Abstract
Gene ytkD of Bacillus subtilis, a member of the Nudix hydrolase superfamily, has been cloned and expressed in Escherichia coli. The purified protein has been characterized as a nucleoside triphosphatase active on all of the canonical ribo- and deoxyribonucleoside triphosphates. Whereas all other nucleoside triphosphatase members of the superfamily release inorganic pyrophosphate and the cognate nucleoside monophosphate, YtkD hydrolyses nucleoside triphosphates in a stepwise fashion through the diphosphate to the monophosphate, releasing two molecules of inorganic orthophosphate. Contrary to a previous report, our enzymological and genetic studies indicate that ytkD is not an orthologue of E. coli mutT.
The Nudix hydrolases are a widely distributed superfamily of enzymes, so named because they catalyze the hydrolysis of nucleoside diphosphates linked to some other moiety, x (4). The superfamily may be classified into subfamilies dependent on the structure of the substrates hydrolyzed. These include nucleoside and deoxynucleoside triphosphates; ADP-ribose; sugar nucleotides, such as ADP-glucose or GDP-mannose; dinucleoside polyphosphates; coenzymes, such as NADH and coenzyme A (CoA); and CDP-choline, among others. Members of the family can be recognized by a highly conserved signature sequence of amino acids, the Nudix box, which is Gx5Ex7REUxEExGU, where U represents a bulky hydrophobic amino acid, usually Ile, Leu, or Val, and x is any amino acid. A recent BLAST (2) search has revealed over 1,900 open reading frames from more than 370 species, distributed in all three kingdoms and ranging in complexity from viruses to humans. The founding member of the Nudix superfamily, Escherichia coli MutT, is a nucleoside triphosphatase thought to prevent the high mutation frequency seen in mutT cells by sanitizing the nucleotide pool of a mutagenic nucleoside triphosphate (5), reputedly 8-oxo-dGTP (14).
Recently, ytkD, a Nudix hydrolase gene of Bacillus subtilis (13), was cloned and expressed, and the purified protein was characterized as a highly specific nucleoside triphosphatase with 400-fold greater activity on 8-oxo-dGTP than on dGTP. Based on its Nudix hydrolase signature sequence, its 8-oxo-dGTPase activity, and its ability to complement an E. coli mutT mutant strain, it was considered an orthologue of E. coli mutT and was named mutTA (19). These results were surprising to us for two reasons. First, for all orthologues of MutT measured, including E. coli (6, 14), Streptococcus pneumoniae (7), and human (16), dGTP is an excellent substrate. In fact, the E. coli and human enzymes have a higher Vmax on dGTP than on 8-oxo-dGTP. Second, there is very little amino acid identity between YtkD and E. coli MutT, whereas there is a high degree of identity between YtkD and a protein that was recently characterized from Bacillus cereus (20). This is illustrated in Fig. 1, in which YtkD is aligned separately with E. coli MutT or B. cereus NP_834487. Not counting the amino acids in the Nudix box that are highly conserved in almost all Nudix hydrolases, there is 8% amino acid identity between YtkD and MutT and 59% identity between YtkD and NP_834487. Because we already had preliminary evidence that the B. cereus enzyme was a nonspecific nucleoside triphosphatase active on dGTP, we deemed it important to try to rationalize the lack of dGTPase activity reported for YtkD (19) with our expectation that YtkD was an orthologue of the B. cereus nucleoside triphosphatase. Accordingly, we have cloned and expressed ytkD and purified and partially characterized the enzyme. On the basis of its substrate specificity, the products of the reaction, and the failure of the gene to complement an E. coli mutT mutant, we believe that YtkD is not an orthologue of MutT but is, in fact, an unusual nucleoside triphosphatase member of the Nudix hydrolase superfamily.
FIG. 1.
Amino acid sequence alignment of B. subtilis YtkD with E. coli MutT (A) or B. cereus NP_834487 (B). The Nudix box is underlined, and the conserved amino acids representing the Nudix hydrolase signature sequence are in boldface. Asterisks mark amino acid identities.
MATERIALS AND METHODS
Bacterial strains, plasmids, and DNA.
E. coli K-12 strains TX48, wild-type parent, and SB3, a mutT1 transductant of TX48, were as described previously (5). Plasmid pET-24a(+) (Kmr) and E. coli strains HMS174(DE3) and DH5α were from Novagen (Madison, Wis.) and Stratagene (La Jolla, Calif.), respectively, and plasmid pTrc99A (Ampr) was from Amersham Pharmacia Biotech (Piscataway, N.J.). pTrcMutT, which is pTrc99A containing the mutT gene, was constructed in this laboratory. The genomic DNA of Bacillus subtilis strain 168 was from the American Type Culture Collection (ATTC; Manassas, Va.), and the PCR primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, Iowa).
Materials.
Purified MutT nucleoside triphosphate pyrophosphohydrolase was prepared as described previously (1). Sephadex G-100 was from Amersham Pharmacia Biotech, and the restriction enzymes, PCR kits, calf intestinal alkaline phosphatase, and yeast inorganic pyrophosphatase were from Stratagene. The canonical nucleotide substrates and common chemicals were from Sigma-Aldrich Chemical Co. (St. Louis, Mo.) or Life Technologies Inc. (Rockville, Md.), and 8-oxo-dGTP was from TriLink Biotechnologies (San Diego, Calif.). Protein assay reagent was from Bio-Rad Laboratories (Hercules, Calif.).
Cloning of the gene ytkD from B. subtilis strain 168.
Standard cloning technology was used to clone ytkD. Briefly, gene ytkD was amplified directly from the genomic DNA of B. subtilis strain 168 using the DNA PCR. NdeI and BamHI restriction sites were incorporated at the start and end of the gene, respectively, and the amplified DNA was purified, digested with NdeI and BamHI, and ligated into pET24a(+) to place the gene under transcriptional control of the T7 lac promoter. The resultant plasmid, pETytkD, was transformed into E. coli DH5α for storage and into HMS174(DE3) for protein expression. For complementation studies, the gene ytkD was excised from pETytkD with XbaI and SalI and was ligated into the corresponding sites of pTrc99A, putting ytkD under the control of the trc promoter. The resulting plasmid, pTrcytkD, was transformed into the mutT strain SB3.
Growth and expression of HMS174:pETytkD.
One colony of the expression strain of HMS174:pETytkD was inoculated into 40 ml of Luria-Bertani medium containing 30 μg of kanamycin/ml, incubated at 37°C on a rotary shaker over night, transferred to 2 liters of the same medium, and grown to an A600 of 0.8. The culture was derepressed by the addition of isopropyl-β-d-thio-galactopyranoside to a concentration of 0.5 mM and was grown for an additional 3 h, after which the cells were harvested by centrifugation, washed in buffered isotonic saline solution, and frozen at −80°C.
Purification of the enzyme YtkD.
The frozen cells were suspended in 2.5 volumes of buffer A (50 mM Tris-Cl [pH 7.5], 1 mM EDTA, 0.1 mM dithiothreitol), and the cell extract (Fraction I) containing the expressed protein was collected by centrifugation. Streptomycin sulfate (10% in buffer A) was added to a final concentration of 1.5% to precipitate nucleic acids, which were removed by centrifugation, and the supernatant, Fraction II, was brought to 60% saturation with ammonium sulfate. The precipitate was collected and dissolved in a minimal volume of buffer A (Fraction III) and was chromatographed on a Sephadex G-100 gel filtration column equilibrated and eluted with buffer A containing 200 mM sodium chloride. The fractions were located by A280 absorbance and identified by their migration on an sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. Those containing the purified protein were pooled and stored at −80°C (Fraction IV).
Enzyme assays.
The standard reaction mixture was in 50 μl of a solution containing 50 mM Tris-Cl (pH 8.5), 5 mM Mg2+, 2 mM substrate, 0.5 U of yeast inorganic pyrophosphatase for substrates such as (deoxy)nucleoside triphosphates and their derivatives (or 4 U of alkaline phosphatase for all other substrates), and 0.1 to 2 mU of purified enzyme. The solution was incubated at 37°C for 15 min and stopped by the addition of 250 μl of 4 mM EDTA (or a Norit suspension to remove unreacted triphosphates), and the liberated inorganic orthophosphate was assayed by the colorimetric procedure of Fiske and SubbaRow (11) as modified by Ames and Dubin (3). A unit of enzyme hydrolyzes 1 μmol of substrate per min.
Product determination.
The products of the reactions were analyzed by high-performance liquid chromatography (HPLC) as follows. The standard reaction mixture (minus inorganic pyrophosphatase and alkaline phosphatase) was scaled up fivefold to 250 μl. Fifty microliters of the reaction mixture was withdrawn and stopped by adding 55 μl of 25 mM EDTA at 0, 5, 15, 30, and 45 min. Each of the fractions was injected into a 250- by 4.6-mm C18 column (YMC Inc.) and eluted with a buffer containing 12.5 mM citric acid, 25 mM sodium acetate, 30 mM NaOH, and 10 mM acetic acid at a flow rate of 1 ml/min. Peaks were visualized with UV optics and were identified by their elution time.
Complementation test for the mutT mutator phenotype.
Gene ytkD was cloned into pTrc99A and transformed into E. coli strain SB3 lacking a functional mutT gene. The mutation frequency of this construct was compared to that of TX48 (the wild-type parental strain of SB3), to that of SB3 transformed with pTrcMutT, and to that of SB3 transformed with plasmid pTrc99A lacking the ytkD insert. Mutation frequencies were calculated from the number of colonies resistant to nalidixic acid or streptomycin as previously described (18).
RESULTS AND DISCUSSION
Gene cloning, expression, and enzyme purification.
Cloning of ytkD as described in Materials and Methods went without incident, and the plasmid insert was sequenced and found to be identical to that reported. The protein expressed well, was soluble, and, as with some other Nudix hydrolases, was extractable into buffer merely by subjecting the cells to a freeze-thaw cycle (8, 9, 12, 17, 18). This left the bulk of the cellular proteins behind, thereby considerably simplifying the purification. The procedure described in Materials and Methods led to a highly enriched fraction estimated to be over 90% pure with the expected mobility on an SDS-PAGE gel (Fig. 2).
FIG. 2.
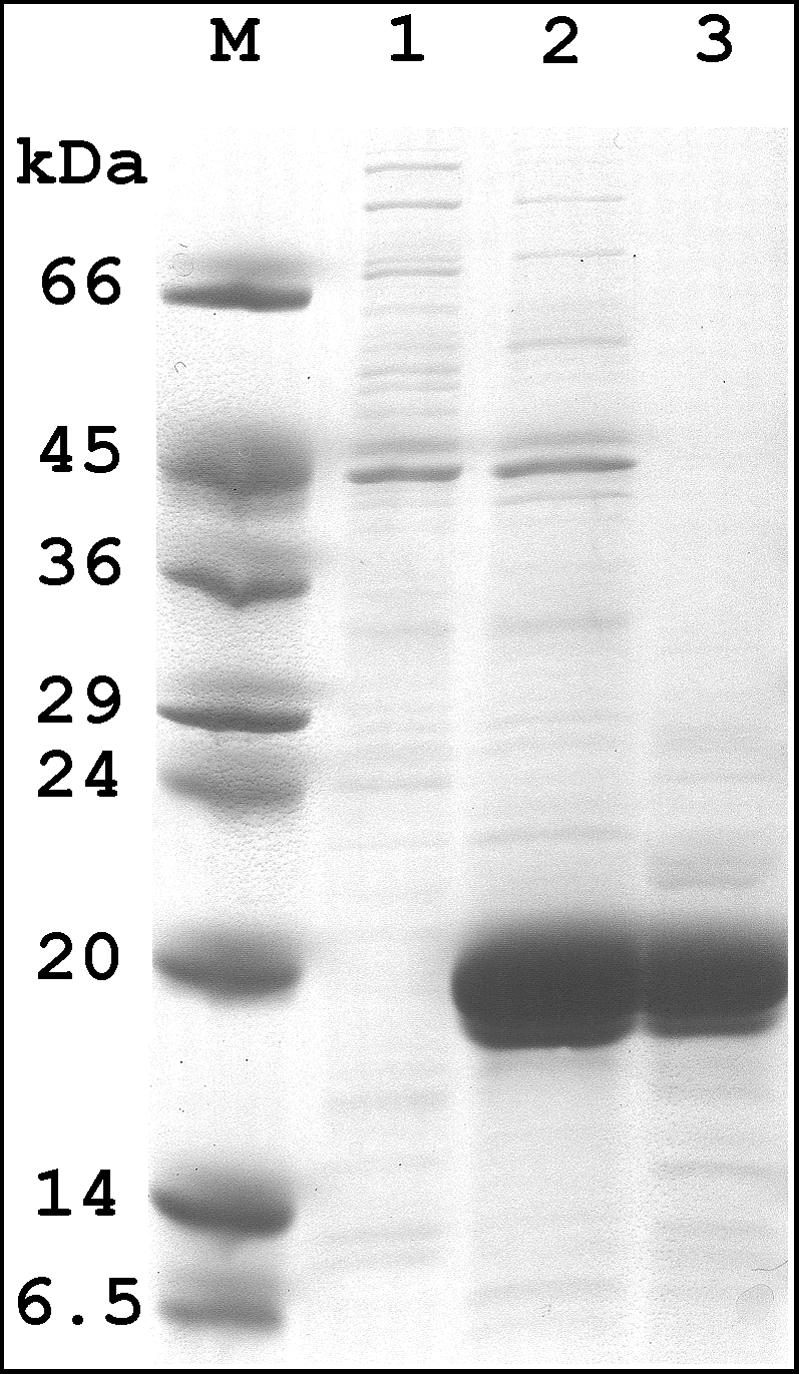
Expression and purification of B. subtilis YtkD. Shown is a polyacrylamide gel (10 to 20%) containing 1% SDS. Lane M, protein standards with the indicated molecular masses. Lanes 1 and 2, equal volumes of extracts from induced cells containing pET24a and pETytkD, respectively. Lane 3, the same number of units of enzyme as for lane 2.
Identification of enzyme activity and substrate specificity.
A unifying feature of the Nudix hydrolase superfamily of enzymes is their preference for nucleoside diphosphate derivatives. When faced with a potentially new enzyme, without any recognizable amino acid landmarks external to the Nudix box relegating it to one of the known subfamilies, we systematically tested a large number of biologically significant compounds containing a nucleoside diphosphate moiety. We used a simple assay based on cleavage of the pyrophosphate linkage (see Materials and Methods). In the case of YtkD, however, its high level of amino acid identity to the B. cereus protein NP_834487, which was previously identified as a nucleoside triphosphatase (20), intimated that it might have a similar activity. We therefore tested a number of nucleoside triphosphates as potential substrates as well as some nucleoside diphosphate derivatives acted on by other Nudix hydrolases. The data in Table 1 clearly demonstrate that the enzyme is active on all of the common nucleoside triphosphates as well as on the analogue, 8-oxo-dGTP. Because it was found that YtkD hydrolyses nucleoside triphosphates in a stepwise fashion without the intermediate formation of PPi (see below), yeast inorganic pyrophosphatase was omitted from the standard assay. Also listed in Table 1 are several substrates of other Nudix hydrolases that show little or no hydrolysis with YtkD. The low but significant rate of hydrolysis of Ap4A [adenosine (5′)-tetraphospho (5′)-adenosine] might be explained by its accommodation in the active site of the enzyme as a derivative of ATP. Kinetic data for three of the substrates are listed in Table 2. Especially noteworthy is the relatively high rate of hydrolysis of dGTP, about 75% of 8-oxo-dGTP. In fact, the catalytic efficiency (kcat/Km) is actually higher for dGTP. This is in contrast to results of a previous report (19), indicating a 400-fold higher rate of hydrolysis of 8-oxo-dGTP over dGTP. At present, we have no satisfactory explanation for the conflicting data. The only obvious difference between the two enzyme preparations was the use of a histidine-tagged protein by the previous authors to facilitate purification, whereas we have worked with the native enzyme. Because only results with (d)GTP and 8-oxo-(d)GTP were reported in the earlier paper (19), we cannot compare the two enzyme preparations with respect to the other canonical (deoxy)nucleoside triphosphates.
TABLE 1.
Substrate specificitya
| Substrate | Amt of substrate hydrolyzed (nmol) | Relative activity (%) |
|---|---|---|
| dATP | 49 | 100 |
| 8-oxo-dGTP | 33 | 69 |
| dGTP | 29 | 59 |
| dCTP | 27 | 56 |
| dUTP | 26 | 54 |
| dTTP | 22 | 45 |
| ATP | 35 | 71 |
| CTP | 24 | 50 |
| UTP | 19 | 40 |
| GTP | 13 | 26 |
| Ap4Ab | 10 | 20 |
| ADP-ribose, GDP-mannose, UDP-glucose, NADH, FAD, CDP-choline | <5 | <10 |
Rates of substrate hydrolysis were measured by using the standard assay described in Materials and Methods.
Ap4A is adenosine (5′)-tetraphospho-(5′)-adenosine.
TABLE 2.
Kinetic analysis of YtkD triphosphatasea
| Substrate | Vmax (U/mg) | kcat (s−1) | Km (mM) | kcat/Km (s−1M−1) |
|---|---|---|---|---|
| dATP | 4.7 | 1.5 | 0.89 ± 0.27 | 1.6 × 103 |
| 8-oxo-dGTP | 3.1 | 0.96 | 0.43 ± 0.17 | 2.4 × 103 |
| dGTP | 2.4 | 0.74 | 0.16 ± 0.015 | 4.6 × 103 |
Vmax and Km were obtained from a nonlinear regression analysis. One unit of enzyme catalyzes the hydrolysis of 1 μmol of substrate/min.
Products of the reaction.
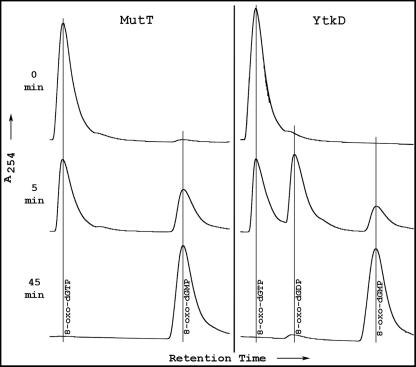
In addition to the original MutT dGTPases (6, 7, 14), several Nudix hydrolases active on different nucleoside triphosphates have been characterized. These include dATPase (18), CTPase (17), riboTTPase, (21), 3-amino-ATPase (10), and dUTPase (C. A. Dunn, D. Smith, and M. J. Bessman, unpublished data). Without exception, they catalyze the following type of reaction: nucleoside triphosphate + H2O → nucleoside monophosphate + PPi (equation 1). These enzymes are pyrophosphohydrolases that liberate inorganic pyrophosphate and the cognate nucleoside monophosphate. However, when the products of the YtkD-catalyzed hydrolysis were analyzed, we were surprised to find that Pi, not PPi, was released. An example is shown in Fig. 3, where the hydrolyses of 8-oxo-dGTP, catalyzed by YtkD and E. coli MutT, are compared. MutT shows a time-dependent conversion of the triphosphate to the monophosphate, without the intermediate formation of diphosphate. No Pi appeared during the course of the reaction, indicating that 8-oxo-dGDP was not transiently formed and rapidly converted to the monophosphate (data not shown). On the other hand, YtkD catalyzes the stepwise conversion of 8-oxo-dGTP to the monophosphate with the intermediate formation of 8-oxo-dGDP. This pathway is seen with all the triphosphates listed in Table 1. The inclusion of yeast inorganic pyrophosphatase in the reaction mixture produced no additional Pi, indicating that PPi was not formed during the reaction. Thus, the reaction catalyzed by YtkD may be written as follows: nucleoside triphosphate + 2H2O → nucleoside monophosphate + 2Pi (equation 2). As with most Nudix hydrolases, the enzyme has an alkaline pH optimum of 8.5 to 9. A divalent cation, preferably Mg2+, is required for activity, with Mn2+ about 15% as effective.
FIG. 3.
Product formation by E. coli MutT and B. subtilis YtkD nucleoside triphosphatases. Standard reaction mixtures (minus inorganic pyrophosphatase) were scaled up fivefold. At the indicated times, aliquots were applied to a C18 reverse-phase column and were analyzed by high-performance liquid chromatography.
This is the first report of a nucleoside triphosphatase member of the Nudix hydrolase superfamily with phosphohydrolase rather than pyrophosphohydrolase activity. However, there is preliminary evidence that at least three of the 21 Nudix hydrolases of Deinococcus radiodurans (DR0274, 0603, and 1025) are involved in nucleoside triphosphatase-catalyzing reactions according to equation 2 (W.-L. Xu, J.-Y. Shen, C. A. Dunn, and M. J. Bessman, FASEB J. 17[part 1, suppl.]:A574, 2003).
Complementation studies.
It has been reported that B. subtilis ytkD is an orthologue of E. coli mutT, and it complements SB3, an E. coli mutT1 mutant (19). Because the enzymatic properties of purified YtkD described above are not characteristic of previously studied MutT preparations, we deemed it necessary to determine whether our cloned gene could complement SB3(mutT1), the same strain we had sent to Ramírez et al. (19) for use in their experiments. The data in Table 3 show clearly that, whereas the plasmid pTrcMutT containing the mutT gene completely suppresses the mutator phenotype of SB3(mutT1), pTrcYtkD has no significant effect on the mutation frequency. Ramírez et al (19) report an approximately 10-fold decrease in spontaneous mutation frequency in the SB3 mutator strain transformed with ytkD and a 10-fold increase in spontaneous mutation frequency in a ΔytkD B. subtilis construct compared to that of its wild-type parent (19). These are unusually low effects compared to the 500- to 10,000-fold influence on mutation frequency seen with E. coli mutT. Again, we have no satisfactory explanation for the discrepancy in the results from the two laboratories.
TABLE 3.
Noncomplementation of mutT1 by ytkD
| Bacterial strain | Plasmid | Mutation frequenciesa
|
|
|---|---|---|---|
| Streptomycin resistant | Nalidixin resistant | ||
| TX48 (wild type)b | <1 | <1 | |
| SB3 (mutT1) | pTrc99A | 1997 ± 873 | 624 ± 49 |
| SB3 (mutT1) | pTrcMutT | <1 | <1 |
| SB3 (mutT1) | pTrcYtkD | 2526 ± 770 | 529 ± 228 |
Number of colonies resistant to streptomycin (120 μg/ml) or nalidixic acid (25 μg/ml) per 109 cells. Values are the means and average deviations of the means of three determinations.
TX48 is the wild-type parent of SB3 (5).
As pointed out in the introduction, from an examination of its amino acid sequence, its substrate specificity, the products of the reaction it catalyzes, and its failure to complement an E. coli mutT1 strain, we conclude that YtkD is not an orthologue of E. coli MutT. Instead, YtkD is the first example of a Nudix hydrolase nucleoside triphosphatase degrading ribo- and deoxyribonucleoside triphosphates in a stepwise fashion from the diphosphate to the monophosphate. What is its physiological role? It has been previously pointed out that the Nudix hydrolases function as cellular surveillance enzymes that are involved in removing potentially toxic substances, such as ADP-ribose or modified nucleotide derivatives; regulating cell signaling molecules, such as dinucleoside polyphosphates; or modulating the accumulation of metabolic intermediates, such as nucleotide sugars or NADH, during the cell cycle (4). YtkD might play a role in preventing deleterious imbalances in nucleoside triphosphate pool sizes. Assuming that the Km values we measured for dATP and dGTP are representative of the other canonical nucleoside triphosphates, they are slightly above the measured pool sizes of the individual nucleoside triphosphates in E. coli (15) and thus are poised to prevent large increases in the individual nucleotides. How this relates to B. subtilis is conjectural, but in this respect it would be of interest to examine the ytkD deletion mutant (19) for its effect on the physiology of B. subtilis.
Acknowledgments
This work was supported by U.S. Public Health Service grant GM-18649 from the National Institute of General Medical Studies.
Footnotes
Publication 1528 of the McCollum-Pratt Institute.
REFERENCES
- 1.Abeygunawardana, C., D. J. Weber, D. N. Frick, M. J. Bessman, and A. S. Mildvan. 1993. Sequence-specific assignments of the backbone 1H, 13C and 15N resonances of the MutT enzyme by heteronuclear multidimensional NMR. Biochemistry 32:13071-13080. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames, B. N., and D. T. Dubin. 1960. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J. Biol. Chem. 235:769-775. [PubMed] [Google Scholar]
- 4.Bessman, M. J., D. N. Frick, and S. F. O'Handley. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271:25059-25062. [DOI] [PubMed] [Google Scholar]
- 5.Bhatnagar, S. K., and M. J. Bessman. 1988. Studies on the mutator gene, mutT of Escherichia coli: molecular cloning of the gene, purification of the gene product, and identification of a novel nucleoside triphosphatase. J. Biol. Chem. 263:8953-8957. [PubMed] [Google Scholar]
- 6.Bhatnagar, S. K., L. C. Bullions, and M. J. Bessman. 1991. Characterization of the MutT nucleoside triphosphatase of Escherichia coli. J. Biol. Chem. 266:9050-9054. [PubMed] [Google Scholar]
- 7.Bullions, L. C., V. Méjean, J. P. Claverys, and M. J. Bessman. 1994. Purification of the MutX protein of Streptococus pneumoniae, a homologue of Escherichia coli mutT: identification of a novel catalytic domain for nucleoside triphosphate pyrophosphohydrolase activity. J. Biol. Chem. 269:12339-12344. [PubMed] [Google Scholar]
- 8.Conyers, G. B., and M. J. Bessman. 1999. The gene ialA, associated with the invasion of human erythrocytes by Bartonella bacilliformis, designates a nudix hydrolase active on dinucleoside 5′-polyphosphates. J. Biol. Chem. 274:1203-1206. [DOI] [PubMed] [Google Scholar]
- 9.Dunn, C. A., S. F. O'Handley, D. N. Frick, and M. J. Bessman. 1999. Studies on the ADP-ribose pyrophosphatase subfamily of the nudix hydrolases and tentative identification of trgB, a gene associated with tellurite resistance. J. Biol. Chem. 274:32318-32324. [DOI] [PubMed] [Google Scholar]
- 10.Espinosa, J. C., J. A. Tercero, M. A. Rubio, and A. Jimenez. 1999. The pur7 gene from the puromycin biosynthetic pur cluster of Streptomyces alboniger encodes a nudix hydrolase. J. Bacteriol. 181:4914-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiske, C. H., and Y. SubbaRow. 1925. The colorimetric determination of phosphorous. J. Biol. Chem. 66:375-400. [Google Scholar]
- 12.Frick, D. N., B. D. Townsend, and M. J. Bessman. 1995. A novel GDP-mannose mannosyl hydrolase shares homology with the MutT family of enzymes. J. Biol. Chem. 270:24086-24091. [DOI] [PubMed] [Google Scholar]
- 13.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Dusterhoft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S. Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Henaut, H. Hilbert, S. Holsappel, S. Hosono, M. F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klaerr-Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S. M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauel, C. Medigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'Reilly, K. Ogawa, A. Ogiwara, B. Oudega, S. H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B. S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H. F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 14.Maki, H., and M. Sekiguchi. 1992. MutT protein specifically hydrolyzes a potent mutagenic substrate for DNA synthesis. Nature 355:273-275. [DOI] [PubMed] [Google Scholar]
- 15.Mathews, C. K. 1972. Biochemistry of deoxribonucleic acid-defective amber mutants of bacteriophage T4. J. Biol. Chem. 247:7430-7438. [PubMed] [Google Scholar]
- 16.Mo, J.-Y., H. Maki, and M. Sekiguchi. 1992. Hydrolytic elimination of a mutagenic nucleotide, 8-oxodGTP, by human 18-kilodalton protein: sanitization of nucleotide pool. Proc. Natl. Acad. Sci. USA 89:11021-11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Handley, S. F., C. A. Dunn, and M. J. Bessman. 2001. Orf135 from Escherichia coli is a nudix hydrolase specific for CTP, dCTP, and 5-methyl-dCTP. J. Biol. Chem. 276:5421-5426. [DOI] [PubMed] [Google Scholar]
- 18.O'Handley, S. F., D. N. Frick, L. F. Bullions, A. S. Mildvan, and M. J. Bessman. 1996. Escherichia coli orf17 codes for a nucleoside triphosphate pyrophosphohydrolase member of the MutT family of proteins. J. Biol. Chem. 271:24649-24654. [DOI] [PubMed] [Google Scholar]
- 19.Ramírez, M. I., F. X. Castellanos-Juárez, R. E. Yasbin, and M. Pedraza-Reyes 2004. The ytkD (mutTA) gene of Bacillus subtilis encodes a functional antimutator 8-oxo-(dGTP/GTP)ase and is under dual control of sigma A and sigma F RNA polymerases. J. Bacteriol. 186:1050-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu, W.-L., C. A. Dunn, C. R. Jones, G. D'Souza, and M. J. Bessman. 2004. The 26 nudix hydrolases of Bacillus cereus, a close relative of Bacillus anthracis. J. Biol. Chem. 279:24861-24865. [DOI] [PubMed] [Google Scholar]
- 21.Xu, W.-L., J.-Y. Shen, C. A. Dunn, and M. J. Bessman. 2003. A new subfamily of the nudix hydrolase superfamily active on 5-methyl-UTP (riboTTP) and UTP. J. Biol. Chem. 278:37492-37496. [DOI] [PubMed] [Google Scholar]