already suggested tmRNA and rnpB (10, 14). The low level of sRNAs in B. burgdorferi could be related to the lack of both the RNA-binding protein Hfq and the RNase E protein. Our results suggest a tight relationship among these three components.
MATERIALS AND METHODS
B. burgdorferi growth conditions.
B. burgdorferi B31 (3) bacteria were grown in BSK-II medium (2) supplemented with 6% rabbit serum at 34°C until they reached mid-exponential or stationary growth phase at ∼4.3 × 107 or ∼1.2 × 108 cells/ml, respectively. The bacteria had previously been passaged in culture medium no more than nine times and were counted using a light microscope and a Petroff-Hausser counting chamber.
Computer analysis.
Intergenic sequences of ≥150 bp were identified on both the chromosome and the plasmids of the B. burgdorferi B31 genome (10). Sequences from 329 regions (96 regions from the chromosome and 233 regions from the plasmids) were retrieved and compared to the NCBI Microbial Genome Database by using the BLAST program (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). Nucleotide sequence homologies were determined by performing a nucleotide-nucleotide BLAST (BLASTN) search on the NCBI website (www.ncbi.nlm.nih.gov/BLAST) or a CMR BLAST search on the TIGR website (http://tigrblast.tigr.org/cmr-blast/). The multiple-sequence alignment program ClustalW at the European Bioinformatics Institute website (http://www.ebi.ac.uk/clustalw/) was used for sequence alignments.
Determination of plasmid profile.
Total DNA of B. burgdorferi B31 spirochetes was prepared using a Wizard Genomic DNA purification kit (Promega), and the plasmid content was investigated by PCR as described elsewhere (8).
Northern blot analysis.
Total RNA was isolated from in vitro-cultured B. burgdorferi by use of an Ultraspec-II RNA isolation system (Biotecx Laboratories) according to the manufacturer's instructions with minor changes. Briefly, 500 ml of Borrelia culture was pelleted by centrifugation at 8,000 × g at 4°C. The bacteria were resuspended in 6 ml of Ultraspec-II RNA reagent, and 0.2 ml of chloroform was added per ml of Ultraspec-II RNA reagent. RNA in the aqueous phase was bound to RNATack resin and eluted with RNase-free water and incubation at 37°C for 30 s. The amount of RNA was determined by measuring the absorbance at 260 nm, and then the RNA was stored in 100-μl aliquots at −80°C. Twenty micrograms of total RNA per lane was separated in a 1 or 1.5% (wt/vol) agarose-formaldehyde gel in 1 × HEPES (20 mM Na-HEPES, 5 mM Na-acetate, 1 mM EDTA) (100 V, 3 h) and transferred by capillary force to a Hybond-N nylon membrane (Amersham Pharmacia Biotech) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The filter was air dried for 1 h, and RNA was cross-linked to the membrane by use of a UV Stratalinker 1800 (Stratagene). RNA blots were stored dry at −20°C until hybridization.
PCR fragments or oligonucleotides were used as radioactive probes for the Northern blots and were generated with [γ-32P]dATP (∼6,000 Ci/mmol; Amersham Biosciences). Primers specific for the DNA sequences of interest are shown in Table 1. Target sequences were amplified by PCR using Taq polymerase (Roche) with genomic DNA as a template. PCR products larger than 100 bp were purified with a QIAquick PCR purification kit (QIAGEN) or a High Pure PCR product purification kit (Roche). Smaller fragments were precipitated and washed with ethanol. Probes were end labeled with γ-32P by use of polynucleotide kinase (Roche). RNA blots were placed in a hybridization oven, prehybridized for 2 h, and then hybridized with the radiolabeled probes at 42°C overnight in Rapid-Hyb buffer (Amersham Biosciences). Membranes were washed once with 0.1% sodium dodecyl sulfate (SDS) in 2× SSC at room temperature for 10 min and then washed once again with 0.1% SDS in 0.5× SSC at 45°C for 5 min. The washed membranes were then placed on autoradiography film at −80°C. Blots that had been probed previously were stripped by washing them twice with boiling 0.1% SDS.
TABLE 1.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| d08d09-F | ATTATCATCTAACACAAATCTCTGC |
| d08d09-R | GTAAACCTTTTTTATAGATTAAGTCTG |
| d08d09-R_2 | TGCAGAGATTTGTGTTAGATGATAATCTAC |
| e02e03-F | AATCTTTTCCATATATTATTAATATTAAC |
| e02e03-R | GAAAGTTATTCTATCAACCTATTTATC |
| i24i25-F | TTTGTTTAGTTAAAATGATATAGGGC |
| i24i25-R | TACTAAAACTTTTTCATTATGGGGC |
| i24i25-R_2 | TTTATAAGTGGAATAGAACTTTAAATGC |
| j22j23-F | TAAAGCTTTATTTTTGCTTGACATGC |
| j22j23-R | CTTTAAAGAGAATATTTATAAGCAAATG |
| k02.1k05-F | CTTATATTTATTCAAAAATTTGAAAAAATG |
| k02.1k05-R | TTAATGTCTTTTTTTTAAAACCTAGATC |
| p27p28-F | AATAATGAGATAAAAGCTAGCACAAC |
| p27p28-R | CTTATCTCCAGTACAATATGTTGAG |
| BBH13-BBH14_1 | CATAAATCAAAGCCAAATATCGGAAC |
| BBH13-BBH14_2 | AAGATAAATTGAAATTATGAGCTTTAGC |
| BBK31-BBK32_1 | ACTTTGTTCACCCTCTTGATAGCAC |
| BBK31-BBK32_2 | TAATAAACTTTTCTTATCAAAAAGAGGC |
| BBQ80-BBQ81_1 | CAAATGTAGAATAGTAAAATATTGGAAG |
| BBQ80-BBQ81_2 | AGTTCTATTAATAAGATAGATTACACTAG |
| BB0402-BB0403_1-F | TGTAAAAGTTATATTTTGATTTTTCAATAC |
| BB0402-BB0403_2-R | TAATTAAATAAATAAAAATACTTATTCAATAC |
| BB0564-BB0565_1-F | TTTTATATTAAATTTAATTTAACCTACAAC |
| BB0564-BB0565_2-R | AAATTAGAAGTTAATAAAATTTAATTTACC |
| BB0577-BB0578_1-F | AATTAAATTAAATTAAATTAAATGAGGAG |
| BB0577-BB0578_2-R | TAAGCTTTCATCTTAAACCATAAAACC |
| BB0607-BB0608_1-F | TTAACCAATATTATTGAATTTATTGAATG |
| BB0607-BB0608_2-R | ATAACTTTCCTTTGATTAAATTAAATCC |
| SRP1-1 | TTCGAATCCCATTCTCTCCGATTC |
| SRP1-2 | AAAGCAAAACATTGCATTTGGGTTC |
| rnpB-F | CATAATGCTAGGTAATGCCTAGG |
| rnpB-R | GAGTTCTGTACTATGCCATCATC |
| rnpB1-1 | ATTATTATAAATATTATAGGAGGGATATG |
| rnpB1-2 | GTTTGTATTAATAAATATAAGTATATAGC |
| rnpB3_rnpB6-1 | AAGTAAAAGTGGTTTAGTTTACTGGAG |
| rnpB3_rnpB6-2 | TTTATGTAGTCCAGTAAACTAAACCAC |
| rnpB7-1 | TTTGGAGAATATATGATAATCTCTGAAC |
| rnpB7-2 | TATAGTCATTAGCTAAATCTAATATATTC |
| rnpB8-1 | GTTAAATATTACTCGAATGGCTGTAC |
| rnpB8-2 | TTTTGGGATAGAATTGTAGATGTTCC |
| rnpB9-1 | AATATTACTTTGAGAATTTTCATTTGGG |
| rnpB9-2 | TAGGGCAAGTTGCTTTATATATACTC |
| rnpB10-1 | GTTTAATCATAGTTTACAATTCCCGC |
| rnpB10-2 | TAAACTTGATTACATTTGAGTTCTCG |
| tmRNA-1-F | TTTCAGTCAAATCCAAAACATCCCC |
| tmRNA-2-R | TAATAATTGCTACTGCAACCCATGC |
| tmRNA1-1 | ATTCGTTGATCATAATTAATTGATATTG |
| tmRNA1-2 | TATAAAAATTCCAAAAATATGCAAGTTG |
| tmRNA2-1 | AATTGTCTCCATTTATTATGTAGTTGG |
| tmRNA2-2 | AATTCCAAATGGGAATATTTCTCGAC |
| tmRNA4-1 | AATAAAGCCCTTAAAATGGTTTTACAG |
| tmRNA4-2 | ACATAATTATTTAACATTTCAAAATATTGC |
| tke8-1 | TATAAATCTAAATAAATCTAATTTACAAATAG |
| tke8-2 | ATATAATTTCACATTAACGTTATTAAAAGC |
RESULTS
Direct homology approach.
As a first approach to identify sRNAs in B. burgdorferi, we analyzed the genome for homologs to the 62 sRNAs hitherto found in E. coli. Except for tmRNA and rnpB, we observed only one E. coli sRNA (IS063) (5), which displayed a low level of homology to an intergenic sequence of B. burgdorferi B31. To determine if this sRNA was also represented in B. burgdorferi, total RNA was extracted from spirochetes at either the logarithmic or stationary phase of growth and separated on a formaldehyde-agarose gel. There were no complementary RNA species detected by Northern hybridization of total RNA when a probe specific to the intergenic region harboring the postulated IS063 homolog was used (Table 2 and data not shown).
TABLE 2.
Overview of putative sRNAs in B. burgdorferi identified with the different methods used
| Method and genetic materiala | Locationb | Coordinatesc | Testingd by:
|
Test result | Commentg | |
|---|---|---|---|---|---|---|
| PCRe | Oligonucleotidef | |||||
| Homology approach | ||||||
| IS063 | cp26 | 11741-12013 | T | NT | − | |
| Comparative genomics approach | ||||||
| BB0260-BB0261 | Chromosome | 273648-273853 | NT | T | − | |
| BB0564-BB0565 | Chromosome | 577188-577394 | T | NT | − | Same as region complementary to rpoS |
| BBD08-BBD09 | lp17 | 4806-5057 | T | T | − | |
| BBE02-BBE03 | lp25 | 4157-4418 | T | NT | − | |
| BBP27-BBP28 | cp32-1 | 17061-17231 | T | NT | + | Untranslated mRNA from BBP27 or BBP28 |
| BBK02.1-BBK05 | lp36 | 3771-4901 | NT | T | − | |
| BBK14-BBK15 | lp36 | 9017-9372 | T | T | + | tmRNA4 located within this region |
| BBK31-BBK32 | lp36 | 20170-20388 | T | NT | + | Large band, probably untranslated mRNA of adjacent gene |
| BBI24-BBI25 | lp28-4 | 14442-15210 | T | NT | − | |
| BBH13-BBH14 | lp28-3 | 10517-10817 | T | NT | − | |
| BBL35-BBL36 | cp32-8 | 22692-23305 | T | NT | − | rnpB1 located within this region |
| BBQ80-BBQ81 | lp56 | 48627-49046 | T | NT | − | |
| Ortholog-based search strategy | ||||||
| BB0402-BB0403 | Chromosome | 415794-415845 | T | NT | − | |
| BB0564-BB0565 | Chromosome | 577188-577394 | T | NT | − | Complementary to rpoS upstream region |
| BB0577-BB0578 | Chromosome | 590936-591187 | T | T | + | |
| BB0607-BB0608 | Chromosome | 634465-634580 | T | NT | − | |
| Annotation-based search strategy | ||||||
| srp | Chromosome | 482285-482439 | T | NT | + | Overlaps with tRNA_Ser-3 |
| rnpB1 | cp32-8 | 22959-23354 | T | NT | − | |
| rnpB2 | Chromosome | 750818-751169 | T | T | + | Previously annotated rnpB |
| rnpB3 | lp21 | 4744-5129 | NT | T | + | |
| rnpB4 | lp21 | 5431-5816 | NT | T | + | |
| rnpB5 | lp21 | 13049-13432 | NT | T | + | 376 additional homologs on lp21 |
| rnpB6 | lp21 | 14051-14436 | NT | T | + | |
| rnpB7 | lp28-3 | 26447-26836 | T | NT | − | |
| rnpB8 | lp38 | 29152-29520 | T | NT | − | |
| rnpB9 | lp28-4 | 2288-2668 | T | NT | − | |
| rnpB10 | lp28-4 | 9445-9849 | T | NT | − | |
| tmRNA1 | cp32-9 | 28239-28454 | T | T | − | |
| tmRNA2 | lp17 | 542-886 | T | T | − | Two copies inside BBQ88 and BBH02 |
| tmRNA3 | Chromosome | 46691-47049 | T | T | + | Previously annotated tmRNA |
| tmRNA4 | lp36 | 9099-9314 | T | T | + | Same as BBK14-BBK15 |
Homologous or annotated sRNA or intergenic region as described in the text.
Designations are of plasmids.
According to http://www.tigr.org.
Testing for presence of sRNA with Northern blotting experiments. T, tested; NT, not tested.
32P-labeled PCR fragments comprising the specific sRNA as intergenic region.
32P-labeled oligonucleotides showing complementarity to the specific sRNA or intergenic region.
Specific comments relative to extra copies, homologs, complementary regions, etc., of particular sRNAs.
Comparative genomics approach.
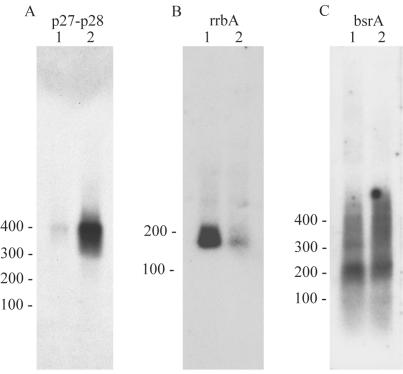
In 2001, Wassarman et al. (23) identified several new sRNAs in E. coli by a homology-based approach that utilizes the high sequence conservation among homologous sRNAs between different species. We adopted this method and applied it with minor modifications to B. burgdorferi. Briefly, intergenic sequences of ≥150 bp (329 regions) were identified on both the chromosome and the plasmids of B. burgdorferi B31 MI (TIGR sequence database). These sequences were compared to microbial genomes at the NCBI website by use of the BLAST program (see above). Regions displaying sufficient homology (giving homology scores of 3 or higher on a scale of 1 to 5) were investigated further. sRNA candidates were considered only if they showed significant levels of homology with sequences in other species and if these homologous sequences were located within intergenic regions in those organisms (see Table S1 in the supplemental material). Our results indicated that only a few intergenic sequences could fulfill the above-described criteria for further analysis. If the region displaying the highest level of homology was situated at a distance of approximately 30 bases from the 5′ or 3′ end, the intergenic sequence was not considered further, since it probably contained an untranslated region of adjacent genes. Consequently, the two intergenic sequences displaying the highest levels of homology to other organisms proved to be the 5′ ends of adjacent genes (see Table S1 in the supplemental material). Intergenic sequences found among the 21 plasmids showed, in many cases, very high homology to other sequences in both B. burgdorferi and other species. However, these sequences were mostly localized intragenically on other plasmids and were not considered further. A compilation of all homologous sequences is found in Table S1 in the supplemental material, together with homology scores and an explanation of why certain sequences were omitted. To determine whether the remaining intergenic sequences (both plasmid and chromosome borne) could harbor sRNAs, Northern blotting was performed with total RNA extracted from exponential- and stationary-phase-grown B. burgdorferi B31. 32P-labeled oligonucleotides and/or 32P-labeled PCR fragments were used as probes (Table 1). No new sRNA was detected in the chromosome of B. burgdorferi by the comparative genomics method, although our positive controls (the previously suggested sRNAs rnpB and tmRNA) could be detected by both labeled oligonucleotides and PCR fragments (Fig. 1 and Table 2). In additional control experiments, we clearly detected the transcript encoding the channel-forming protein P13 (Fig. 1) (18, 19). Plasmid-borne intergenic sequences showing very high homology to other intergenic sequences on plasmids and to sequences in other species did not reveal any new sRNA species (see Table S1 in the supplemental material). One exception was the hybridization with the intergenic sequence between BBP27 and BBP28, located on circular plasmid cp32-1, that gave a signal with the RNA isolated from the stationary phase. However, the size of the fragment suggested that it was the untranslated 5′ region of one of the adjacent genes, BBP27 (483 bp) or BBP28 (447 bp), instead of a novel sRNA (Fig. 2A and Table 2). In conclusion, the comparative genomics method did not reveal any new sRNA in B. burgdorferi.
FIG. 1.
Northern blot analysis of RNA from B. burgdorferi B31. RNA samples (20 μg/lane) were separated by electrophoresis and hybridized with probes specific for p13, rnpB, or tmRNA, respectively. RNA from spirochetes is shown at mid-exponential growth phase (lanes 1) and stationary growth phase (lanes 2). Molecular sizes in base pairs are indicated on the left.
FIG. 2.
Northern blot analysis of RNA from B. burgdorferi B31. RNA samples (20 μg/lane) were separated by electrophoresis and hybridized with probes specific to BBP27-BBP28 (A), rrbA (dsrA ortholog) (B), and bsrA (C). In each panel, RNA from spirochetes is shown at mid-exponential growth phase (lane 1) and stationary growth phase (lane 2). Molecular sizes in base pairs are indicated to the left of each panel.
Ortholog-based search strategy.
The low level of sRNA homology found between E. coli and B. burgdorferi (see above) caused us to undertake a different strategy to identify sRNAs. We reasoned that mRNA targets regulated by an sRNA in E. coli could also be regulated by the same sRNA mechanism in B. burgdorferi, although the sequences of the sRNAs of these two species would differ. If such a prediction was accurate, the sRNAs would be detected by searching for RNA sequences complementary to the target mRNAs. One obvious candidate was rpoS, which is regulated by at least three different antisense sRNAs in E. coli (20). When searching for RNA sequences complementary to rpoS in B. burgdorferi, we found four good candidates that were located in intergenic regions on the chromosome (Table 2). No expression was detected from three of the candidates (BB0402-BB0403, BB0564-BB0565, and BB0607-BB0608). However, when analyzing the intergenic region of BB0577-BB0578, we identified an RNA fragment (rrbA for the rpoS regulator in Borrelia A) with a size of approximately 170 bases (Fig. 2B) that was more highly expressed in exponential-phase-grown cells than in stationary-phase-grown cells. In a manner similar to that for rpoS, other mRNA targets were analyzed for complementarity against sequences located within intergenic regions, but no significant results that could suggest other putative sRNAs were found (data not shown).
Annotation-based search strategy.
During the course of this work, 13 new potential sRNAs, in addition to tmRNA and rnpB, were annotated at the TIGR B. burgdorferi website (see above). The annotation of the potential sRNAs suggested that they were homologs of either tmRNA or rnpB (3 or 9 of the 13 new potential sRNAs, respectively) or to be the 4.5 sRNA (srp), which is part of the signal recognition particle. By our analyses, however, neither the postulated tmRNA homologs nor those of rnpB exhibited any great sequence homology to tmRNA or rnpB (maximum sequence homology, 54 or 45%, respectively) (Table 2).
Additionally, not only were the sRNA homologs rnpB3, rnpB4, rnpB5, and rnpB6 very homologous (at least 96%) to each other, but also a new homology search of the B. burgdorferi genome by use of the BLAST program, with rnpB3 as the query sequence, revealed 376 other very homologous sequences located between positions 3618 and 14605 on linear plasmid lp21. This region is part of the large, repeated DNA sequence of lp21, whose functions remain unknown. We were not able to amplify a single PCR fragment by using primers for rnpB3, probably due to the many homologous sequences. Therefore, we used 32P-labeled oligonucleotides (rnpB3_rnpB6-1 and rnpB3_rnpB6-2), which revealed numerous fragments of different sizes by Northern blotting that could correspond to functional sRNAs (see Fig. S1 in the supplementary material). However, the complexity of this region made it difficult to determine the exact locations and sizes of these sRNAs. We were not able to detect any RNA fragment for rnpB1, rnpB7, rnpB8, rnpB9, or rnpB10 when using 32P-labeled PCR fragments (Table 2).
Among the newly annotated tmRNAs, we could identify migrating RNA fragments of the size expected for tmRNA4 (∼220 nucleotides [nt]) when we used 32P-labeled PCR fragments (Fig. 2C). RNA fragments were also identified when we probed for tmRNA1 and tmRNA2, but the sizes of these RNA molecules suggested that they were parts of the adjacent genes rather than independent RNA molecules (Table 2 and data not shown).
Although we identified an RNA fragment when a Northern filter was hybridized with a 32P-labeled PCR fragment constituting the annotated srp sRNA, the size of this fragment (∼90 nt) and the 23-nt overlap with tRNA Ser-3 suggested rather that it constituted the tRNA Ser-3 in B. burgdorferi (data not shown).
B. burgdorferi lacks Hfq as well as RNase E.
A possible explanation for the low abundance of sRNAs could be that the RNA-binding protein Hfq is absent in B. burgdorferi. We therefore undertook an extensive in silico search for Hfq or Hfq-like proteins in B. burgdorferi B31. A recent report (22) suggested that the Hfq protein could be very diverse in different species. We therefore used that report's search criteria (sequence length, similarity, motif, and pattern) together with the sequences of recently discovered Hfq homologs reported therein to analyze the B. burgdorferi genome. These searches did not reveal any Hfq-like proteins. Our closest match corresponded to RpsL, a ribosome protein, emphasizing the low probability of an Hfq protein existing in B. burgdorferi.
The function of RNase E has been conserved in both eukaryotes and prokaryotes (6). It is also intimately involved in the turnover of sRNAs that bind to their respective target mRNAs (13). In order to analyze its status in B. burgdorferi, we performed a homology search by use of the BLAST program with the E. coli protein sequence as a query, but, surprisingly, we were not able to identify the RNase E protein in B. burgdorferi.
DISCUSSION
By using four different approaches, we have identified two new sRNAs in the gram-negative pathogenic bacterium B. burgdorferi. We believe that the different approaches tested (direct homology, comparative genomics, and ortholog- and annotation-based search strategies) would be able to identify many of the sRNAs present in B. burgdorferi. This interpretation is justified by the finding of several RNA structures (mainly 5′ untranscribed sequences) by these methods (see Table S1 in the supplemental material). Moreover, the apparent overlap among the methods used, where the same intergenic region was in many cases identified with different methods (Table 2), further strengthens this argument.
One of the sRNAs found, rrbA, may bind to rpoS and control its expression, since it shows complementarity to the rpoS upstream region. In the case of E. coli, the upstream region of rpoS forms a secondary structure masking the Shine-Dalgarno site, and the binding of dsrA to this upstream region discloses the rpoS Shine-Dalgarno site, allowing translation (12). The expression pattern of rrbA suggests another mechanism (Fig. 2B). Since rrbA is mostly expressed in logarithmic growth phase and only slightly expressed in stationary phase (Fig. 2), it probably negatively controls rpoS expression by an antisense mechanism. Such a mechanism would prevent the expression of rpoS at an inappropriate moment (logarithmic growth phase). Clearly, further work is required to disclose the exact function of rrbA. Three other intergenic regions displaying complementarity to the rpoS upstream region were identified, but we were not able to detect any expression of sRNAs from these regions. However, we cannot exclude the possibility that sRNAs would be expressed from these intergenic regions under other conditions of growth, as has been shown for the OxyS sRNA in E. coli (1).
Another sRNA identified was tmRNA4, which originally was suggested to constitute another tmRNA molecule in B. burgdorferi. As tmRNA and tmRNA4 are very different in length and their overall identity is rather low, we suggest that the tmRNA4 RNA species should rather be named “bsrA,” for Borrelia small RNA A, until the function of this RNA species has been established. Although the function of bsrA remains unknown, it could act as an antisense RNA, since several mRNA sequences show good complementarity to bsrA (data not shown).
Linear plasmid lp21 contains a large, repeated DNA sequence. The function of this sequence is presently unknown, but our data suggest that multiple RNA species with different sizes are expressed from this region (see Fig. S1 in the supplemental material).
Interestingly, another gram-negative bacterium harboring a few functional sRNAs, Rickettsia conorii (7), also seems to be devoid of Hfq and Hfq-like proteins, as suggested by extensive homology searches (data not shown). The function of Hfq has previously been shown to be crucial for the activities of sRNAs (16, 24), and the apparent lack of Hfq in both B. burgdorferi and R. conorii underscores the tight relationship between the function of Hfq and the presence of sRNAs. Due to the lack of several metabolic pathways, both B. burgdorferi and R. conorii require complex media containing exogenous amino acids and vitamins to grow. It is tempting to speculate that the lack of such metabolic pathways makes the function of sRNAs redundant, since many sRNAs hitherto characterized have been involved in metabolism. These small-genome bacterial pathogens have probably lost Hfq and functional sRNAs during evolution, either because the regulatory role of the sRNAs has been replaced by other regulatory components (i.e., proteins) or because the (metabolic) pathway they control in other organisms does not exist in either B. burgdorferi or R. conorii.
We cannot exclude the possibility that the putative sRNA candidates that we have analyzed may be expressed under other conditions (i.e., low temperature, low oxygen, etc.). However, we believe that the two conditions tested reflect the most relevant environments that B. burgdorferi encounters during growth in human blood.
Another interesting feature of B. burgdorferi is that it is most likely devoid of RNase E, a protein present in several other bacterial species (6). In E. coli, RNase E is responsible for the degradation of target mRNAs when they have been bound by sRNAs (13). Also, RNase E and Hfq compete for the same binding sites on the RNA (9, 15). Our finding that both Hfq and RNase E seem to be absent in a strain harboring only a few sRNAs is interesting and requires further work.
Supplementary Material
Acknowledgments
We thank May Ali for critically reading the manuscript.
S.B. was supported by Swedish Research Council grant 07922, by Swedish Council for Forestry and Agricultural Research grant 23.0161, and by Swedish Strategic Research Foundation grant A301:93/01/02. J.J. was supported by Swedish Research Council grant 15144, by the Wenner-Gren Foundation, and by Umeå University.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altuvia, S., D. Weinsterin-Fischer, A. Zhang, L. Postow, and G. Storz. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90:43-53. [DOI] [PubMed] [Google Scholar]
- 2.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 3.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 4.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 5.Chen, S., E. A. Lesnik, T. A. Hall, R. Sampath, R. H. Griffey, D. J. Ecker, and L. B. Blyn. 2002. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. BioSystems 65:157-177. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, S. N., and K. J. McDowall. 1997. RNase E: still a wonderfully mysterious enzyme. Mol. Microbiol. 23:1099-1106. [DOI] [PubMed] [Google Scholar]
- 7.Davids, W., H. Amiri, and S. G. Andersson. 2002. Small RNAs in Rickettsia: are they functional? Trends Genet. 18:331-334. [DOI] [PubMed] [Google Scholar]
- 8.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folichon, M., V. Arluison, O. Pellegrini, E. Huntzinger, P. Régnier, and E. Hajnsdorf. 2003. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 31:7302-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. C. Venter, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 11.Geissmann, T. A., and D. Touati. 2004. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 23:396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massé, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattsson, J. G., S. G. Svärd, and L. A. Kirsebom. 1994. Characterization of the Borrelia burgdorferi RNase P RNA gene reveals a novel tertiary interaction. Mol. Biol. 241:1-6. [DOI] [PubMed] [Google Scholar]
- 15.Moll, I., T. Afonyushkin, O. Vytvytska, V. R. Kaberdin, and U. Bläsi. 2003. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9:1308-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Møller, T., T. Franch, P. Højrup, D. R. Keene, H. P. Bächinger, R. G. Brennan, and P. Valentin-Hansen. 2002. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9:23-30. [DOI] [PubMed] [Google Scholar]
- 17.Møller, T., T. Franch, C. Udesen, K. Gerdes, and P. Valentin-Hansen. 2002. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 16:1696-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noppa, L., Y. Östberg, M. Lavrinovicha, and S. Bergström. 2001. P13, an integral membrane protein of Borrelia burgdorferi, is C-terminally processed and contains surface-exposed domains. Infect. Immun. 69:3323-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Östberg, Y., M. Pinne, R. Benz, P. Rosa, and S. Bergström. 2002. Elimination of channel-forming activity by insertional inactivation of the p13 gene in Borrelia burgdorferi. J. Bacteriol. 184:6811-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Repoila, F., N. Majdalani, and S. Gottesman. 2003. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the rpoS paradigm. Mol. Microbiol. 48:855-861. [DOI] [PubMed] [Google Scholar]
- 21.Storz, G., J. A. Opdyke, and A. Zhang. 2004. Controlling mRNA stability and translation with small noncoding RNAs. Curr. Opin. Microbiol. 7:140-144. [DOI] [PubMed] [Google Scholar]
- 22.Valentin-Hansen, P., M. Eriksen, and C. Udesen. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 51:1525-1533. [DOI] [PubMed] [Google Scholar]
- 23.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9:11-22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.