Abstract
Glucansucrases of oral streptococci and Leuconostoc mesenteroides have a common pattern of structural organization and characteristically contain a domain with a series of tandem amino acid repeats in which certain residues are highly conserved, particularly aromatic amino acids and glycine. In some glucosyltransferases (GTFs) the repeat region has been identified as a glucan binding domain (GBD). Such GBDs are also found in several glucan binding proteins (GBP) of oral streptococci that do not have glucansucrase activity. Alignment of the amino acid sequences of 20 glucansucrases and GBP showed the widespread conservation of the 33-residue A repeat first identified in GtfI of Streptococcus downei. Site-directed mutagenesis of individual highly conserved residues in recombinant GBD of GtfI demonstrated the importance of the first tryptophan and the tyrosine-phenylalanine pair in the binding of dextran, as well as the essential contribution of a basic residue (arginine or lysine). A microplate binding assay was developed to measure the binding affinity of recombinant GBDs. GBD of GtfI was shown to be capable of binding glucans with predominantly α-1,3 or α-1,6 links, as well as alternating α-1,3 and α-1,6 links (alternan). Western blot experiments using biotinylated dextran or alternan as probes demonstrated a difference between the binding of streptococcal GTF and GBP and that of Leuconostoc glucansucrases. Experimental data and bioinformatics analysis showed that the A repeat motif is distinct from the 20-residue CW motif, which also has conserved aromatic amino acids and glycine and which occurs in the choline-binding proteins of Streptococcus pneumoniae and other organisms.
The glucansucrases (E.C. 2.4.1.5; commonly named glucosyltransferases [GTF] or dextransucrases) are extracellular enzymes from oral streptococci, Leuconostoc mesenteroides, and certain lactobacilli that catalyze the transfer of glucosyl units from the cleavage of sucrose to a growing glucan chain (11, 13, 26, 34). The enzymes are capable of synthesizing α-1,2, α-1,3, α-1,4, and α-1,6 linkages between the glucose units and are sometimes given trivial names according to their product, e.g., dextransucrase, mutansucrase, and alternansucrase. The streptococcal glucans are a key factor in the sucrose-dependent accumulation of mutans group streptococci on tooth surfaces and subsequent human dental caries formation (11), while dextransucrase from Leuconostoc has an important industrial application in the manufacture of dextran and other products with commercial potential (23, 37).
All glucansucrases possess a common pattern of structural organization. They are of high molecular mass, ranging from 160 to 313 kDa, and have a signal sequence followed by a variable stretch of approximately 200 amino acids and a highly conserved catalytic core region of about 900 amino acids that is a cyclically permuted version of the (β/α)8 barrel found in the amylase superfamily (33). Most glucansucrases have a C-terminal domain comprising about one-third of the protein, which characteristically contains a series of tandem amino acid repeats. The exception is DsrE, which has a repeat domain located centrally between two catalytic domains (8). A number of different types of repeating units have been identified in the primary sequence of glucansucrases and are termed A, B, C, and D repeats (5). These vary in length from 20 to 48 amino acids, but in all cases certain residues are highly conserved, particularly aromatic amino acids and glycine. However, the 33-residue A repeat, first identified in GtfI of Streptococcus downei by visual inspection of aligned sequences (16), was the only repeat found in all GTF (34, 44). Its existence as a distinctive motif was recently confirmed by analysis of 16 glucansucrase sequences with the MEME/MAST motif discovery tool, which uses statistical modeling techniques to automatically find and describe repeated motifs in a set of sequences (42).
The repeat domain is not required for the catalytic activity of glucansucrases, though in a number of instances it has been shown to influence the rate of reaction, possibly by removing the growing glucan chain from the active site (25, 30, 32). In a number of the streptococcal GTF, the repeat domain has been experimentally demonstrated to bind glucans such as dextran and has been identified as a glucan binding domain (GBD) (1, 16, 24, 28, 46). The A repeat motif is also found in several proteins that do not have glucansucrase activity: the GbpA glucan binding protein of Streptococcus mutans (4), the GbpD glucan binding lipase of S. mutans and Streptococcus sobrinus (42), and the Dei dextranase inhibitor of S. sobrinus (43), all of which bind dextran. It has also recently been identified near the N termini of some glucansucrases (22). However, it is not yet clear whether all proteins with a repeat-containing region really bind glucans, even though the term GBD has been widely used. In addition, there are unanswered questions about the relationship of the A repeat motif to the CW, or cell wall binding motif, which also has conserved aromatic amino acids and glycine and which is found in the choline-binding proteins of Streptococcus pneumoniae, the toxins of Clostridium difficile, and some other surface-associated proteins (17, 47). There is a lack of information on how the similarities in sequence reflect the specificity of these putative binding domains. This paper examines the distribution of A repeats in a range of glucansucrases and characterizes the glucan binding by representative recombinant proteins. The critical importance of conserved aromatic residues is demonstrated, and the specificity of binding is discussed with regard to the relationship between A and CW repeats.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. mutans strain UA159 was grown in Todd-Hewitt broth (Oxoid Ltd., Hampshire, United Kingdom) supplemented with 5% yeast extract. The cultures were incubated at 37°C in candle jars. L. mesenteroides NRRL B-1355, B-512F, and B-1299 strains were provided by the National Center for Agricultural Utilization Research stock culture collection in Peoria, Ill. Cells were grown at 30°C on standard medium as previously described (14). E. coli XL1Blue was used in all cloning and protein expression procedures. E. coli cultures were grown in Luria-Bertani or 2× yeast extract-tryptone (YT) broth containing ampicillin (100 μg/ml) where required. For solid media, bacteriological agar (agar no. 1; Oxoid Ltd.) was added at a final concentration of 1.5%.
Bioinformatics.
Sequences were retrieved from the GenBank database. Sequences examined (accession numbers are in parentheses) were those for S. mutans GtfB (AAA88588), GtfC (AAA88589), GtfD (AAA26895), GbpA (A37184), and GbpD (AAN58492); Streptococcus gordonii GtfG (AAC43483); S. downei GtfI (AAC41412) and GtfS (AAA63063); Streptococcus sobrinus GtfT (D13928), GtfU (AB089438), and Dei (L34406); Streptococcus salivarius GtfJ (CAA77900), GtfK (CAA77901), GtfL (AAC41412), and GtfM (AAC41413); Streptococcus oralis GtfR (BAA95201); and L. mesenteroides Asr (CAB76565), DsrA (JC5473), DsrB (AAB95453), DsrS (I09598), and DsrE (CAD22883). The sequence set was submitted to the MEME motif-searching program (2, 3), available at http://meme.sdsc.edu/meme/website/, to identify the consensus repeat sequence. Individual repeats were compiled and subjected to multiple alignment with the CLUSTALW program (10), available at http://www.ebi.ac.uk/clustalw/.
Molecular biology techniques.
Standard DNA manipulations were carried out by protocols described by Sambrook and Russell (39). Restriction enzymes were obtained from New England Biolabs and used according to manufacturer's instructions. Large-scale plasmid extractions for DNA sequencing were carried out with the Plasmid Midi-Kit (QIAGEN). DNA sequencing was carried out at the Molecular Biology Unit, University of Newcastle, by Thermosequenase and dye terminator chemistry on an ABI377 sequencer (Amersham). DNA sequences were assembled and analyzed with OMIGA, version 2.0, software (Oxford Molecular). PCRs were carried out with the high-fidelity, premixed Extensor Long PCR Master Mix (ABgene) without oil overlays, under cycling conditions recommended by the manufacturer, on a GeneAmp9700 thermal cycler (Applied Biosystems). All PCR primers were custom synthesized by Genset (Paris, France). PCR products for cloning were separated by electrophoresis and purified from the agarose gel with the Qiaex II kit (QIAGEN). Electrophoresis of proteins and Western blotting were carried out by standard protocols described by Sambrook and Russell (39).
Cloning and expression of His-tagged proteins.
Clones for expression of six-His-tagged GBD of S. downei GtfI and the full-length S. mutans GbpD have been described previously (41, 42). GbpA from S. mutans (4) and the GBD of GtfS from S. downei (19) were cloned by PCR amplification of the GBD-coding regions with primers that incorporated BamHI and HindIII sites to enable in-frame cloning into pQE30 (QIAGEN) to give an N-terminal six-His tag. The clones were sequenced to confirm that no misincorporation had occurred. For expression of proteins, E. coli XL1Blue competent cells were transformed with expression plasmids. Single colonies were used to inoculate 2× YT medium and shaken overnight at 37°C. The culture was used to inoculate 5 ml of fresh 2× YT medium and allowed to grow for 1 h at 37°C. Expression of tagged protein was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 37°C. The cells were harvested by centrifugation and resuspended in 1 ml of B-PER protein extraction reagent (Pierce) containing 2 mg of lysozyme/ml and frozen at −20°C overnight. After thawing on ice the lysates were cleared by centrifugation and retention of supernatant. The supernatants were stored at 4°C until necessary.
Site-directed mutagenesis of GBD1A.
Mutagenesis was performed with pGBD1A, a six-His-tagged truncated version of the GBD of S. downei GtfI that has only four A repeats (41). Mutations leading to single amino acid changes were generated by the two-step, overlap extension PCR method (21) using pGBD1A as the template. The end primers were used to incorporate BamHI (forward primer) and HindIII (reverse primer) restriction sites to enable cloning into the pQE30 vector (QIAGEN). The fragments were cloned in frame such that the encoded protein was fused to a six-His tag similarly to the unmutated “wild-type” GBD1A. After cloning, the inserts were sequenced to confirm that the desired mutation had been generated and that no undesired mutations had been generated owing to misincorporation during PCR. Expression of each of the mutant proteins was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) before use in the binding experiments.
Binding assays.
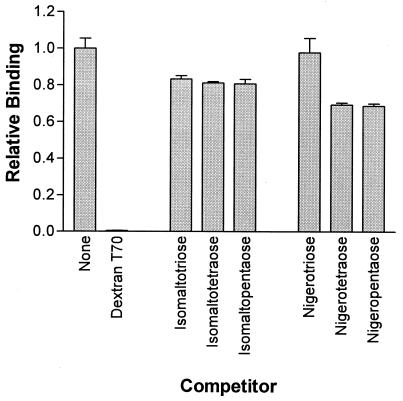
To measure the binding of dextran by recombinant proteins, the microtiter plate assay utilizing the Ni affinity of the histidine tag to immobilize the protein was performed (41, 42). Cleared lysates of induced E. coli cells carrying the overexpression plasmids or vector controls were prepared as described above, and 50 μl of cleared lysate was added to Ni-nitrilotriacetic acid (NTA)-coated 96-well HiSorb plates (QIAGEN) in phosphate-buffered saline (PBS) containing 0.05% (vol/vol) Tween 20 (PBST) to a final volume of 200 μl and incubated overnight at 4°C. Such lysates contain an excess of tagged protein and saturate the binding capacity of the plates (41, 42). The protein solutions were removed, and the wells were washed four times for 1 min with PBST. Two hundred microliters of a 100-μg/ml solution of biotin-dextran (biotinylated dextran T70; Fluka) in PBS-0.2% bovine serum albumin (BSA) was added and incubated for 10 min. After being washed as before, wells were incubated with 200 μl of a 1/20,000 dilution of Extravidin-alkaline phosphatase conjugate (Sigma) in PBS-0.2% BSA for 30 min and washed again. One hundred microliters of phosphatase substrate solution (1 mg of p-nitrophenylphosphate Sigma 104 substrate/ml, 28 mM NaHCO3, 22 mM NaCO3, 5 mM MgCl2) was added, and color change was monitored by readings at 405 nm in a Titertek Multiskan MCC 340 plate reader. For competition studies as shown in Fig. 3, 200 μl of competitor solution at 100 μg/ml was added and the plates were incubated at room temperature for 10 min prior to the addition of 200 μl of biotin-dextran at 1 μg/ml. Isomaltosaccharides (3 to 5 glucose units, α-1,6 linked) were from commercial sources, and nigerooligosaccharides (3 to 5 glucose units, α-1,3 linked) were kindly provided by H. Mukasa (36).
FIG. 3.
Competition with biotinylated dextran for binding to GBD0 by unlabeled dextran, isomaltosaccharides, and nigerooligosacharides.
Determination of binding affinity.
Cleared lysates from cells induced for expression of binding domains of GtfI and GtfS and full-length GbpA and GbpD were prepared as described above. A control lysate from E. coli containing pQE30 was also prepared. Fifty microliters of cleared lysate was added to Ni-NTA-coated 96-well HiSorb plates (QIAGEN), and the plates were incubated overnight at 4°C. Two wells were coated with pQE30 lysate, and each of the protein expression lysates was used to coat 12 wells. The protein solutions were removed, and the wells were washed four times for 1 min with PBST. Two hundred microliters of PBS-0.2% BSA containing biotin-dextran solution (Fluka) at 100 μM, 10 μM, 1 μM, 100 nM, 10 nM, or 1 nM was added to the sample wells. Two wells per protein were treated with each concentration of biotin-dextran. Two hundred microliters of biotin-dextran at 100 μM only was added to the wells treated with the control pQE30 lysate. These wells were used as the zero control. After incubation for 10 min at room temperature and washing with PBST as before, wells were incubated with 200 μl of a 1/20,000 dilution of Extravidin-alkaline phosphatase conjugate (Sigma) in PBS-0.2% BSA for 30 min and washed again. One hundred microliters of phosphatase substrate solution (1 mg of p-nitrophenylphosphate Sigma 104 substrate/ml, 28 mM NaHCO3, 22 mM NaCO3, 5 mM MgCl2) was added, and color change was monitored by readings at 405 nm in a Titertek Multiskan MCC 340 plate reader. Data were plotted with the GraphPad Prism, version 3, program. The Kd values for binding were determined by linear regression using the one-site binding equation within the program (http://www.graphpad.com/prism/Prism.htm).
Electroblotting and detection with biotin-dextran.
S. mutans samples were prepared from a culture supernatant concentrated 40-fold with a 10-kDa-cutoff ultrafiltration membrane. L. mesenteroides samples were prepared from a 10-fold-concentrated culture by resuspension the cell pellet in 20 mM sodium acetate buffer at pH 5.4. Zymography to detect glucansucrase activity was performed as previously described (16) with 3 mU of glucansucrase activity loaded on SDS-PAGE gel. One glucansucrase unit is defined as the enzyme quantity that releases 1 μmol of fructose per min at 30°C, pH 5.4, and 100 g of sucrose/liter. The same samples were run in parallel for biotin-dextran detection on SDS-PAGE gel (about 50 mU of glucansucrase activity) and electroblotted onto nitrocellulose membranes. The membranes were kept overnight in 20 mM PBS, pH 7.3, containing 3% (wt/vol) BSA (initial heat shock fraction; Sigma) to block nonspecific binding and washed three times for 15 min in PBST. One hundred micrograms of biotin-dextran (Fluka)/ml in PBS-0.2% BSA was then added. After incubation at room temperature with shaking, the membranes were washed in PBST for 45 min with three buffer changes and then incubated for 1 h at room temperature with a 1:20,000 dilution of Extravidin-alkaline phosphatase conjugate (Sigma) in PBS-0.2% BSA. Proteins that bound to dextran were revealed by a 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium color reaction with reagents supplied by Zymed (San Francisco, Calif.) according to the manufacturer's instructions. In some experiments, dextran T70 or alternan was labeled with biocytin hydrazide (7).
RESULTS
Identification of a consensus A repeat.
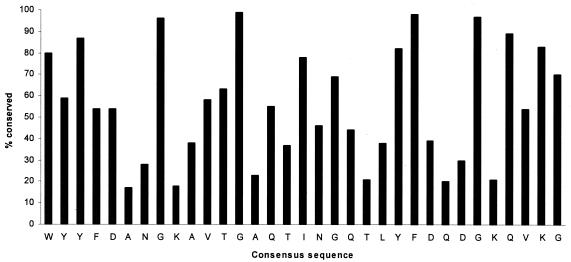
The 20 proteins examined are all concerned with the synthesis or binding of α-1,3 and/or α-1,6-linked glucans. As determined by the MEME program, they contain a total of 96 copies of a conserved 33-amino-acid motif corresponding to the A repeat. Seventy-six of these motif units can be directly aligned without gaps by CLUSTALW, and the consensus is shown in Fig. 1, which also illustrates the extent of conservation of the individual residues. Most striking are the almost universally conserved glycines and the aromatic residues tryptophan, tyrosine, and phenylalanine. The remaining 20 A repeats also contain these highly conserved residues but have 1 to 3 additional residues in the positions shown as having least conservation in Fig. 1. The consensus sequence is WYYFDANGKAVTGAQTINGQTLYFDQDGKQVKG.
FIG. 1.
Conservation of individual residues in the 33-amino-acid A repeat derived from CLUSTAL alignment. The consensus sequence is shown at the bottom.
The A repeat can be presented in Prosite notation as [WYFTLR]-[YFLARGQV]-[YFRGQVAML]-x (4, 5)-[GYVFT]-x (4)-G-x (9)-[YLFHA]-[FY]-x (3, 4)-[GS]-x-[QMAELY]-[VIALMTF]-[KRYVTL]-[GNDEAHS].
Binding affinity of streptococcal GBD.
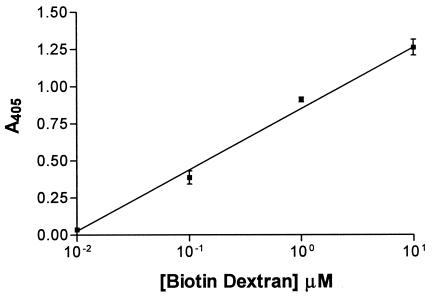
The GBD of S. downei GtfI and GtfS, as well as GbpA and GbpD of S. mutans, were expressed in E. coli as recombinant proteins with N-terminal six-His tags, which allowed them to be immobilized on Ni-NTA-coated microtiter trays and used in an enzyme-linked immunosorbent assay-type assays to measure binding of biotinylated dextran. Figure 2 shows the equilibrium binding by the GBD from GtfI (GBD0), with a linear plot over a 1,000-fold concentration range. The other proteins tested also gave linear plots. Assays using biotin-dextran concentrations covering a range of 1 nM to 100 μM allowed calculation of the dissociation constant Kd for the GBD from GtfI as 5.9 × 10−7 M. The corresponding values for the GBD from GtfS, GbpA, and GbpD were 11.2 × 10−7, 4.5 × 10−7, and 3.2 × 10−7 M, respectively. Addition of unlabeled dextran T70 to compete with the biotinylated dextran showed a stoichiometric relationship (i.e., an equimolar concentration of competitor gave a 50% reduction in the absorbance reading). This indicated that the biotin moieties had little or no effect on interaction of the dextran moiety with the binding domain and that biotin-dextran competes for the same sites as unlabeled dextran.
FIG. 2.
Kinetics of binding of biotinylated dextran to the recombinant binding domain of S. downei GtfI (GBD0).
Specificity of binding domain of S. downei GtfI.
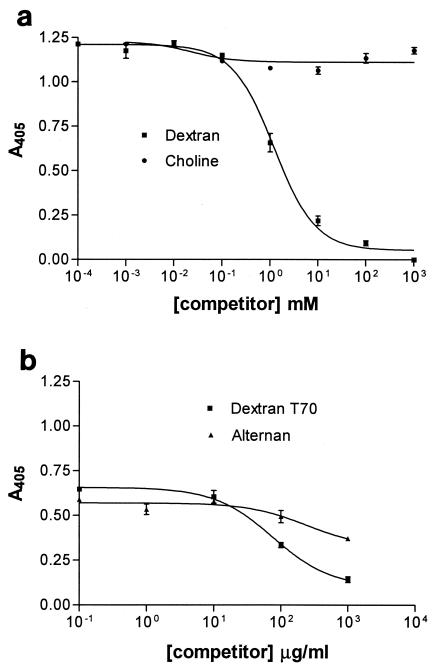
GtfI of S. downei binds to dextran or to mutan, and these can be used as affinity absorbents to purify the enzyme (16). To explore the specificity of binding, a range of potential ligands were tested for the ability to compete with binding by biotinylated dextran to the recombinant GBD of GtfI, with the competitors in a 100-fold excess over the labeled dextran. Unlabeled dextran was an efficient inhibitor, and there was also some inhibition by oligosaccharides of the α-1,6-linked isomaltose series and the α-1,3-linked nigerose series (Fig. 3). It was not possible to test the α-1,3-linked polymer mutan in this assay, due to its insolubility. To investigate the perceived relationship between the A repeat and the choline-binding CW repeat, the ability of choline to compete with biotin-dextran for binding by the GtfI binding domain was tested (Fig. 4a). Choline was unable to displace biotin-dextran, even at concentrations 3 orders of magnitude higher. In contrast, alternan, which contains alternating α-1,3 and α-1,6 linkages, was able to compete with biotin-dextran (Fig. 4b). Thus proteins containing the A repeat have a specificity of binding distinct from that of proteins containing the CW repeat.
FIG. 4.
Competition with biotin-dextran for the binding domain of S. downei GtfI (GBD0) by unlabeled dextran T70, choline, and alternan. The concentration of biotinylated dextran was kept constant at 1 mM (a) or 100 μg/ml (b). The competitors choline or alternan were added after a 10-min preincubation with biotin-dextran and washed after a further 10-min incubation. Detection and washing steps were carried out as described in Materials and Methods. In panel b, the units for the ligands are micrograms per milliliter because the molecular weight distribution of the alternan is unknown.
Binding by Leuconostoc glucansucrases.
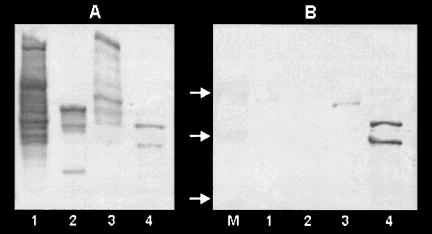
A sensitive method for detecting the ability to bind dextran is the use of biotinylated dextran to detect proteins electroblotted onto nitrocellulose (42). Figure 5A shows the migration of glucansucrases from three strains of L. mesenteroides and S. mutans UA159, detected by staining the glucan formed during incubation in sucrose. Probing with biotinylated dextran (Fig. 5B) showed that neither the alternansucrase and dextransucrases of strain B-1355 (234 and 165 kDa respectively; lane 1), nor the dextransucrase of B-512F (174 kDa; lane 2), nor the high-molecular-weight DsrE dextransucrase of B-1299 (313 kDa; lane 3) bound dextran. The only Leuconostoc enzyme to bind was the 194-kDa DsrB dextransucrase of B-1299, whereas strong binding was shown by the S. mutans GTF (lane 4). S. mutans glucan binding proteins GbpA and GbpD, which also bind biotin-dextran (42), are of lower molecular weight and are not shown on this gel. In a parallel experiment where the blot was probed with biotinylated alternan, alternansucrase and dextransucrases showed no labeling, whereas the S. mutans proteins did bind the label (not shown).
FIG. 5.
SDS-PAGE analysis of cell extracts from L. mesenteroides and S. mutans strains. (A) Zymogram of active glucansucrases after SDS-PAGE separation and incubation in sucrose. (B) The same samples electroblotted and developed with biotin-dextran to detect glucan binding proteins. Lane 1, L. mesenteroides NRRL B-1355; lane 2, L. mesenteroides NRRL B-512F; lane 3, L. mesenteroides NRRL B-1299; lane 4, S. mutans UA159; lane M, molecular mass ladder of 250, 150, and 100 kDa (arrows).
Contribution of conserved amino acids to binding by GBD of S. downei GtfI.
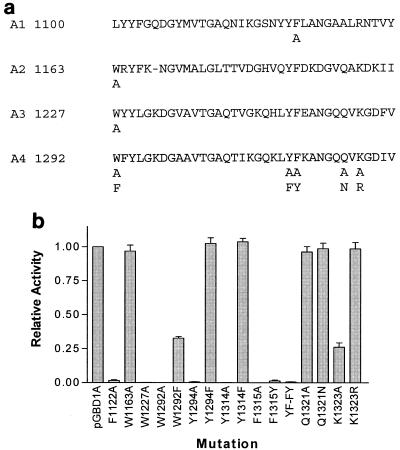
The terminal A repeat of GBD1A (which corresponds to repeat A4 in GTFI) was selected as a target for site-directed mutagenesis because removal of this repeat results in a marked reduction in binding capacity (41). Figure 6a shows the residues that were altered, and Fig. 6b summarizes the results. Conversion of the first tryptophan to alanine (W1292A) or of either the conserved tyrosine or phenylalanine to alanine (Y1314A and F1315A) resulted in complete loss of binding capacity. Twenty-five percent of the binding was retained when the tryptophan was converted to another aromatic amino acid (W1292F), and the conserved tyrosine could also be replaced by phenylalanine without any loss of function (Y1314F). However, it was not possible to replace the conserved phenylalanine and retain function (F1315Y). A double-mutant protein where YF was changed to FY was also without binding activity, confirming the essential function of F1315. It is striking that this phenylalanine residue is conserved in all except one of the 94 repeats examined; in the single exception it was replaced by tyrosine. The two other highly conserved amino acids examined by mutagenesis were glutamine 1321 and lysine 1323. Despite the fact that the former is conserved in 89% of the aligned A repeats, it could be converted to alanine or asparagine (Q1321A and Q1321N) without affecting function. The lysine, however, could satisfactorily be replaced by arginine, but a basic residue seems to be essential because replacement by alanine (K1323A) gave a drop in binding. The functional importance of a basic lysine or arginine is supported by the fact that, in 91% of the A repeats, one or the other of these residues was present in this position.
FIG. 6.
(a) A repeats present in recombinant GBD1A, which was derived by truncating the GBD of S. downei GtfI (41). Amino acid substitutions introduced by mutagenesis are indicated below the main sequence. (b) Relative binding of biotinylated dextran by GBD1A and specific mutant forms. YF-FY, residues 1314 and 1315.
The question of whether all of the A repeats contribute to binding was not examined thoroughly, but mutations F1123A in repeat A1 and W1227A in repeat A3 resulted in a loss in binding whereas mutation W1163A in repeat 2 did not, suggesting that all the repeats do not contribute equally to the overall binding of dextran or that corresponding residues in different repeats may differ in their contributions.
DISCUSSION
The analysis of 20 proteins previously known to contain A repeats has allowed recognition of the 33-residue consensus sequence and information on the extent to which different residues are conserved. The A repeats occur in all of the glucansucrases from streptococci and Leuconostoc for which the sequences are available. However, not all glucansucrases have the A repeats because they are not found in glucansucrases of lactobacilli, although these may contain other repeat motifs (26), nor are they found in amylosucrase of Neisseria polysaccharea (12). The consensus motif will be of value in scanning genomes for related proteins, particularly for gene products that are not glucansucrases, and its use has led to the discovery of a novel glucan-binding lipase in S. mutans (42).
The phenomenon of glucan binding is considered to be an important feature that contributes to dental plaque formation and the virulence of the oral streptococci associated with dental caries (11). There has thus been interest in the molecular basis of binding, and it has repeatedly been shown that truncation of the C-terminal repeat-containing domain affects binding. Also, antibodies directed against the domain can inhibit attachment to surfaces and have the potential for preventing dental caries (5). The work presented here extends this by site-directed mutagenesis of specific conserved residues within the A repeat and shows the crucial importance of the aromatic residues that may be able to “stack” with sugar units and the conserved polar residues (K and R) that may allow hydrogen bonding with hydroxyl residues. It has been well established in other classes of proteins, such as cellulases and chitanases, that the disposition of aromatic residues is the main determinant of strength and specificity of binding to polysaccharides. The functional importance of the conserved glycines was not explored by mutagenesis, but these residues are thought to be responsible for the high content of the β-sheet and the flexibility of the repeat domain demonstrated by circular dichroism (20, 24, 31).
GtfS, GtfI, GbpA, and GbpD all had a Kd for dextran binding in the 10−7 M range, similar to values from other reports (5) and indicating that the A repeats within these proteins have similar affinities for binding. However, these values were calculated assuming a simple, single-binding-site model. The data indicated that, in the binding domain of GtfS at least, there may be heterogeneity in the binding site. The cause of this heterogeneity is unknown but may be due to subtle differences in sequence and protein folding. The similarity in binding affinity, despite the fact that GtfS synthesizes α-1,6 linkages whereas GtfI synthesizes primarily α-1,3 links, is interesting.
Comparison of the primary sequences of streptococcal and Leuconostoc glucansucrases shows that they are very similar in the number and arrangement of A repeats within the C-terminal domain (34). Experiments in which this domain was truncated have also shown similar effects in all the glucansucrases examined, with a reduction in the rate of enzyme activity and a loss of stimulation by exogenous dextran, but no influence on the specificity of the catalytic reaction (1, 25, 27, 30, 32). There is, however, a great difference in the ability to bind glucan. We were unsuccessful in our attempts to express isolated Leuconostoc GBD in E. coli, but, as illustrated in Fig. 5, most of the Leuconostoc glucansucrases do not bind biotinylated dextran under conditions when S. mutans GTF bind strongly. This lack of binding by Leuconostoc dextransucrases and alternansucrase has been confirmed by a microtiter tray assay and by attempts to purify these enzymes by affinity chromatography on dextran or mutan by methods effective with streptococcal proteins (38; our unpublished observations). It can thus be concluded that, while A repeats are necessary for binding by the streptococcal GTF (and also GbpA, GbpD, and Dei), the mere presence of A repeats in a sequence may not justify its designation as a GBD.
The molecular basis for the difference in binding between streptococcal and Leuconostoc glucansucrases remains unknown but is presumably due to as yet unrecognized subtle differences in sequence. We can only speculate as to the functional importance of the difference in binding. Insofar as it is possible to compare the catalytic properties of GTF and dextransucrases described in different reports, they seem to be remarkably similar (34). The difference may thus not relate to the involvement of the C-terminal repeat domain in glucan synthesis, but rather to benefits to streptococci of having GTF bound to the glucans dextran and mutan in dental plaque. GTF and the glucan binding proteins GbpA and GbpD bind to both these glucans (24, 38, 42). In this paper we show that both α-1,6- and α-1,3-linked oligosaccharides, as well as alternan, which contains both types of linkage, can compete with dextran for binding to GtfI of S. downei (Fig. 4). The oligosaccharides compete poorly with dextran, presumably because they are below the optimal size for binding (25). The inherent flexibility of the domain thus allows recognition of different types of glucan; note, however, that no binding to other α-linked glucans, such as amylose or amylopectin, is found. It is, however, important to recognize the possibility that other macromolecules that have not been experimentally investigated may be bound. It will be of particular interest to explore whether the repeat domains in Leuconostoc might be responsible for the surface location of these enzymes (50) and interaction with molecules such as lipoteichoic acid (LTA). For example, it is known that another class of repeat motif is involved in binding the internalin molecule to surface LTA in Listeria monocytogenes (9).
Repetitive CW motifs that resemble A repeats in having conserved glycines and aromatic residues are found in choline-binding proteins of S. pneumoniae and its bacteriophages; toxins of Clostridium difficile and Clostridium sordellii that recognize carbohydrate targets in the gut epithelium (35); and surface-associated proteins from Erysipelothrix rhusiopathiae, Clostridium acetobutylicum, Peptostreptococcus micros, and Lactococcus lactis (47). The best-characterized of these are the choline-binding proteins, so we investigated the capacity of the binding domain derived from S. downei GtfI to bind choline. There is no evidence for any binding of choline whatever (Fig. 4). Furthermore, streptococcal GTF and GBP are not retained by DEAE-cellulose, a substrate commonly used for affinity purification of choline-binding proteins (40). Conversely, choline-binding proteins have not been reported to be retarded by chromatography on Sephadex, which serves as an affinity matrix for GTF and GBP. There is thus no experimental evidence for considering those proteins that contain A repeats to be functionally related to choline-binding proteins containing CW repeats. The similarity in sequence between glucan-binding and choline-binding proteins, as well as the toxins of C. difficile, originates from the observation that all contained tandem repeats with conserved glycines and aromatic residues and were believed to have binding functions. Evidence for structural similarity also came from immunological cross-reaction and the fact that an antibody against a synthetic peptide based on a consensus conserved motif could react with both C. difficile toxin and S. mutans GbpA (48). Numerous attempts have been made to align the various types of repeats, based on the conserved glycines and the critical YF dyad (18, 45, 47, 49). Automatic sequence alignment programs, such as those used to compile the Pfam and CDART (conserved domain architecture retrieval tool) databases (6, 29) also conflate proteins with A and CW repeats (PF01473 and COG5263, respectively). We believe that the current database resources widely used for automated bioinformatics annotation of genomes thus have a clear potential to give rise to erroneous conclusions about the relationship and functional activity of repeat-containing domains because the analysis fails to take into account two crucial factors: (i) the consensus A repeat is 33 residues whereas the consensus CW repeat is 20 residues long and (ii) CW tandem repeats are almost always contiguous whereas A repeats have variable spacing but always have intervening nonconserved regions.
The structure of LytA autolysin of S. pneumoniae, which contains six CW repeats, has recently been elucidated (15). It has a novel solenoid fold with six β-hairpins, where the apex of the hairpin is glycine and the aromatic residues interact with choline. This structure is dependent on the folding of the consecutive 20-residue CW repeats, and it is difficult to see how the larger, widely spaced A repeats could be constrained into the same fold. It has so far proved impossible to obtain crystals of an A repeat domain but comparison of a three-dimensional structural model with LytA would be extremely interesting and would provide valuable insights into the relationship between primary sequence, structure, and function of these intriguing molecules.
Acknowledgments
This work was supported by the Wellcome Trust Project grant 060993 and Biomedical Collaboration grant 056112/Z/98/Z.
REFERENCES
- 1.Abo, H., T. Matsumura, T. Kodama, H. Ohta, K. Fukui, K. Kato, and H. Kagawa. 1991. Peptide sequences for sucrose splitting and glucan binding within Streptococcus sobrinus glucosyltransferase (water-insoluble glucan synthetase). J. Bacteriol. 173:989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 3.Bailey, T. L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48-54. [DOI] [PubMed] [Google Scholar]
- 4.Banas, J. A., R. R. B. Russell, and J. J. Ferretti. 1990. Sequence analysis of the gene for the glucan-binding protein of Streptococcus mutans Ingbritt. Infect. Immun. 58:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banas, J. A., and M. M. Vickerman. 2003. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral Biol. Med. 14:89-99. [DOI] [PubMed] [Google Scholar]
- 6.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayer, E. A., H. Ben-Hur, and M. Wilchek. 1988. Biocytin hydrazide—a selective label for sialic acids, galactose, and other sugars in glycoconjugates using avidin-biotin technology. Anal. Biochem. 170:271-281. [DOI] [PubMed] [Google Scholar]
- 8.Bozonnet, S., M. Dols-Laffargue, E. Fabre, S. Pizzut, M. Remaud-Simeon, P. Monsan, and R. M. Willemot. 2002. Molecular characterization of DSR-E, an alpha-1,2 linkage-synthesizing dextransucrase with two catalytic domains. J. Bacteriol. 184:5753-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, L., S. Dramsi, P. Dehoux, H. Bierne, G. Lindahl, and P. Cossart. 1997. InIB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol. Microbiol. 25:285-294. [DOI] [PubMed] [Google Scholar]
- 10.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colby, S. M., and R. R. B. Russell. 1997. Sugar metabolism by mutans streptococci. J. Appl. Microbiol. 83:S80-S88. [DOI] [PubMed] [Google Scholar]
- 12.De Montalk, G. P., M. Remaud-Simeon, R. M. Willemot, V. Planchot, and P. Monsan. 1999. Sequence analysis of the gene encoding amylosucrase from Neisseria polysaccharea and characterization of the recombinant enzyme. J. Bacteriol. 181:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devulapalle, K. S., and G. Mooser. 1994. Subsite specificity of the active site of glucosyltransferases from Streptococcus sobrinus. J. Biol. Chem. 269:11967-11971. [PubMed] [Google Scholar]
- 14.Dols, M., M. R. Simeon, R. M. Willemot, M. R. Vignon, and P. F. Monsan. 1997. Structural characterization of the maltose acceptor-products synthesized by Leuconostoc mesenteroides NRRL B-1299 dextransucrase. Carbohydr. Res. 305:549-559. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Tornero, C., A. Ramon, C. Fernandez-Cabrera, G. Gimenez-Gallego, and A. Romero. 2002. Expression, crystallization and preliminary X-ray diffraction studies on the complete choline-binding domain of the major pneumococcal autolysin. Acta Crystallogr. D Biol. Crystallogr. 58:556-558. [DOI] [PubMed] [Google Scholar]
- 16.Ferretti, J. J., M. L. Gilpin, and R. R. B. Russell. 1987. Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinus MFe28. J. Bacteriol. 169:4271-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia, J. L., A. R. Sanchez-Beato, F. J. Medrano, and R. Lopez. 1998. Versatility of choline-binding domain. Microb. Drug Resist. 4:25-36. [DOI] [PubMed] [Google Scholar]
- 18.Giffard, P. M., and N. A. Jacques. 1994. Definition of a fundamental repeating unit in streptococcal glucosyltransferase glucan-binding regions and related sequences. J. Dent. Res. 73:1133-1141. [DOI] [PubMed] [Google Scholar]
- 19.Gilmore, K. S., R. R. B. Russell, and J. J. Ferretti. 1990. Analysis of the Streptococcus downei gtfS gene, which specifies a glucosyltransferase that synthesizes soluble glucans. Infect. Immun. 58:2452-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas, W., R. MacColl, and J. A. Banas. 1998. Circular dichroism analysis of the glucan binding domain of Streptococcus mutans glucan binding protein-A. Biochim. Biophys. Acta 1384:112-120. [DOI] [PubMed] [Google Scholar]
- 21.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain-reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 22.Janecek, S., B. Svensson, and R. R. B. Russell. 2000. Location of repeat elements in glucansucrases of Leuconostoc and Streptococcus species. FEMS Microbiol. Lett. 192:53-57. [DOI] [PubMed] [Google Scholar]
- 23.Joucla, G., T. Brando, M. Remaud-Simeon, P. Monsan, and G. Puzo. 2004. Capillary electrophoresis analysis of gluco-oligosaccharide regioisomers. Electrophoresis 25:861-869. [DOI] [PubMed] [Google Scholar]
- 24.Kingston, K. B., D. M. Allen, and N. A. Jacques. 2002. Role of the C-terminal YG repeats of the primer-dependent streptococcal glucosyltransferase, GtfJ, in binding to dextran and mutan. Microbiology 148:549-558. [DOI] [PubMed] [Google Scholar]
- 25.Konishi, N., Y. Torii, T. Yamamoto, A. Miyagi, H. Ohta, K. Fukui, S. Hanamoto, H. Matsuno, H. Komatsu, T. Kodama, and E. Katayama. 1999. Structure and enzymatic properties of genetically truncated forms of the water-insoluble glucan-synthesizing glucosyltransferase from Streptococcus sobrinus. J. Biochem (Tokyo) 126:287-295. [DOI] [PubMed] [Google Scholar]
- 26.Kralj, S., G. H. van Geel-Schutten, H. Rahaoui, R. J. Leer, E. J. Faber, M. J. van der Maarel, and L. Dijkhuizen. 2002. Molecular characterization of a novel glucosyltransferase from Lactobacillus reuteri strain 121 synthesizing a unique, highly branched glucan with α-(1→4) and α-(1→6) glucosidic bonds. Appl. Environ. Microbiol. 68:4283-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kralj, S., G. H. van Geel-schutten, M. van der Maarel, and L. Dijkhuizen. 2003. Efficient screening methods for glucosyltransferase genes in Lactobacillus strains. Biocatal. Biotransform. 21:181-187. [Google Scholar]
- 28.Lis, M., T. Shiroza, and H. K. Kuramitsu. 1995. Role of C-terminal direct repeating units of the Streptococcus mutans glucosyltransferase-S in glucan binding. Appl. Environ. Microbiol. 61:2040-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monchois, V., M. Arguello-Morales, and R. R. B. Russell. 1999. Isolation of an active catalytic core of Streptococcus downei MFe28 GTF-I glucosyltransferase. J. Bacteriol. 181:2290-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monchois, V., J. H. Lakey, and R. R. B. Russell. 1999. Secondary structure of Streptococcus downei GTF-1 glucansucrase. FEMS Microbiol. Lett. 177:243-248. [DOI] [PubMed] [Google Scholar]
- 32.Monchois, V., A. Reverte, M. Remaud-Simeon, P. Monsan, and R. M. Willemot. 1998. Effect of Leuconostoc mesenteroides NRRL B-512F dextransucrase carboxy-terminal deletions on dextran and oligosaccharide synthesis. Appl. Environ. Microbiol. 64:1644-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monchois, V., M. Vignon, P. C. Escalier, B. Svensson, and R. R. B. Russell. 2000. Involvement of Gln937 of Streptococcus downei GTF-I glucansucrase in transition-state stabilization. Eur. J. Biochem. 267:4127-4136. [DOI] [PubMed] [Google Scholar]
- 34.Monchois, V., R. M. Willemot, and P. Monsan. 1999. Glucansucrases: mechanism of action and structure-function relationships. FEMS Microbiol. Rev. 23:131-151. [DOI] [PubMed] [Google Scholar]
- 35.Moncrief, J. S., and T. D. Wilkins. 2000. Genetics of Clostridium difficile toxins. Curr. Top. Microbiol. Immunol. 250:35-54. [DOI] [PubMed] [Google Scholar]
- 36.Mukasa, H., A. Shimamura, and H. Tsumori. 2000. Nigerooligosaccharide acceptor reaction of Streptococcus sobrinus glucosyltransferase GTF-I. Carbohydr. Res. 326:98-103. [DOI] [PubMed] [Google Scholar]
- 37.Remaud-Simeon, M., C. Albenne, G. Joucia, E. Fabre, S. Bozonnet, S. Pizzut, P. Escalier, G. Potocki-Veronese, and P. Monsan. 2003. Glucansucrases: structural basis, mechanistic aspects, and new perspectives for engineering. Oligosaccharides Food Agric. 849:90-103. [Google Scholar]
- 38.Russell, R. R. B. 1979. Glucan-binding proteins of Streptococcus mutans serotype c. J. Gen. Microbiol. 112:197-201. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sanchez-Puelles, J. M., J. M. Sanz, J. L. Garcia, and E. Garcia. 1992. Immobilization and single-step purification of fusion proteins using DEAE-cellulose. Eur. J. Biochem. 203:153-159. [DOI] [PubMed] [Google Scholar]
- 41.Shah, D. S. H., and R. R. B. Russell. 2002. Glucan binding domain of streptococcal glucosyltransferases. Biologia 57:129-136. [Google Scholar]
- 42.Shah, D. S. H., and R. R. B. Russell. 2004. A novel glucan-binding protein with lipase activity from the oral pathogen, Streptococcus mutans. Microbiology 150:1947-1956. [DOI] [PubMed] [Google Scholar]
- 43.Sun, J. W., S. Y. Wanda, A. Camilli, and R. Curtiss III. 1994. Cloning and DNA sequencing of the dextranase inhibitor gene (dei) from Streptococcus sobrinus. J. Bacteriol. 176:7213-7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vickerman, M. M., M. C. Sulavik, J. D. Nowak, N. M. Gardner, G. W. Jones, and D. B. Clewell. 1997. Nucleotide sequence analysis of the Streptococcus gordonii glucosyltransferase gene, gtfG. DNA Seq. 7:83-95. [DOI] [PubMed] [Google Scholar]
- 45.von Eichel-Streiber, C., M. Sauerborn, and H. K. Kuramitsu. 1992. Evidence for a modular structure of the homologous repetitive C-terminal carbohydrate-binding sites of Clostridium difficile toxins and Streptococcus mutans glucosyltransferases. J. Bacteriol. 174:6707-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong, C., S. A. Hefta, R. J. Paxton, J. E. Shively, and G. Mooser. 1990. Size and subdomain architecture of the glucan-binding domain of sucrose:3-α-d-glucosyltransferase from Streptococcus sobrinus. Infect. Immun. 58:2165-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wren, B. W. 1991. A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol. Microbiol. 5:797-803. [DOI] [PubMed] [Google Scholar]
- 48.Wren, B. W., R. R. B. Russell, and S. Tabaqchali. 1991. Antigenic cross-reactivity and functional inhibition by antibodies to Clostridium difficile toxin A, Streptococcus mutans glucan-binding protein, and a synthetic peptide. Infect. Immun. 59:3151-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yother, J., and D. E. Briles. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J. Bacteriol. 174:601-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zahnley, J. C., and M. R. Smith. 2000. Cellular association of glucosyltransferases in Leuconostoc mesenteroides and effects of detergent on cell association. Appl. Biochem. Biotechnol. 87:57-70. [DOI] [PubMed] [Google Scholar]