Abstract
Oral streptococci such as Streptococcus gordonii are facultative anaerobes that initiate biofilm formation on tooth surfaces. An isolated S. gordonii::Tn917-lac biofilm-defective mutant contained a transposon insertion in an open reading frame (ORF) encoding a homolog of NosX of Ralstonia eutropha, a putative maturation factor of nitrous oxide reductase. Located downstream are two genes, qor1 and qor2, predicted to encode two putative NADPH quinone oxidoreductases. These three genes are cotranscribed, forming a putative oxidative stress response (osr) operon in S. gordonii. Inactivation of nosX, qor1, or qor2 resulted in biofilm-defective phenotypes. Expression of nosX, measured by the β-galactosidase activity of the nosX::Tn917-lac mutant, was growth-phase dependent and enhanced when grown under aerobic conditions or in the presence of paraquat. Real-time reverse transcription-PCR revealed that nosX-specific mRNA levels were increased approximately 8.4 and 3.5 fold in biofilm-derived cells grown on plastic and glass, respectively, when compared to planktonic cells. Expression of nosX increased 19.9 fold in cells grown under aerated aerobic conditions and 4.7 fold in cells grown under static aerobic conditions. Two ORFs immediately adjacent to the osr operon encode a putative NADH oxidase (Nox) and a putative thiol-specific antioxidant enzyme (AhpC, for alkyl hydroperoxide peroxidase C). Expression of nox and ahpC was also significantly increased in cells grown under aerated and static aerobic conditions when compared to anaerobic conditions. In addition, nox expression was increased in biofilm cells compared to planktonic cells. These genes may be part of an island that deals with oxidoreductive response, some of which may be important in S. gordonii biofilm formation.
Oral biofilms are complex, highly organized bacterial communities which contribute significantly towards the development of dental caries and periodontal disease. Formation of oral biofilms is initiated by the attachment of primary colonizers to saliva-conditioned oral surfaces. The facultative anaerobe, Streptococcus gordonii, is one of the pioneer colonizers of tooth surfaces and a species from the mitis streptococcal group (21), which constitutes a majority of the cultivable bacteria found in dental plaque (12). Subsequent attachment of other bacteria and growth result in microcolonies that increase in size and the eventual establishment of a complex oral biofilm, consisting of an ordered community of heterogeneous microbial species. The growth of this multilayered, cellular, matrix-embedded community and its eventual dispersal and spread require the coordinated expression of an array of genes (42, 49). Isolation and analysis of biofilm-defective mutants have resulted in the identification of a number of biofilm-associated genetic loci in oral bacteria (26). These include genetic competence (25, 27, 60), quorum sensing and signaling (4, 5, 14, 24, 27, 32, 57, 60), cell wall biosynthesis (27, 57, 60), exopolysaccharide synthesis (20), stress response (23, 28), nutritional and growth phase adaptation (9, 29, 57), and specific bacterial surface proteins (13, 22, 27, 41, 57).
The transition from a planktonic existence to sessile growth occurs primarily in response to environmental cues. Oral biofilms are highly structured, have distinct architecture, and are capable of adapting to abrupt changes in environmental conditions such as O2 levels, nutrient availability, and acidification (24, 57). For example, oxygen levels in the environment have been shown to affect biofilm formation of oral streptococci on abiotic surfaces (6).
Although often considered to be relatively anaerobic, oral biofilms have active oxygen metabolism and biofilm bacteria, including anaerobes, and have developed defenses against oxidative stress (30). In subgingival biofilms, the measured residual oxygen levels were sufficient to allow for oxygen metabolism by organisms considered to be extremely anaerobic, such as Treponema denticola, which metabolize oxygen by means of NAD (or NADH) oxidases and produce the protective enzymes superoxide dismutase and NADH peroxidase (30).
Oral streptococci are facultative anaerobes found mainly in supragingival biofilms. The oxidative capabilities of streptococci have been observed, such as the aerobic pathway for glucose oxidation in Streptococcus agalactiae (33). An unavoidable by-product of the aerobic lifestyle is oxidative stress, as O2− and H2O2 are generated by the auto-oxidation of components of the respiratory chain. Both aerobic and anaerobic bacteria have adaptive responses to elevated levels of oxidative stress, probably by sensing the increased levels of reactive oxygen species and by transducing the signal into increased expression of defense activities (50). Exposure to reactive oxygen intermediates can damage proteins, nucleic acids, and cells (50). To counter this, bacterial cell membranes constitutively express enzymes that detoxify the reactive oxygen species and repair the damage caused. Quinones are widely distributed in nature and play vital roles in management of oxidative stress and gene regulation (47). Ubiquinone is a bacterial respiratory quinone that functions as a redox mediator in aerobic respiration and retains antioxidant activity only in its reduced state. In Escherichia coli, a soluble quinone oxidoreductase (Qor) complexed with NADPH maintains ubiquinone in its reduced state, thereby promoting its antioxidant function (47).
A number of oxidative stress response systems have been identified in streptococci. In Streptococcus pyogenes, the PerR regulator, which is involved in the regulation of oxidative stress responses and iron homeostasis, also contributes to virulence (40). Two distinct oxygen-inducible NADH oxidases have been identified in Streptococcus mutans (17). LuxS-mediated quorum sensing in S. mutans has recently been shown to be involved in the regulation of oxidative stress and acid tolerance as well as biofilm formation (57). Inactivation of msrA in S. gordonii increased sensitivity to hydrogen peroxide, suggesting a role for streptococcal MsrA, a methionine sulfoxide reductase homolog, in protecting against oxidative stress (54).
This study reports the isolation and characterization of a S. gordonii Tn917-lac biofilm-defective mutant, which harbors a unique transposon insertion at a locus we designated nosX, part of a putative oxygen stress response (osr) operon. The osr operon consists of three genes, nosX and two Qor genes, the qor1 and qor2 genes. Results from the present study suggest that these genes are regulated by environmental oxygen levels, are likely to be involved in bacterial response to oxidative stress, and probably play a significant role in the development of biofilms.
MATERIALS AND METHODS
Bacteria, media, and chemicals.
S. gordonii Challis 2, the rifamycin-resistant (500 μg/ml) strain of S. gordonii Challis (27), was used as the parent strain. Unless otherwise indicated, bacteria were subcultured and maintained routinely on brain heart infusion (BHI) agar (BBL; Becton Dickinson, Cockeysville, Md.) or Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) supplemented with 0.2% yeast extract (THBYE) at 37°C under anaerobic conditions (VWR [Plainfield, N.J.] anaerobic chamber). Kanamycin or erythromycin was included in the medium when appropriate at a final concentration of 350 or 10 μg per ml, respectively. All chemicals were purchased from Sigma (St. Louis, Mo.) or Fisher Scientific (Pittsburgh, Pa.). All enzymes for DNA manipulations were purchased from Promega (Madison, Wis.) or Fisher Scientific unless stated otherwise. All primers used are listed in Table 1.
TABLE 1.
Oligonucleotide primers used in this studya
| Primer | Nucleotide sequence (5′ to 3′) (enzyme) | Location | Application | Amplification target (bp) |
|---|---|---|---|---|
| SSP-917B | AAC TGT ACC ACT AAT AAC TCA CAA T | Tn917-lac | PCR | Tn917-lac 5′ flanking region |
| PBSSK3 | PBSSK3: GTA AAA CGA CGG CCA GT | pBluescript | PCR | |
| Kan1 MluI | CGA CGC GTC GCC GTC GAT ACT ATG TTA T (MluI) | pSF151 | Mutagenesis | kan (1,073) |
| Kan2 SalI | CGG TCG ACC GAT CAG AGT ATG GAC AGT T (SalI) | pSF151 | Mutagenesis | |
| NosX1 | CGG GTA CCC GGG TCT TCC AAG TTC TTA G (KpnI) | nox | Mutagenesis of nosX | nosX 5′ flanking region (972) |
| NosX2 | CGA CGC GTC GGA ATG GAT CTA GAA GTG A (MluI) | nosX | Mutagenesis of nosX | |
| NosX3 | CGG TCG ACC GCG TTA TTA CAG AAG ATA ATC (SalI) | nosX | Mutagenesis of nosX | nosX 3′ flanking region (998) |
| NosX4 | CGC TCG AGC GAG CCA TTC AAG TAC AGA T (XhoI) | qor1 | Mutagenesis of qor1 | |
| O1P1 | CGG GTA CCC GGT GAT AAA AGT GGA TCT C (KpnI) | nosX | Mutagenesis of qor1 | qor1 5′ flanking region (1,062) |
| O1P2 | CGA CGC GTC GAA AGT GTT TTT GCA TGT A (MluI) | qor1 | Mutagenesis of qor1 | |
| O1P3 | CGG TCG ACC GGT ATC TTC GTG AAG ATT A (SalI) | qor1 | Mutagenesis of qor1 | qor1 3′ flanking region (858) |
| O1P4 | CGC TCG AGC GCA TAT CGA TTT GGT TAA C (XhoI) | qor2 | Mutagenesis of qor1 | |
| O2P1 | CGG GTA CCC GAT TAC TGT CCA TCC AAG A (KpnI) | qor1 | Mutagenesis of qor2 | qor2 5′ flanking region (1,018) |
| O2P2 | CGA CGC GTC GAT GAT TGA TCG TAG TTT G (MluI) | qor2 | Mutagenesis of qor2 | |
| O2P3 | CGG TCG ACC GTG GAG GTC ATG GAG AC (SalI) | qor2 | Mutagenesis of qor2 | qor2 3′ flanking region (698) |
| O2P4 | CGC TCG AGC GAG CTG CAG TGT TTT TAA C (XhoI) | ahpC | Mutagenesis of qor2 | |
| O2P1 | See above | nosX | RT-PCR | nosX-qor2 (1,018) |
| O2P2 | See above | qor2 | RT-PCR | |
| O1P3 | See above | qor1 | RT-PCR | qor1-qor2 (858) |
| O1P4 | See above | qor2 | RT-PCR | |
| NosX RT5′ | ATT GGC TTT AAA GAT GCC AGA GT | nosX | Real-time RT-PCR | nosX (101) |
| NosX RT3′ | GAA CGG GCA GTC GGA TCT AAT | nosX | Real-time RT-PCR | |
| O1RT5′ | TTG CCA TTG TTG GAA CGA ACT | qor1 | Real-time RT-PCR | qor1 (108) |
| O1RT3′ | CCT TGA TTT CAA CAA GTT CAA TTT C | qor1 | Real-time RT-PCR | |
| O2RT5′ | TTG CTA AAC GTG TAC CAC CTC AA | qor2 | Real-time RT-PCR | qor2 (111) |
| O2RT3′ | CGT TGC GAA GTG TTC CGA TA | qor2 | Real-time RT-PCR | |
| Nox RT5′ | AAC CAA GGA AAG AGA TGT TTG A | nox | Real-time RT-PCR | nox (109) |
| Nox RT3′ | GGT GCT AAC CAC GCA GGT A | nox | Real-time RT-PCR | |
| AhpC RT5′ | TTT CTT ACA TGG CCC ACA CCA | ahpC | Real-time RT-PCR | |
| AhpC RT3′ | TGA AGT CTA TGG ACG GAA AAA CTG T | ahpC | Real-time RT-PCR | ahpC (98) |
| 23S rRNA RT5′ | ACTGCAATGTGGACTCAGAATTTAT | 23S rRNA | Real-time RT-PCR | 23S rRNA gene (90) |
| 23S rRNA RT3′ | TACAGAATCTATTTAAAATACGAGGCTCT | 23S rRNA | Real-time RT-PCR |
Engineered restriction sites not present in the template sequence are in boldface type. The corresponding restriction enzyme is shown in parentheses to the right of the primer sequence.
Tn917-lac mutagenesis of S. gordonii.
S. gordonii Challis 2 was mutagenized with pTV32OK (10), and a lacZ fusion library was generated and screened for biofilm-deficient mutants as described previously (27). Chromosomal DNA isolated from S. gordonii Challis 2, the biofilm-deficient mutant was digested with HindIII and transferred onto a nitrocellulose membrane, and Southern hybridization with a digoxigenin-labeled pTV32-OK probe was performed as described previously (28).
Localization of transposon insertion site and sequence analyses.
The location of the transposon insertion was determined by sequence analysis of the region flanking the transposon as described previously (27). Briefly, pBluescript vector and chromosomal DNA from the mutant were digested with HindIII, purified, ligated, and used as a PCR template to amplify the Tn917 junctional region. The sequence obtained from the PCR product was compared with sequences in GenBank using the BLASTX and TBLASTN programs (1) to identify homologous bacterial sequences. Amino acid sequence alignments and phylogenetic analyses were performed and analyzed with the AlignX program in Vector NTI (Informax, Inc., Bethesda, Md.).
Biofilm assays.
The in vitro biofilm formation assays of S. gordonii Challis 2 and the biofilm-defective Challis::Tn917-lac mutant were performed with biofilm medium (BM) as previously described (27, 28). In addition to the microtiter plate biofilm assay, biofilm formation of S. gordonii Challis 2 and the biofilm-defective mutant was assayed on borosilicate glass coverslips suspended in BM in 50-ml Falcon tubes. The biofilm bacteria present on the glass coverslip were visualized directly by phase-contrast microscopy. Images were captured with a Nikon Coolpix 4500 digital camera, as described previously (28).
Biofilm formation was also assessed with a flow cell system (36) and visualized by confocal scanning laser microscopy (CSLM). A glass flow cell (BioSurface Technologies Corp., Bozeman, Mont.) containing a flow channel, 0.2 mm deep by 11.4 mm wide by 40.6 mm long, was used. The substratum consisted of a microscope glass coverslip. An overnight BHI culture was washed twice and resuspended in BM. An inoculum from this suspension was introduced in a 250-μl volume (2 × 107 cells) and allowed to bind to the surface by inversion of the flow cell for 15 min without flow, after which BM was pumped through the flow cell channel at a rate of 200 μl per min. After bacteria were grown in the flow cell at 37°C for 18 h, CSLM was used to examine the spatial organization of the biofilm formed in situ. After staining with the LIVE/DEAD bacLight bacterial viability stain according to the manufacturer's instructions (Molecular Probes, Eugene, Oreg.), cells were visualized with an air objective (magnification, ×20) with a TCS SP2 confocal microscope (Leica LaserTechnik GmbH, Heidelberg, Germany) equipped with an argon-krypton laser. Images were collected and analyzed with Image-Pro Plus computer software (Media Cybernetics, Silver Spring, Md.).
Image analysis of each strain was performed using five image stacks acquired from random positions of the flow channel, approximately 5 mm from the inlet. Images were acquired at 5-μm intervals down through the biofilm; therefore, the number of images in each stack varied according to the thickness of the biofilm. The data were processed using COMSTAT software (16). The parameters chosen to characterize the biofilm structures are total biomass, average thickness, area occupied, substratum coverage, and roughness coefficient of the biofilm.
Reverse transcription-PCR (RT-PCR) of S. gordonii Challis 2 RNA.
To characterize the transcription of the S. gordonii nosX gene, RT-PCR was performed using total RNA extracted from S. gordonii Challis 2 grown to mid-log phase (A600 of 0.3 to 0.4). RNA was purified from bacterial cells with an RNeasy minikit (QIAGEN) and DNase I was treated as previously described (28). Primers O2P1 and OP2, specific for an intergenic region that spans nosX to qor2, were used (Table 1). RT-PCR was performed with the Access RT-PCR system (Promega) under the conditions described previously (28). The RT-PCR products were visualized after 1% agarose gel electrophoresis. Strain Challis 2 DNA was used as the positive control, and the negative controls without reverse transcriptase were used to ensure that there was no contaminating DNA in each DNase I-treated RNA sample.
Construction and characterization of nosX, qor1, and qor2 mutants.
PCR ligation mutagenesis with vectorless intermediates was used to construct nosX, qor1, and qor2 deletion mutants (28). A kanamycin resistance gene, kan, amplified from plasmid pSF151 (51), was used as the antibiotic marker insert (Table 1). PCR products of the 5′- and 3′-flanking region and the kan cassette were ligated and used for transformation of S. gordonii Challis 2 as described previously (28). The segment of nosX which encodes residues 30 to 299 of the predicted 320-amino-acid NosX on the chromosome of S. gordonii was replaced with kan to generate a mutant with a nosX::Kanr allele. The two downstream genes, qor1 (residues 28 to 173 of the predicted 201-amino-acid sequence) and qor2 (residues 25 to 381 of the predicted 414-amino-acid sequence), were also replaced with kan to generate mutants with either a qor1::Kanr or qor2::Kanr allele, respectively. Transformants were plated on BHI agar containing 350 μg of kanamycin per ml and incubated at 37°C anaerobically for 3 to 5 days. RT-PCR was performed as described above with RNA isolated from nosX::Tn917-lac, nosX::Kanr, and qor1::Kanr mutants to determine whether these strains possessed a polar or a nonpolar mutation. S. gordonii Challis 2 RNA was used as a control.
The growth rates of S. gordonii Challis 2 strain and nosX::Tn917-lac, nosX::Kanr, qor1::Kanr, and qor2::Kanr mutants were assessed by inoculating the strains from an overnight THBYE culture into fresh THBYE (10 ml) and growing them at 37°C under anaerobic and static aerobic conditions. Bacterial growth was quantified by recording the absorbance at 600 nm at regular intervals over 24 h.
A disk diffusion assay modified from Vriesema et al. (54) was used to determine the susceptibility of bacteria to H2O2. Briefly, 100 μl of bacteria (106 CFU/ml) grown to exponential phase in BHI was added to 2 ml of liquid 0.7% BHI agar at 45°C and layered onto BHI plates (prewarmed to 37°C in an incubator). An 8-mm filter paper disk containing 10 μl of H2O and 1, 2, or 4 M H2O2 was placed on the plate. The plates were incubated overnight at 37°C, and the zone of growth inhibition was measured. All assays were performed in triplicate, and results were reproducible within a range of ±1 mm. No growth inhibition was seen in disks containing H2O. To assess the sensitivity of bacteria to the superoxide-generating agent paraquat, overnight cultures were inoculated in fresh BM containing 0.1 mM paraquat (40). After 72 h of incubation at 37°C, bacterial growth, recorded as absorbance at 575 nm, was expressed as a percentage of the growth in BM with no paraquat. The ability of S. gordonii Challis 2, nosX::Tn917-lac, nosX::Kanr, qor1::Kanr, and qor2::Kanr strains to form biofilms was assessed on a microtiter plate, on a glass surface using phase-contrast microscopy, and in a flow cell system with CSLM as described above.
Expression of nosX in different environmental conditions.
To study the regulation of nosX expression, β-galactosidase activity of the biofilm-defective S. gordonii nosX::Tn917-lac mutant was determined by a fluorimetric assay (28) using 4-methylumbelliferyl-β-d-galactoside (MUG). The effect of growth phase on the expression of nosX was examined by measuring the β-galactosidase activity of the nosX::Tn917-lac mutant grown in THBYE under anaerobic conditions over 24 h. To determine the effects of various substrates on nosX expression, cells were grown in BM under the conditions described previously (28), in BM under static aerobic conditions, and in BM under anaerobic conditions in the presence of oxidizing agents (paraquat and H2O2), reducing agents (cysteine hydrochloride and dithiothreitol), FeCl3, thiamine pyrophosphate, and thiamine monophosphate.
Real-time quantitative RT-PCR.
To independently and directly assess transcriptional changes of nos, qor1, and qor2 genes in various environmental conditions, real-time quantitative RT-PCR was performed with the ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, Calif.) as described previously (14). The primer pairs used (Table 1) were developed from the S. gordonii genome database with Primer Express (Applied Biosystems) for uniformity in amplicon size (approximately 100 bp) and melting temperature. Expression levels of the nosX, qor1, qor2, nox (NADH oxidase), and ahpC (alkyl hydroperoxide peroxidase C) genes were examined. RNA was extracted from S. gordonii Challis 2 biofilm and planktonic cultures and from cells grown under aerated aerobic (on a shaker-incubator at 250 rpm), static aerobic, and anaerobic conditions. Total RNA was extracted from cells grown in BM to mid-log phase as described previously (28). RNA was also isolated from the four mutant strains and Challis 2 grown to mid-log phase in BM under anaerobic conditions. Three independent RNA preparations from three separate experiments of each particular growth condition were used for real-time PCR analysis. RNA concentrations were normalized by using amplification of the 23S rRNA gene of S. gordonii as an internal standard. For each RT-PCR, cDNA synthesis and PCR amplification were performed in a two-step reaction mixture. RT was performed with DNaseI-treated RNA with the Taqman RT reagent kit (Applied Biosystems). Each 50-μl reaction mixture containing 5.5 mM MgCl2, 500 μM each deoxynucleoside triphosphate, 0.4 U of RNase inhibitor/μl, 1.25 U of reverse transcriptase/μl, 2.5 μM reverse primer, and 10 ng of total RNA in a 1× RT buffer was incubated at 48°C for 30 min, followed by 95°C for 5 min. Real-time PCR amplifications were then performed in triplicate with the SYBR Green PCR master mix (Applied Biosystems) according to the manufacturer's instructions in 50-μl reaction mixtures that contained 1× SYBR Green I PCR master mix, 300 nM forward primer, 50 nM additional reverse primer, and 5 μl of cDNA template. PCR conditions included an initial denaturation at 95°C for 10 min, followed by a 50-cycle amplification, with each cycle consisting of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. Primer pairs were checked for primer-dimer formation by omitting the RNA template in the two-step RT-PCR. As an additional control for each primer pair and each RNA sample, the cDNA synthesis reaction was carried out without reverse transcriptase to identify contamination of RNA samples by residual genomic DNA.
Ten-fold serial dilutions of cDNA were employed to generate standard curves to determine the sensitivity of the assay for the detection of each gene. The value used for quantitation and comparison was the threshold cycle (CT), defined as the number of cycles required to cross the midpoint of the detectable amplification curve, which was normalized to a passive reference dye (carboxy-X-rhodamine) included in each reaction. Real-time PCR analysis was performed on three independent RNA preparations from three separate cultures. For each target gene, the CT values for RNA prepared from different growth conditions were compared. Expression levels for each gene in each environment are presented as relative fold induction. Student's t test with an alpha level of 0.05 was used to calculate the significance of the difference between the mean expression levels of a particular gene in two different growth conditions or strains.
Sequences.
DNA and protein sequences used in this study were retrieved from the National Center for Biotechnology Information genomic BLAST pages that contain sequences of completed and unfinished microbial genomes (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi).
RESULTS
Identification and characterization of an S. gordonii biofilm-defective mutant.
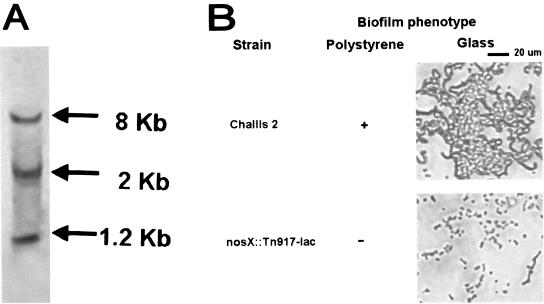
A biofilm-defective mutant was identified after screening an in-frame lacZ-fusion library generated by Tn917-lac mutagenesis of S. gordonii Challis 2 for mutants with altered ability to form biofilms. Southern hybridization with a Tn917-lac probe confirmed that a single transposon insertion had occurred in the biofilm-defective Tn917-lac mutant (Fig. 1A). The microtiter plate biofilm assay showed that biofilm formation of the nosX::Tn917-lac mutant (A575 ± standard deviation, 0.72 ± 0.32) was reduced by 77% when compared to the S. gordonii Challis 2 strain (A575 ± standard deviation, 4.63 ± 0.05). The mutant also displayed a biofilm-deficient phenotype on borosilicate glass (Fig. 1B), as the mutant was not able to form a biofilm over time.
FIG. 1.
(A) Southern hybridization of HindIII-digested DNA from Tn917-lac biofilm-defective S. gordonii nosX::Tn917-lac mutant. A digoxigenin-labeled pTV32-OK containing Tn917-lac (which has two HindIII sites) was used as the probe. Three hybridizing bands were produced in HindIII-digested DNA, demonstrating the presence of a single transposon insertion. (B) Biofilm phenotypes of S. gordonii Challis 2 and nosX::Tn917-lac mutant as determined by the polystyrene microtiter plate assay and direct observation of biofilms formed on borosilicate glass surfaces by phase-contrast microscopy. Cells were grown in BM for 24 h at 37°C under anaerobic conditions.
The nucleotide sequence of the region 5′ to the transposon insertion was used in sequence similarity BLASTN searches of the S. gordonii genome database (http://www.tigr.org). The transposon insertion was located immediately downstream of the start codon of an open reading frame (ORF) which encodes a protein homologous to a Ralstonia eutropha NosX (GenBank accession no. AAP86003). NosX, a homolog of ApbE from Salmonella enterica serovar Typhimurium (3), is one of seven putative maturation factors (NosX) of nitrous oxide reductase in Pseudomonas putida (58). This gene was not identified when Tn916 mutagenesis was used in a previous screening of biofilm-defective mutants of S. gordonii Challis 2 (27).
Genetic organization and phylogenetic analyses of the putative osr operon.
Located downstream of the transposon insertion locus, nosX, are two ORFs that encode proteins homologous to Qors, which we designated qor1 and qor2 based on sequence homology. Immediately downstream of qor2 is a palindromic sequence (TAAGTAAAAAAAGAAT), which begins with the qor2 stop codon (TAA). Another palindrome (ACCTTTAAAATTTAATTTACCA) was identified 216 bp upstream from the start codon of nosX.
NosX.
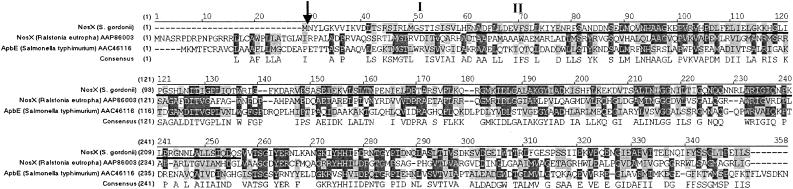
The 960-bp nosX ORF encodes a 320-amino-acid sequence with a predicted molecular mass of 35.7 kDa. The homologs to NosX that were found are NosX of R. eutropha (accession no. AAP86003), NosX of P. putida (58), NosX of Rhizobium meliloti (8), and ApbE of S. enterica serovar Typhimurium (3). NosX also has homology in the C-terminal domain with RnfF of Rhodobacter capsulatus, an iron-sulfur secretory membrane protein with a signal sequence of 44 amino acids involved in electron transport to nitrogenase (45). The N-terminal portion of R. capsulatus RnfF contains a cysteine motif (C-X3-C-X-C-X2-C) typical of 4Fe-4S proteins. In contrast, another homolog, NosX of R. meliloti, is a peripheral membrane protein involved in N2O reduction that contains only one cysteine residue, lacks cysteine motifs, and is not an iron-sulfur protein (8). Similarly, S. gordonii NosX does not contain any cysteine residues and therefore is probably not an Fe-S protein. S. gordonii NosX does not have a transmembrane helix, a secretory leader sequence, or a lipoprotein peptide signal cleavage sequence, indicating it is not a lipoprotein (Fig. 2). The N-terminal of NosX does not contain the conserved twin-arginine type of signal peptide found in the NosX of Paracoccus denitrificans (43), suggesting it is not a metalloprotein.
FIG. 2.
Alignments of the deduced amino acid sequences of S gordonii nosX with homologs in R. eutropha (accession no. AAP86003) and S. enterica serovar Typhimurium (accession no. AAC46116) with the AlignX program of Vector NTI (Informax). Amino acids that are identical and conserved are highlighted in dark grey and light grey, respectively. A black arrow marks the position of the Tn917-lac insertion in the nosX::Tn917-lac mutant. Residues homologous to a NADH-ubiquinone oxidoreductase chain 2 (I) and NADH dehydrogenase (II) signature sequences are underlined.
S. gordonii NosX has an NADH-ubiquinone oxidoreductase chain 2 signature sequence (SIRLMGSTISISVL), at residues 17 to 30 (Fig. 2). A conserved RNA structure, known as the thi box, is highly conserved in the 5′ regions of thiamine biosynthetic and transport genes of both gram-positive and gram-negative bacteria and is involved in regulation of thiamine biosynthetic gene expression (34). Since apbE, a nosX homolog in S. enterica, was originally proposed as being involved in thiamine biosynthesis, we examined whether a thi box was present in the promoter region of nosX in S. gordonii. No thi box homolog was found in the 5′ region of nosX, qor1, or qor2. Although the function of S. gordonii NosX is currently unknown, it may be involved in the maturation of the oxidoreductases encoded by qor1 and qor2.
Qor1.
The two ORFs located downstream of nosX encode proteins that are homologous to the NAD(P)H Qor family of reductases. The 603-bp qor1 starts 20 bp downstream of the nosX stop codon and is predicted to encode a 201-amino-acid peptide with a predicted molecular mass of 22.6 kDa. The homologs of Qor1 found are YieF of E. coli, a hypothetical protein which contains an NADPH-dependent reductase protein motif, and two Qors, MdaB of Helicobacter pylori (55) and Nqr of the plant Arabidopsis thaliana (48).
Amino acid residues 2 to 167 of S. gordonii Qor1 were homologous to NADPH-dependent flavin mononucleotide (FMN) reductases. FMN reductases, such as SsuE of E. coli (53), catalyze the reaction NAD(P)H + FN = NAD(P) + FMNH (2). Also present are NAD(P)H dehydrogenase (quinone) motifs at residues 63 to 108 and 141 to 191 and a possible transmembrane helix at residues 83 to 100.
Qor2.
The qor2 ORF was 1,242 bp in length, starting 18 bp downstream of the qor1 stop codon. It encodes a deduced 414-amino-acid peptide with a predicted molecular mass of 45.7 kDa. Qor2 also shares significant homology with the oxidoreductases Qor and YieF of E. coli and Nqr of A. thaliana. Amino acid residues 1 to 163 of S. gordonii Qor2 were homologous to the NADPH-dependent FMN reductases, whereas residues 181 to 371 were homologous to oxidoreductases.
In many bacteria, qor genes are unique in the genome, an exception being Staphylococcus aureus, which possesses two qor genes located adjacent to each other that share 26.2% identity (31). Two qor genes are also present in S. gordonii, which encode the highly homologous proteins Qor1 and Qor2. Qor1 and Qor2 share 19.0% identity and 28.2% similarity; however, this homology is limited to the N terminus of Qor2 (Fig. 3). Qor2 is homologous to YieF of E. coli and two Qors, MdaB of H. pylori (55) and Nqr of the plant A. thaliana (48). Similar to QOR of E. coli, Qor1 of S. gordonii contains the Qor nucleotide-binding fingerprint motif AXXGXXG (Fig. 3), which is the result of the insertion of an alanine residue in the fingerprint region of the nucleotide-binding domain (52). On the other hand, S. gordonii Qor2 contains an alcohol dehydrogenase-type NAD(P)H-binding motif, GXGXXG (37) but does not possess the Qor nucleotide-binding motif (Fig. 3). Likewise, one of the two Qors in S. aureus, QorA, contains an NAD(P)H-binding motif, while the second downstream Qor homolog contains an alcohol dehydrogenase-type NAD(P)H-binding motif sequence (31). A copper/zinc superoxide dismutase motif identified in Qor2 (residues 373 to 406) is not found in Qor1, suggesting that Qor2 may be involved in degradation of oxygen radicals. In addition, there are two His residues at the N-terminal of Qor2 that are absent from Qor1, which may be a putative metal-binding site (Fig. 3).
FIG. 3.
Alignments of the deduced amino acid sequences of S. gordonii oxidoreductases Qor1 and Qor2. Amino acids that are identical and conserved are highlighted in dark grey and light grey, respectively. A Qor nucleotide-binding motif present in Qor1 is boxed. In Qor2, an NAD(P)H binding motif (I) and an NADP binding motif (II) are underlined, while two His residues are marked with an asterisk.
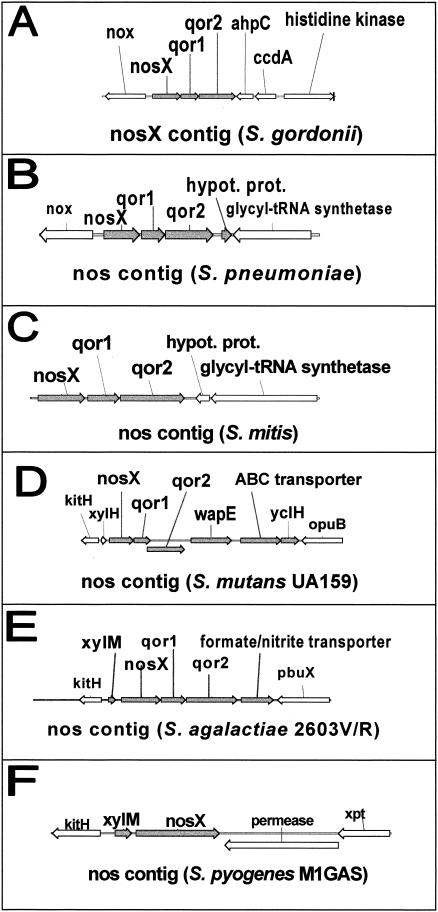
The organization of the putative osr operon in S. gordonii was compared to homologs in other streptococci (Fig. 4). The nosX genes in the mitis group (S. gordonii, Streptococcus pneumoniae, and Streptococcus mitis), S. mutans, and S. agalactiae are contiguous with qor1 and qor2, whereas only the nosX gene was identified in S. pyogenes. In S. mutans, S. agalactiae, and S. pyogenes, an additional ORF located upstream of nosX is transcribed in the same direction and encodes an oxalocrotonate tautomerase.
FIG. 4.
Organization of the putative nos operon in different streptococci, as determined by sequences from their completed and unfinished microbial genome databases (http://www.ncbi.nih.gov). The nosX genes in the mitis group (S. gordonii, S. pneumoniae, and S. mitis), S. mutans, and S. agalactiae are contiguous with qor1 and qor2, whereas only the nosX gene was identified in S. pyogenes. In S. mutans, S. agalactiae, and S. pyogenes, an additional ORF located upstream of nosX is transcribed in the same direction and encodes an oxalocrotonate tautomerase. ORFs transcribed in the same direction as nosX are shown in grey.
NADH oxidase.
Located upstream of nosX is a divergently oriented ORF with a start codon 249 bp upstream from the start codon of nosX. This ORF was designated nox, as it encodes a putative 458-amino acid protein that is highly homologous to Nox2 of S. mutans (75.4% identity, 84.5% similarity) (17) and Nox of S. pneumoniae, a soluble flavoprotein that reoxidizes NADH and reduces O2 to H2O (88.9% identity, 95% similarity) (2). Nox2, one of two NADH oxidases in S. mutans, is an H2O-forming oxidase that catalyzes the four-electron reduction of O2 by NADH and is induced under aerobic conditions (17). Inactivation of nox in S. pneumoniae attenuated its virulence and persistence in mice, suggesting that NADH oxidase activity is important for the regulation virulence, possibly by sensing and adapting to O2 concentration in the environment (2). Nox could be involved in the defense of S. pneumoniae against oxidative stress by reducing potentially dangerous oxygen to water or by regulating competence to allow the capture of DNA as a source of nucleotides and DNA fragments for the repair of O2-induced chromosomal damage (2).
AhpC.
Another ORF oriented in the opposite direction was identified downstream of qor2, with a stop codon 53 bp from the stop codon of qor2. It encodes a 186-amino-acid protein homologous to the thiol-specific antioxidant protein/alky hydroperoxide peroxidase C (AhpC) family of proteins (7), and was therefore designated AhpC. The thiol-specific antioxidant/AhpC family is a type of peroxidase that provides defense against oxidative stress by reducing hydroperoxides, by using thioredoxin and other thiol-containing reducing agents (19, 38). The deduced S. gordonii AhpC amino acid sequence is homologous to AhpC of Bacillus cereus (GenBank accession no. NP_979929) and AhpC of S. mutans (38).
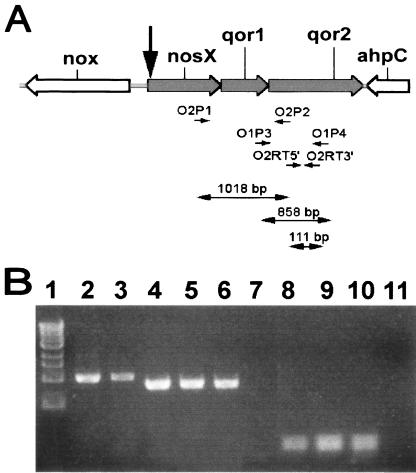
RT-PCR with RNA from the Challis 2 strain using primers O2P1 and O2P2 (Table 1), which are specific for a 1,018-bp region that spans the nosX and qor2 intergenic region produced a product of the predicted size (Fig. 5B, lane 3). Fidelity of the primers used was confirmed by PCR with Challis 2 DNA as the template (Fig. 5B, lane 2). These results demonstrate that nosX, qor1, and qor2 are cotranscribed as a single operon. The two adjacent genes, which are the putative nox and ahpC genes, are divergently transcribed. Therefore, the putative osr operon in S. gordonii consists of the three ORFs, nosX, qor1, and qor2.
FIG. 5.
RT-PCR analysis of RNA extracted from S. gordonii Challis 2 and nosX::Tn917-lac strains. (A) The organization of nos and adjacent genes, location of primers, and predicted size of successfully amplified RT-PCR products are shown. See Table 1 for all primers used. The black arrow denotes the position of the Tn917-lac insertion in the biofilm-defective mutant. (B) RT-PCR analysis of transcripts of the ORFs within the putative nos operon. The total RNA extracted from the Challis 2, nosX::Tn917-lac, nosX::Kanr, and qor1::Kanr strains was used as the template. Challis 2 DNA was used as the positive control to verify the fidelity of the primers used. Lanes: 1, 1-kb DNA marker; 2, Challis 2 DNA with primers O2P1 and O2P2 (spans nosX-qor2; 1,018 bp); 3, Challis 2 RNA with primers O2P1 and O2P2; 4, Challis 2 DNA with primers O1P3 and O1P4 (spans qor1-qor2; 858 bp); 5, Challis 2 RNA with primers O1P3 and O1P4; 6, nosX::Kanr RNA with primers O1P3 and O1P4; 7, nosX::Tn917-lac RNA with primers O1P3 and O1P4; 8, Challis 2 DNA with primers O2RT5′ and O2RT3′ (within qor2; 111 bp); 9, Challis 2 RNA with primers O2RT5′ and O2RT3′; 10, qor1::Kanr RNA with primers O2RT5′ and O2RT3′; 11, nosX::Tn917-lac RNA with primers O2RT5′ and O2RT3′.
Expression of the nosX gene.
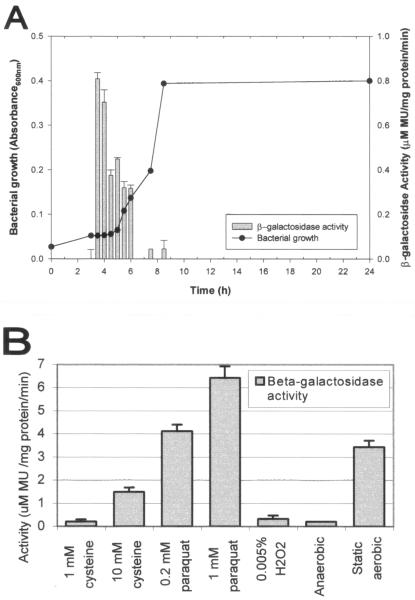
The effect of growth phase and various environmental conditions on the expression of nosX was examined by measuring the β-galactosidase activity of the S. gordonii nosX::Tn917-lac. When grown under anaerobic conditions, the expression of nosX was only observed during early to mid-exponential growth phase in S. gordonii (Fig. 6A). The expression of nosX was significantly increased in the presence of paraquat and cysteine (Fig. 6B). Expression of nosX was also upregulated when cells were grown as a biofilm. Exogenous FeCl3 at a concentration of 0.5 μM or higher has been shown to correct the growth defect associated with mutation of apbE, the nosX homolog in S. enterica serovar Typhimurium (46). In contrast, no effect on nosX expression in the presence of FeCl3 (0.05, 0.1, and 0.5 mM) was observed in S. gordonii. Dithiothreitol (1 and 0.1 mM), 10 mM thiamine, 10 mM thiamine monophsophate, and 10 mM thiamine pyrophosphate also did not significantly affect nosX expression. All other conditions tested did not have a significant effect on expression (data not shown). These include BM adjusted to various pHs or supplemented with zinc (1 mM), manganese (1 mM), 10% clarified whole human saliva, 10% human serum, 0.8% (wt/vol) sugar (fructose, galactose, glucose, lactose, maltose, mannose, sucrose, mannitol, sorbitol, or xylitol), or 100 mM amino acid (alanine, arginine, aspartate, cysteine, glutamine, glutamate, glycine, histidine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tyrosine, or valine).
FIG. 6.
Effect of different growth conditions on nosX expression. The biofim-deficient nosX::Tn917-lac mutant was grown at 37°C under anaerobic conditions unless otherwise specified. The β-galactosidase activities of the nosX-lacZ transcriptional fusion were quantified with MUG (28). All assays were performed in triplicate, and mean values and standard deviations of the β-galactosidase activities are shown. MU, 4-methyl-umbelliferone. (A) Relation between growth phase and nosX expression. The nosX::Tn917-lac mutant was grown in THBYE. Growth and β-galactosidase activity at various times over 24 h are shown. (B) Effects of different concentrations of various substrates on nosX expression. The nosX::Tn917-lac mutant was grown in BM with different substrates or under static anaerobic conditions as indicated.
Mutational analysis of nosX, qor1, and qor2 mutants.
To confirm the biofilm-defective phenotype identified in S. gordonii nosX::Tn917-lac, an insertional inactivation of nosX was constructed by allelic exchange. The qor1 and qor2 genes were also inactivated in the Challis 2 strain to investigate their role in biofilm formation and oxygen tolerance. Mutants with either a qor1::Kanr or qor2::Kanr allele were also generated by the same method. PCR confirmed that integration of the kan resistance gene in nosX, qor1, or qor2 in the chromosome had occurred as predicted (data not shown).
RT-PCR with primers spanning the qor1 and qor2 intergenic region successfully obtained a PCR product of the predicted size from RNA isolated from S. gordonii Challis 2 but not from nosX::Tn917-lac RNA (Fig. 5B, lanes 5, 7, and 11), indicating that the transposition resulted in a polar nosX::Tn917-lac mutation. RT-PCR results confirmed that the nosX and qor1 mutations constructed were nonpolar (Fig. 5B, lanes 6 and 10). Fidelity of the primers used was confirmed by PCR with Challis 2 DNA as the template (Fig. 5B, lanes 4 and 8). RT-PCR with Challis 2 RNA as the template (Fig. 5B, lanes 5 and 9) verified the presence of the transcript in S. gordonii under the conditions used for RNA extraction.
Biofilm formation of nosX::Tn917-lac (A575 ± standard deviation, 0.72 ± 0.32), nosX::Kanr (A575 ± standard deviation, 1.05 ± 0.12), qor1::Kanr (A575 ± standard deviation, 2.90 ± 0.18) and qor2::Kanr (A575 ± standard deviation, 1.64 ± 0.83) mutants were at levels all lower than that of the Challis 2 strain (A575 ± standard deviation, 4.63 ± 0.05). When compared to the Challis 2 strain, the polar mutation in nosX::Tn917-lac resulted in a 84% reduction in biofilm formation, while inactivation of nosX, qor1, and qor2 resulted in 77, 37, and 65% reduction in biofilm formation, respectively. The most significant reductions in biofilm formation were observed when either the whole operon or nosX was inactivated. These results suggest that all three genes may be involved in biofilm formation of S. gordonii.
Growth assays.
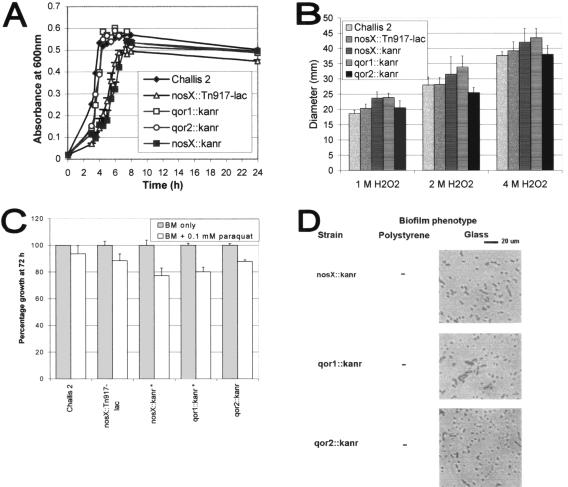
Under static aerobic conditions, two of the mutant strains, nosX::Tn917-lac and nosX::Kanr, grew at a slower rate than the Challis 2 strain (Fig. 7A), indicating that nosX and qor1 probably play a role in the growth of S. gordonii in the presence of higher levels of O2. However, the final growth yields of both mutants after 24 h were not significantly different from that of Challis 2. It is likely that there is minimal oxygen diffusion during static growth into the cultures and that the culture may become relatively anaerobic. Therefore, the differences observed in the growth rate of these two mutants appear to be an extension of the lag period.
FIG. 7.
Oxidative stress assays. (A) Growth curves of S. gordonii Challis 2, nosX::Tn917-lac, nosX::Kanr, qor1::Kanr, and qor2::Kanr strains in 10 ml of THBYE broth over 24 h at 37°C under static aerobic conditions. Growth was measured as absorbance at 600 nm. All assays were performed in triplicate, and mean values and standard deviations are shown. (B) A disk diffusion assay was performed to compare the H2O2 sensitivity of S. gordonii Challis 2, nosX::Tn917-lac, nosX::Kanr, qor1::Kanr, and qor2::Kanr mutant strains. All assays were performed in triplicate, and mean values and standard deviations are shown. (C) A growth assay was performed to compare the sensitivity of S. gordonii Challis 2 and nosX::Tn917-lac, nosX::Kanr, qor1::Kanr, and qor2::Kanr mutant strains to paraquat. Cells were grown in 100 μl of BM under anaerobic conditions in the presence of 1 mM paraquat. The bacterial growth (A575) at 72 h was measured and expressed as the percentage of growth relative to BM without paraquat. Strains marked with an asterisk (nosX::Kanr and qor1::Kanr) denote significant difference and when compared to the Challis 2 strain were significantly lower than Challis 2 (P < 0.05). All assays were performed in triplicate, and mean values and standard deviations are shown. (D) Biofilm phenotypes of S. gordonii nosX::Kanr, qor1::Kanr, and qor2::Kanr strains were determined by the polystyrene microtiter plate assay; direct observation of biofilms formed on borosilicate glass surfaces was compared by phase-contrast microscopy. Cells were grown in BM for 24 h at 37°C under anaerobic conditions.
Results from sequence analysis suggest that the putative osr operon may be involved in tolerance to oxidative stress. The paraquat and H2O2 sensitivity of the Challis 2 and mutant strains nosX::Tn917-lac, nosX::Kanr, qor1::Kanr, and aqor2::Kanr were compared (Fig. 7B and C). Although the zones of inhibition produced by H2O2 in the mutant strains were slightly higher than those of the Challis 2 strain, the differences were not statistically significant. The percentages of growth of nosX::Kanr and qor1::Kanr in the presence of 0.1 mM paraquat were significantly lower than in the Challis 2 strain (P < 0.05). These results suggest the possible involvement of nosX and qor1 in oxidative stress tolerance.
Phase-contrast and confocal microscopy analysis of biofilms.
In addition to the microtiter plate biofilm assay, biofilm formation of nosX::Kanr, qor1::Kanr, and qor2::Kanr on borosilicate glass coverslips was examined 24 h after inoculation. The ability of these strains to form biofilms was confirmed by direct visualization of the biofilm formation on the glass surfaces using phase-contrast microscopy (Fig. 7D). After incubation for 24 h, the Challis 2 strain formed large clusters of cells that were interspersed with sparsely covered areas. A number of dense microcolonies were also observed. In contrast, significantly fewer cells from the nosX, qor1, and qor2 mutant strains had attached, forming a scattered pattern that was markedly different from that of the Challis 2 biofilm, with large areas that were devoid of cells. The biofilm phenotypes of the nosX, qor1, and qor2 mutants suggest that these genes are important for biofilm formation.
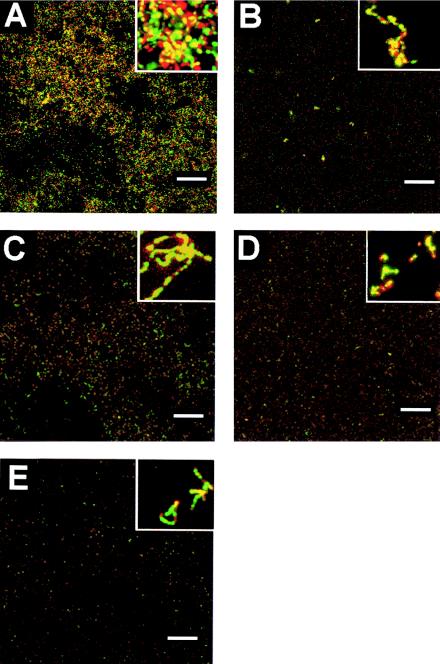
Confocal microscopy was carried out to assess whether the mutant strains could form biofilms in a flow cell system and how these biofilms differ when compared to the S. gordonii Challis 2 strain. The biofilms formed by the mutant strains appeared to contain significantly fewer cells than biofilms formed by Challis 2 (Fig. 8). The Challis 2 biofilm was characterized as a thick, compact biofilm containing dense clusters of cells. In contrast, the mutant biofilms contained sparse clusters made up of a much smaller number of cells. Similar amounts of live (green) to dead (red) bacteria were observed in the biofilms of all the S. gordonii strains examined.
FIG. 8.
Confocal scanning laser microscopic analysis of S. gordonii biofilms in a flow-cell system. S. gordonii Challis 2 (A), nosX::Tn917-lac (B), nosX::Kanr (C), qor1::Kanr (D), and qor2::Kanr (E) strains were grown in BM in a BST FC71 flow cell unit (flow rate, 180 μl/min; initial inoculum, A660 = 0.1) over 18 h at 37°C. Live cells were stained with SYTO-9 (green), and dead cells were stained with propidium iodide (red); cells were then examined by confocal microscopy. A representative image from a set of randomly selected x-y stacks is shown for each strain. Bars, 100 μm. The small inset in each panel represents a 10× magnification of the panel.
To determine the spatial architecture of the mutant biofilms versus the Challis 2 biofilm, quantitative analysis of the biofilms was performed using the COMSTAT program (16). Results from the COMSTAT analysis of confocal images using five separate image stacks at 5-μm intervals from different areas of the biofilm are shown in Table 2. Student's t tests performed to compare values obtained for all four mutant strains with those of the S. gordonii Challis 2 strain found significant differences in all the six parameters examined. The total biomass of the Challis 2 strain is significantly higher than nosX::Tn917-lac, nosX::Kanr, qor1::Kanr, and qor2::Kanr, with a total biomass that ranged from 1.4 to 5.1 times that present in the biofilms formed by the four mutants. The average thickness of Challis 2 biofilms was significantly higher than nosX::Tn917-lac, nosX::Kanr, qor1::Kanr, and qor2::Kanr biofilms. The average area occupied by Challis 2 was approximately four times higher than the mutant strains. The roughness coefficient of Challis 2 (Ra = 1.93) was significantly lower than the mutant strains (Ra = 1.99). These results indicate that there were significant differences in the total biomass, average thickness, area occupied, substratum coverage, and roughness coefficient of biofilms formed by the mutant strains when compared to the parent strain.
TABLE 2.
COMSTAT analysis of CSLM images of biofilms formed by S. gordonii strains in a flow cell systema
| Strain | Total biomass (μm3/μm2) | Avg thickness (μm) | Area occupied (%) | Substratum coverage (%) | Roughness coefficient (Ra) |
|---|---|---|---|---|---|
| Challis 2 | 0.61 ± 0.02 | 0.50 ± 0.04 | 1.98 ± 0.65 | 1.83 ± 0.21 | 1.93 ± 0.02 |
| nosX::Tn917-lac | 0.44 ± 0.05 | 0.41 ± 0.05 | 0.52 ± 0.01 | 0.53 ± 0.02 | 1.99 ± 0.00 |
| nosX::Kanr | 0.24 ± 0.04 | 0.22 ± 0.04 | 0.52 ± 0.02 | 0.53 ± 0.02 | 1.99 ± 0.00 |
| qor1::Kanr | 0.14 ± 0.02 | 0.12 ± 0.02 | 0.52 ± 0.01 | 0.52 ± 0.00 | 1.99 ± 0.00 |
| qor2::Kanr | 0.12 ± 0.02 | 0.10 ± 0.02 | 0.52 ± 0.00 | 0.52 ± 0.00 | 1.99 ± 0.00 |
The values shown are the means and standard deviations obtained from image stacks of five different areas of the biofilm after 18 h. Student's t tests were performed to compare values obtained for each mutant strain with those of the S. gordonii Challis 2 strain. All P values were <0.05. Substratum coverage represents the percentage of glass surface covered by bacteria, whereas area occupied indicates the average area occupied by cells throughout the thickness of the biofilm.
Real-time RT-PCR.
The sensitivity of real-time PCR and its high-throughput potential will facilitate individual statistical determination and quantitation of any transcriptional changes of each biofilm-associated gene in response to particular changes in environmental conditions observed in MUG assays. The induction of nosX, nox, and ahpC genes in sessile cells relative to expression in planktonic cells is shown in Table 3. There were significant increases in nosX-specific mRNA levels in sessile cells grown as biofilms on both glass and plastic surfaces. When compared to planktonic cells, levels of nosX-specific mRNA were increased approximately eightfold in biofilm-derived cells grown on a polystyrene surface and approximately threefold in biofilm-derived cells grown on a borosilicate glass surface. Compared to the levels in planktonic cells, the levels of nox (NADH oxidase) gene transcription were increased approximately 7-fold in sessile cells grown on polystyrene and approximately 10-fold in biofilm-derived cells grown on borosilicate glass. On the other hand, the mRNA levels of ahpC in sessile and planktonic cells were not significantly different. The mRNA levels of qor1 and qor2 in S. gordonii Challis 2 grown under these conditions were also examined, and expression levels of these two genes were similar to those of nosX (data not shown).
TABLE 3.
Quantification of nosX, nox, and ahpC transcript levels by real-time RT-PCRa
| Gene | Growth condition | Fold change | P value |
|---|---|---|---|
| nosX | Biofilm on plastic | 8.4 | 0.001* |
| nosX | Biofilm on glass | 3.5 | 0.04* |
| nox | Biofilm on plastic | 7.0 | 0.03* |
| nox | Biofilm on glass | 9.9 | 0.003* |
| ahpC | Biofilm on plastic | 1.2 | 0.9 |
| ahpC | Biofilm on glass | 1.1 | 0.4 |
Change in abundance of S. gordonii Challis 2 mRNA from biofilm cells relative to planktonic cells when grown in BM under anaerobic conditions. The data are presented as the relative expression normalized to that of the endogenous reference, 23S rRNA. Values of >1 reflect a relative increase in Challis 2 expression levels in biofilm cells compared to planktonic cells; values of <1 reflect a relative decrease. An asterisk (*) indicates changes with statistical significance (P value of <0.05). Each data point and statistical significance are derived from three experiments with three independently isolated RNA preparations. The threshold cycle (CT) used for quantitation and comparison among the samples was identified at the threshold fluorescence of 0.2. A standard curve, generated by 10-fold serial dilutions of cDNA, shows the amplification values of 23S rRNA for four concentrations of cDNA, indicating a log-linear relationship in the cDNA range tested (data not shown).
The relative abundance of nosX-, nox-, and ahpC-specific mRNA in S. gordonii Challis 2 cells grown under anaerobic, static, and aerated aerobic conditions was examined. The induction of these genes relative to expression in anaerobic BM cultures is shown in Table 4. There were increased levels of nosX transcripts from cells grown under both aerated and static aerobic conditions. When compared to cells grown under anaerobic conditions, there was a 20-fold increase in nosX expression in cells grown under aerated conditions (at 250 rpm), while cells grown under static aerobic conditions had a 4.7-fold increase in nosX expression. Compared to cells grown under anaerobic conditions, the mRNA levels of the nox (NADH oxidase gene) were increased approximately 39-fold in cells grown under aerated aerobic conditions and increased approximately 10-fold in cells grown under static aerobic conditions. The mRNA levels of the ahpC gene were increased approximately fivefold in cells grown under aerated aerobic conditions and increased approximately fourfold in cells grown under static aerobic conditions.
TABLE 4.
Quantification of nosX, nox, and ahpC transcript levels by real-time RT-PCRa
| Gene | Growth condition/strainb | Fold change | P value |
|---|---|---|---|
| nosX | Static aerobic | 4.7 | 0.0003* |
| nosX | Aerated aerobic | 19.9 | 0.02* |
| nosX | qor1::Kanr mutantb | 7.5 | 0.003* |
| nosX | qor2::Kanr mutantb | 0.7 | 0.4 |
| nox | Static aerobic | 9.7 | 0.002* |
| nox | Aerated aerobic | 38.9 | 0.0003* |
| nox | nosX::Kanr mutantb | 1.0 | 1.0 |
| nox | qor1::Kanr mutantb | 26.6 | 0.005* |
| nox | qor2::Kanr mutantb | 1.1 | 0.7 |
| ahpC | Static aerobic | 3.8 | 0.02* |
| ahpC | Aerated aerobic | 4.7 | 0.01* |
| ahpC | nosX::Kanr mutantb | 0.7 | 0.9 |
| ahpC | qor1::Kanr mutantb | 2.8 | 0.01* |
| ahpC | qor2::Kanr mutantb | 1.5 | 0.4 |
Change in abundance of S. gordonii mRNA when cultured in BM under different conditions, relative to expression levels when grown under anaerobic conditions. The data are presented as the relative expression normalized to that of the endogenous reference, 23S rRNA. Values of >1 reflect a relative increase in Challis 2 mRNA abundance compared to BM under anaerobic conditions, values of <1 reflect a relative decrease. An asterisk (*) indicates changes with a statistical significance (P value of <0.05). Each data point and statistical significance are derived from three experiments using three independently isolated RNA preparations.
The relative expression in the mutant strain compared to Challis 2 when grown in BM under anaerobic conditions.
The mRNA levels of nosX, nox, and ahpC in mutant cells were compared to the Challis 2 cells (Table 4). When compared to the Challis 2 strain, there was a significant increase in the expression of nosX in the qor1::Kanr mutant but not in the qor2::Kanr mutant. Expression levels of nox and ahpC in the qor1::Kanr mutant were also significantly higher than in Challis 2. On the other hand, there was no significant difference in the expression of nox or ahpC in the nosX::Kanr or the qor2::Kanr mutants when compared to the Challis 2 strain. These observations suggest that compensatory interactions may exist between qor1 and the two adjacent genes, nox and ahpC.
These results suggest that expression of nosX, qor1, qor2, nox, and ahpC genes are tied to oxygen levels whereas expression of nosX, qor1, qor2, and nox genes are tied to a sessile mode of growth. The relative increases in the expression of nosX, qor1, qor2, nox, and ahpC genes observed during aerobic conditions suggest that this region constitutes an island responsible for dealing with oxidative stress that may also be associated with biofilm formation. As real-time quantitative RT-PCR is based on the amplification of mRNA, results obtained will only reflect transcriptional processes; extension of the results to the protein level is beyond the scope of this study.
DISCUSSION
S. gordonii, a facultative anaerobe present in the oral cavity, is a pioneer colonizer of tooth surfaces that plays a vital role in the formation of oral biofilms. In this study, we have identified a putative osr operon in S. gordonii consisting of nosX, qor1, and qor2 genes after a biofilm-defective S. gordonii::Tn917-lac mutant was isolated and characterized. A recent study reported that nosX is a component of Cu-S center assembly factors in P. putida that are required for the activity of bacterial nitrous oxide reductase (58). Although the function of S. gordonii NosX is currently unknown, it may be involved in the maturation of the oxidoreductases Qor1 and/or Qor2.
Qors found in bacteria are involved in the respiratory chain and are important factors in the response to oxidative stress (18, 52). The expression of QorA, an NADPH-dependent Qor in S. aureus, which catalyzes a one-electron reduction of quinone, is enhanced by oxidative stress (31). MdaB, an NADPH quinone reductase in H. pylori, has been shown to play an important role in managing oxidative stress and colonization of the mouse stomach (55).
Mutagenesis of each of the three ORFs, nosX, qor1, and qor2, resulted in biofilm-defective phenotypes when examined in a polystyrene microtiter plate biofilm assay, on borosilicate glass, and in a flow cell system. Quantitative analysis of the biofilm structure also confirmed that the biofilms formed by nosX, qor1, and qor2 mutants were significantly different than those formed by the Challis 2 biofilm.
The expression of nosX in S. gordonii is growth phase dependent and induced by aerobic growth and paraquat. Paraquat isa redox compound that is reduced by low-potential electron donors inside the bacterial cell and then oxidized by molecular oxygen, thereby generating superoxide in the cytoplasm (15). The toxicity of paraquat depends upon its entry into cells, which leads to increased intracellular production of the superoxide radical O2−. The observation that the nosX::Tn917-lac fusion was induced by paraquat but not H2O2 suggests that external H2O2 has no significant effect on nosX expression in S. gordonii. This heterogeneity in oxidative stress susceptibility has also been observed in a S. pyogenes mutant where the perR gene, which is involved in oxidative stress response and iron homeostasis, was inactivated (40). It may be due to the cells producing enough superoxide dismutase to handle moderate levels of oxidative stress in the form of intracellular O2− generated by paraquat.
In this study, significant increases were observed in the abundance of mRNA encoding NosX in RNA derived from biofilm cells grown on both glass and plastic surfaces. When flanking genes of the osr operon were examined, the expression of nox was increased in biofilm cells, but ahpC expression levels were similar in both biofilm and planktonic cells. These results provide further evidence that the osr operon and nox gene play a role in biofilm formation. A Nox homolog in S. pneumoniae is involved in modulating the level of genetic transformability and virulence of S. pneumoniae in response to oxygen concentration (11). A recent study that examined S. mutans protein expression during the initial stage of biofilm formation identified an NADH oxidase as being enhanced 2.1-fold in biofilm cells compared to planktonic cells (56). Examination of the S. mutans UA159 genome identified two NADH oxidases, which correspond to the H2O2-forming oxidase (Nox1) that functions as part of an alkyl hydroperoxide reductase system and the H2O-forming NADH oxidase (Nox2), which plays an important role in aerobic energy metabolism in S. mutans (17).
Expression of both nox and ahpC was significantly increased in cells grown under aerated and static aerobic conditions when compared to anaerobic conditions. Most thiol peroxidase genes, such as the bcp gene of E. coli, are inducible by oxidative stress (19), and results from this study also indicate that the ahpC gene is inducible in response to oxygen stress. The nosX, nox, and ahpC genes responded to oxidative stress, lending support to the postulated role of the putative osr operon, nox, and ahpC in oxidative stress response.
When nosX or the osr operon was inactivated, aerobic growth was significantly reduced. Growth was not significantly affected when either qor1 or qor2 was inactivated, which may be due to compensation by other proteins involved in oxygen stress resistance function. In S. mutans, the antioxidant Dpr had increased production upon mutation of other antioxidant-encoding genes (59). We observed that inactivation of qor1 increased the expression of nosX, nox, and ahpC in S. gordonii, lending support to the hypothesis that compensatory interactions exist among S. gordonii genes involved in oxidative stress response.
The transition from a planktonic to a sessile existence may have to respond and adapt to changes in environmental factors, such as oxygen gradients. Genes involved in resistance to oxygen are regulated by environmental oxygen levels and appear to be important in bacterial biofilm formation. Disruption of the genes in the putative osr operon also affected the biofilm formation of S. gordonii. In S. mutans, both oxidative stress tolerance and biofilm formation are regulated by LuxS-mediated quorum sensing (57). A recent study found that the transcriptional regulator SinR controls the maturation of Agrobacterium tumefaciens and proposed that a signal cascade responsive to oxygen limitation activates sinR expression in response to decreased oxygen levels and influences the biofilm formation of A. tumefaciens (39). Schembri et al. (44), in a study of the global gene in E. coli biofilms using microarray analysis, reported that putative genes encoding oxidoreductases and cytochrome terminal oxidases were up-regulated in biofilm cells when compared to planktonic cells. In S. gordonii, the putative osr operon, nox, and ahpC appear to be part of an island responsible for dealing with oxidative stress. These genes might modulate biofilm formation by playing a role in maintaining a reduced environment. Taken together, these observations support the hypothesis that a number of S. gordonii genes involved in bacterial defense against oxidative stress are linked in biofilm formation. In contrast, oral streptococcal biofilm cells grown in tryptone-yeast extract-sucrose broth had demonstrated repressed respiration and NADH oxidase activity when compared with cells from aerobic culture (35). These differences may be due to the differences in the media used. On the other hand, both respiration and NADH oxidase activity of S. mutans were greatly enhanced in aerobic growth, while minor effects were observed for S. gordonii (35).
The mechanism by which inactivation of the putative osr operon affects S. gordonii biofilm formation is not entirely clear. Although oral biofilms are often considered relatively anaerobic, active oxygen metabolism is present in both supragingival and subgingival biofilms, which necessitates the development of defenses against oxidative stress (30). In summary, the present study specifically implicates the transcription of nosX, qor1, and qor2 in S. gordonii biofilm formation. Results from this study suggest genes involved in bacterial defense against oxidative stress also play a significant role in the development of biofilms. Further characterization of the putative osr operon will provide insight into the molecular mechanisms of biofilm formation and oxygen tolerance of oral streptococci.
Acknowledgments
This work was supported by U.S. Public Health Service grant RO1-DE13328 from the National Institute of Dental and Craniofacial Research.
Special thanks go to Z. Skobe and Elke Pravda at the Biostructure Core Facility in The Forsyth Dental Institute for assistance with confocal microscopy.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Auzat, I., S. Chapuy-Regaud, G. Le Bras, D. Dos Santos, A. D. Ogunniyi, I. Le Thomas, J.-R. Garel, J. C. Paton, and M.-C. Trombe. 1999. The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol. Microbiol. 34:1018-1028. [DOI] [PubMed] [Google Scholar]
- 3.Beck, B. J., and D. M. Downs. 1998. The apbE gene encodes a lipoprotein involved in thiamine synthesis in Salmonella typhimurium. J. Bacteriol. 180:885-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhagwat, S. P., J. Nary, and R. A. Burne. 2001. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205:225-230. [DOI] [PubMed] [Google Scholar]
- 5.Blehert, D. S., R. J. Palmer, Jr., J. B. Xavier, J. S. Almeida, and P. E. Kolenbrander. 2003. Autoinducer 2 production by Streptococcus gordonii DL1 and the biofilm phenotype of a luxS mutant are influenced by nutritional conditions. J. Bacteriol. 185:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burne, R. A., Y. Y. Chen, and J. E. Penders. 1997. Analysis of gene expression in Streptococcus mutans in biofilms in vitro. Adv. Dent. Res. 11:100-109. [DOI] [PubMed] [Google Scholar]
- 7.Chae, H. Z., K. Robison, L. B. Poole, G. Church, G. Storz, and S. G. Rhee. 1994. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc. Natl. Acad. Sci. USA 91:7017-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, Y.-K., W. A. McCormick, and R. J. Watson. 1997. A new nos gene downstream from nosDFY is essential for dissimilatory reduction of nitrous oxide by Rhizobium (Sinorhizobium) meliloti. Microbiology 143:2817-2834. [DOI] [PubMed] [Google Scholar]
- 9.Chen, W., R. J. Palmer, and H. K. Kuramitsu. 2002. Role of polyphosphate kinase in biofilm formation by Porphyromonas gingivalis. Infect. Immun. 70:4708-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cvitkovitch, D. G., J. A. Gutierrez, J. Behari, P. J. Youngman, J. E. Wetz, P. J. Crowley, J. D. Hillman, L. J. Brady, and A. S. Bleiweis. 2000. Tn917-lac mutagenesis of Streptococcus mutans to identify environmentally regulated genes. FEMS Microbiol. Lett. 182:149-154. [DOI] [PubMed] [Google Scholar]
- 11.Echenique, J. R., S. Chapuy-Regaud, and M.-C. Trombe. 2000. Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol. Microbiol. 36:688-696. [DOI] [PubMed] [Google Scholar]
- 12.Frandsen, E. V., V. Pedrazzoli, and M. Kilian. 1991. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol. Immunol. 6:129-133. [DOI] [PubMed] [Google Scholar]
- 13.Froeliger, E. H., and P. Fives-Taylor. 2001. Streptococcus parasanguis fimbria-associated adhesin Fap1 is required for biofilm formation. Infect. Immun. 69:2512-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilmore, K. S., P. Srinivas, D. R. Akins, K. L. Hatter, and M. S. Gilmore. 2003. Growth, development, and gene expression in a persistent Streptococcus gordonii biofilm. Infect. Immun. 71:4759-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassett, D. J., B. E. Britigan, T. Svendsen, G. M. Rosen, and M. S. Cohen. 1987. Bacteria form intracellular free radicals in response to paraquat and streptonigrin. Demonstration of the potency of hydroxy radicals. J. Biol. Chem. 262:13404-13408. [PubMed] [Google Scholar]
- 16.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 17.Higuchi, M., Y. Yamamoto, L. B. Poole, M. Shimada, Y. Sato, N. Takahashi, and Y. Kamio. 1999. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J. Bacteriol. 181:5940-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaiswal, A. K. 2000. Regulation of genes encoding NAD(P)H:quinone oxidoreductases. Free Radic. Biol. Med. 29:254-262. [DOI] [PubMed] [Google Scholar]
- 19.Jeong, W., M.-K. Cha, and I.-H. Kim. 2000. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J. Biol. Chem. 275:2924-2930. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan, J. B., M. F. Meyenhofer, and D. H. Fine. 2003. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J. Bacteriol. 185:1399-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawamura, Y., X.-G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S RNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 22.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 23.Lemos, J. A., T. A. Brown, Jr., and R. A. Burne. 2004. Effects of relA on key virulence properties of planktonic and biofilm populations of Streptococcal mutans. Infect. Immun. 72:1431-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Y.-H., P. C. Lau, N. Tang, G. Svensater, R. P. Ellen, and D. G. Cvitkovitch. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184:6333-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Y.-H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loo, C. Y. 2003. Oral streptococcal genes that encode biofilm formation, p. 189-211. In M. Wilson and D. Devine (ed.), Medical implications of biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 27.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loo, C. Y., K. Mitrakul, I. B. Voss, C. V. Hughes, and N. Ganeshkumar. 2003. Involvement of the adc operon and manganese homeostasis in Streptococcus gordonii biofilm formation. J. Bacteriol. 185:2887-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loo, C. Y., K. Mitrakul, I. B. Voss, C. V. Hughes, and N. Ganeshkumar. 2003. Involvement of an inducible fructose phosphotransferase operon in Streptococcus gordonii biofilm formation. J. Bacteriol. 185:6241-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marquis, R. E. 1995. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J. Ind. Microbiol. 15:198-207. [DOI] [PubMed] [Google Scholar]
- 31.Maruyama, A., Y. Kumagai, K. Morikawa, K. Taguchi, H. Hayashi, and T. Ohta. 2003. Oxidative-stress-inducible qorA encodes an NADPH-dependent quinone oxidoreductase catalyzing a one-electron reduction in Staphylococcus aureus. Microbiology 149:389-398. [DOI] [PubMed] [Google Scholar]
- 32.Merritt, J., F. Qi, S. D. Goodman, M. H. Anderson, W. Shi. 2003. Mutation of luxS affects biofilm formation in Streptococcus mutans. Infect. Immun. 71:1972-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mickelson, M. N. 1967. Aerobic metabolism of Streptococcus agalactiae. J. Bacteriol. 94:184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miranda-Rios, J., M. Navarro, and M. Soberon. 2002. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc. Natl. Acad. Sci. USA 98:9736-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen, P. T., J. Abranches, T.-N. Phan, and R. E. Marquis. 2002. Repressed respiration of oral streptococci grown in biofilms. Curr. Microbiol. 44:262-266. [DOI] [PubMed] [Google Scholar]
- 36.Palmer, R. J., Jr., and D. E. Caldwell. 1995. A flow-cell for the study of plaque removal and regrowth. J. Microbiol. Methods 24:171-182. [Google Scholar]
- 37.Persson, B., J. S. Zigler, Jr., and H. Jornvall. 1994. A super-family of medium-chain dehydrogenases/reductases (MDR). Sub-lines including zeta-crystallin, alcohol and polyol dehydrogenases, quinone oxidoreductase enoyl reductases, VAT-1 and other proteins. Eur. J. Biochem. 226:15-22. [DOI] [PubMed] [Google Scholar]
- 38.Poole, L. B., M. Higuchi, M. Shimada, M. L. Calzi, and Y. Kamio. 2000. Streptococcus mutans H2O2-forming NADH oxidase is an alkyl hydroperoxide reductase protein. Free Radic. Biol. Med. 28:108-120. [DOI] [PubMed] [Google Scholar]
- 39.Ramey, B. E., A. G. Matthysse, and C. Fuqua. 2004. The FNR-type transcriptional regulator SinR controls maturation of Agrobacterium tumefaciens biofilms. Mol. Microbiol. 52:1495-1511. [DOI] [PubMed] [Google Scholar]
- 40.Ricci, S., R. Janulczyk, and L. Bjorck. 2002. The regulator PerR is involved in oxidative stress response and iron homeostasis and is necessary for full virulence of Streptococcus pyogenes. Infect. Immun. 70:4968-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogers, J. D., R. J. Palmer, Jr., P. E. Kolenbrander, and F. A. Scannapieco. 2001. Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation. Infect. Immun. 69:7046-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saunders, N. F., J. J. Hornberg, W. N. Reijnders, H. V. Westerhoff, S. de Vries, and R. J. van Spanning. 2000. The NosX and NirX proteins of Paracoccus denitrificans are functional homologs: their role in maturation of nitrous oxide reductase. J. Bacteriol. 182:5211-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schembri, M. A., K. Kjaergaard, K., and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 45.Schmehl, M., A. Jahn, A. Meyerzu Vilsendorf, S. Hennecke, B. Masepohl, M. Schuppler, M. Marxer, J. Oelze, J., and W. Klipp. 1993. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 241:602-615. [DOI] [PubMed] [Google Scholar]
- 46.Skovran, E., and D. M. Downs. 2003. Lack of the apbC and apbE protein results in a defect in Fe-S cluster metabolism in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soballe, B., and R. K. Poole. 1999. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 145:1817-1830. [DOI] [PubMed] [Google Scholar]
- 48.Sparla, F., G. Tedeschi, P. Pupillo, and P. Trost. 1999. Cloning and heterologous expression of NAD(P)H:quinone reductase of Arabidopsis thaliana, a functional homologue of animal DT-diaphorase. FEBS Lett. 463:382-386. [DOI] [PubMed] [Google Scholar]
- 49.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 50.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 51.Tao, L., D. J. LeBlanc, and J. J. Ferretti. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120:105-110. [DOI] [PubMed] [Google Scholar]
- 52.Thorn, J. M., H. D. Barton, N. E. Dixon, D. L. Ollis, and K. J. Edwards. 1995. Crystal structure of Eschericha coli QOR quinone oxidoreductase complexed with NADPH. J. Mol. Biol. 249:785-799. [DOI] [PubMed] [Google Scholar]
- 53.van Der Ploeg, J. R., R. Iwanicka-Nowicka, T. Bykowski, M. M. Hryniewicz, and T. Leisinger. 1999. The Escherichia coli ssuEADCB gene cluster is required for the utilization of sulfur from aliphatic sulfonates and is regulated by the transcriptional activator Cb1. J. Biol. Chem. 274:29358-29365. [DOI] [PubMed] [Google Scholar]
- 54.Vriesema, A. J., J. Dankert, and S. A. Zaat. 2000. A shift from oral to blood pH is a stimulus for adaptive gene expression of Streptococcus gordonii CH1 and induces protection against oxidative stress and enhanced bacterial growth by expression of msrA. Infect. Immun. 68:1061-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, G., and R. J. Maier. 2004. An NADPH quinone reductase of Helicobacter pylori plays an important role in oxidative stress resistance and host colonization. Infect. Immun. 72:1391-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welin, J., J. C. Wilkins, D. Beighton, and G. Svensater. 2004. Protein expression by Streptococcus mutans during initial stage of biofilm formation. Appl. Environ. Microbiol. 70:3736-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wunsch, P., M. Herb, H. Wieland, U. M. Schiek, and W. G. Zumft. 2003. Requirements for CuA and Cu-S center assembly of nitrous oxide reductase deduced from complete periplasmic enzyme maturation in the nondenitrifier Pseudomonas putida. J. Bacteriol. 185:887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamamoto, Y., M. Higuchi, L. B. Poole, and Y. Kamio. 2000. Identification of a new gene responsible for the oxygen tolerance in aerobic life of Streptococcus mutans. Biosci. Biotechnol. Biochem. 64:1106-1109. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, Y., Y. Lei, A. Khammanivong, and M. C. Herzberg. 2004. Identification of a novel two-component system in Streptococcus gordonii V288 involved in biofilm formation. Infect. Immun. 72:3489-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]