Abstract
Site-directed mutagenesis experiments combined with fluorescence microscopy shed light on the role of Escherichia coli FtsW, a membrane protein belonging to the SEDS family that is involved in peptidoglycan assembly during cell elongation, division, and sporulation. This essential cell division protein has 10 transmembrane segments (TMSs). It is a late recruit to the division site and is required for subsequent recruitment of penicillin-binding protein 3 (PBP3) catalyzing peptide cross-linking. The results allow identification of several domains of the protein with distinct functions. The localization of PBP3 to the septum was found to be dependent on the periplasmic loop located between TMSs 9 and 10. The E240-A249 amphiphilic peptide in the periplasmic loop between TMSs 7 and 8 appears to be a key element in the functioning of FtsW in the septal peptidoglycan assembly machineries. The intracellular loop (containing the R166-F178 amphiphilic peptide) between TMSs 4 and 5 and Gly 311 in TMS 8 are important components of the amino acid sequence-folding information.
The wall peptidoglycan is a bacterium-specific polymer that preserves cell integrity and plays an important role in bacterial morphogenesis. To allow bacterial cell growth and division, two morphogenetic networks channel peptidoglycan assembly into wall expansion and septum formation in a cell cycle-dependent fashion. The morphogenetic protein machineries responsible for maintenance of the cell's cylindrical shape during cell elongation and for septation during cell division involve a set of network-specific proteins (2, 16, 38, 42).
In Escherichia coli, the first morphogenetic network includes class B penicillin-binding protein 2 (PBP2) and the RodA protein, which are responsible for wall elongation and maintenance of the rod shape. They are encoded by the pbpA and rodA genes, respectively, which form an operon with the dacA gene, encoding the monofunctional PBP5 in the 14-min region of the chromosome (49, 50). Inactivation of RodA or PBP2 leads to the formation of spherical cells. MreB, MreC, and MreD also belong to this morphogenetic network (53). If MreB is not produced, the cells become spherical (54). MreB is an actin-like protein (52) that polymerizes into fibrous spirals at the inner face of the membrane in E. coli and Bacillus subtilis (25, 48). This protein is required for proper chromosome segregation (32). In Caulobacter crescentus, MreB seems to spatially coordinate the activities of the cell wall assembly proteins (17).
Formation of the septum in E. coli requires class B PBP3 (also called FtsI) and FtsZ, FtsA, ZipA, FtsK, FtsQ, FtsL, FtsB, FtsW, and FtsN, which constitute the second morphogenetic network. The majority of the genes encoding these proteins lie in the 2-min region of the chromosome and form the dcw (division cell wall) operon. Inactivation of one gene inhibits septation and leads to filamentous growth (4, 36). To initiate cell division, the GTP-binding tubulin-like FtsZ protein forms an intracellular ring at the division site (34, 35). The Z-ring serves as a cytoskeletal scaffold for the recruitment of at least 11 proteins of the cell division machinery, called the divisome. FtsA, an actin-like protein, and ZipA bind directly to the C terminus of FtsZ (1, 43). Then, FtsK, FtsQ, and FtsL, together with FtsB (previously called YgbQ), FtsW, PBP3, and FtsN, appear sequentially at the division site (4, 6, 7, 40). FtsQ, FtsB, and FtsL form a protein subcomplex before their localization to the division site. It is not known if the downstream proteins are components of this assemblage (5). The transmembrane protein FtsW is involved in the recruitment of PBP3, which catalyzes peptidoglycan cross-linking and presumably mediates protein-protein interactions critical for septal peptidoglycan synthesis (37, 40).
In B. subtilis, a third morphogenetic system that is specifically activated during sporulation for the spore cortex formation exists. It includes the class B PBP called SpoVD and the SpoVE protein, which are related to PBP3 and FtsW, respectively, and whose encoding genes are also part of the dcw cluster. Disruption of SpoVE causes a complete block of spore cortex formation but does not interfere with vegetative growth (21). Development of the spore cortex in SpoVD mutants is severely affected (8).
RodA, FtsW, and SpoVE are polytopic membrane proteins which share 30% identity (23, 26). Most of the conserved residues are located in the C-terminal part of the proteins. They belong to the SEDS family (for shape, elongation, division, and sporulation) (22). These proteins appear to work in coordination with one class B PBP to catalyze peptidoglycan polymerization during the cell cycle. Their mechanism of action is not known.
FtsW is present in virtually all bacteria that have a peptidoglycan cell wall. It is an essential cell division protein (3). In addition to its role in the localization of PBP3, FtsW may have other functions. It is present at the interface between the cytoplasm and the periplasm and is made of 10 transmembrane segments (TMSs) (33). The protein may serve to integrate signals between the cytoplasmic (FtsZ, FtsA, ZipA, and FtsK) and the periplasmic (FtsK, FtsQ, FtsL, FtsB, PBP3, and FtsN) components of the divisome (36). It was proposed that FtsW might be involved in the translocation of the lipid-linked peptidoglycan precursor through the cytoplasmic membrane, on the basis of the twofold accumulation of lipid-linked precursor in cells overproducing PBP3 and FtsW (39). Less is known about how it is targeted to the septum, how it recruits PBP3, and how it functions in cell division.
In the experiments reported here, the roles of several domains of FtsW were investigated by site-directed mutagenesis. The functionality and the localization of the modified proteins were studied by complementation assays and fluorescence microscopy. The results allowed the identification of distinct roles for several regions of the protein. The periplasmic loop from residues P368 to P375 plays an important role in the septal recruitment of PBP3, the E240-A249 periplasmic amphiphilic segment appears to be a key element in the functioning of FtsW in the septal peptidoglycan assembly machineries, and the R166-F178 intracellular amphiphilic peptide seems to be involved in the conformation of FtsW.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, and media.
Bacterial strains and plasmids are described in Table 1. Oligonucleotides were from Amersham Pharmacia Biotech. The rich media used were Luria-Bertani (LB) medium, LB medium without NaCl and containing 0.1% glucose and 20 mg of thymine/liter (LBΔNaCl), and LB medium containing 5 g of NaCl/liter instead of 10 g of NaCl/liter (T4). Minimal medium contained 6.33 g of K2HPO4 · 3H2O, 2.95 g of KH2PO4, 1.05 g of (NH4)2SO4, 0.10 g of MgSO4 · 7H2O, 0.28 mg of FeSO4 · 7H2O, 7.1 mg of Ca(NO3)2 · 4H2O, 4 mg of thiamine, 4 g of glucose, and 50 μg of lysine, each per liter (pH 7.0). The media were supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), chloramphenicol (30 μg/ml), thymine (20 μg/ml), and/or IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) when appropriate.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genetic marker(s) or features | Construction, source, or reference |
|---|---|---|
| Strains | ||
| JLB17 | F−thr trp his thy ara lac gal xyl mtl rspL tonA ftsW(Ts) | 24 |
| LMC500 | F−araD139 Δ(argF-lac)U169 deoC1 flbB5301 ptsF25 rbsR relA1 rpsL150 lysA1 | 51 |
| Plasmids | ||
| pBAD/His A | araBAD promoter, N-terminal His tag, Ampr | Invitrogen |
| pRU277 | ftsW under the control of the lac promoter-operator, Ampr | Aventis |
| pET28a(+) | T7 lac promoter-operator, N-terminal His tag, Kanr | Novagen |
| pMCL210 | lac promoter-operator, p15A origin, Cmr | 41 |
| pDSW234 | gfp-ftsl under the control of a weakened trc promoter, Ampr | 55 |
| pDML2400 | pET28a-ftsW(wild-type) | This study |
| pDML2401 | pET28a-ftsW(R172S) | This study |
| pDML2402 | pET28a-ftsW(R243Q) | This study |
| pDML2403 | pET28a-ftsW(R402Q) | This study |
| pDML2404 | pET28a-ftsW(P368A) | This study |
| pDML2405 | pET28a-ftsW(P375A) | This study |
| pDML2406 | pET28a-ftsW(P368A P375A) | This study |
| pDML2407 | pMCL210-ftsW(wild-type) | This study |
| pDML2408 | pMCL210-ftsW(R172S) | This study |
| pDML2409 | pMCL210-ftsW(R243Q) | This study |
| pDML2410 | pMCL210-ftsW(R402Q) | This study |
| pDML2411 | pMCL210-ftsW(P368A) | This study |
| pDML2412 | pMCL210-ftsW(P375A) | This study |
| pDML2413 | pMCL210-ftsW(P368A P375A) | This study |
| pDML2414 | gfp-ftsW(wild-type) derived from pDSW234 | This study |
| pDML2415 | pDML2414-ftsW(R172S) | This study |
| pDML2416 | pDML2414-ftsW(R243Q) | This study |
| pDML2417 | pDML2414-ftsW(R402Q) | This study |
| pDML2418 | pDML2414-ftsW(P368A) | This study |
| pDML2419 | pDML2414-ftsW(P375A) | This study |
| pDML2420 | pDML2414-ftsW(P368A P375A) | This study |
| pDML2421 | pMCL210-ftsW(L164 HA) | This study |
| pDML2422 | pMCL210-ftsW(E293 HA) | This study |
| pDML2424 | pMCL210-ftsW(R411 HA) | This study |
| pDML2425 | pDML2414-ftsW(G311D) | This study |
Recombinant plasmids.
For construction of the FtsW mutants, pRU277 bearing the ftsW gene served as the template for site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene) using the oligonucleotides (mutated codons are underlined) 5′-GTAAAGGCGACGAAGTATCTAATAACCTGCG-3′ and 5′-CGCAGGTTATTAGATACTTCGTCGCCTTTAC-3′ (mutation R172S), 5′-CCGAACCGTACCAAATCCGCCGTGTTAC-3′ and 5′-GTAACACGGCGGATTTGGTACGGTTCGG-3′ (mutation R243Q), 5′-GGGGATGTTAGCGACCAAAGG-3′ and 5′-CCTTTGGTCGCTAACATCCCC-3′ (mutation P368A), 5′-GACATTGGCGCTGATCAGTTACG-3′ and 5′-CGTAACTGATCAGCGCCAATGTC-3′ (mutation P375A), and 5′-GTATTGATTATGAAACGCAGCTGGAGAAAG-3′ and 5′-CTTTCTCCAGCTGCGTTTCATAATCAATAC-3′ (mutation R402Q). The mutated genes were identified by DNA sequencing. The StyI-HindIII fragments containing the R243Q, R402Q, P368A, P375A, and P368A P375A mutations or the BglII-StyI fragment containing the R172S mutation were then replaced with the corresponding unmodified segment of pRU277. The BglII-HindIII segments carrying ftsW or modified ftsW were then excised from pRU277 or recombinant pRU277 plasmids and ligated to the BamHI and HindIII sites of pET28a(+). The resulting plasmids, pDML2400, pDML2401, pDML2402, pDML2403, pDML2404, pDML2405, and pDML2406, encoded His-tagged FtsW, His-tagged FtsW R172S, His-tagged FtsW R243Q, His-tagged FtsW R402Q, His-tagged FtsW P368A, His-tagged FtsW P375A, and His-tagged FtsW P368A P375A, respectively. In these constructions, the modified ftsW genes were under the control of the T7 lac promoter-operator.
For complementation assays, the genes encoding the His-tagged (wild-type and modified) FtsW proteins were inserted into pMCL210 under the control of the lac promoter-operator. The ftsW gene was amplified by PCR using pDML2400 as the template, 5′-GGATCCCTGAAGGAGATATACCATGGGCAGC-3′ as the sense primer, and 5′-GAAGCTTATCATCGTGAACCTCGTACAAAC-3′ as the antisense primer. (BamHI and HindIII sites, respectively, are underlined. Bold characters indicate a stop codon, a ribosome binding site, and a start codon embedded in the sense primer.) The amplified product was cloned in the pGEM-T Easy vector (Promega), and the nucleotide sequence was analyzed to detect any error. The BamHI-HindIII fragment containing the ftsW gene was then inserted into the corresponding sites of pMCL210, giving rise to pDML2407. The NruI-HindIII fragments containing the R172S, R243Q, R402Q, P368A, P375A, and P368A P375A mutations were then excised from pDML2401, pDML2402, pDML2403, pDML2404, pDML2405, and pDML2406 and exchanged with the unmodified segment of pDML2407, giving rise to pDML2408, pDML2409, pDML2410, pDML2411, pDML2412, and pDML2413, respectively.
Construction of GFP fusions.
The ftsW gene was amplified by PCR using pDML2400 as the template, 5′-CACGAATTCAACAACAACCGTTTATCTCTCCCTCGCCTGAAAATGC-3′ as the sense primer, and 5′-GCCGCAAGCTTATCATCGTGAACCTC-3′ as the antisense primer. The purified PCR fragment was digested by EcoRI and HindIII (respective underlined sites) and ligated to the same sites of pDSW234. The EcoRI-NruI DNA segment was completely sequenced. The NruI-HindIII DNA fragment was then replaced by the NruI-HindIII DNA fragment from the original pDML2400 to create pDML2414. The gene fusion was under the control of the trc promoter weakened in the −35 region and encoded FtsW fused to the C-terminal end of the green fluorescent protein (GFP). To generate GFP-FtsW mutants, the NruI-HindIII fragments containing the R172S, R243Q, and R402Q mutations or the SnaBI-HindIII fragments containing the P368A, P375A, and P368A P375A mutations were excised from pDML2401, pDML2402, pDML2403, pDML2411, pDML2412, and pDML2413 and exchanged with the unmodified segment of pDML2414, giving rise to pDML2415, pDML2416, pDML2417, pDML2418, pDML2419, and pDML2420, respectively. The ftsW(Ts) gene was amplified by PCR using the chromosomal DNA from E. coli JLB17 as the template, 5′-GAAACTATGGAACAGGCGATGC-3′ as the sense primer, and 5′-GTTCCGCCTGCCATCACC-3′ as the antisense primer. The purified PCR fragment was completely sequenced and inserted into pGEM-T Easy to produce pGEM-ftsWts. The NruI-SalI fragment containing the G311D mutation was excised from pGEM-ftsWts and exchanged with the unmodified fragment from pDML2414 to create pDML2425.
Construction of HA epitope-tagged FtsW derivatives.
Epitope-tagged FtsW derivatives were generated by introduction of a nonapeptide (YPYDVPDYA) from the hemagglutinin (HA) of the influenza virus into loops 4/5 and 7/8 of FtsW by use of the method described by Kamman et al. (27). Three primers were used in two successive PCRs with pDML2407 as the template. In the first reaction, we used the sense primer 5′-CACGAATTCAACAACAACCGTTTATCTCTCCCTCGCCTGAAAATGC-3′ and the antisense primer 5′-GCCTTTACCACCGCGTAATCTGGTACGTCGTACGGATACAGATATTGGC-3′ to insert the epitope (underlined) between L164 and V165 or the antisense primer 5′-GTCAGTGTGCGCGCCGCGTAATCTGGTACGTCGTACGGATATTCCGGCAGATA-3′ to insert the epitope (underlined) between E293 and A294. The reaction products were then used as the sense primer in combination with the antisense primer 5′-GCCGCAAGCTTATCATCGTGAACCC-3′, which hybridizes with the 3′ end of ftsW. The amplified products were digested with NruI and XcmI (loop 4/5) or XcmI and HindIII (loop 7/8) and were used to replace the corresponding fragments of pDML2407. The modified regions were sequenced. The resulting plasmids were named pDML2421 and pDML2422 and encoded His-tagged FtsW(L164 HA) or His-tagged FtsW(E294 HA). Insertion of the epitope in the C-terminal loop of FtsW between R411 and G412 was carried out using pDML2407 as the template, 5′-CACGAATTCAACAACAACCGTTTATCTCTCCCTCGCCTGAAAATGC-3′ as the sense primer, and 5′-GATAAGCTTATCATCGTGAACCCGCGTAATCTGGTACGTCGTACGGATATCGTACAAACG-3′ as the antisense primer. (The segment encoding the epitope is underlined.) The amplified fragment digested by SalI and HindIII was ligated to the same site of pDML2400 to create pDML2423. After the modified segment was sequenced, the SnaBI-HindIII fragment was excised from pDML2423 and inserted in the corresponding site of pDML2407, giving rise to pDML2424, which encodes His-tagged FtsW(R411 HA).
Complementation assay and immunolabeling.
E. coli JLB17 was electrotransformed with the appropriate vectors. Transformants were inoculated in 25 ml of LBΔNaCl and grown overnight at 28°C. Subsequently, the culture was diluted 1:20 and grown to an A450 of 0.5. A sample of the culture was fixed for microscopy. The culture was diluted 1:4 in prewarmed medium at 42°C and grown at this temperature for 2 h for a complementation assay and for two mass doublings for immunolabeling before harvest and fixation of the cells for microscopy.
Membrane preparation and detection of FtsW derivatives.
The level of production of GFP-FtsW (wild type and mutants) was estimated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) of crude extracts prepared as follows. The cells from 1 ml of cultures at an A600 of 1 were suspended in 66 μl of 10 mM Tris-HCl (pH 8.0)-10% glycerol-10% ethylene glycol-0.5 M NaCl. Samples were incubated at room temperature for 20 min in the presence of denaturing buffer (0.06 M Tris-HCl [pH 6.8], 1% SDS, 9.5% glycerol, 3.3% mercaptoethanol, 0.002% bromophenol blue) before electrophoresis. Fluorescence detection was done directly on the gel in an FX molecular imager (Bio-Rad Laboratories) with the parameter set designed for fluorescein isothiocyanate detection (excitation by 488-nm-wavelength Ar ion laser with a 515- to 545-nm band-pass emission filter).
Fixation and permeabilization.
Due to the salt sensitivity of the cells, a procedure adapted from the work of den Blaauwen et al. was used for the fixation and permeabilization of the cells (10). Cells were fixed in 2.8% formaldehyde and 0.04% glutaraldehyde in growth medium for 15 min at room temperature. The cells were collected by centrifugation at 8,000 × g for 5 min, washed once in low-salt phosphate buffer (PBΔS; 27 mM KCl, 10 mM Na2HPO4 · 2H2O, 2 mM KH2PO4 [pH 7.2]), and subsequently incubated in 0.025% Triton X-100 in PBΔS for 15 min at room temperature. All subsequent centrifugations were performed at 4,500 × g. The cells were washed three times in PBΔS and incubated in PBΔS containing 100 μg of lysozyme/ml and 5 mM EDTA for 15 min at room temperature. Finally, the cells were washed three times in PBΔS.
Microscopy and image analysis.
The JLB17 cells were immunolabeled as described previously (11), with the modifications that phosphate-buffered saline was replaced with PBΔS in all steps and the cells were labeled with affinity-purified polyclonal antibodies against PBP3 (37) and goat anti-rabbit conjugated with Alexa 546 (Molecular Probes). Subsequently, the cells were suspended in water and immobilized on agarose slides as described by Koppelman et al. (31). They were photographed with a cooled Princeton or Photometrics CoolSNAP fx charge-coupled-device camera (Roper Scientific) mounted on an Olympus BX60 microscope through a UPlanFI 100×, 1.3 oil immersion objective, in both phase contrast and fluorescence (with a U-MNB filter cube [for GFP] with a 470- to 490-nm band-pass excitation filter and a >515-nm long-pass emission filter or with a U-MNG filter cube [for Alexa 546] with a 530- to- 550-nm band-pass excitation filter and a >590-nm long-pass emission filter). Images were taken using the public domain program OBJECT-IMAGE 2.09 (http://simon.bio.uva.nl/object-image.html) by N. Vischer (University of Amsterdam), which is based on NIH IMAGE by W. Rasband. The photographs were stacked, and the length of each cell was measured in the phase-contrast image as described previously (11).
Prediction of interaction sites.
Interaction sites were predicted by the method described by Gallet et al. (18), on the basis of plots of mean hydrophobic moment versus mean hydrophobicity, constructed according to the algorithm of Eisenberg et al. (14). These values were calculated with a 7-residue window and a gyration angle of 100° (i.e., the angle between two consecutive residues observed in the direction of the helix axis), corresponding to an α-helix conformation. The consensus hydrophobicity scale (15) was used for the calculations. The mean α-helical hydrophobic moment and the mean hydrophobicity were assigned to the central amino acid of the window. Five amphiphilic peptides, designated A through E, exhibiting mean hydrophobicities lower than −0.5 and mean hydrophobic moments higher than 0.25, were detected (see Fig. S1 in the supplemental material). Segments C and D have five and four heptapeptide windows, respectively, in which the central amino acid residue is characterized by a mean hydrophobic moment greater than 0.5. These segments are, thus, more amphiphilic and presumably have more interaction potentiality than segments A, B, and E, which have no or only one heptapeptide window with a mean hydrophobic moment of greater than 0.5.
RESULTS
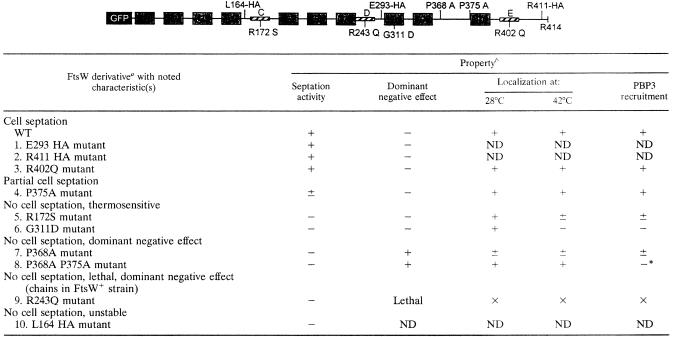
The topology of E. coli FtsW has been deduced from the study of truncated FtsW fused to reporter proteins and from comparison with the topology of Streptococcus pneumoniae FtsW (19, 33). As shown in Fig. 1, the model contains 10 TMSs, a large periplasmic loop between TMSs 7 and 8 (loop 7/8), and both N- and C-terminal ends in cytoplasm. To investigate the role of FtsW, attempts were carried out to identify and characterize the regions of the protein involved in its localization, in the recruitment of PBP3, and in its cell division activity. For this purpose, derivatives altered at specific sites in putative protein-protein interacting segments and in conserved residues or modified by the insertion of a small peptide were constructed. They were fused to a His tag or to the GFP. The effects of the modifications were studied by analyzing the level of production of the derivatives compared to that of wild-type FtsW. The activity of the FtsW derivatives in cell division was analyzed by a complementation assay with E. coli JLB17 producing chromosomal, thermosensitive FtsW (24). The localization of the derivatives was examined by fluorescence microscopy using E. coli JLB17 or in E. coli LMC500, which produces chromosomal wild-type FtsW. The modified FtsW derivatives are shown in Table 2. On the basis of the effects of the mutations introduced into the ftsW gene, the modified FtsW derivatives fall into six groups (see below).
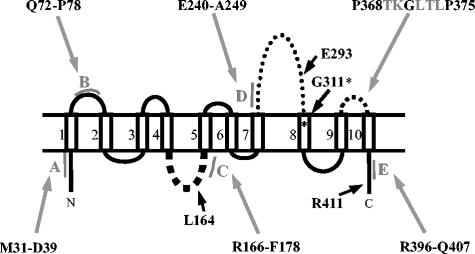
FIG. 1.
Membrane topology of E. coli FtsW. The localizations of amphiphilic segments A through E are shown in gray in the model. Black arrows indicate the localizations of HA inserts and Gly 311*, which is modified into Asp in E. coli JLB17. The N-terminal end contains 46 residues, loop 1/2 contains 19 residues, loop 4/5 contains 14 residues, loop 7/8 contains 66 residues, loop 9/10 contains 10 residues, and the C-terminal end contains 20 residues.
TABLE 2.
Properties of FtsW mutants in E. coli JLB17 grown in LBΔNaCl mediuma
In the FtsW diagram above the table, TMs and putative protein-protein interaction sites are represented by numbered gray and striped boxes, respectively. Amino acid substitutions are shown. For mutant derivatives 1, 2, and 10, GFP is replaced by a His tag. WT, wild type.
+, presence of activity (septation, localization, or PBP3 recruitment) or dominant negative effect; −, absence of activity or dominant negative effect; ±, weak activity; ND, not determined; ×, could not be done.
Characterization of the ftsW(Ts) gene of E. coli JLB17 used in complementation.
The ftsW(Ts) gene of E. coli JLB17 was amplified by PCR. The DNA fragment obtained from 10 independent amplifications was sequenced. The results revealed that the ftsW(Ts) gene contained one transition mutation that changed Gly 311 located in TMS 8 into Asp (Fig. 1). Gly 311 is conserved in 24 out of 25 FtsW sequences. The ftsW(Ts) gene of E. coli JLB17 thus encodes the FtsW G311D mutant.
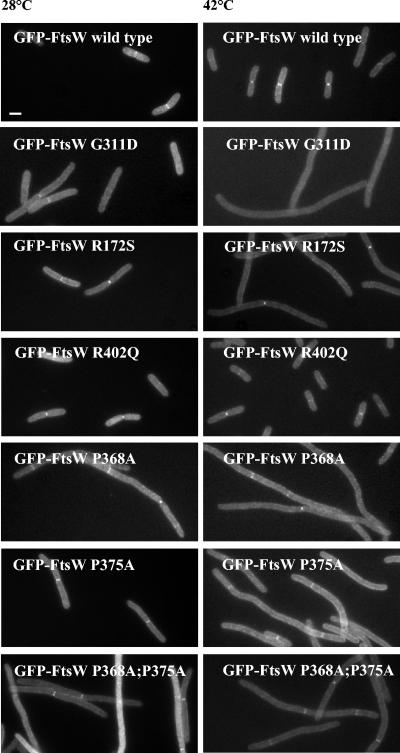
To determine the effect of the mutation on the targeting of FtsW to the division site, thermosensitive FtsW G311D was fused to the C-terminal end of the GFP and placed under the control of a weakened trc promoter (55). E. coli JLB17 was used as a host to produce GFP-FtsW G311D (Table 2, derivative 6) from pDML2425 and GFP-FtsW from pDML2414 (Table 1). Crude extracts from transformants grown in LBΔNaCl (for JLB17) at 28 and 42°C without IPTG were analyzed by SDS-PAGE and fluorescence detection. GFP-FtsW G311D was produced at both temperatures in similar or slightly larger amounts than the wild-type fusion (Fig. 2). Both GFP fusion proteins (wild type and mutant) were susceptible to proteolysis; one of the breakdown products corresponded to the GFP. The cells were also collected, fixed with cross-linking agents, and examined by fluorescence microscopy (Fig. 3). A faint GFP-FtsW G311D fluorescent signal was detected at the midcell region at 28°C but not at 42°C, whereas the wild-type fusion was found at the septal site at both temperatures (Fig. 3). Thus, thermosensitive GFP-FtsW G311D was not functional, because it failed to localize to the division site at 42°C.
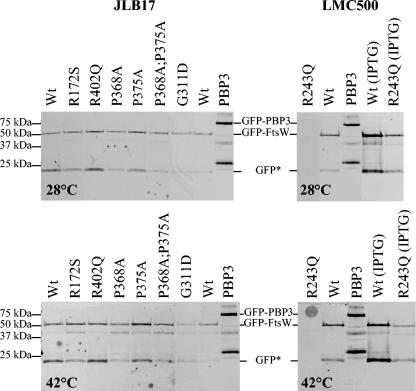
FIG. 2.
Expression levels of GFP-FtsW fusions. Fluorescence detection after SDS-PAGE of membranes isolated from E. coli JLB17 grown exponentially in LBΔNaCl in the absence of IPTG at 28 and 42°C and from E. coli LMC500 grown in minimal medium in the absence of IPTG at 28 and 42°C or grown in LB medium with 1 mM IPTG at 28 and 42°C. Each loaded sample corresponded to 250 μl of the cell culture at an A600 of 1. GFP-PBP3 was used as a reference and contained some degradation products. GFP*, a GFP-FtsW breakdown product with a molecular mass similar to that of GFP.
FIG. 3.
Complementation activity and localization of GFP-FtsW fusions in E. coli JLB17. The cells were harvested and fixed after exponential growth in LBΔNaCl in the absence of IPTG at 28°C and after 2 h of growth at 42°C. See Materials and Methods for details on fluorescence microscopy. Bar, 1 μm.
Role of putative protein-protein interacting segments.
Hydrophobic moments can be used to detect peptide segments of a protein that are capable of forming strongly amphiphilic α-helices or β-sheets (14, 15). Amphiphilic peptide segments were shown to have protein-protein interaction potentials (9). By analyzing the hydrophobic moment plot of a database of 50 known interaction sites, 66% of the experimental interacting residues were predicted (18). This approach was applied to PBP3 and allowed the identification of regions with different functionalities (37). In an attempt to identify regions of FtsW possibly involved in protein interaction, the hydrophobic moment plot of the 414 amino acid residues of the protein was therefore calculated (see Fig. S1 in the supplemental material), which allowed the detection of five amphiphilic peptides, A through E. On the basis of the model of FtsW shown in Fig. 1, segments A and E are located in the cytosolic N and C ends of the protein, segment B is located in periplasmic loop 1/2, segment C is in intracellular loop 4/5, and segment D is in periplasmic loop 7/8. We decided to characterize segments C and D, which are more amphiphilic and presumably have more interaction potentiality than the others (see Fig. S1 in the supplemental material), and intracellular segment E. In order to determine the role of these putative protein interaction domains, mutations that decrease their amphiphilicity were selected (see Fig. S1 in the supplemental material). Arg 172 in cytoplasmic R166-F178 segment C was replaced by Ser, Arg 243 in periplasmic E240-A249 segment D was replaced by Gln, and Arg 402 in intracellular R396-Q407 segment E was replaced by Gln. These FtsW mutants are characterized below.
Intracellular R166-F178 segment C.
E. coli JLB17 was used as the host of pDML2415 bearing the gene encoding GFP-FtsW R172S (Table 2, derivative 5). The cells were grown in LBΔNaCl without IPTG. Figure 2 shows that the level of production of GFP-FtsW R172S was similar to that of the wild-type fusion protein at 28 and 42°C. As shown in Fig. 3, at the permissive temperature, cells expressing GFP-FtsW R172S grew as slightly longer rods than those expressing the wild-type fusion. GFP-FtsW R172S fluorescent signals were present in the midcell region. The cells producing the mutant grew as filaments at 42°C, and one fluorescent ring that was sometimes faint was observed at potential division sites. A similar morphology was observed with His-tagged FtsW R172S (data not shown). Calculation of the number of GFP-FtsW R172S rings per micrometer of JLB17 cell length indicated that localization of the FtsW mutant was reduced at 42°C in comparison to its localization at 28°C, suggesting that this mutant was thermosensitive (Table 3). Thus, the mutation in segment C did not affect the localization activity of the protein but affected its thermosensitivity.
TABLE 3.
Localization of GFP-FtsW derivatives and PBP3 relative to JLB17 cell lengtha
| GFP-FtsW derivative | 28°C
|
42°C
|
||||
|---|---|---|---|---|---|---|
| Length (μm) (mean ± SD) | No. of protein rings/μm (no. of cells)
|
Length (μm) (mean ± SD) | No. of protein rings/μm (no. of cells)
|
|||
| GFP-FtsW | PBP3 | GFP-FtsW | PBP3 | |||
| No GFP-FtsW | 5.66 ± 1.50 | 0 (604) | 0.05 (449) | 11.99 ± 4.61 | 0 (456) | 0 (12) |
| WTb | 5.35 ± 1.36 | 0.05 (410) | 0.06 (408) | 5.19 ± 2.30 | 0.08 (426) | 0.07 (367) |
| R172S mutant | 6.49 ± 2.20 | 0.07 (572) | 0.08 (163) | 17.17 ± 7.88 | 0.035 (164) | 0.035 (164) |
| P368A mutant | 13.25 ± 8.91 | 0.04 (150) | 0.03 (91) | 32.40 ± 19.92 | 0.02 (20) | 0.03 (37) |
| P375A mutant | 4.60 ± 1.10 | 0.06 (401) | 0.07 (319) | 7.95 ± 3.68 | 0.07 (471) | 0.07 (211) |
| P368A P375A mutant | 11.60 ± 6.54 | 0.06 (323) | 0.01 (122) | 18.64 ± 13.94 | 0.04 (144) | 0.01 (84) |
| R402Q mutant | 3.65 ± 0.87 | 0.06 (407) | 0.06 (441) | 5.90 ± 2.29 | 0.08 (421) | 0.08 (361) |
JLB17 cells were grown in LBΔNaCl, and measurements were taken at 28°C and after two mass doublings at 42°C.
WT, wild type.
Periplasmic E240-A249 segment D.
Attempts to transform E. coli JLB17 with plasmid pDML2409, encoding His-tagged FtsW R243Q, or pDML2416, encoding GFP-FtsW R243Q (Table 2, derivative 9), failed. The only transformants that were isolated contained a plasmid with mutations or deletions. The FtsW R243Q protein thus appeared to be lethal for E. coli JLB17. By contrast, E. coli LMC500 was successfully transformed with these plasmids. The cells grew as a mix of chains and normal cells in LBΔNaCl without IPTG at 28 or 42°C, whereas in the presence of IPTG, all transformants grew as long chains of rods that were prone to lysis (Fig. 4). Similar morphologies were observed when the cells grew in minimal medium. The R243Q mutant was thus dominant negative in LMC500 at both temperatures. Figure 2 shows that the mutant protein was detected only in small amounts in cells growing in the presence of IPTG and that its production level was slightly lower at 42°C than at 28°C.
FIG. 4.
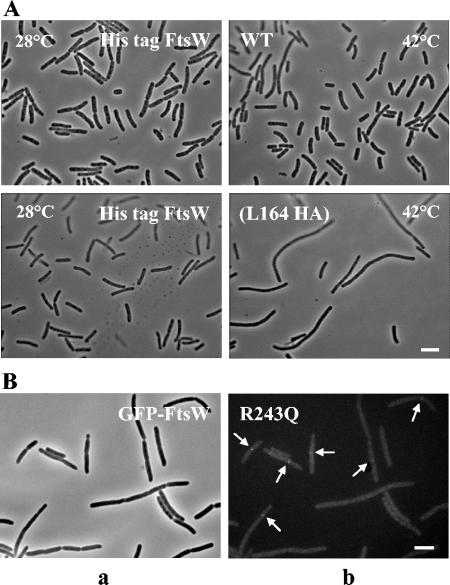
(A) Complementation activity of His-tagged FtsW(L164 HA) in E. coli JLB17. See the legend to Fig. 3 for growth conditions. The cells were observed by phase-contrast microscopy. Bar, 5 μm. (B) Localization of GFP-FtsW R243Q (arrows) in E. coli LMC500. Cells were harvested and fixed after two generation times of exponential growth at 28°C in TY medium in the absence of IPTG. Panel a is a phase-contrast micrograph, and panel b is a fluorescence micrograph. Bar, 1 μm.
As shown in Fig. 4B, a very faint fluorescent GFP-FtsW R243Q signal was observed at the septa of some cells grown in a TY rich medium without IPTG at both temperatures, indicating septal localization of the mutant protein. Hence, segment D appears to play a key role in the formation of septa and the separation of the cells.
Intracellular R396-Q407 segment E.
The R402Q mutation did not affect the production of GFP-FtsW R402Q (Table 2, derivative 3), its in vivo activity as measured by complementation in E. coli JLB17, or its localization to the division site as examined in E. coli JLB17 at 28 or at 42°C and in E. coli LMC500 at 28°C (Fig. 2 and 3).
Insertion of a nonapeptide from the HA of the influenza virus in loops 4/5 and 7/8 and the C terminus.
A small antigenic peptide epitope (YPYDVPDYA) containing part of the HA of the influenza virus was inserted in loops 4/5 and 7/8 and in the C-terminal end of the protein in order to investigate the structural and/or functional role of these regions. Using the program PEPTIDE STRUCTURE (12), insertion sites were chosen outside amphiphilic segments and conserved amino acid residues so that the epitope could be accessible to the anti-HA antibody (see Fig. S2 in the supplemental material). The epitope was inserted between L164 and V165 in loop 4/5, between E293 and A294 in loop 7/8, and between R411 and G412 in the C-terminal end (Fig. 1). His-tagged FtsW(E293 HA) (Table 1, derivative 1) and His-tagged FtsW(R411 HA) (Table 2, derivative 2) were expressed as revealed by immunoblotting and were functional as examined by complementation in E. coli JLB17 (data not shown). In contrast, His-tagged FtsW(L164 HA) (Table 2, derivative 10) could not be produced; the protein was very unstable, prone to proteolysis, and, as expected, lacked complementation activity (Fig. 4A and data not shown). These results suggest that loop 4/5 was important for proper folding of FtsW.
Role of the conserved amino acid residues P368 and P375 in periplasmic loop 9/10.
Comparison of 24 amino acid sequences of FtsW found in databases shows the presence of conserved amino acid residues in the C-terminal region of the protein (see Fig. S2 in the supplemental material). We focused on the P368TKGLTLP375 segment located in periplasmic loop 9/10 and which contained three strictly conserved residues (shown in bold in Fig. S2 in the supplemental material). In order to determine their role by increasing the flexibility of the loop, the proline residues P368 and/or P375 were changed to Ala (Table 2, derivatives 4, 7, and 8).
E. coli JLB17 transformants expressing the GFP-FtsW P368A (from pDML2418), GFP-FtsW P375A (from pDML2419), and GFP-FtsW P368A P375A (from pDML2420) mutants were grown at 28 and 42°C in LBΔNaCl without IPTG. Figure 2 shows that the three GFP-FtsW mutants were produced in amounts similar to that of the wild-type fusion at both temperatures.
As shown in Fig. 3, the FtsW P368A mutant and the double mutant exhibited a dominant negative effect in a ftsW(Ts) background (the cells grew as filaments), indicating that they each competed with the resident FtsW G311D mutant of E. coli JLB17 at the permissive temperature and prevented it from being functional. By contrast, the GFP-FtsW P375A mutant did not show such an effect and partially complemented ftsW(Ts) at 42°C. Similar results were obtained with cells producing the FtsW derivatives fused to a His tag (data not shown). The three FtsW mutants were recruited to the division site or to some potential division sites of filamentous cells at both temperatures.
The GFP-FtsW P368A mutant and the GFP-FtsW double mutant did not exhibit a strong dominant negative effect in a ftsW+ background, in contrast with the effect observed in a ftsW(Ts) background, but they induced a lengthening of LMC500 cells grown exponentially in LBΔNaCl without IPTG. This result suggests that these FtsW mutants did not compete as well with the wild-type resident FtsW mutant. At 42°C, only cells producing GFP-FtsW P368A were longer (Table 4), suggesting that the mutation of Pro 375 could cause thermosensitivity of the protein. Alteration of loop 9/10 thus did not affect the localization activity but affected another function of the protein.
TABLE 4.
Phenotypes of E. coli LMC500 producing FtsW derivatives with lesions in periplasmic loop 9/10
| GFP-FtsW derivative | Avg cell length (μm) ata:
|
|
|---|---|---|
| 28°C | 42°C | |
| WTb | 4.7 ± 1.1 (309) | 4.2 ± 1.6 (321) |
| P368A mutant | 9.5 ± 2.5 (175) | 8.5 ± 6.4 (202) |
| P375A mutant | 6.6 ± 1.6 (242) | 4.6 ± 2.6 (514) |
| P368A P375A mutant | 7.7 ± 3.5 (292) | 4.4 ± 2.3 (311) |
The numbers in parentheses are the total numbers of cells.
WT, wild type.
Recruitment of PBP3 by FtsW mutants.
To assess whether FtsW mutants were affected in their ability to recruit PBP3 to the cell division site, its localization was examined in ftsW(Ts) JLB17 transformants grown to exponential phase in LBΔNaCl at 28°C without IPTG. Samples from one part of cells were collected and fixed. The samples were imaged for the determination of the length of the cells and the GFP localization. Subsequently, the cells were permeabilized and labeled with anti-PBP3 antibodies and secondary antibodies labeled with the Alexa 546 fluorophore (see Materials and Methods). The rest of the cells were diluted four times in LBΔNaCl and incubated at 42°C for two mass doublings. The imaging and analysis procedure was repeated. The number of GFP-FtsW rings and PBP3 rings per micrometer of cells was calculated. The results are shown in Table 3. PBP3 localized at 28°C in JLB17 but did not localize at 42°C as expected, since FtsW G311D did not localize at the nonpermissive temperature. For each transformant except the one producing the GFP-FtsW P368A P375A mutant, the number of PBP3 fluorescent signals per micrometer of JLB17 was similar to that of the GFP-FtsW, GFP-FtsW R172S, GFP-FtsW P368A, GFP-FtsW P375A, or GFP-FtsW R402Q rings, indicating that these FtsW mutants were able to recruit PBP3 to the division site. In contrast, although it localized reasonably well to the potential division sites in filaments at both temperatures, the GFP-FtsW P368A P375A mutant failed to target a significant amount of PBP3 to these sites. This result suggests that at the permissive temperature, the FtsW double mutant competed with the resident FtsW G311D mutant for the preassembled division complex or recruiting factor, preventing PBP3 localization at the septum. At 42°C, the number of FtsW double mutant rings decreased, suggesting that the protein was thermosensitive as previously shown. Thus, the GFP-FtsW P368A P375A mutant is not functional because it cannot recruit PBP3 at the division site.
DISCUSSION
The FtsW protein is an essential component of the divisome. It is a late recruit to the division site and is required to target PBP3 to the midcell region. The results presented in this work allow the identification of distinct regions performing specific functions. Three regions of the protein appear to play an essential role in septum assembly.
Periplasmic loop 9/10 is involved in the recruitment of PBP3 at the division site.
The sequence of periplasmic loop 9/10 is well conserved among the known FtsW proteins. Replacement of the conserved Pro 368 residue and of both conserved Pro 368 and Pro 375 residues by Ala gives rise to proteins (Table 2, derivatives 7 and 8) that are not functional, have dominant negative effects, and are able to localize. However, both proteins differ in their abilities to recruit PBP3 to the division site. The P368A modification still allows the recruitment of PBP3, while the P368A P375A double mutation does prevent PBP3 targeting to the division site. Loop 9/10 thus appears to play a key role in the recruitment of PBP3 to the divisome. PBP3 recruitment may be mediated by protein-protein interaction. Results obtained by using an E. coli two-hybrid system showed that FtsW and PBP3 interact (13). On the other hand, it was shown that the structural determinants required to target PBP3 to the septum are present in the first 56 residues of PBP3 (44, 56). They might partly interact with loop 9/10 of FtsW.
E240-A249 segment D located in periplasmic loop 7/8 is a key element in FtsW functioning.
Substitution of Gln for Arg 243 to decrease the amphiphilicity of segment D gives rise to a protein (Table 2, derivative 9) that is not functional in vivo, that has a dominant negative effect at 28 and 42°C in a ftsW+ strain, that is produced in small amounts, and that localizes to the division site. In contrast to those of other ftsW mutants which grew as filaments, the ftsW+ cells producing the FtsW R243Q mutant grew as chains that were prone to lysis. This phenotype is similar to that of amiA, amiB, and amiC deletion mutants (20). The gene encoding FtsW R243Q is lethal for E. coli JLB17, producing thermosensitive FtsW G311D because the plasmid encoding the mutant could not be transformed to JLB17. One explanation is that in JLB17, the FtsW R243Q mutant could much better compete with the resident FtsW G311D mutant than with the wild-type protein in E. coli LMC500, preventing it from being functional. Segment D thus appears to be a key element in the functioning of FtsW, presumably by providing an essential interacting site with other components of the peptidoglycan assembly divisome machineries.
Replacement of Pro 253 close to segment D by Leu blocks cell division (28). Proline residues frequently occur near protein-protein interaction sites (30). Alteration of this Pro residue may disturb the structure of the amphiphilic segment D and thus the functioning of FtsW at the division site. However, the P253L mutation is not as deleterious as the R243Q mutation. By contrast, insertion of the HA epitope between E293 and A294 in loop 7/8 (Table 2, derivative 1) has no deleterious effect on the activity of the protein, showing that the overall structure of FtsW was not significantly altered.
Cytoplasmic loop 4/5 and TMS 8 are important determinants for the conformation of FtsW.
R166-F178 segment C is located in cytoplasmic loop 4/5. Decreasing the amphiphilicity of segment C by replacing Arg 172 with Ser results in a protein mutant (Table 2, derivative 5) that is inactive in cell division and that is thermosensitive but that localizes at the midcell region and still recruits PBP3 at the division site. In a study by Khattar et al. (29), the replacement of Glu 170 by Lys in segment C, which also decreases its amphiphilicity, causes a thermosensitive early block of cell division and makes the mutant strain hypersensitive to the overexpression of ftsZ at the permissive temperature (28). Thus, segment C is not involved in FtsW recruitment or PBP3 targeting but seems to be involved in the conformation of FtsW. The protein might be stabilized presumably through interaction of segment C with a cytoplasmic component of the division machinery.
Insertion of an antigenic peptide epitope containing part of the HA of the influenza virus at position 164 in cytoplasmic loop 4/5 is not tolerated. The protein (Table 2, derivative 10) loses its in vivo activity and is so unstable that it cannot be isolated from the producing transformants. Replacement of Pro 181 located in TMS 5 by Leu prevents cell division (28). This alteration could modify the packing in the membrane and thus alter the conformation of loop 4/5. Thus, the conformation of this loop seems to be essential for the structural integrity of the protein.
Modification of Gly 311 into Asp in TMS 8 seems to alter the structure of the protein, which becomes thermosensitive (Table 2, derivative 6). TMS 8 might be required for the correct conformation of FtsW to allow its localization. G311 in TMS 8 is conserved and is part of the GXXX(G/A) motif that is known to drive association between TMSs by favoring hydrogen bond interactions (45, 46, 47). The possibility that a dimer is formed or that an interaction with TMS of another protein occurs could not be excluded. Thus, TMS 8 seems to be an important determinant of the conformation of FtsW.
Supplementary Material
Acknowledgments
This work was supported in part by the Belgian program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Services Fédéraux des Affaires Scientifiques, Techniques et Culturelles (PAI no. P5/33), the Fonds de la Recherche Fondamentale Collective (contract 2.4521.01), the Actions de Recherche Concertées (grant 03/08-297), a Vernieuwingsimpuls grant (016.001.024; T.D.B.) from The Netherlands Organization for Scientific Research (NWO), and a European Commission grant (LSMH-CT-2003-503; COBRA). S.P., A.P., and B.W. were fellows of the Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture, Brussels. A.T. is a research director at INSERM (Paris, France). R.B. is a research director at the Fonds National de la Recherche Scientifique, Brussels.
We thank D. S. Weiss (University of Iowa, Iowa City) for the gift of plasmid pDSW234 and M. Marrec-Fairley, B. Joris, J. Coyette, and J. M. Frère for fruitful discussions.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Addinall, S. G., and J. Lutkenhaus. 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J. Bacteriol. 178:7167-7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begg, K. J., A. Takasuga, D. H. Edwards, S. J. Dewar, B. G. Spratt, H. Adachi, T. Ohta, H. Matsuzawa, and W. D. Donachie. 1990. The balance between different peptidoglycan precursors determines whether Escherichia coli cells will elongate or divide. J. Bacteriol. 172:6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle, D. S., M. M. Khattar, S. G. Addinall, J. Lutkenhaus, and W. D. Donachie. 1997. ftsW is an essential cell-division gene in Escherichia coli. Mol. Microbiol. 24:1263-1273. [DOI] [PubMed] [Google Scholar]
- 4.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol. 5:553-557. [DOI] [PubMed] [Google Scholar]
- 5.Buddelmeijer, N., and J. Beckwith. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 52:1315-1327. [DOI] [PubMed] [Google Scholar]
- 6.Buddelmeijer, N., N. Judson, D. Boyd, J. J. Mekalanos, and J. Beckwith. 2002. YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc. Natl. Acad. Sci. USA 99:6316-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. C., and J. Beckwith. 2001. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol. Microbiol. 42:395-413. [DOI] [PubMed] [Google Scholar]
- 8.Daniel, R. A., S. Drake, C. E. Buchanan, R. Scholle, and J. Errington. 1994. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J. Mol. Biol. 235:209-220. [DOI] [PubMed] [Google Scholar]
- 9.De Loof, H., M. Rosseneu, R. Brasseur, and J. M. Ruysschaert. 1986. Use of hydrophobicity profiles to predict receptor binding domains on apolipoprotein E and the low density lipoprotein apolipoprotein B-E receptor. Proc. Natl. Acad. Sci. USA 83:2295-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.den Blaauwen, T., A. Lindqvist, J. Löwe, and N. Nanninga. 2001. Distribution of the Escherichia coli structural maintenance of chromosomes (SMC)-like protein MukB in the cell. Mol. Microbiol. 42:1179-1188. [DOI] [PubMed] [Google Scholar]
- 11.den Blaauwen, T., M. E. G. Aarsman, N. O. E. Vischer, and N. Nanninga. 2003. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol. Microbiol. 47:539-547. [DOI] [PubMed] [Google Scholar]
- 12.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Lallo, G., M. Fagioli, P. Gheraldini, and L. Paolozzi. 2003. Use of a two-hybrid system to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353-3359. [DOI] [PubMed] [Google Scholar]
- 14.Eisenberg, D., R. M. Weiss, and T. C. Terwilliger. 1982. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299:371-374. [DOI] [PubMed] [Google Scholar]
- 15.Eisenberg, D., E. Schwarz, M. Komaromy, and R. Wall. 1984. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179:125-142. [DOI] [PubMed] [Google Scholar]
- 16.Errington, J., R. A. Daniel, and D.-J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figge, R. M., A. V. Divakaruni, and W. J. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321-1332. [DOI] [PubMed] [Google Scholar]
- 18.Gallet, X., B. Charloteaux, A. Thomas, and R. Brasseur. 2000. A fast method to predict protein interaction sites from sequences. J. Mol. Biol. 302:917-926. [DOI] [PubMed] [Google Scholar]
- 19.Gérard, P., T. Vernet, and A. Zapun. 2002. Membrane topology of the Streptococcus pneumoniae FtsW division protein. J. Bacteriol. 184:1925-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidrich, C., M. F. Templin, A. Ursinus, M. Merdanovic, J. Berger, H. Schwarz, M. A. de Pedro, and J. V. Holtje. 2001. Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41:167-178. [DOI] [PubMed] [Google Scholar]
- 21.Henriques, A. O., H. de Lencastre, and P. J. Piggot. 1992. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie 74:735-748. [DOI] [PubMed] [Google Scholar]
- 22.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda, M., T. Sato, M. Wachi, H. K. Jung, F. Ishino, Y. Kobayashi, and M. Matsuhashi. 1989. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 171:6375-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishino, F., H. K. Jung, M. Ikeda, M. Doi, M. Wachi, and M. Matsuhashi. 1989. New mutations fts-36, lts-33, and ftsW clustered in the mra region of the Escherichia coli chromosome induce thermosensitive cell growth and division. J. Bacteriol. 171:5523-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 26.Joris, B., G. Dive, A. Henriques, P. J. Piggot, and J. M. Ghuysen. 1990. The life-cycle proteins RodA of Escherichia coli and SpoVE of Bacillus subtilis have very similar primary structures. Mol. Microbiol. 4:513-517. [DOI] [PubMed] [Google Scholar]
- 27.Kamman, M., J. Laufs, J. Schell, and B. Gronenborn. 1989. Rapid insertion mutagenesis of DNA by polymerase chain reaction (PCR). Nucleic Acids Res. 17:5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khattar, M. M., S. G. Addinall, K. H. Stedul, D. S. Boyle, J. Lutkenhaus, and W. D. Donachie. 1997. Two polypeptide products of the Escherichia coli cell division gene ftsW and a possible role for FtsW in FtsZ function. J. Bacteriol. 179:784-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khattar, M. M., K. J. Begg, and W. D. Donachie. 1994. Identification of FtsW and characterization of a new ftsW division mutant of Escherichia coli. J. Bacteriol. 176:7140-7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kini, R. M., and J. H. Evans. 1995. A hypothetical structural role for proline residues in the flanking segments of protein-protein interaction sites. Biochem. Biophys. Res. Commun. 212:115-1124. [DOI] [PubMed] [Google Scholar]
- 31.Koppelman, C. M., M. E. G. Aarsman, J. Postmus, E. Pas, A. O. Muijsers, D. J. Scheffers, N. Nanninga, and T. den Blaauwen. 2004. R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling, and is essential for cell division. Mol. Microbiol. 51:645-657. [DOI] [PubMed] [Google Scholar]
- 32.Kruse, T., J. Moller-Jensen, A. Lobner-Olesen, and K. Gerdes. 2003. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 22:5283-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lara, B., and J. A. Ayala. 2002. Topological characterization of the essential Escherichia coli cell division protein FtsW. FEMS Microbiol. Lett. 216:23-32. [DOI] [PubMed] [Google Scholar]
- 34.Löwe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203-206. [DOI] [PubMed] [Google Scholar]
- 35.Lutkenhaus, J., and S. G. Addinall. 1997. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66:93-116. [DOI] [PubMed] [Google Scholar]
- 36.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 37.Marrec-Fairley, M., A. Piette, X. Gallet, R. Brasseur, H. Hara, C. Fraipont, J. M. Ghuysen, and M. Nguyen-Distèche. 2000. Differential functionalities of amphiphilic peptide segments of the cell-septation penicillin-binding protein 3 of Escherichia coli. Mol. Microbiol. 37:1019-1031. [DOI] [PubMed] [Google Scholar]
- 38.Matsuhashi, M., M. Wachi, and F. Ishino. 1990. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res. Microbiol. 141:89-103. [DOI] [PubMed] [Google Scholar]
- 39.Matsuhashi, M. 1994. Utilization of lipid-linked precursors and the formation of peptidoglycan in the process of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and the mechanism of their regulation. New Compr. Biochem. 27:55-71. [Google Scholar]
- 40.Mercer, K. L., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano, Y., Y. Yoshida, Y. Yamashita, and T. Koga. 1995. Construction of a series of pACYC-derived plasmid vectors. Gene 162:157-158. [DOI] [PubMed] [Google Scholar]
- 42.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piette, A., C. Fraipont, T. den Blaauwen, M. E. G. Aarsman, S. Pastoret, and M. Nguyen-Distèche. 2004. Structural determinants required to target penicillin-binding protein 3 to the septum of Escherichia coli. J. Bacteriol. 186:6110-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russ, W. P., and D. M. Engelman. 2000. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296:911-919. [DOI] [PubMed] [Google Scholar]
- 46.Senes, A., M. Gerstein, and D. M. Engelman. 2000. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J. Mol. Biol. 296:921-936. [DOI] [PubMed] [Google Scholar]
- 47.Senes, A., I. Ubarretxena-Belandia, and D. M. Engelman. 2001. The Cα-H…O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc. Natl. Acad. Sci. USA 98:9056-9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shih, Y. L., T. Le, and L. Rothfield. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl. Acad. Sci. USA 100:7865-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spratt, B. G. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 72:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoker, N. G., J. K. Broome-Smith, A. Edelman, and B. G. Spratt. 1983. Organization and subcloning of the dacA-rodA-pbpA cluster of cell shape genes in Escherichia coli. J. Bacteriol. 155:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taschner, P. E., P. G. Huls, E. Pas, and C. L. Woldringh. 1988. Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J. Bacteriol. 170:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van den Ent, F., and J. Löwe. 2000. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 19:5300-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wachi, M., M. Doi, Y. Okada, and M. Matsuhashi. 1989. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J. Bacteriol. 171:6511-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wachi, M., and M. Matsuhashi. 1989. Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J. Bacteriol. 171:3123-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss, D. S., J. C. Chen, J.-M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wissel, M. C., and D. S. Weiss. 2004. Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J. Bacteriol. 186:490-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.