Abstract
Background
Manganese peroxidase (MnP) of white rot basidiomycetes, an extracellular heme enzyme, is part of a peroxidase superfamily that is capable of degrading the different phenolic compounds. Ganoderma, a white rot basidiomycete widely distributed worldwide, could secrete lignin-modifying enzymes (LME), including laccase (Lac), lignin peroxidases (LiP) and MnP.
Results
After the selection of a G. lucidum strain from five Ganoderma strains, the 1092 bp full-length cDNA of the MnP gene, designated as G. lucidum MnP (GluMnP1), was cloned from the selected strain. We subsequently constructed an eukaryotic expression vector, pAO815:: GlMnP, and transferred it into Pichia pastoris SMD116. Recombinant GluMnP1 (rGluMnP1) was with a yield of 126 mg/L and a molecular weight of approximately 37.72 kDa and a specific enzyme activity of 524.61 U/L. The rGluMnP1 could be capable of the decolorization of four types of dyes and the degradation of phenol. Phenol and its principal degradation products including hydroquinone, pyrocatechol, resorcinol, benzoquinone, were detected successfully in the experiments.
Conclusions
The rGluMnP1 could be effectively expressed in Pichia pastoris and with a higher oxidation activity. We infer that, in the initial stages of the reaction, the catechol-mediated cycle should be the principal route of enzymatic degradation of phenol and its oxidation products. This study highlights the potential industrial applications associated with the production of MnP by genetic engineering methods, and the application of industrial wastewater treatment.
Electronic supplementary material
The online version of this article (doi:10.1186/s12896-017-0338-5) contains supplementary material, which is available to authorized users.
Keywords: Ganoderma lucidum, Yeast expression system, Manganese peroxidase, Degradation, Phenolic compound
Background
Manganese peroxidase (MnP) (E.C. 1.11.1.13. Mn2+: H2O2 oxidoreductases) belongs to the family of oxidoreductases, to be specifically those actions on peroxide as acceptor (peroxidases), is an extracellular hemeprotein which catalyze the H2O2-dependent oxidation of lignin-derivatives based polymers [1]. MnP is a specific enzyme that can oxidize Mn2+ to Mn3+, which diffuses from the enzyme surface and in turn oxidizes the phenolic substrate, including lignin model compounds and some organic pollutants [2]. In nature, MnP catalyzes plant lignin de-polymerization as component of ligninolytic enzymes complex. So it is one of the most common lignin degradation enzymes and has great application potential in the field of agriculture for degradation of some cellulose, hemicellulose and lignin, etc. To protect the environment, it was widely used in many industrial fields for degradation some recalcitrant organic pollutants such as polycyclic aromatic hydrocarbons, chlorophenols, industrial dyes and nitroaromatic compounds, which are very harmful to human health [3]. Recently, more and more attention has been paid to the value of bioremediation of this enzyme.
MnP was first discovered in Phanerochaete chrysosporium [4] and seems to be the most ubiquitous ligninolytic enzyme among white-rot fungi. At present, it has been purified and characterized from various white rot fungi [5–11]. Properties and application on MnPs isolated from different sources had been investigated widely. Much previous research has suggested that some azo dyes could be efficiently degraded by the purified MnPs, which were isolated from P. chrysosporium, Lentinula edodes, Trametes versicolor, Dichomitus squalens, Stereum ostrea, Irpex lacteus and etc. [3, 12–16]. However, many factors influenced the application of MnP, which include slow fungal growth rate, accumulation of extracellular polysaccharides, similar chromatographic properties of MnP and laccase, and etc. [17]. Therefore, searching for new MnP from widely distributed worldwide and fast fungal growth rate is essential for the application of MnP in industrial and agricultural productions, and environmental protection.
Ganoderma, a white rot basidiomycete widely distributed worldwide, can be cultivated on various substrates by different cultivation model, and could secrete lignin-modifying enzymes (LME), including laccase (Lac), lignin peroxidases (LiP) and MnP. Because of the rapid growth rate and extensive decolorization on solid media, Ganoderma is suitable for a wide range of applications in the field of environment and biotechnology; previous publications had reported that several species of Ganoderma can produce high amounts of MnP enzymes in solid or liquid cultures [2]. However, as we know, few studies focused their attention on the evaluation of the capability of purified and heterologous expression MnP tolerating different for dyes or other industrial pollutants. In the previous publications, most of them mainly focused on inducing secretion of MnP from different Ganoderma, and their potential uses in decolorization of textile effluents [18] and degrades β-carotene from G. applanatum under alkaline conditions [19].
In the present study, the possible difference of various G. lucidum strain for production of the MnPs was investigated using a qualitative plate assay method by using O-methoxyphenol as a color indicator. The fungal colony showing the largest zone of decolorization was selected for cloning the MnP1 cDNA sequence, and then an expression vector, pAO815:: GluMnP1, was constructed and transferred into P. pastoris SMD1168H by electroporation-mediated transformation. The expression products were demonstrated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting. We also carried out a preliminary exploration on the ability of rGluMnP1 to biodegrade four dyes and phenol, and infer a probable degradation route of phenolic compounds, which should be taken into account in producing and designing a related industrial wastewater treatment process. This study provides a production strategy for MnP and will aid our understanding of the role of fungal MnP oxidation in biodegradation and bioremediation.
Results
Selection of the strain from various G. species
The ability of producing lignin-degrading enzymes of five species of G. lucidum strains was measured by comparing the diameter of the colony and reddish brown circles. The results showed that the ratio of diameter of reddish brown circles and the diameter of fungal colony was the largest when G. lucidum 00679 was cultured for 7 days (Fig. 1). In order to better understand the lignin-degrading enzyme from G. lucidum, we tested the action of manganese peroxidase. At initial concentrations of 1.2 mM NH4 2+ and 3 μM Mn2+, extracellular production of MnP and Lac began by day 4, with maximum levels of 1003 U/L Lac on day 14 and 57 U/L MnP on day 16. At a higher initial Mn2+ concentration of 200 μM, MnP and Lac production also began at day 4 with more MnP produced. The maximum level of Lac was less. No LiP was detected. The results showed that initial concentrations of 1.2 mM NH4 2+ and 200 μM Mn2+, extracellular production of MnP of G. lucidum 00679 with maximum levels, reached 670 (U/L) (Table 1). Despite significant differences in enzyme production, cultures at both Mn2+ concentrations rapidly colorful reaction in the PDA-O-methoxyphenol plate, with no difference in the ratio of diameter of reddish brown circles (Fig. 2). As a result, the fungal colony of G. lucidum 00679 for highest decolorization zone was chosen for the further study.
Fig. 1.
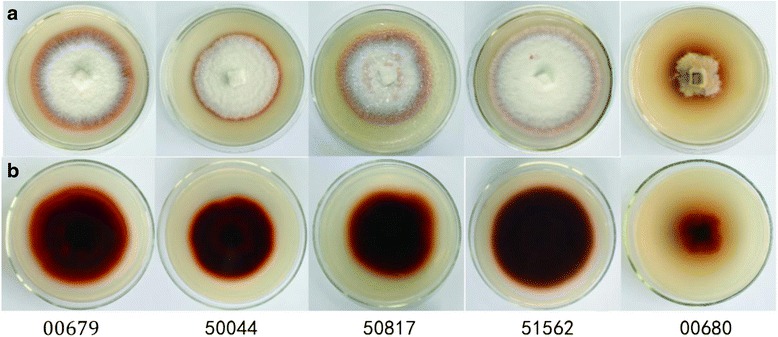
Decolorization of O-methoxyphenol with five G. lucidum strains. G. lucidm 00679, 50044, 50817, 51562 and 00680 was cultured on PDA medium for 7 d, and then was taken photographs. a displayed on the front of the petri dish, and (b) displayed the reverse side of the petri dish
Table 1.
MnP, Lac and Lip production by N-limited and N-rich batch cultures at 3 μM and 200 μM Mn2+
| enzymes | 1.2 mM NH4 2+ | 12 mM NH4 2+ | ||
|---|---|---|---|---|
| 3 μM Mn2+ | 200 μM Mn2+ | 3 μM Mn2+ | 200 μM Mn2+ | |
| Maximum MnP (U/L) | 57 | 670 | 33 | 72 |
| Maximum Lac (U/L) | 1003 | 69 | 890 | 230 |
| Maximum LiP (U/L) | undetected | undetected | undetected | undetected |
Fig. 2.
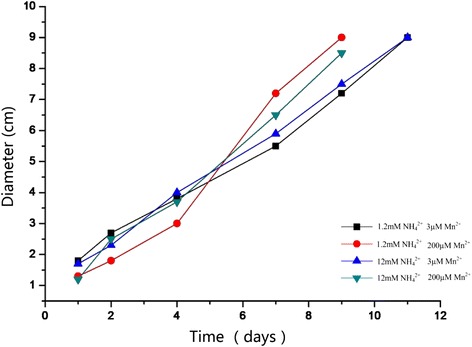
Diameters of colored red-brown circled with G. lucidum 00679 by N-limited and N-rich cultures at 3 μM and 200 μM Mn2+
Decolorization of four dyes by the culture supernatants of G. lucidum strains
The results showed that G. lucidum 00679 could efficiently decolorize these four dyes. Drimaren Blue CL-BR, Drimaren Yellow X-8GN, Drimaren Red K-4Bl in the aqueous solutions (500 mg/L) were respectively decolorized up to 92.8, 90.2 and 70.1% by G. lucidum 00679 within 72 h. Disperse Navy Blue HGL in the aqueous solution (500 mg/L) could be decolorized up to 93.4% by G. lucidum 00679 within 12 h. MnP, Lac, and LiP activities were assayed in the supernatant medium before and after decolorization. Extracellular MnP activities were significantly induced by 278.1, 300.9, 259.3 and 191.3% respectively after decolorization of four dyes by G. lucidum 00679. Less lac and nor LiP was detected during the decolorization process. Induction in MnP activity during the decolorization process suggested that MnP was involved in the decolorization of these four dyes.
Isolation and Sequence Characterization of the MnP gene
Based on total RNA isolated from the mycelia of G. lucidum, degenerate primers MnPF1 and MnPR1 [see Additional file 1] were used to specifically amplify a 461 bp core fragment using a method of the one-step real-time reverse transcriptase-PCR (RT-PCR) [see Additional file 2A]. A BLAST search showed that the PCR core fragment was homologous to MnP genes from other white rot fungi species (data not shown). The 5′ and 3′-ends fragments (222 and 870 bp, respectively) were amplified by 5′ RACE [see Additional file 2 B] and 3′ RACE [see Additional file 2C], based on the 461 bp core fragments. The core fragment, and the 3′- and 5′-ends fragments were assembled using Vector NTI Suite 10 and the deduced full-length GluMnP1 cDNA sequence obtained was confirmed by sequencing. The full-length cDNA of GluMnP1 was 1,341 bp [see Additional file 2D], comprising a 70 bp 5′-untranslated region, an ORF of 1095 bp and a 176 bp 3′-untranslated region.
Sequence analysis confirmed isolation of a full-length cDNA of GlMnP1 encoding a protein of 364 amino acids, with a calculated molecular mass of 37.7 kDa and isoelectric point (pI) of 4.43. Amino acids of GlMnP1 involved in aromatic substrate oxidation on the distal side of the heme, Ca2+ side binding residues, heme pocket residues and Mn2+ binding site. These features suggested that GlMnP1 encoded a probable manganese peroxidase. A database search with Blastx (https://blast.ncbi.nlm.nih.gov/Blast.cgi) showed that there was a relatively high similarity between GluMnp1 and other MnPs from strains, such as GluMnP, GapMnP, GfoMnP, and GauMnP. A number of gaps and insertions were made in the sequences to optimize the alignment. The percentages of identity among GluMnP, GauMnP, and GfoMnP were 98, 88 and 87%, respectively, suggesting they were closely related to each other [see Additional file 3]. Amino acids of GluMnP1 involved in aromatic substrate oxidation were first analyzed and compared with various other plants and fungi by bioinformatics analysis [see Additional file 3]. Amino acids involved in aromatic substrate oxidation [see Additional file 3A] on the distal side of the heme, Ca2+ side binding residues [see Additional file 3C], heme pocket residues [see Additional file 3H] and Mn2+ binding site [see Additional file 3, M] were conserved in the MnP sequences from G. lucidum as well as in peroxidase sequences from various other plants and fungi. The deduced sequence contained eight cysteines [see Additional file 3C], which probably form four disulfide bonds in the mature protein.
Heterologous expression of GluMnP1 gene in P. pastoris
The presence of GluMnP1 in the transformants was confirmed by PCR (Fig. 3). SDS-PAGE analysis after Coomassie Brilliant Blue R-250 staining indicated that rGluMnP1 could be efficiently expressed in P. pastoris cells (Fig. 4a). The theoretical mass of the target rGluMnP1 protein was 38 KDa, and the mass of the rGluMnP1 protein after glycosylation modification was higher than the theoretical value.
Fig. 3.
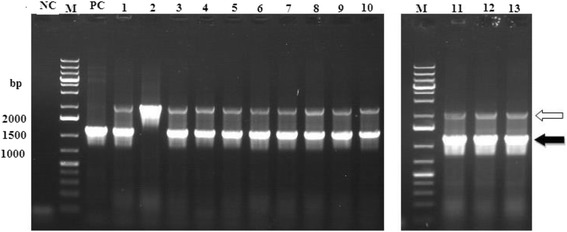
Electrophoresis of PCR amplification of GluMnP1 from pAO815::GluMnP1 Lane M: DNA marker DL 10000; lane NC: negative control; lane PC: positive control; Lane 1–13: selected transformants. The hollow arrow showed the DNA bands of AOX gene from yeast (about 2200 bp). The solid arrow showed the DNA bands of the GluMnP1 gene plus a part of vector sequences (about 1300 bp)
Fig. 4.
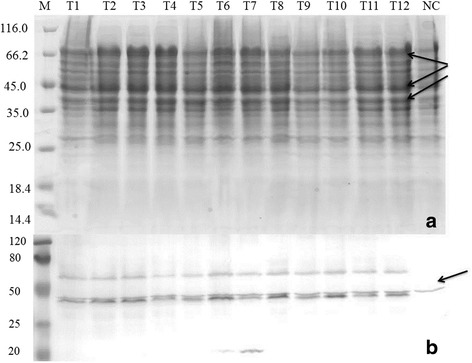
SDS-PAGE (a) and western blot analysis (b) of positive clones of recombinant Ganoderma MnP after 2 days of induction (a), lane M: protein marker; lanes 1–13: recombinant plasmid pAO815::MnP; Lane NC: negative control. Arrows showed the expressed bands. b, lane M: protein marker; lanes 1–13: re-pAO815-MnP; lane NC: negative control; lane PC: positive control. An arrow indicated the target band
The size of expressed protein was analyzed by western blotting of samples from a three-day fermentation of the yeast. The results of western blot analysis showed that the target protein of GluMnP1 from G. lucidum was heterologously expressed in P. pastoris (Fig. 4b).
Analysis of enzyme yield and activity
The content of total soluble protein was determined approximately 1258 mg/L using the Bradford Protein Assay Kit. The density of protein bands was detected using the software Bandscan 5.0 (Glyko, Novato, USA) and the rGluMnP1 protein was estimated to account for about 10% of total soluble protein. Therefore, the yield of rGluMnP1 produced by the yeast transformants reached roughly 126 mg/L.
Methanol was used to induce expression in the P. pastoris transformants in BMMY medium. After fragmenting of P. pastoris and centrifugation, the highest rGluMnP1 activity in the culture supernatant of total protein extracted from P. pastoris transformants reached about 524.61 U/L after 48 h of incubation.
Decolorization of four dyes using the rGluMnP1
The decolorization experiments were performed with crude protein extracts in 50 mM sodium malonate (pH 4.5) and 500 mg/L of dyes in a final volume of 1 mL at 25 °C. The results showed that the maximum decolorization rates of the four dyes all reached 70% (Fig. 5), indicating that rGluMnP1 had a higher decolorizing ability. Reaction containing MnSO4 and H2O2 could only decolorize four dyes by about 49% in 15 min. However, rGluMnP1 could decolorize Drimaren Red K-4Bl by more than 62% after 15 min. The decolorization rates of the four dyes increased quickly at the start of the reaction, but increased more slowly after 30 min.
Fig. 5.
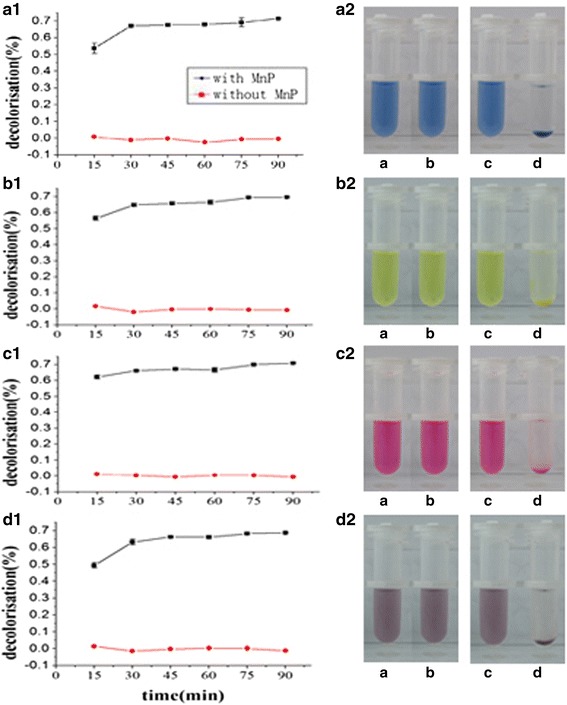
Time course and visual effect of decolorization of four dyes by the crude enzymatic solution. A1, B1, C1 and D1 show the in vitro decolorization rate of four dyes over time by consumption of crude rGluMnP1, which included Drimaren Blue CL-BR (A1), Drimaren Yellow X-8GN (B1), Drimaren Red K-4Bl (C1) and Disperse Navy Blue HGL (D1). The black line in each image shows the decolorization rate after treatment by rGluMnP1; the red line shows the results for the negative control. A2, B2, C2 and D2 showed the visual decolorization effect of four dyes by control (untransformed yeast) and crude rGluMnP1 solutions (transformed yeast). (a) and (b) show the visual decolorization effects before/after treated by the untransformed yeast, (c) and (d) show the visual decolorization effects before/after treated by the yeast transformants. Yeasts were broken to prepare the crude enzyme solutions. The reactions were carried out in a 2 mL EP tube
HPLC-based analysis of the degradation rate of phenol and the principal degradation products
Based on the establishment of the standard curves of phenol and the principal degradation products (data not shown), the regression equations of calibration curves and their coefficients were calculated as follows: for phenol, Y = 6046.4X + 21365 (R2 = 0.9998); for hydroquinone, Y = 11297X − 3710.4 (R2 = 0.9999); for pyrocatechol, Y = 13955X − 20052 (R2 = 0.9999). The high performance liquid chromatography (HPLC) analysis results showed that rGluMnP1 solutions can degrade phenol in aqueous solution effectively and the degradation products contained hydroquinone and pyrocatechol at least (Fig. 6). Furtherly, the contents of phenol in the samples dealt with by the crude enzymes solutions at the concentration of 5, 10 and 15% were about 88.914 ± 0.958, 84.642 ± 1.478 and 84.258 ± 1.613 μg/mL, respectively. The degradation rates of phenol in aqueous solution treated by 5, 10 and 15% crude enzymes solutions were 7.262 ± 0.999%, 8.079 ± 1.605% and 4.873 ± 1.821%, respectively. The results suggested that optimum concentration of crude enzymes solutions was about 10% under the present conditions.
Fig. 6.
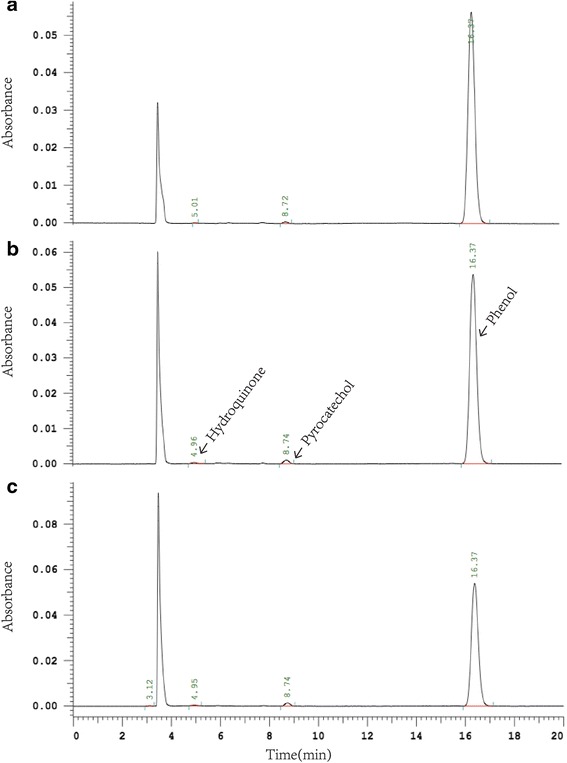
HPLC chromatography of phenol and the main degradation products. HPLC analysis of the phenol (retention time = 16.37 ± 0.1 min) and its main degradation products of hydroquinone (retention time = 4.96 ± 0.1 min), pyrocatechol (retention time = 8.74 ± 0.1 min). The aqueous solutions of phenol were treated by 5% (Fig. 6a, upper spectrum), 10% (Fig. 6b, middle spectrum) and 15% (Fig. 6c, lower spectrum) rGlMnP1 enzyme solutions, respectively
The HPLC analysis results of the likely oxidation products of phenol showed that among the four possible degradation products including hydroquinone, pyrocatechol, resorcinol and benzoquinone, only two chemical compounds, hydroquinone and pyrocatechol were determined in the treated sample, the others were not determined by HPLC (Fig. 6). The contents of hydroquinone and pyrocatechol in the sample treated by crude enzymes solutions at the concentration of 5, 10 and 15% were found to be 0.359 ± 0.053, 0.517 ± 0.028, 0.503 ± 0.124 μg/mL and 1.640 ± 0.047, 2.014 ± 0.123, 2.180 ± 0.137 μg/mL, respectively.
Discussion
For screening process on the ability of producing LMEs from different G. lucidum strains by comparing the diameter of the colony and reddish brown circles, based on previous theory, the smaller the ratio is, the stronger the strains’ ability of producing lignin-degrading enzymes is [20]. As a result, G. lucidum 00679 selected for the further study should be correct. Previous studies had demonstrated that Mn peroxidase production was controlled by the concentration of Mn2+ [21]. At higher Mn2+ concentrations, production of MnP increased and that of laccase decreased, but the rate or number of decolozations was unaffected [22]. In addition, the nitrogen source and its concentration were found to influence MnP production [23]. In order to obtain more information about the MnP from G. lucidum, Mn and nitrogen concentration were routine used to test the effects of MnP production and activity in the present study.
White rot fungi have been widely studied over the last 30 years because they could release LMEs and had a high capacity for biodegradation of environmental pollutants [24]. Most white-rot basidiomycetes are capable of degrading or oxidizing a range of aromatic organic compounds with the aid of certain enzymes, such as LiP, MnP and other versatile peroxidases [25]. To date, more than thirty enzymes have been isolated and investigated from at least a dozen Ganoderma species. MnP, as one of major fungal oxidative enzymes, plays a key role in enzymatic degradation of phenolic compounds in vitro. There are many reports concerning the decolorization of wastewater from dyeing factories [2, 26]. However, high-level expression of MnP must be taken into account before it can be used commercially. MnP expression level in some isolates is too low for industrial application. Heterologous expression in P. pastoris could meet these requirements, because it enhanced the expression levels by 10-, 100-, or even 1000-fold compared with the natural host [27].
G. lucidum contained 7 peroxidases genes in its genome, was the third largest number of peroxidases, which may suggest its strong ligninolytic ability [28]. In order to better use Ganoderma MnP for the degradation of different phenolic compounds, the MnP gene of and its full-length cDNA were successfully cloned and characterized from G. lucidum 00679. In the process of yeast expression, the complexity of the yeast intracellular proteins meant that the target protein would migrate with other proteins, possibly resulting in no significant band being observed on SDS-PAGE; however, as the recombinant expressed protein is expressed at a higher level than similar sized endogenous proteins, an individual protein band was observed that was not present in the negative control, indicating expression of the target protein (Fig. 4). For the yield or enzymatic activity of protein expression, compared with the native host, recombinant fungi and yeast strains could produce from 5 to 100 mg/L rMnP [29]. Comparing with the results of previous studies [30], in the present study, the highest rGluMnP1 activity in the culture supernatant of total protein extracted from P. pastoris transformants reached about 524.61 U/L at 25 °C and pH 4.5.
Peroxidases are hemoproteins that catalyze reactions in the presence of hydrogen peroxide. MnPs have a reaction mechanism that starts with enzyme oxidation by H2O2 to an oxidized state during the catalytic cycle [31]. The degradation mechanism of LMEs has been studied extensively using different white rot fungi [32, 33]. Zhang et al. (1999) demonstrated that MnP plays an important role in the decolorization of cotton bleaching effluent by an unidentified white-rot fungus, while there was no obvious role for LiP in this decolorization [34]. In other words, the relationship between these fungal oxidative enzymes in the decolorization process is not clear. However, it is likely that the main enzymes that decolorize different dyes are not the same [35, 36]. In 2005, Champagne and Ramsay approved that the combination of the MnP and Lac had an additive effect, and that MnP was the principal active enzyme in the reaction process [14]. Although H2O2 or −OH may oxidize a dye without an enzyme, the decolorization of dyes mainly relies on Mn3+ acting on the organic acid compounds, which were also demonstrated in this study. In addition, based on the opinions of previous literature [37], a crude enzymatic solution was used to decolorize the four dyes in the present study, which could narrow the high costs associated with enzyme purification.
The above analysis had proved that the hydroxyl radical played a major role in phenol degradation. Resorcinol and benzoquinone were not found in the probable oxidation products due to the in perfect reaction system or in appropriate reaction conditions. We infer that the results are relevant to the phenol structure. The electronic arc in phenolic hydroxyl oxygen atom can have p-π conjugation with π electronic in the phenyl ring which made phenyl ring had more negative charge on the ortho- and/or para- in phenolic hydroxyl. So it is not difficult to produce hydroquinone and pyrocatechol by hydroxyl radical attack. The carboxylation of the phenyl ring is the first step of phenolic compounds degradation. Subsequently, the phenyl ring was open for the formation of carboxyl aromatic ring, and finally was completely mineralized to carbon dioxide and water (Fig. 7) [38]. Phenol intermediate products and its degradation route were needed for further investigation and analysis.
Fig. 7.
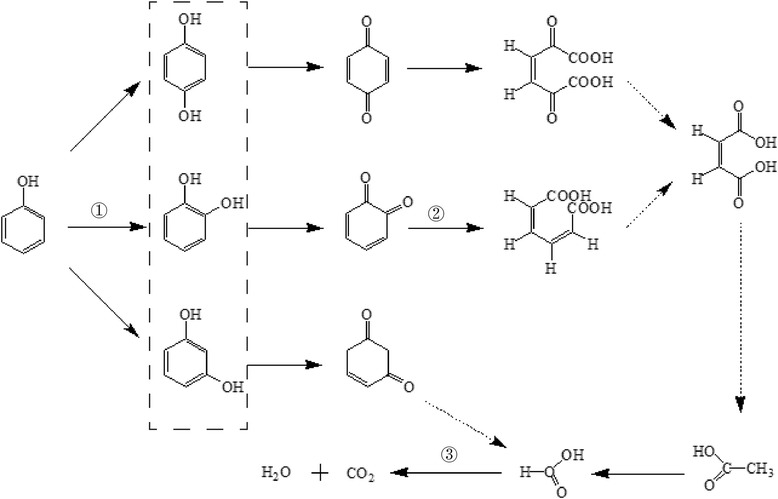
Pictorial scheme of the enzymatic degradation route of phenol. ① Hydroxylation of benzene formed the dihydroxybenzene and quinones. ② Dihydroxybenzene and quinones dehydrogenated and opened loop to form carboxylic acids. ③ Carboxylic acids mineralized to carbon dioxide and water. Dashed frame represented the compounds that were detected in this experiment
Conclusions
In this study, we found Ganoderma strains with a capacity of the decolorisation of four types of dyes, cloned a MnP gene from G. lucidum 00679 and expressed this gene in the methylotrophic yeast P. pastoris that produced an intracellular rGluMnP1 with a stable and active form. The higher oxidation capacity of the recombinant proteins was established by using the enzymes for the decolorization of four dyes and the degradation of phenol, in which phenol and the main degradation products were especially confirmed by HPLC. From this result, we inferred that the degradation of phenolic compounds may relate to the phenol structure. In the initial stages of the reaction, this catechol-mediated cycle should be the principal route of enzymatic degradation of phenol and its oxidation products. In summary, the rGluMnP1 showed a great potential for the enzymatic degradation of industrial dyes and phenolic compounds.
Methods
Strains, plasmids and media
G. lucidum strain 51562, 50044, 00679, 50817, 00680 were purchased from the Agricultural Culture Collection of China (ACCC) (Beijing, China). Escherichia coli DH5α were preserved by the Plant Biotechnology Research Center, School of Agriculture and Biology, Shanghai Jiao Tong University (Shanghai, China). P. pastoris strain SMD1168H and the pAO815 yeast expression vectors were purchased from Invitrogen (San Diego, CA, USA).
Potato dextrose agar (PDA) medium was used to culture Ganoderma species. Four different kinds of media, yeast extract peptone dextrose (YPD) medium, minimal dextrose (MD) medium, buffered minimal glycerol-complex (BMGY) medium and buffered methanol-complex (BMMY) medium, were used to culture P. Pastoris [29, 37].
Preparation and selection of fungal strains
The culture of G. lucidum mycelia was based on those described in the previous literature [39]. To select the suitable strain, stock cultures of different G. lucidum strains (51562, 50044, 00679, 50817, and 00680) were maintained in slant tubes at 4 °C on improved PDA medium (potato 200 g/L, dextrose 20 g/L, MgSO4 · 7H2O g/L, KH2PO4 2.5 mg/L, vitamin B1 10 mg/L, agar 20 g/L). Stock cultures were transferred onto agar plates containing the improved PDA medium and allowed to incubate for 5 d at 28 °C. Subsequently, agar blocks of the same size with the activated mycelia were cut from the edges of the growing colonies on the agar plates covered by the mycelia. Cut cultures were then transferred onto the Petri dishes containing the improved PDA medium containing 1 g/L O-methoxyphenol and allowed to incubate at 28 °C for 7 days. The diameters of the respective colonies and the decolorized zones were observed on the 13th day [21].
To screen the Mn influence on the MnP production, the mycelia suspension (0.5 mL) was added into 500 mL Erlenmeyer flasks containing 200 mL of liquid PDA medium. Two media, N-rich (12 mM ammonium tartrate) and N-limited (1.2 mM ammonium tartrate) PDA media with 3 μM and 200 μM Mn2+, were established by adding appropriate amounts of MnSO4 · H2O. Furtherly, 30 mL portions of inoculum were inoculated into 200 mL of medium in 500 mL Erlenmeyer flasks, then the cultures were incubated at 28 °C with 200 rpm. 1 g/L O-methoxyphenol was added to flasks on day 0. All batch experiments in the current study were done in duplicate; results were reported as the average of analyses of triplicate sample. For decolorization of four dyes by the culture supernatants, the culture supernatants prepared from G. lucidum 00679 were used to decolorize four dyes. The assays were performed at 28 °C. The reaction mixture in a total volume of 1 mL contained (final concentration): dyes (Drimaren Blue CL-BR, Drimaren Yellow X-8GN, Drimaren Red K-4Bl and Disperse Navy Blue HGL: 500 mg/L) and 100 μL culture supernatant.
Cloning and Expression of MnP
Total RNA was extracted from 1.0 g of freshly harvested G. lucidum mycelia using a TIANGEN RNA prep pure plant kit (Tiangen Biotech Co. Ltd., Beijing, China). Total RNA was reverse-transcribed into cDNA using the PrimeScript®RT Master Mix Perfect Real Time, according to the manufacturer’s instructions (TaKaRa Biotechnology Co., Ltd., Dalian, China). The core fragment of MnP gene was cloned according to standard protocols of the one step R-T PCR kit (AMV) (TaKaRa, Dalian, China) using forward primer MnPF1 and reverse primer MnPR1 that were designed according to the conserved regions of the MnP gene of Ganoderma sp., such as G. lucidum (ACA48488), G. formosanum (ABB77243), G. applanatum (BAA88392) and G. australe (ABB77244) deposited in GenBank. Following this step, 5′- and 3′-ends fragments were conducted using SMART technology (SMARTTM RACE cDNA Amplification Kit) to produce a full-length MnP sequence. Two gene-specific primers 3GlMnPF1 and 3GlMnPF2 were used only for 3′-ends fragments of GlMnP, and two gene-specific primers 5GlMnPR1 and 5GlMnPR2 for 5′-ends fragments. All amplified PCR products were purified, sub-cloned with the pMD 18-T vector system (TaKaRa, Dalian, China) and then sequenced. By aligning and assembling the products of the 3′-ends fragments, 5′-ends of the fragments and the core fragment, the full-length MnP sequence of G. lucidum was deduced and subsequently amplified using primers GlMnPFullF1 and GlMnPFullR1. All the primers [see Additional file 1] employed in PCR amplification were synthesized by the Shanghai Sangon Biotech Co. Ltd. (Shanghai, China).
After digestion with HindIII and EcoRI, digested products encoding GluMnP1 gene were sub-cloned directly into vector pAO815 that was predigested with the same restriction enzymes. The ligation products were transformed into E. coli strain DH5α and transformants were confirmed by PCR. The resulting recombinant plasmid, designated as pAO815::GluMnP1, was then sequencing.
Competent P. pastoris cells were prepared using the Invitrogen EasySelecte™ Pichia Expression Kit (Invitrogen), according to manufacturer’s instructions. 80 μL of P. pastoris cells were transformed with 20 μL of pAO815::GluMnP1, previously linearized with Pol, as described in the instruction of Multi-Copy Pichia Expression Kit (Invitrogen, Carlsbad, USA). Transformed clones were selected on MD (Mininal Dextrose) medium with ampicillin at 0.5, 1.0 and 2.0 mg/mL. Genomic DNA was extracted from P. pastoris using the Yeast DNA Isolation Kit (Sangon, Shanghai, China) according to the manufacturer’s instructions. The transformants were further confirmed by PCR amplification of the GluMnP1 gene, using the same primers used to clone it and with the AOX forward and reverse primers supplied with the kit. P. pastoris SMD1168 was used as a positive control and sterile water was used as a control.
Induction time screening and western blot analysis
In order to find out the optimum time points and analyze the time-course of expression, 1 mL cultures were induced to express rGluMnP1 by the addition heme to 1 mM and MnSO4 to 0.5 mM and supplemented methanol to 1% each 24 h until the transformants OD600 reached to 5. Samples of the cultures were taken out at 24, 48 and 72 h and disrupted using a high-pressure homogenizer (APV-2000, Germany). The culture supernatant was then harvested by centrifugation. The expression product was extracted using a commercial Kit (BSP013; Sangon), and resuspended in 100 μL of 2 × SDS-PAGE sample buffer and boiled for 5 min at 95 °C. The samples were prepared according to the previous description, and analyzed by 15% SDS-PAGE and stained with Coomassie Brilliant Blue R-250 [40].
Estimation of total protein and determination of enzyme activities
After being induced and cultured for 48 h, the recombinant P. pastoris transformants were harvested by centrifugation (4000 × g for 5 min at 4 °C). Cells were then resuspended in PBS buffer and disrupted by a high pressure homogenizer. After centrifugation, the supernatant was collected and determination of the protein content was based on a modified Bradford method [41]. To quantitatively analyze the relative concentrations of the expressed rGlMnP1 in cell supernatants was from the densitometry of the bands using the software BandScan 5.0 (Glyko, Novato, USA).
MnP activity was estimated by monitoring the oxidation of Mn2+ to Mn3+ at 270 nm using a UV/Visible spectrophotometer (DU 800, Beckman, USA), according to the Wariishi’s method [42]. The enzyme reaction system contained 0.5 mL sodium malonate (100 mM, pH 4.5), 0.1 mL of MnSO4 (10 mM) and 0–50 μL crude enzyme. The mixtures were incubated in 1.5 mL centrifuge tubes at 28 °C for 30 min. The reaction was started by adding 10 μL H2O2 (10 mM). The absorbance was immediately measured at 270 nm after the reactions were initiated using the addition of H2O2 to a concentration of 10 μM at room temperature. Both Lac and LiP activities were monitored as previously described [43].
Decolorization of dyes by the rGluMnP1
Four dyes, Drimaren Blue CL-BR, Drimaren Yellow X-8GN, Drimaren Red K-4Bl and Disperse Navy Blue HGL, were used for enzymatic decoloration treatment. Decolorization reactions were carried out at room temperature. The disappearance of the dyes was determined spectrophotometrically (Evolution 300UV-VIS spectrophotometer, Thermo Scientific) by measuring the absorbance at the wavelength of the maximum absorbance for 150 mg/L of each dye [13]. The maximum absorbance of the four dyes are 590 nm, 425 nm, 540 nm, 570 nm respectively. Typically, 0.4 mL dye (500 mg/L) and 0.1 mL crude enzyme were added to 0.39 mL sodium malonate buffer (100 mM, pH 4.5) containing 0.1 mL of MnSO4 (1 mM). The buffer was used as a negative control. Reactions were initiated by adding 10 μL of H2O2 (100 μM) to the reaction mixture. Decolorization was followed spectrophotometrically by a microplate reader (Power wave XS, Bio-tek) using the maximum absorbance curves recorded under these conditions. The decolorizing change of each dye was calculated every 15 min for 90 min. The decolorization percentage was calculated using a method described in the previous literature [44]. Decolorization percentage A% = (A0-A)/A0 × 100% (A0-initial absorbance; A-final absorbance). All of the experiments were performed using three replicates and were repeated at least twice. The data presented in the text correspond to the mean values.
HPLC-based analysis of phenol degradation rate and its oxidation products
The analysis of phenol and its oxidation products were carried out with a HPLC (Waters Corporation, Milford, MA, USA). Phenol, hydroquinone, pyrocatechol, resorcinol, benzoquinone and water were all the HPLC-grade and purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). The standard phenolic compounds including phenol, hydroquinone, pyrocatechol, resorcinol, benzoquinone solvent were consecutively injected five times to draw calibration curves. The injection volume was 2, 4, 8, 16 and 22 mL, respectively.
The oxidation reaction was carried out as previously described using 50 mL EP tubes [45]. A solution of 500 mg/L of simulation phenol-containing wastewater was made up with dissolving 1.00 g phenol at the constant volume of 2000 mL distilled water. The solution was sterilized at 121 °C for 20 min and then cooled to 30 °C. 5 mL phenol solutions were added into 50 mL EP tubes and add 5, 10 and 15% rGluMnP1 crude enzymes. Reactions are completely at 210 rpm/min and 28 °C for more than 24 h. The determination condition of the samples was set as follows: the column was YMC-pack ODS-A 250 × 4.60 mm S-5.0 μm; the mobile phase adopted in the analysis consists of methanol and water was in the ratio 30:70 (v/v). The separation was conducted in isocratic elution at a flow rate of 0.8 ml/min. The detection wavelength of photo-diode array was set at 280 nm; the column temperature was 30 °C. The injection volume was 20 μL and a data acquisition time of 20 minutes was used [46]. The degradation rate of phenol was calculated according to the following formula: Degradation rate (100%) = (C0-C)/C0 × 100%. In the formula, C0 was the concentration of phenol in the control group; C was the concentration of residual phenol in the treated phenol aqueous solution.
All experiments were performed at least twice using three replicates. The data presented in the text corresponded to the mean values.
Acknowledgments
HPLC analysis was performed in Plant Biotechnology Research Center, Shanghai Jiao Tong University.
Funding
This study was funded by the National Natural Science Foundation of China (No: 30771500) and Tibet Shenglong Industry Co., Ltd (No: 2013310031001210).
Availability of data and materials
The datasets generated during and analysed during the current study are available in the NCBI Short Read Archive repository (under accession number KR106991) and in Additional files.
Author contributions
XWZ acquired the funding and designed the whole study. XHB supervised the research group. HX, MYG and YHG performed all the experiments and analyzed data. HX collected the data and wrote the manuscript. MYG was responsible for the drawing. YHG contributed to preparation for the experiment. XWZ and XHB revised and enhanced the paper. All authors reviewed and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- BMGY
Buffered minimal glycerol-complex
- BMMY
Buffered methanol-complex
- GluMnP1
MnP1 from G. lucidum
- Lac
Laccase
- LiP
Lignin peroxidase
- LMEs
Lignin-modifying enzymes
- MD
Minimal dextrose
- MnP
Manganese peroxidase
- PDA
Potato dextrose agar
- rGluMnP1
Recombinant GluMnP1
- SDS-PAGE
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- YPD
Yeast extract peptone dextrose
Additional files
Primers used in this study. (DOC 30 kb)
Electrophoresis photos of GluMnP1 gene cloning from G. lucidum 00679. (DOC 79 kb)
Multiple alignment of the amino acid sequences of GluMnP1 from G. lucidum 00679. (DOC 45 kb)
Contributor Information
Hui Xu, Email: gedawawa@163.com.
Meng-Yuan Guo, Email: 971276109@qq.com.
Yan-Hua Gao, Email: g.ryan@sjtu.edu.cn.
Xiao-Hui Bai, Phone: +86-21-34205778, Email: xhbai@sjtu.edu.cn.
Xuan-Wei Zhou, Phone: +86-021-34203714, Email: xuanweizhou@sjtu.edu.cn.
References
- 1.Gold MH, Alic M. Molecular biology of the lignin-degrading basidiomycete Phanerochaete chrysosporium. Microbiol Rev. 1993;57:605–22. doi: 10.1128/mr.57.3.605-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou XW, Cong WR, Su KQ, Zhang YM. Ligninolytic enzymes from Ganoderma spp: Current status and potential applications. Crit Rev Microbiol. 2013;39:416–26. doi: 10.3109/1040841X.2012.722606. [DOI] [PubMed] [Google Scholar]
- 3.Qin X, Zhang J, Zhang X, Yang Y. Induction, purification and characterization of a novel manganese peroxidase from Irpex lacteus CD2 and its application in the decolorization of different types of dye. PLoS One. 2014;9:e113282. doi: 10.1371/journal.pone.0113282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paszczyński A, Huynh VB, Crawford R. Enzymatic activities of an extracellular, manganese-dependent peroxidase from Phanerochaete chrysosporium. FEMS Microbiol Lett. 1985;29:37–41. doi: 10.1111/j.1574-6968.1985.tb00831.x. [DOI] [Google Scholar]
- 5.Tello M, Corsini G, Larrondo LF, Salas L, Lobos S, Vicuña R. Characterization of three new manganese peroxidase genes from the ligninolytic basidiomycete Ceriporiopsis subvermispora. Biochim Biophys Acta. 2000;1490:137–44. doi: 10.1016/S0167-4781(99)00227-4. [DOI] [PubMed] [Google Scholar]
- 6.Maeda Y, Kajiwara S, Ohtaguchi K. Manganese peroxidase gene of the perennial mushroom Elfvingia applanata: cloning and evaluation of its relationship with lignin degradation. Biotechnol Lett. 2001;23:103–9. doi: 10.1023/A:1010341232388. [DOI] [Google Scholar]
- 7.Johansson T, Nyman PO, Cullen D. Differential regulation of mnp2, a new manganese peroxidase-encoding gene from the ligninolytic fungus Trametes versicolor PRL 572. Appl Microbiol Biotechnol. 2002;68:2077–80. doi: 10.1128/AEM.68.4.2077-2080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lankinen P, Hildén K, Aro N, Salkinoja-Salonen M, Hatakka A. Manganese peroxidase of Agaricus bisporus: grain bran-promoted production and gene characterization. Appl Microbiol Biotechnol. 2005;66:401–7. doi: 10.1007/s00253-004-1731-2. [DOI] [PubMed] [Google Scholar]
- 9.Hildén K, Martinez AT, Hatakka A, Lundell T. The two manganese peroxidases Pr-MnP2 and Pr-MnP3 of Phlebia radiata, a lignin-degrading basidiomycete, are phylogenetically and structurally divergent. Fungal Gen Biol. 2005;42:403–19. doi: 10.1016/j.fgb.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Sakamoto Y, Nakade K, Nagai M, Uchimiya H, Sato T. Cloning of Lentinula edodes lemnp2, a manganese peroxidase that is secreted abundantly in sawdust medium. Mycoscience. 2009;50:116–22. doi: 10.1007/S10267-008-0463-Z. [DOI] [Google Scholar]
- 11.Dong YC, Dai YN, Xu TY, Cai J, Chen QH. Biodegradation of chestnut shell and lignin-modifying enzymes production by the white-rot fungi Dichomitus squalens, Phlebia radiata. Bioproc Biosyst Eng. 2014;37:755–64. doi: 10.1007/s00449-013-1045-9. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez CS, Domínguez A, Sanromán A. Production of manganese-dependent peroxidase in a new solid-state bioreactor by Phanerochaete chrysosporium grown on wood shavings. Application to the decolorization of synthetic dyes. Folia Microbiol. 2002;47:417–21. doi: 10.1007/BF02818701. [DOI] [PubMed] [Google Scholar]
- 13.Boer CG, Obici L, de Souza CG, Peralta RM. Decolorization of synthetic dyes by solid state cultures of Lentinula (Lentinus) edodes producing manganese peroxidase as the main ligninolytic enzyme. Biores Technol. 2004;94:107–12. doi: 10.1016/j.biortech.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 14.Champagne PP, Ramsay JA. Contribution of manganese peroxidase and laccase to dye decoloration by Trametes versicolor. Appl Microbiol Biotechnol. 2005;69:276–85. doi: 10.1007/s00253-005-1964-8. [DOI] [PubMed] [Google Scholar]
- 15.Susla M, Novotný C, Erbanová P, Svobodová K. Implication of Dichomitus squalens manganese-dependent peroxidase in dye decolorization and cooperation of the enzyme with laccase. Folia Microbiol. 2008;53:479–85. doi: 10.1007/s12223-008-0075-1. [DOI] [PubMed] [Google Scholar]
- 16.Praveen K, Usha KY, Viswanath B, Reddy BR. Kinetic properties of manganese peroxidase from the mushroom Stereum ostrea and its ability to decolorize dyes. J Microbiol Biotechnol. 2012;22:1540–8. doi: 10.4014/jmb.1112.12011. [DOI] [PubMed] [Google Scholar]
- 17.Périé FH, Sheng D, Gold MH. Purification and characterization of two manganese peroxidase isozymes from the white-rot basidiomycete Dichomitus squalens. Biochim Biophys Acta. 1996;1297:139–48. doi: 10.1016/S0167-4838(96)00096-9. [DOI] [PubMed] [Google Scholar]
- 18.Iqbal HMN, Asgher M. Decolorization applicability of sol-gel matrix immobilized manganese peroxidase produced from an indigenous white rot fungal strain Ganoderma lucidum. BMC Biotechnol. 2013;13:56. doi: 10.1186/1472-6750-13-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanfermann I, Linke D, Nimtz M, Berger RG. Manganese peroxidases from Ganoderma applanatum degrade β-carotene under alkaline conditions. Appl Biochem Biotechnol. 2015;175:3800–12. doi: 10.1007/s12010-015-1548-8. [DOI] [PubMed] [Google Scholar]
- 20.Hatvani N, Mécs I. Effect of the nutrient composition on the dye decolorization and extracellular enzyme production by Lentinus edodes on solid medium. Enzyme Microb Tech. 2002;30:381–6. doi: 10.1016/S0141-0229(01)00512-9. [DOI] [Google Scholar]
- 21.Perez J, Jeffries TW. Roles of manganese and organic acid chelators in regulating lignin degradation and biosynthesis of peroxidases by Phanerochaete chrysosporium. Appl Environ Microbiol. 1992;58:2402–9. doi: 10.1128/aem.58.8.2402-2409.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swamy J, Ramsay JA. Effects of Mn2+ and NH4+ concentrations on laccase and manganese peroxidase production and Amaranth decoloration by Trametes versicolor. Appl Microbiol Biotechnol. 1999;51:391–6. doi: 10.1007/s002530051408. [DOI] [Google Scholar]
- 23.Martínez MJ, Ruiz-Dueñas FJ, Guillén F, Martínez AT. Purification and catalytic properties of two manganese peroxidase isoenzymes from Pleurotus eryngii. Eur J Biochem. 1996;237:424–32. doi: 10.1111/j.1432-1033.1996.0424k.x. [DOI] [PubMed] [Google Scholar]
- 24.Camarero S, Martínez MJ, Martínez AT. Understanding lignin biodegradation for the improved utilization of plant biomass in modern biorefineries. Biofue Bioprod Bior. 2014;8:615–25. doi: 10.1002/bbb.1467. [DOI] [Google Scholar]
- 25.Fu Y, Viraraghavan T. Fungal decolorization of dye wastewaters: a review. Bioresource Technol. 2001;79:251–62. doi: 10.1016/S0960-8524(01)00028-1. [DOI] [PubMed] [Google Scholar]
- 26.Kariminiaae-Hamedaani HR, Sakurai A, Sakakibara M. Decolorization of synthetic dyes by a new manganese peroxidase-producing white rot fungus. Dyes Pigments. 2007;72:157–62. doi: 10.1016/j.dyepig.2005.08.010. [DOI] [Google Scholar]
- 27.Daly R, Hearn MT. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J Mol Recognit. 2005;18:119–38. doi: 10.1002/jmr.687. [DOI] [PubMed] [Google Scholar]
- 28.Liu D, Gong J, Dai W, Kang X, Huang Z, Zhang HM, Liu W, Liu L, Ma J, Xia Z, Chen Y, Chen Y, Wang D, Ni P, Guo AY, Xiong X. The genome of Ganoderma lucidum provides insights into triterpenes biosynthesis and wood degradation. PLoS One. 2012;7:e36146. doi: 10.1371/journal.pone.0036146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang F, Kongsaeree P, Charron R, Lajoie C, Xu H, Scott G, Kelly C. Production and separation of manganese peroxidase from heme amended yeast cultures. Biotechnol Bioeng. 2008;99:540–9. doi: 10.1002/bit.21590. [DOI] [PubMed] [Google Scholar]
- 30.Gu L, Lajoie C, Kelly C. Expression of a Phanerochaete chrysosporium manganese peroxidase gene in the yeast Pichia pastoris. Biotechnol Progr. 2003;19:1403–9. doi: 10.1021/bp025781h. [DOI] [PubMed] [Google Scholar]
- 31.Chacko JT, Subramaniam K. Enzymatic degradation of azo dyes-a review. Int J Environ Sci. 2011;1:1250–60. [Google Scholar]
- 32.Goszczynski S, Paszczynski A, Pasti-Grigsby M, Crawford R, Crawford D. New pathway for degradation of sulfonated azo dyes by microbial peroxidases of Phanerochaete chrysosporium and Streptomyces chromofuscus. J Bacteriol. 1994;176:1339–47. doi: 10.1128/jb.176.5.1339-1347.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardin IR, Cao H, Wilson SS, Akin DE. Decolorization of textile wastewater by selective Fungi. Text Chem Color Am D. 2000;32:38–42. [Google Scholar]
- 34.Zhang FM, Knapp JS, Tapley KN. Decolourisation of cotton bleaching effluent with wood rotting fungus. Water Res. 1999;33:919–28. doi: 10.1016/S0043-1354(98)00288-7. [DOI] [Google Scholar]
- 35.Wesenberg D, Kyriakides I, Agathos SN. White-rot fungi and their enzymes for the treatment of industrial dye effluents. Biotechnol Adv. 2003;22:161–87. doi: 10.1016/j.biotechadv.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 36.Levin L, Papinutti L, Forchiassin F. Evaluation of Argentinean white rot fungi for their ability to produce lignin-modifying enzymes and decolorize industrial dyes. Bioresource Technol. 2004;94:169–76. doi: 10.1016/j.biortech.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Jiang F, Kongsaeree P, Schilke K, Lajoie C, Kelly C. Effects of pH and temperature on recombinant manganese peroxidase production and stability. Appl Biochem Biotechnol. 2008;146:15–27. doi: 10.1007/s12010-007-8039-5. [DOI] [PubMed] [Google Scholar]
- 38.Qiu YL, Chen L, Ma JH, Zhao JF. Analysis of phenol intermediate products and investigation into its degradation route resulted from photocatalytic oxidation. Sichuan Environment. 2005;24:5–8. [Google Scholar]
- 39.Zhou XW, Li QZ, Zhao JY, Tang KX, Lin J, Yin YZ. Comparison of rapid DNA extraction methods applied to PCR identification of medicinal mushroom Ganoderma spp. Prep Biochem Biotechnol. 2007;37:369–80. doi: 10.1080/10826060701593282. [DOI] [PubMed] [Google Scholar]
- 40.Li QZ, Wang XF, Chen YY, Lin J, Zhou XW. Cytokines expression induced by Ganoderma sinensis fungal immunomodulatory proteins (FIP-gsi) in mouse spleen cells. Appl Biochem Biotechnol. 2010;162:1403–13. doi: 10.1007/s12010-010-8916-1. [DOI] [PubMed] [Google Scholar]
- 41.Carlsson N, Borde A, Wölfel S, Åkerman B, Larsson A. Quantification of protein concentration by the Bradford method in the presence of pharmaceutical polymers. Anal Biochem. 2011;411:116–21. doi: 10.1016/j.ab.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 42.Wariishi H, Valli K, Gold MH. Manganese (II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators. J Biol Chem. 1992;267:23688–95. [PubMed] [Google Scholar]
- 43.Chi YJ, Yan HB. Detection on laccase, manganese peroxidase and lignin peroxidase in ligninolytic enzymes of Pleurotus dyamor. Sci Silvae Sin. 2009;45:154–8. [Google Scholar]
- 44.Mielgo I, López C, Moreira M, Feijoo G, Lema J. Oxidative degradation of azo dyes by manganese peroxidase under optimized conditions. Biotechnol Prog. 2003;19:325–31. doi: 10.1021/bp020136w. [DOI] [PubMed] [Google Scholar]
- 45.Yu LY, Ma HJ, Su MM, Zhou JZ. Content detection of phenol during the degradation process by yeasts. Environ Pollut Cont. 2013;35:60–7. [Google Scholar]
- 46.Fan CM, Sun YP. Simultaneous determination of phenol and its photo-catalyzed degradation intermediates by HPLC. J Instrumental Analysis. 2000;19:48–50. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and analysed during the current study are available in the NCBI Short Read Archive repository (under accession number KR106991) and in Additional files.


