Abstract
The results reported here have identified yueB as the essential gene involved in irreversible binding of bacteriophage SPP1 to Bacillus subtilis. First, a deletion in an SPP1-resistant (pha-2) strain, covering most of the yueB gene, could be complemented by a xylose-inducible copy of yueB inserted at amyE. Second, disruption of yueB by insertion of a pMutin4 derivative resulted in a phage resistance phenotype regardless of the presence or absence of IPTG (isopropyl-β-d-thiogalactopyranoside). YueB homologues are widely distributed in gram-positive bacteria. The protein Pip, which also serves as a phage receptor in Lactococcus lactis, belongs to the same family. yueB encodes a membrane protein of ∼120 kDa, detected in immunoblots together with smaller forms that may be processed products arising from cleavage of its long extracellular domain. Insertional inactivation of yueB and the surrounding genes indicated that yueB is part of an operon which includes at least the upstream genes yukE, yukD, yukC, and yukBA. Disruption of each of the genes in the operon allowed efficient irreversible adsorption, provided that yueB expression was retained. Under these conditions, however, smaller plaques were produced, a phenotype which was particularly noticeable in yukE mutant strains. Interestingly, such reduction in plaque size was not correlated with a decreased adsorption rate. Overall, these results provide the first demonstration of a membrane-bound protein acting as a phage receptor in B. subtilis and suggest an additional involvement of the yukE operon in a step subsequent to irreversible adsorption.
The interaction of a bacteriophage with the bacterial surface (adsorption) is the first step in the infection process. This step involves recognition of and binding to one or more cell envelope constituents and leads to ejection of the phage DNA from the capsid. It has long been appreciated that adsorption can proceed in two steps, one reversible step followed by irreversible commitment (1). Irreversible adsorption and ejection of DNA are possibly different descriptions of the same phenomenon, but this has not really been clarified. Similarly, DNA ejection from the capsid and injection into the cytoplasm are at best difficult to separate in vivo, except in those cases where injection itself proceeds in distinct phases, as in T5 (13).
In the gram-negative world, several phage receptors have been identified, and in a number of cases (e.g., T4, T5, and T7 of Escherichia coli), the adsorption and injection processes are now beginning to be understood at a detailed molecular level (14, 18, 26). Much less is known about the first steps of infection for phages preying on gram-positive bacteria. Early work has established that glucosylated polyglycerol phosphate, the major and essential teichoic acid in Bacillus subtilis, serves as a receptor for several phages, including φ29, φ25, and SP01 (24, 29, 34, 36). Mutations leading to a lack of glucosylation of this polymer block the phage adsorption process and plaque-forming ability. More recently, selection for B. subtilis mutants resistant to bacteriophage φ3T led to the identification of another type of cell wall receptor, a minor cell wall anionic polymer, poly-(glucosyl-N-acetylgalactosamine 1-phosphate) (6).
Interestingly, no membrane protein has yet been implicated in phage binding to B. subtilis, in contrast to the evidence from gram-negative systems. Nevertheless, phage infection of B. subtilis protoplasts from both wild-type and phage-resistant strains has been reported, suggesting the presence of membrane-linked receptors (9, 10). In Lactococcus lactis, Geller et al. have established that phages of the c2 group adsorb first reversibly to a cell wall polymer and then irreversibly to a membrane protein, which was designated Pip (for phage infection protein) (7). To our knowledge, this is the only instance where a membrane receptor for a phage infecting a gram-positive bacterium has been documented.
Twenty years ago, a B. subtilis phage resistance mutation (pha-2) blocking irreversible binding of phage SPP1 was described (29). pha-2 mutants retained the capacity to adsorb phages at a wild-type rate, but phage-host complexes could be readily separated by dilution, showing the process to be wholly reversible. The mutation was found to be specific for SPP1 and related phages and was mapped, by PBS1-mediated transduction, in the vicinity of the ald-1 marker (29, 30).
Based on the map position of the pha-2 locus and the availability of the complete sequence of the B. subtilis genome, we have now attempted to identify the gene required for SPP1 irreversible adsorption. The experiments described here identify yueB, an orthologue of pip, as the key gene involved in irreversible adsorption of SPP1. yueB and upstream genes are organized as an operon, the products of which may contribute additionally to efficient SPP1 infection. Nonetheless, only YueB appears to be required for irreversible binding of the phage. This is the first demonstration of a membrane-bound protein acting as a phage receptor in B. subtilis. Furthermore, it strengthens the notion that YueB/Pip-like proteins, which are widely distributed among gram-positive species, may be generally used as bacteriophage receptors.
MATERIALS AND METHODS
Bacterial strains, phages, plasmids, and growth conditions.
The bacterial strains, plasmids, and phages used in this work are listed in Table 1. E. coli and B. subtilis strains were usually grown in Luria-Bertani (LB) medium (27) with aeration at 37°C, except for E. coli transformants harboring pMutin4 derivatives, which were selected and propagated at 30°C. Agar was added to LB medium to a final concentration of 0.7 or 1.5% (wt/vol) in order to obtain top or bottom LB plates, respectively. Media were supplemented with 10 mM CaCl2 for phage propagation. E. coli strains carrying vectors or recombinant plasmids were grown in the presence of ampicillin (100 μg/ml) and/or kanamycin (40 μg/ml), while neomycin (7.5 μg/ml) or erythromycin (0.5 μg/ml) was used for the selection of B. subtilis transformants. B. subtilis transformants expressing β-galactosidase were selected in X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-supplemented (0.02%, wt/vol) LB plates. SPP1 and SP01 propagation was as described by Santos et al. (29).
TABLE 1.
Bacterial strains, phages, and plasmids used in this work
| Bacterial strains, phages, and plasmids | Genotype or relevant features | Source or reference |
|---|---|---|
| E. coli strains | ||
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′[traD36 proAB+lacIqlacZΔM15] | Stratagene |
| MC1061 | araD139 Δ(ara leu)7697 ΔlacX74 galU galK hsdR2 strA mcrA mcrB1 | 3 |
| CG61 | BL21 derivative carrying pGP1-2 | 31 |
| CG65 | CG61 derivative carrying pCSJ65 | This work |
| B. subtilis strainsa | ||
| L16601 | B. subtilis 168; wild-type strain; SPP1 indicator strain | 16 |
| CSJ1 | pha-2 Δ6 | This work |
| CSJ2 | pha-2 | This work |
| CSJ3 | yueBΩpCSJ42; yueB conditional mutant | This work |
| CSJ4 | CSJ1 derivative with gene eryR inserted in the amyE locus | This work |
| CSJ6 | CSJ4 derivative with yueB inserted in the amyE locus | This work |
| CBM6 | yukEΩpCBM6 | This work |
| CBM7 | yukDΩpCBM7 | This work |
| CBM8 | yukCΩpCBM8 | This work |
| CBM9 | yukBAΩpCBM9 | This work |
| CBM10 | yueBΩpCBM10 | This work |
| CBM11 | yueCΩpCBM11; yueC conditional mutant | This work |
| B. subtilis phages | ||
| SPP1 | 25 | |
| SP01 | 20 | |
| Vectors and plasmids | ||
| pIVEX2.3d | Expression vector; allows production of His-tagged fusion proteins; Ampr | Roche Applied Science |
| pGR40 | Integration vector; allows insertion of genes under the control of PxylA in the amyE locus of B. subtilis; Ampr Neor | G. Real and A. O. Henriques, unpublished data |
| pMutin4 | Integration vector used for gene inactivation; Ampr Eryr | 32 |
| pCSJ65 | pIVEX2.3d derivative expressing YueB780 | This work |
| pCSJ42 | pMutin4 derivative carrying a PCR product covering the RBSb and 5′ end of yueB | This work |
| pCBM6 | pMutin4 derivative carrying a PCR product internal to yukE | This work |
| pCBM7 | pMutin4 derivative carrying a PCR product internal to yukD | This work |
| pCBM8 | pMutin4 derivative carrying a PCR product internal to yukC | This work |
| pCBM9 | pMutin4 derivative carrying a PCR product internal to yukBA | This work |
| pCBM10 | pMutin4 derivative carrying a PCR product internal to yueB | This work |
| pCBM11 | pMutin4 derivative carrying a PCR product covering the RBS and 5′ end of yueC | This work |
All B. subtilis strains used in this work are L16601 derivatives.
RBS, ribosome binding site.
General recombinant-DNA techniques.
B. subtilis genomic DNA was obtained from 4-ml culture samples. Cells were recovered in 0.2 ml of TEG buffer (27) supplemented with 2 mg of fresh lysozyme/ml, and after an incubation period of 10 min at 37°C, the cells were lysed by the addition of 0.6 ml of GES-β (23). The lysate was then extracted twice with a phenol-chloroform-isoamyl alcohol mixture (25:24:1) and once with a chloroform-isoamyl alcohol solution (24:1) before DNA precipitation with 2-propanol. DNA was finally resuspended in Tris-EDTA buffer (27) supplemented with 10 μg of RNase A/ml. Restriction endonuclease digestions, DNA ligations, and conventional agarose gel electrophoresis were performed essentially as described by Sambrook and Russel (27). PCR amplification of DNA fragments was carried out in a RoboCycler Gradient 96 thermocycler (Stratagene) using Taq DNA polymerase (Invitrogen), ExTaq long-range polymerase (TaKaRa Biomedicals), or Pfu polymerase (Stratagene) as recommended by the suppliers. All the oligonucleotides used in this work were purchased from Invitrogen and are listed in Table 2. When needed, DNA products resulting from restriction endonuclease cleavage or PCR amplification were extracted from agarose gels and purified using the JETquik gel extraction spin kit (Genomed). Highly specific PCR products were directly purified from the PCR mixtures using the JETquik PCR purification spin kit (Genomed). Plasmid DNA extraction and purification were carried out using the JETquik miniprep extraction kit (Genomed). Transformations of E. coli and B. subtilis with recombinant plasmids were performed essentially as described by Chung et al. (4) and Yasbin et al. (35), respectively.
TABLE 2.
Oligonucleotides used in this work
| Oligo-nucleotide | Sequence (5′→3′)a |
|---|---|
| yueB-4 | GAGTGCTccatggCAGAACAACG |
| yueB-5 | CACTCAGcccgggGGTACCAGACAGCCC |
| yueB-25 | TATactagtCTGAAAGGGAGAGTGCTAGATG |
| yueB-2 | TTAtctagaTCACGCTTCATACGTTTCATCGC |
| yueB-22 | ATGgaattcTGAAAGGGAGAGTGCTAGATG |
| yueB-23 | GAAggatccATTCGAGCGTTGCCTTTTGAG |
| yukE-3 | TGAgaattcTGGCAGGATTAATTCGTGTCAC |
| yukE-4 | TGggatccGAAGGTTTGAGCTGCTCGTATT |
| yukD-3 | ACAgaattcTTGAAACATTATAACGGCAGTG |
| yukD-4 | CACggatccCTTGTTCACCACTCTGATCC |
| yukD-5 | TCTgaattcTGAAAAGCATGTGGGAAGGTG |
| yukD-6 | TCAggatccCTGCCGTTATAATGTTTCAAATC |
| yukC-3 | TCAgaattcAATCGATGTGACGGATGATGA |
| yukC-4 | GTCggatccAAACGCAAAAGCGCCGTCAA |
| yukBA-3 | TCAgaattcAAATCTGTCTAACCTCCCGTC |
| yukBA-7 | GAGggatccACAATTTCATCCCGATTTCCCG |
| yueB-27 | TGCgaattcACAGAACAACGAAAAAGCTTGAT |
| yueC-4 | GTAgaattcTGGGCACTTGTTTACAATGGG |
| yueC-3 | TGAggattcCAAATCTTTTATCAGCTCGGCG |
| pMutin-1 | TTCTACATCCAGAACAACCTC |
| lacZR1 | GTGCTGCAAGGCGATTAAGTT |
Boldface lowercase letters indicate endonuclease restriction sites.
yueB insertion in the B. subtilis amyE locus.
A DNA fragment corresponding to the yueB coding sequence and its translation signals was amplified by PCR (primer pair yueB-25/yueB-2 [Table 2]), double digested with SpeI and XbaI, and ligated to the equally digested vector pGR40 (G. Real and A. O. Henriques, unpublished data). pGR40 carries the 5′ and 3′ portions of the B. subtilis amyE gene flanking a neomycin resistance gene and an upstream xylose-inducible promoter (PxylA). In the described procedure, yueB is placed under the control of PxylA and upstream of the Neor marker. The amyE segments are oriented in the direction opposite to yueB. B. subtilis CSJ4 competent cells were directly transformed with the ligation mixture, and integrants resulting from the double crossover between the amyE segments were selected by marker replacement (neomycin resistance and erythromycin sensitivity; CSJ4 carries an erythromycin resistance gene inserted at amyE, which is lost upon recombination).
Inactivation of coding sequences in the B. subtilis genome.
DNA fragments ranging from ∼150 to 400 bp and corresponding to internal regions of the open reading frames (ORFs) yukE, yukD, yukC, yukBA, and yueB were PCR amplified (using primer pairs yukE-3/yukE-4, yukD-3/yukD-4, yukC-3/yukC-4, yukBA-3/yukBA-7, and yueB-27/yueB-23, respectively), double digested with EcoRI and BamHI, and ligated to similarly digested pMutin4 (33). The resulting recombinant plasmids were obtained in E. coli strain MC1061 (Table 1) and then used to transform B. subtilis L16601. The integrants (CBM6, CBM7, CBM8, CBM9, and CBM10 [Table 1]) resulting from Campbell-type recombination between the chromosomal loci and the cloned sequences were selected for erythromycin resistance and blue color in X-Gal-LB plates. Gene disruption resulting from each integration event was confirmed by PCR using pMutin4-specific primers (pMutin-1 and lacZR1) and primers flanking the target loci. A similar strategy was used to construct yueB and yueC conditional mutants. In these cases, the PCR products (obtained with primer pairs yueB-22/yueB-23 and yueC-4/yueC-3, respectively) covered the translation signals and the 5′ end of each ORF. In the resulting strains (CSJ3 and CBM11 [Table 1]), the genes are separated from their natural transcriptional elements and placed under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter Pspac.
B. subtilis RNA extraction and Northern blot analysis.
B. subtilis total RNA was prepared from 20-ml culture samples (optical density at 600 nm [OD600], 0.8) using the TRIzol reagent (Invitrogen) according to the supplier's instructions, except that the cells were recovered in TE buffer supplemented with 10 mg of fresh lysozyme/ml, incubated for 5 min at 37°C, and lysed by the addition of 5 ml of the reagent. RNA molecules were separated in 1% RNase-free agarose gels prepared in NBC buffer (50 mM boric acid, 1 mM sodium citrate, 5 mM NaOH, pH 7.5) containing 1% formaldehyde and 0.25 μg of ethidium bromide/ml and were run in the same buffer. Before gel loading, RNA samples (10 to 15 μg in up to 5 μl) were supplemented with 2 μl of 10× NBC buffer, 3 μl of 37% formaldehyde, and 10 μl of formamide, followed by a denaturing step of 5 min at 65°C. Standard RNA markers were from Invitrogen. Northern blotting was carried out essentially as described by Sambrook and Russel (27), with 32P-labeled probes prepared with a Ready-To-Go DNA Labeling Beads (minus dCTP) kit (Amersham Pharmacia Biotech). After RNA integrity was checked by staining the RNA-carrying nylon membranes with a solution of 0.03% (wt/vol) methylene blue-0.3 M sodium acetate (32), the membranes were incubated in a prehybridization solution (5× Denhardt's solution, 6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.5% sodium dodecyl sulfate [SDS], and 0.1 mg of salmon sperm single-stranded DNA/ml) for 60 min at 68°C. The prehybridization solution was then replaced by the hybridization solution (2× Denhardt's solution, 6× SSC, and 0.1% SDS). After the addition of the denatured 32P-labeled probes, hybridization occurred overnight at 60°C. Unattached probes were discarded with two washes of 10 min each at 60°C with a 5× SSC-0.1% SDS solution, followed by two to four washes of 10 min each with a 2× SSC-0.1% SDS mixture at room temperature. After a brief wash with 2× SSC, the membranes were autoradiographed.
Determination of SPP1 adsorption constants.
One-milliliter samples of B. subtilis cultures grown at an OD600 of 0.8 were supplemented with 15 mM MnCl2 and maintained at 37°C. Mn2+ ions fulfill the role of Ca2+ in phage adsorption, with the advantage of inhibiting further B. subtilis growth and SPP1 intracellular development (28). The cells were infected with ∼107 PFU/ml and diluted 100-fold in TBT buffer (2) supplemented with 10% chloroform at different times after infection. After being vigorously vortexed for 5 s, the mixtures were allowed to equilibrate for 5 min at room temperature and were centrifuged (15,000 × g; 5 min), and the supernatants were recovered for the enumeration of free phages (unadsorbed plus reversibly bound virions). For evaluation of irreversible-adsorption rates, phage titers in the supernatants were plotted as a function of time, and the slopes of the resulting curves were determined [ln (P0/P)/Δt, where P0 is the initial phage titer and P is the phage titer in the supernatant after a Δt period]. Adsorption constants (Kads) are the ratio between adsorption rates and bacterial cell mass, here expressed as optical density values (OD600).
Measurements of β-galactosidase activity.
β-Galactosidase activity was determined essentially as described by Miller (17) from 1- or 10-ml samples of liquid cultures of B. subtilis strains grown to late log phase.
Production of B. subtilis membrane vesicles and protein extracts.
Cell cultures (200 ml; OD600, 0.8) were collected by centrifugation and concentrated 50-fold in prechilled lysis buffer (50 mM Tris · Cl, 300 mM NaCl, pH 8.0) supplemented with 2 mg of lysozyme/ml, 1 mM phenylmethylsulfonyl fluoride, and 5% (vol/vol) Complete Mini EDTA-free Protease Inhibitor Cocktail (Roche Applied Science). The cells were then disrupted by at least three freeze-thaw cycles (liquid nitrogen and a 37°C water bath) and homogenized with an ultrasonic processor (UP 200 S; Dr. Hielscher, GmbH) by performing three cycles of 45 s each at 60% power and 0.5 pulse, intercalated with 1 min on ice. Following a centrifugation step at 10,000 × g for 10 min at 4°C, the supernatants were further subjected to ultracentrifugation (120,000 × g; 90 min; 4°C). The membrane-enriched pellets were resuspended in 0.2 ml of lysis buffer for immunoblot analysis.
Anti-YueB antibodies and immunoblot analysis.
A rabbit polyclonal serum raised against a hydrophilic portion of YueB (see below) was prepared at the Centre National de la Recherche Scientifique facilities, Gif-sur-Yvette, France. The protein used for antibody production corresponds to a histidine-tagged version of YueB (YueB780) that includes amino acids from positions 30 to 797 of its sequence. To obtain this recombinant protein, the corresponding sequence of yueB, obtained by PCR with the primer pair yueB-4/yueB-5, was cloned into pIVEX2.3d (Table 1) after digestion of both the product and the vector with the enzymes NcoI and XmaI. The resulting plasmid (pCSJ65) was used to transform E. coli strain CG61, which produces T7 RNA polymerase upon temperature upshift (from 28 to 42°C) (31). This constructed strain was used to overproduce YueB780, which was then purified from E. coli extracts by affinity chromatography (C. São-José and S. Lhuillier, unpublished data). For immunoblot analysis, ∼30 μg of total protein was analyzed by electrophoresis on SDS-8% polyacrylamide gel electrophoresis gels (12). The resolved proteins were then blotted onto nitrocellulose membranes (Bio-Rad) using the Trans-Blot SD Transfer Cell Apparatus (Bio-Rad). Detection of protein-anti-YueB780 complexes was carried out with the Chemiluminescence Western Blotting kit (Roche Applied Science), according to the manufacturer's instructions and using a serum dilution of 1:30,000.
Bioinformatic analyses.
Protein homology searches were carried out by PSI-BLAST analysis (http://www.ncbi.nlm.nih.gov). Conserved and functional domains were identified with RPS-BLAST (http://www.ncbi.nlm.nih.gov) and MotifScan (http://hits.isb-sib.ch/cgi-bin/PFSCAN). The public-domain SignalP version 2.0 (http://www.cbs.dtu.dk/services) was used for the prediction of signal peptides and cleavage site positions, while transmembrane segments and membrane topology were inferred from analysis with TMHMM (also available at http://www.cbs.dtu.dk/services). Multiple sequence alignments of protein sequences were performed with CLUSTALW (http://npsa-pbil.ibcp.fr). PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred) was used for prediction of protein secondary structures.
RESULTS
yueB as a putative SPP1 receptor gene.
Mutants of B. subtilis 168 that allowed reversible adsorption but not infection by bacteriophage SPP1 were previously described (29). All mutations were mapped to a single locus, designated pha-2, which was shown to be located near the ald-1 marker, as inferred from the high cotransduction values (ca. 95%) obtained by PBS1-mediated transduction. Irreversible adsorption of SPP1 (impaired in pha-2 strains) was further shown to require a protease-sensitive, heat-labile component associated with protoplasts and membrane fractions (28). Taking advantage of the availability of the genome sequence of B. subtilis (Z99120) (11), we have now examined the region surrounding the gene ald (coding for l-alanine dehydrogenase), looking for genes that might code for membrane proteins. The yueB gene, located ∼9 kb upstream of ald (Fig. 1A) but transcribed in the opposite direction, was identified as a good candidate for a phage receptor gene. Its putative product (1,076 residues; ∼120 kDa) is an orthologue of Pip, a membrane protein previously shown to be required for infection of L. lactis by phage c2 (7, 19). The yueB product is predicted to insert in the membrane through one N-terminal and five C-terminal transmembrane domains (TMDs), with most of its sequence (>800 residues) facing the wall compartment, a membrane topology which is also expected for Pip (Fig. 1B).
FIG. 1.
(A) Gene organization of the B. subtilis chromosome region containing yueB. In databases, yukBA (superscript 1) appears to be divided into two different genes (yukB and yukA) due to a sequencing error (see the text). Inverted repeats with the potential to form stem-loop-type structures (ΔG ≤ −10.5) are indicated by circles. Bs-2 and Bs-9 represent primers used for screening deletion events in pha-2 strains. The region covered by the deletion Δ6 found in strain CSJ1 is boxed. The KpnI restriction sites in boldface were used to clone a PCR product harboring the Δ6 deletion boundaries. The numbers of predicted TMDs of putative membrane products are indicated. (B) Predicted membrane topology of Pip and YueB proteins. Putative TMDs are depicted as black rectangles. Coordinates of the amino acid (aa) residues that are expected to face the cell wall are indicated in parentheses.
As a first attempt to correlate SPP1 resistance with defects in yueB, we screened a collection of pha-2 mutants by PCR, using primers amplifying, in the wild-type genome, the region covering yueB and upstream sequences (Fig. 1A). We observed that for one of the mutants (CSJ1), the amplified product lacked >3 kb of DNA compared to the wild type, indicating the occurrence of a deletion event in the corresponding strain (not shown). The deleted region in strain CSJ1 was delimited in a first step by restriction analysis of the PCR product. Subsequently, the deletion ends were identified at the sequence level by cloning and sequencing a KpnI DNA fragment contained in the PCR product (Fig. 1A). The deletion, which we named Δ6, covers 3,467 bp and seems to result from a recombination event involving the short repeated sequence 5′-GACTGGCG-3′ that flanks the abolished region (with the 5′ end at positions 53159 and 56626 in Z99120). In strain CSJ1 (Δ6), most of yueB and also the 3′ end of the upstream gene are lacking (Fig. 1A). It should be noted that this particular gene (yukA) (21) in fact constitutes a single open reading frame, together with the upstream gene yukB. This conclusion was first suggested by the homology of yukB and yukA products to the N- and C-terminal portions of the same proteins (e.g., the FtsK/SpoIIIE-like protein BH0975 of Bacillus halodurans; NP_241841). Sequencing a PCR product containing the yukB-yukA junction region revealed that a cytosine was missing in the submitted sequence between positions 75648 and 75649 (Z99120), causing a frameshift and the occurrence of a premature stop codon. In this work, the complete gene resulting from the fusion of the previously reported ORFs is designated yukBA.
Ectopic expression of yueB complements the Δ6 mutation with respect to SPP1 sensitivity.
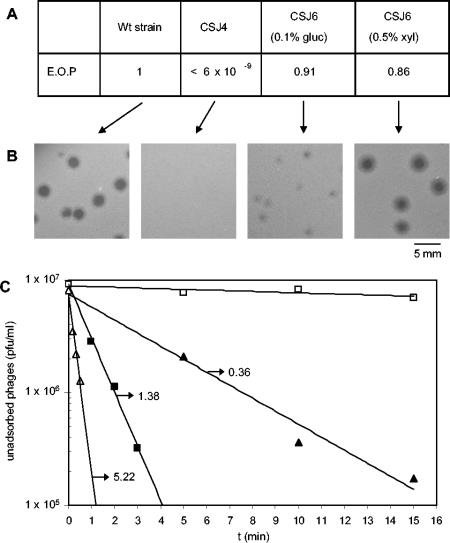
The Δ6 mutation observed in strain CSJ1 supported the involvement of YueB in the irreversible adsorption of SPP1 to B. subtilis cells. However, a possible role of yukBA in the process could not be excluded because the deletion also affected that gene. To ascertain that the CSJ1 resistance phenotype resulted exclusively from the lack of YueB, we tested the complementation of the Δ6 mutation by a xylose-inducible yueB copy inserted at the amyE gene (strain CSJ6; see Materials and Methods). In strain CSJ6, transcription of yueB is expected to be repressed by the native B. subtilis XylR repressor and activated in the presence of xylose. CSJ6 was thus tested for sensitivity to SPP1 infection under repressed (0.1% glucose) or induced (0.5% xylose) conditions. In the presence of xylose, CSJ6 behaved essentially like the wild-type strain, as it allowed normal efficiency of plating and SPP1 plaque morphology (Fig. 2A and B). This result seems to rule out a significant role of yukBA in SPP1 adsorption. SPP1 was also able to plate in CSJ6, even under repressed conditions, although smaller phage plaques were produced. This suggests that leaky expression of yueB can occur in the absence of xylose.
FIG. 2.
Complementation of the Δ6 mutation. (A) Efficiencies of plating (E.O.P) of phage SPP1 in strain CSJ4 (Δ6) and in its derivative, CSJ6 (carrying a xylose-inducible yueB copy in the amyE locus), under repressed (0.1% glucose [gluc]) or induced (0.5% xylose [xyl]) conditions. Wt, wild-type. (B) Phage plaque morphologies in the different strains. (C) Kinetics of SPP1 irreversible adsorption to the wild-type strain (▪), strain CSJ4 (□), and strain CSJ6 under repressed (▴) or induced (▵) conditions. The adsorption constant values (min−1 · UOD−1) obtained from each curve are indicated, except for strain CSJ4, for which no significant SPP1 adsorption could be measured.
To correlate the SPP1 plating phenotypes with YueB receptor activity, SPP1 irreversible adsorption was quantified and expressed as adsorption constants (Kads). These basically reflect the initial rate of irreversible adsorption per unit of cellular optical density (UOD). The Kads value obtained with strain CSJ6 grown in xylose-supplemented medium (5.22 min−1 · UOD−1) was ∼3.5-fold higher than that observed for the wild-type strain (1.38 min−1 · UOD−1) and 14.5-fold higher than that observed with strain CSJ6 grown under repressed conditions (0.36 min−1 · UOD−1). These results (Fig. 2C) are in agreement with plating data, and they further suggest that irreversible adsorption of SPP1 can be modulated by YueB levels (see below).
yueB and surrounding genes: evidence for an operon organization.
The complementation of the Δ6 mutation accomplished by the strategy described above indicates that yueB is a B. subtilis gene essential for SPP1 irreversible adsorption and infection. However, the gene organization around yueB suggests an operon structure, and the complementation experiments alone would not rule out the involvement of other genes of this putative operon in the SPP1 binding and infection process.
Gene organization of the region surrounding yueB and analysis of its nucleotide sequence suggested that at least the genes yukD to yueB, delimited by putative hairpin-like secondary structures (Fig. 1A), could constitute an operon. To test the putative operon organization of yueB and upstream genes, we constructed several strains in which each gene was individually disrupted. For that purpose, PCR products corresponding to parts of the genes yukE to yueC were cloned in pMutin4, an integration vector specially designed for systematic inactivation of coding sequences in the B. subtilis genome (32). This vector has three major properties that make it suitable for our purposes. First, upon integration, the targeted gene becomes disrupted but its 5′ end is transcriptionally fused to spoVG-lacZ, allowing quantification of its native expression. Secondly, strong transcriptional terminators disable native transcription downstream of the integration site. Finally, the presence of the IPTG-inducible promoter Pspac downstream from the terminator region allows the regulated expression of genes downstream from the recombination site, thus bypassing eventual polar effects resulting from the integration event.
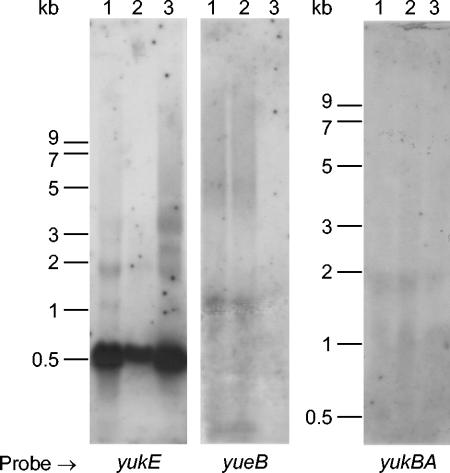
In none of the constructs, except for the one with integration at yueC, could SPP1 plate efficiently in the absence of IPTG (Fig. 3). In contrast, SPP1 could plate at near-wild-type efficiency in all integrants in the presence of IPTG, except for the strain in which yueB was disrupted. Overall, these results indicate that yueB is the sole gene in the examined cluster which is absolutely necessary for plaque formation and that the effect of disrupting upstream genes results from a polar effect on yueB expression. Therefore, these experiments also establish that at least the genes from yukE to yueB are organized as an operon. Interestingly, however, β-galactosidase activity values measured in these strains revealed a much higher expression of the first gene (yukE) than of the others (Fig. 3). In Northern blots, a probe internal to yukE showed a major band with a strong intensity signal ∼600 nucleotides in size, while probes further down the operon failed to detect any clear-cut band. Instead, a smear was apparent, starting at the tops of the gels (Fig. 4), suggesting degradation of a high-molecular-mass mRNA species. Inspection of the region between yukE and yukD showed an inverted-repeat sequence that may form a hairpin secondary structure (Fig. 1A). This structure may function as an attenuator and limit read-through of transcription initiated at the yukF/yukE intergenic region. Secondary structures flanking yukE may also be involved in transcript stabilization (see Discussion).
FIG. 3.
Kads values, efficiencies of plating (E.O.P.), and β-galactosidase activities measured in the wild-type strain (L16601) and in the different integration mutants in the absence or presence of 1 mM IPTG. A schematic representation of the relevant DNA structure of each integrant is provided (genes not drawn to scale). Disrupted ORFs are represented by interrupted white arrows, while intact genes are depicted as grey arrows. pMutin4-derived elements (spoVG-lacZ, transcriptional terminators, and Pspac promoter) are in black. Putative hairpin-like secondary structures are also indicated (open circles). The indicated Kads and β-galactosidase activity values are the averages from at least three independent experiments ± standard deviations of the mean, except under conditions where no accurate Kads measurements (≤0.01) could be performed. E.O.P. values are expressed as the ratio between the phage titer obtained in each mutant strain and that obtained in the wild-type strain (L16601). (*), minute phage plaques could be observed, suggesting an E.O.P almost identical to that observed in the presence (+) of IPTG, but their reduced size did not allow accurate counting.
FIG. 4.
Northern blot analysis of RNA extracted from B. subtilis strains. Lanes: 1, L16601 (wild-type strain); 2, CSJ2 (pha-2 strain); 3, CSJ1 (pha-2 Δ6 strain). Radiolabeled probes correspond to internal PCR products of the indicated genes. The yueB probe is included in the DNA segment deleted in Δ6.
Irreversible adsorption rates versus plaque morphology.
Even though SPP1 could plate efficiently in all but yueB integrants in the presence of IPTG, differences in plaque size were evident and particularly noticeable when yukE was disrupted (Fig. 5). The observed smaller plaques might suggest lower rates of irreversible adsorption and a contribution of other genes in the operon to SPP1 binding properties. However, when adsorption constants were evaluated in liquid medium supplemented with IPTG, all knockouts in genes upstream from yueB resulted in similar Kads values, ∼2- to 3-fold higher than that obtained for the wild-type strain (Fig. 3). Smaller plaques in these cases are thus not correlated with a decrease in irreversible binding. Although plaque size can be influenced by the host metabolism, we found no difference in the growth rates of the strains. Furthermore, the unrelated phage SP01 formed indistinguishable plaques in all integrants in both IPTG-supplemented and unsupplemented media. The reported effect on plaque size is thus specific for SPP1 (see Discussion).
FIG. 5.
SPP1 plaque morphology when plated in the wild-type strain and in the different integration mutants in the absence (−) or presence (+) of 1 mM IPTG.
YueB expression levels correlate with irreversible-adsorption rates.
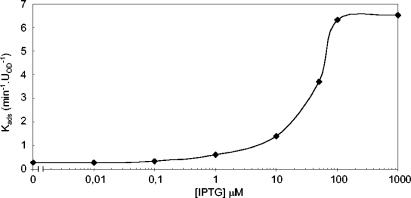
The complementation experiments reported above, as well as the Kads values obtained with the different mutants of the yukE operon grown in IPTG-supplemented medium, suggested a correlation between increased expression of yueB and irreversible-adsorption levels. To directly test this idea, we engineered a strain in which the Pspac promoter was located immediately upstream of yueB. For that purpose, we cloned in vector pMutin4 a PCR product of 409 bp covering the translation signals and the first 391 bp of yueB. The resulting plasmid (pCSJ42) was used to transform the wild-type strain, yielding strain CSJ3. We determined the Kads values for CSJ3 grown in the presence of different IPTG concentrations, and the results show that in the range of 0.1 to 100 μM IPTG, the rate of adsorption increases as the inducer concentration rises (Fig. 6). The irreversible binding of SPP1 to noninduced cultures was also measurable, although very slow, in this case, reflecting some leakiness of the Pspac promoter. At the calculated value of Kads (0.28 min−1 · UOD−1), small plaques were still visible on solid medium (data not shown).
FIG. 6.
SPP1 irreversible-adsorption constants measured with strain CSJ3 (which carries an IPTG-inducible yueB) as a function of IPTG concentration.
YueB species in B. subtilis membrane fractions.
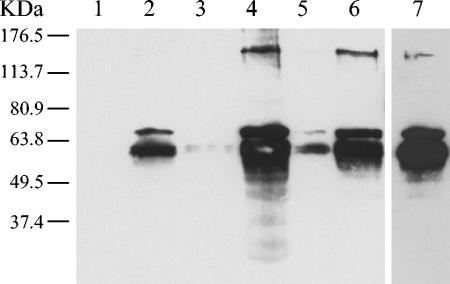
As already mentioned, analysis of the YueB primary sequence clearly indicates that it is an integral membrane protein. We checked YueB production in B. subtilis cells by Western blot analysis using a polyclonal serum raised against its extensive hydrophilic portion (see Materials and Methods) (Fig. 1B). The expected 120-kDa band corresponding to YueB was not detectable in samples from total protein extracts produced from cultures of the wild-type strain (data not shown). This probably results from the low levels of yueB expression inferred from the β-galactosidase activity measurements and Northern blotting results reported above (Fig. 3 and 4). Given its expected membrane location, we sought to detect YueB in extracts enriched for membrane vesicles arising upon culture sonication (see Materials and Methods). Even in these extracts, the expected 120-kDa protein could be easily observed only with material from strains overproducing YueB (CSJ6 in the presence of xylose and CSJ3 grown in the presence of IPTG) (Fig. 7). However, the most obvious signals in all preparations corresponded to two to four bands in the molecular mass range of 50 to 70 kDa (Fig. 7 and results not shown), indicating substantial processing of the SPP1 receptor protein.
FIG. 7.
Western blot analysis of YueB polypeptides in membrane-enriched protein extracts (see Materials and Methods) obtained from different B. subtilis strains. Lanes: 1, CSJ1 (Δ6); 2, L16601 (wild-type strain); 3, CSJ3 (no IPTG); 4, CSJ3 (1 mM IPTG); 5, CSJ6 (0.1% glucose); 6, CSJ6 (0.5% xylose); 7, overexposed lane 2. The polyclonal serum anti-YueB780 was used at a 1:30,000 dilution.
YueB-like proteins in other bacterial species.
A protein database search allowed us to retrieve 56 YueB homologues sharing a membrane topology identical to that predicted for YueB (Fig. 1B). There is little overall similarity among the various proteins at the level of the primary sequence, except in the N-terminal region, where several conserved residues are apparent (Fig. 8). It remains to be established whether such residues play a role in phage binding. The hydrophilic portion between the putative transmembrane domains is predicted to be composed of one or a few long helical structures, as determined by the PSIPRED algorithm. YueB-like proteins seem to be exclusively produced by gram-positive bacterial species and fall into two groups according to their interrelatedness. In fact, some are more closely related to YueB, while others show higher similarity to the product of yhgE, a B. subtilis yueB paralogue. The bacteria listed in Table 3 have one to five copies of these yueB/yhgE-like genes, and some of them are located near homologues of the B. subtilis yuk cluster (yukE to yukBA). We speculate that some of these YueB-like proteins may eventually function as receptors for phages infecting these bacteria, similar to what happens in B. subtilis (this work) and in L. lactis (7).
FIG. 8.
Sequence alignment of a conserved region in YueB-like proteins (bacterial origin provided on the left). A group of 30 sequences, representing the primary structure diversity of this protein family, were compared using the CLUSTALW tool (a representative alignment with only 12 sequences is shown). The positions showing a conservation level of >70% in the 30 sequences analyzed are boxed, while positions with identical, strongly similar, and weakly similar residues are indicated by asterisks, colons, and periods, respectively. The numbers in parentheses refer to the coordinates of each of the regions relative to the whole protein sequence. Predicted secondary structures (α-helices and β-strands) of sequence segments are indicated above the alignment. Protein accession numbers: 1, NP_388897.1; 2, NP_243012.1; 3, NP_267826.1; 4, NP_469996.1; 5, NP_276964.1; 6, NP_781183.1; 7, NP_469397.1; 8, NP_831846.1; 9, NP_692233.1; 10, NP_391064.1; 11, NP_293801.1; 12, NP_765785.1. Bacteria: 1, Bacillus subtilis; 2, Bacillus halodurans; 3, Lactococcus lactis; 4, Listeria innocua; 5, Methanothermobacter thermautotrophicus; 6, Clostridium tetani; 7, L. innocua; 8, Bacillus cereus; 9, Oceanobacillus iheyensis; 10, B. subtilis; 11, Deinococcus radiodurans; 12, Staphylococcus epidermidis.
TABLE 3.
YueB homologues sharing the same predicted membrane topology
| Bacterial species | YueB homologuesa
|
YhgE homologuesa,b
|
Total | ||
|---|---|---|---|---|---|
| No. | No. near yuk-like clusterc | No. | No. near yuk-like clusterc | ||
| Bacillus anthracis | 3 | 0 | 1 | 0 | 4 |
| Bacillus cereus | 3 | 1 | 2 | 0 | 5 |
| Bacillus halodurans | 2 | 1 | 2 | ||
| Bifidobacterium longum | 1 | 0 | 3 | 0 | 4 |
| Clostridium acetobutylicum | 2 | 1 | 2 | 0 | 4 |
| Clostridium perfringens | 1 | 0 | 1 | 0 | 2 |
| Clostridium tetani | 1 | 0 | 1 | ||
| Corynebacterium diphtheriae | 3 | 0 | 3 | ||
| Corynebacterium glutamicum | 1 | 0 | 1 | ||
| Deinococcus radiodurans | 1 | 0 | 1 | ||
| Enterococcus faecalis | 1 | 0 | 1 | ||
| Lactobacillus plantarum | 1 | 0 | 1 | 0 | 2 |
| Lactococcus lactis | 1 | 0 | 1 | 0 | 2 |
| Leuconostoc mesenteroides | 4 | 0 | 4 | ||
| Listeria innocua | 3 | 1 | 3 | ||
| Listeria monocytogenes | 4 | 1 | 4 | ||
| Methanothermobacter thermautotrophicus | 1 | 0 | 1 | ||
| Oceanobacillus iheyensis | 2 | 1 | 2 | ||
| Oenococcus oeni | 1 | 0 | 1 | ||
| Staphylococcus aureus | 1 | 0 | 2 | 1 | 3 |
| Staphylococcus epidermidis | 1 | 0 | 1 | ||
| Streptococcus agalactiae | 1 | 1 | 1 | 0 | 2 |
| Streptococcus mutans | 1 | 0 | 1 | ||
| Streptococcus pyogenes | 1 | 0 | 1 | ||
| Streptomyces avermitilis | 1 | 0 | 1 | ||
| Total | 29 | 7 | 27 | 1 | 56 |
Blast E value ≤ 3 e−04.
B. subtilis YhgE is a YueB paralogue.
Number of members of the YueB family whose genes are located in the vicinity of the B. subtilis yuk cluster (yukE, yukD, yukC, yukBA).
DISCUSSION
In spite of its importance, the interaction of bacterial viruses with the host surface is still a poorly understood process, particularly in gram-positive systems. In the present study, we investigated the nature of the B. subtilis pha-2 locus, previously shown to be required for irreversible adsorption of bacteriophage SPP1 (29).
The results reported in this work have confirmed a hypothesis, based on the map location of pha-2 mutations and bioinformatics analyses, that yueB is the essential gene involved in the irreversible binding of SPP1 to its host. First, a deletion identified in a pha-2 strain and covering most of the yueB gene could be complemented by a xylose-inducible copy of yueB inserted at the amyE locus. Second, disruption of yueB by the insertion of a pMutin4 derivative resulted in a phage resistance phenotype regardless of the presence or absence of IPTG.
By measuring SPP1 adsorption constants in the presence or absence of the inducer in strains similarly disrupted in genes upstream of yueB, we have established that at least the genes yukE, yukD, yukC, yukBA, and yueB are organized as an operon. It should be noted that minute plaques have been observed in the absence of inducer in a yukE disrupted strain. Although this may reflect some leakiness of the Pspac promoter, allowing some expression of yueB, we cannot presently rule out the presence of a weak promoter located upstream of yukD. Interestingly, lacZ fusion data and Northern blotting experiments indicated a much higher level of yukE transcripts than of those of other genes in the operon. A similar observation was made with regard to the gapA operon of B. subtilis, which is involved in the interconversion of triose phosphates of the glycolytic pathway (15). In this case, secondary structures present at both the 3′ and the 5′ ends of gapA appear to confer higher stability on gapA transcripts relative to the remaining genes in the operon. This may apply to yukE as well, since the gene is also delimited by sequences that are likely to form hairpin-like secondary structures (Fig. 1A). Reduced stability of the polycistronic transcript covering yueB and/or limited read-through of transcription beyond the structure found at the 3′ end of yukE may lead to a reduced concentration of YueB in the cell. In fact, we failed to detect the expected 120-kDa form of YueB in immunoblots when whole-cell samples were examined. A corresponding band was nevertheless clearly visible in membrane vesicle preparations, particularly from cells overproducing YueB. Most of the immunoreactive species detected, however, fell in the size range of 50 to 70 kDa in all circumstances. Since a large hydrophilic portion of the protein is expected to face the wall, these lower-molecular-mass products probably reflect processing by cell wall protease(s). It remains an open question whether such processing is a consequence of the extraction procedure or whether it is a natural, biologically relevant event.
By placing the yueB gene under the control of the Pspac promoter present in pMutin4, the irreversible-adsorption rate could be modulated by the IPTG concentration in the medium, reaching values up to fivefold higher than wild-type levels at saturation. Such high adsorption levels had no detectable consequence in terms of SPP1 plating efficiency or plaque morphology. However, smaller plaques have been observed in integrants with genes upstream of yueB disrupted in the presence of IPTG. The small-plaque phenotype was particularly evident when yukE was the insertion target, in spite of the fact that the irreversible-adsorption constant measured in this and other similar cases was about twice as high as the wild-type value. Therefore, although yueB appears to be the only gene in the operon involved in irreversible binding of SPP1, yukE, and possibly downstream genes as well, may be important for a subsequent step in SPP1 infection.
Interestingly, Pallen (22) has recently highlighted the distant similarity between YukE and members of the ESAT-6 family of proteins. In the cases examined, these proteins were detected in culture filtrates, even though they do not possess distinguishable secretion signals. The importance of these small proteins has been recognized in mycobacteria, where they play a fundamental role in virulence and protective immunity (reference 22 and references therein). Given the presence of YukBA homologues near genes encoding members of the ESAT-6 family, and on the basis of homology between YukBA-like proteins and ATPases of the SpoIIIE/FtsK family, Pallen speculated that ESAT-6 protein export could involve a new type of gram-positive bacterial secretion machinery driven by ATP hydrolysis (accomplished by YukBA-related proteins). While our study did not directly address the cellular function of the B. subtilis yukE operon or the subcellular locations of hydrophilic products such as YukE, an additional possibility emerges which seems to us more consistent with the established role of FtsK and SpoIIIE in double-stranded DNA translocation (5). In this view, a putative protein complex formed by yukE operon products could be involved in double-stranded DNA uptake in B. subtilis. The models are not necessarily mutually exclusive if one assumes that energy from ATP hydrolysis carried out by YukBA can be used both for protein export and for DNA import. Our hypothesis is based on the evidence for involvement of the yukE operon products in a step following adsorption, which would likely be DNA injection into the cell. Unlike in competent cells, where uptake of single-stranded DNA occurs, phage DNA enters the cell in double-stranded form. Given the presence of an ftsK-like gene in the operon, one is tempted to speculate that YukBA could play an auxiliary role in driving phage DNA movement across the cell membrane. Even though our experiments indicate that the gene is not essential for infection in solid medium, detailed DNA injection studies are required to provide a measure of relative DNA translocation rates among the various strains. It is becoming clear that in vivo phage DNA injection is not a mere passive process, and in several cases, both phage and host proteins seem to be involved in pulling and pushing the viral DNA inside the cell (8, 18). We speculate that membrane products of the yukE operon (YueB, YukC, and YukBA) could form a complex required both for proper phage binding (involving YueB) and for channeling the ejected DNA through the membrane. Other operon products, such as YukE and YukD, could somehow contribute to the stability or conformation of such a putative complex. Although it is a mere working hypothesis at this stage, the possibility of involvement of yukE operon products in the SPP1 DNA injection process certainly deserves further investigation.
Acknowledgments
The financial support of C. São-José from the Fundação para a Ciência e a Tecnologia, Portugal (grant BPD/9429/2002), is acknowledged.
We thank Gonçalo Real and Adriano Henriques for the kind gift of vector pGR40. The assistance of Sophie Lhuillier (Gif-sur-Yvette, France) in the purification of YueB780 is appreciated. We also thank Ricardo Parreira (IHMT-UNL) for valuable discussions and João Nascimento and Carolina Guerreiro-Pereira, from our laboratory, for commenting on the manuscript. We are particularly indebted to Paulo Tavares (Gif-sur-Yvette, France), who contributed substantially with helpful discussions, critical insight,and encouragement throughout the work.
Footnotes
This paper is dedicated to Hermínia de Lencastre and Luís J. Archer, who first introduced M. A. Santos to the world of phage biology.
REFERENCES
- 1.Adams, M. H. 1959. Bacteriophages. Wiley Interscience, New York, N.Y.
- 2.Biswal, N., A. K. Kleinschmidt, H. C. Spatz, and T. A. Trautner. 1967. Physical properties of the DNA of bacteriophage SP50. Mol. Gen. Genet. 100:39-55. [DOI] [PubMed] [Google Scholar]
- 3.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 4.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Errington, J., J. Bath, and L. J. Wu. 2001. DNA transport in bacteria. Nat. Rev. Mol. Cell Biol. 2:538-545. [DOI] [PubMed] [Google Scholar]
- 6.Estrela, A. I., H. M. Pooley, H. de Lencastre, and D. Karamata. 1991. Genetic and biochemical characterization of Bacillus subtilis 168 mutants specifically blocked in the synthesis of the teichoic acid poly(3-O-β-d-glucopyranosyl-N-acetylgalactosamine 1-phosphate): gneA, a new locus, is associated with UDP-N-acetylglucosamine 4-epimerase activity. J. Gen. Microbiol. 137:943-950. [DOI] [PubMed] [Google Scholar]
- 7.Geller, B. L., R. G. Ivey, J. E. Trempy, and B. Hettinger-Smith. 1993. Cloning of a chromosomal gene required for phage infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 175:5510-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.González-Huici, V., M. Salas, and J. M. Hermoso. 2004. The push-pull mechanism of bacteriophage φ29 DNA injection. Mol. Microbiol. 52:529-540. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson, E. D., and O. E. Landman. 1975. Interaction of protoplasts, L forms, and bacilli of Bacillus subtilis with 12 strains of bacteriophage. J. Bacteriol. 124:445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobson, E. D., and O. E. Landman. 1977. Adsorption of bacteriophages φ29 and 22a to protoplasts of Bacillus subtilis 168. J. Virol. 21:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Letellier, L., L. Plancon, M. Bonhivers, and P. Boulanger. 1999. Phage DNA transport across membranes. Res. Microbiol. 150:499-505. [DOI] [PubMed] [Google Scholar]
- 14.Letellier, L., P. Boulanger, M. de Frutos, and P. Jacquot. 2003. Channeling phage DNA through membranes: from in vivo to in vitro. Res. Microbiol. 154:283-287. [DOI] [PubMed] [Google Scholar]
- 15.Ludwig, H., G. Homuth, M. Schmalisch, F. M. Dyka, M. Hecker, and J. Stulke. 2001. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 41:409-422. [DOI] [PubMed] [Google Scholar]
- 16.Margot, P., and D. Karamata. 1996. The wprA gene of Bacillus subtilis 168, expressed during exponential growth, encodes a cell-wall-associated protease. Microbiology 142:3437-3444. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Molineux, I. J. 2001. No syringes, please: ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 40:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Monteville, M. R., B. Ardestani, and B. L. Geller. 1994. Lactococcal bacteriophages require a host cell wall carbohydrate and a plasma membrane protein for adsorption and ejection of DNA. Appl. Environ. Microbiol. 60:3204-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okubo, S., B. Strauss, and M. Stodolsky. 1964. The possible role of recombination in the infection of competent Bacillus subtilis by bacteriophage deoxyribonucleic acid. Virology 24:552-562. [DOI] [PubMed] [Google Scholar]
- 21.Oudega, B., M. Vandenbol, and G. Koningstein. 1997. A 12 kb nucleotide sequence containing the alanine dehydrogenase at 279° on the Bacillus subtilis chromosome. Microbiology 143:1489-1491. [DOI] [PubMed] [Google Scholar]
- 22.Pallen, M. J. 2002. The ESAT-6/WXG100 superfamily—and a new gram-positive secretion system? Trends Microbiol. 10:209-212. [DOI] [PubMed] [Google Scholar]
- 23.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 24.Pooley, H. M., D. Paschoud, and D. Karamata. 1987. The gtaB marker in Bacillus subtilis 168 is associated with a deficiency in UDP glucose pyrophosphorylase. J. Gen. Microbiol. 133:3481-3493. [DOI] [PubMed] [Google Scholar]
- 25.Riva, S., M. Polsinelli, and A. Falaschi. 1968. A new phage of Bacillus subtilis with infectious DNA having separable strands. J. Mol. Biol. 35:347-356. [DOI] [PubMed] [Google Scholar]
- 26.Rossmann, M. G., V. V. Mesyanzhinov, F. Arisaka, and P. G. Leiman. 2004. The bacteriophage T4 DNA injection machine. Curr. Opin. Struct. Biol. 14:171-180. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Santos, M. A. 1991. Bacteriófagos de Bacillus subtilis do grupo SPP1. Características gerais, especificidade de adsorção e organização genómica. Ph.D. thesis. University of Lisbon, Lisbon, Portugal.
- 29.Santos, M. A., H. de Lencastre, and L. J. Archer. 1983. Bacillus subtilis mutation blocking irreversible binding of bacteriophage SPP1. J. Gen. Microbiol. 129:3499-3504. [DOI] [PubMed] [Google Scholar]
- 30.Santos, M. A., H. de Lencastre, and L. J. Archer. 1984. Homology between phages SPP1, 41c, 22a, ρ15 and SF6 of Bacillus subtilis. J. Gen. Virol. 65:2067-2072. [DOI] [PubMed] [Google Scholar]
- 31.São-José, C., R. Parreira, G. Vieira, and M. A. Santos. 2000. The N-terminal region of the Oenococcus oeni bacteriophage fOg44 lysin behaves as a bona fide signal peptide in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on oenococcal cells. J. Bacteriol. 182:5823-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson, M., J. Doskow, and S. Lindsey. 1991. RNA blots: staining procedures and optimization of conditions. Nucleic Acids Res. 19:679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yasbin, R. E., V. C. Maino, and F. E. Young. 1976. Bacteriophage resistance in Bacillus subtilis 168, W23, and interstrain transformants. J. Bacteriol. 125:1120-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yasbin, R. E., G. A. Wilson, and F. E. Young. 1973. Transformation and transfection in lysogenic strains of Bacillus subtilis 168. J. Bacteriol. 113:540-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young, F. E. 1967. Requirement of glucosylated teichoic acid for adsorption of phage in Bacillus subtilis 168. Proc. Natl. Acad. Sci. USA 58:2377-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]