Abstract
Bacterial populations produce persisters, cells that neither grow nor die in the presence of bactericidal agents, and thus exhibit multidrug tolerance (MDT). The mechanisms of MDT and the nature of persisters have remained elusive. Our previous research has shown that persisters are largely responsible for the recalcitrance of biofilm infections. A general method for isolating persisters was developed, based on lysis of regular cells by ampicillin. A gene expression profile of persisters contained toxin-antitoxin (TA) modules and other genes that can block important cellular functions such as translation. Bactericidal antibiotics kill cells by corrupting the target function (for example, aminoglycosides interrupt translation, producing toxic peptides). We reasoned that inhibition of translation will lead to a shutdown of cellular functions, preventing antibiotics from corrupting their targets, giving rise to MDT persister cells. Overproduction of the RelE toxin, an inhibitor of translation, caused a sharp increase in persisters. Functional expression of a putative HipA toxin also increased persisters, while deletion of the hipBA module caused a sharp decrease in persisters in both stationary and biofilm populations. HipA is thus the first validated persister-MDT gene. We suggest that random fluctuation in the levels of MDT proteins leads to the formation of rare persister cells. The function of these specialized dormant cells is to ensure the survival of the population in the presence of lethal factors.
Bacterial populations exhibit tolerance, an ability to survive killing by bactericidal factors without necessarily expressing a resistance mechanism. The molecular basis of tolerance is unknown. Tolerance to antibiotics is especially significant in survival of bacterial biofilms (15, 32, 51). Biofilms are formed when bacterial cells attach to a surface and grow into a mass encapsulated by an exopolymer matrix (15). Biofilms are responsible for nearly 65% of all human infections in the West (16). These include infections of catheters, orthopedic devices, heart valves, urinary tract infections, and lungs of cystic fibrosis patients (37). It has recently been found that persister cells are largely responsible for the high tolerance of bacterial biofilms to antimicrobials (9, 32, 44). In a variety of bacterial species examined, the level of persisters increased with the density of the culture (28), reaching ∼1% in stationary phase or in a biofilm of Pseudomonas aeruginosa (44), Escherichia coli, and Staphylococcus aureus (A. Spoering and K. Lewis, unpublished data).
Persisters were described in 1944 by Joseph Bigger who noticed that penicillin did not sterilize a culture of Staphylococcus (5). Unlike resistant mutants, persisters are phenotypic variants of the wild type that upon reinoculation produce a culture with a similar amount of persister cells (5, 28). We found that in E. coli, persisters are not formed in early-logarithmic-phase cultures (28). This suggests that persisters are not cells at a particular stage in the cell cycle, as originally suggested by Moyed and Bertrand (34) and that they are not produced in response to antibiotics. Indeed, persisters are rare nongrowing cells preexisting in a population (3). The only reports of genetic effects on the frequency of persisters come from the studies of hipA alleles in the E. coli hipBA operon (6, 7, 34, 35, 42). The fraction of persisters surviving ampicillin treatment is increased in mutant hipA7 cells 1,000 fold (from ∼10−5 to 10−2) compared to the wild type. The hipA7 allele carries two point mutations (29) and confers tolerance to a number of unrelated factors such as cell wall-acting antibiotics, heat, DNA-damaging agents (34), fluoroquinolones (19, 50), and aminoglycoside antibiotics (28). HipB is a transcriptional regulator of the hipBA operon (6), while HipA does not have homology to proteins of known function. It was proposed that the locus carries a toxin-antitoxin (TA) module (20). Similarly to typical TA module products, HipB and HipA form a complex (7); overexpression of HipA is “toxic,” leading to arrest of cell division (19); a hipB mutation could not be obtained due to apparent lethality of free HipA (7); HipB is a repressor of the operon, which is typical for antitoxins; and a homolog of the chromosomal hipBA operon is found on the Rhizobium symbiotic plasmid pNGR234a, where it may play a role in segregation maintenance (20). TA genes were originally identified on plasmids where they constitute a maintenance mechanism (21, 23). Typically, the toxin is a protein that inhibits an important cellular function such as translation or replication, and forms an inactive complex with the antitoxin. The toxin is stable, while the antitoxin is degradable. If a daughter cell does not receive a plasmid after segregation, the antitoxin level decreases due to proteolysis, leaving a toxin that either kills the cell or inhibits propagation. TA modules are also commonly found on bacterial chromosomes, but their role is largely unknown. The E. coli MazEF chromosomal TA module was proposed to serve as a programmed cell death mechanism (24, 41). However, it was reported recently that MazF and an unrelated toxin, RelE, do not actually kill cells but induce stasis by inhibiting translation, a condition that can be reversed by expression of corresponding antitoxins (13, 38). It was also suggested that MazF and RelE act as attenuators of the stringent response (11, 38).
Deletion of the putative TA module hipBA was reported to have no effect on persister formation in a growing culture (7). Consequently, these studies have not received much attention. The common interpretation is that the hipA7 allele creates an unnatural product that interferes with some unidentified components responsible for persister formation. The nature of persisters has remained elusive. Here, we report a gene expression profile of persister cells and suggest that persister formation is dependent on chromosomally encoded TA proteins, including the proteins of the putative TA module gene hipBA, and other proteins that can inhibit important cellular functions, leading to multidrug tolerance (MDT).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are described in Table 1. Luria-Bertani (LB) broth and LB agar media were used for culturing. The medium was supplemented with 75 μg of diaminopimelic acid/ml when culturing strains HM21 and HM22. Unless indicated otherwise, cells were grown by dilution of overnight cultures 1:1,000 in 12 to 25 ml of LB and incubation in 125-ml baffled culture flasks (Belco) for 2.5 h with aeration (250 rpm) at 37°C. Overnight cultures were made by dilution of thawed cells from an 8% dimethyl sulfoxide stock (−80°C) and incubation in LB medium with aeration for 16 to 20 h. For persister isolation experiments, E. coli HM22 was diluted into two 3-liter baffled culture flasks (Belco), each containing 600 ml of LB broth for a total of 1.2 liter for each replicate, and incubated for ∼2.5 h prior to isolation. For susceptibility measurements, HM21 and HM22 strains were cultured for 16 to 18 h, reaching stationary state, prior to testing. For toxin induction and protection studies, cells were cultured in LB containing 50 μg of chloramphenicol/ml and 100 μg of ampicillin/ml, and inducers were added at appropriate times.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Reference |
|---|---|---|
| Strains | ||
| MG1655 | Wild-type E. coli | 5 |
| MC1000 | Δ(ara-leu) Δlac rpsL150 | 6 |
| MG1 | ΔrelBE derivative of MC1000 | 7 |
| EMG2 | Wild-type E. coli K-12 CGSC# 4401 | 8 |
| KL310 | ΔrelBEF derivative of EMG2 | This work |
| KL312 | ΔhipBA derivative of EMG2 | This work |
| AT984 | F+dapA | 9 |
| HM21 | AT984 zde-264::Tn10 | 10 |
| HM22 | AT984 zde-264::Tn10 hipA7 | 10 |
| KL313 | ΔhipBA derivative of HM22 | This work |
| Plasmids | ||
| pKD13 | Template plasmid aphA | 1 |
| pKD46 | λ Red recombinase expression plasmid bla | 1 |
| pCP20 | pSC101rep (Ts) bla cat | 1 |
| pBAD33 | P15 cat araC pBAD | 11 |
| pKP219 | Mini-R1 bla lacIq pA1/O4/O3 | 12 |
| pKP3033 | pKP219 pA1/O4/O3::opSD::relB | 13 |
| pKP3035 | pBAD33 pBAD::relE | 13 |
Strain construction.
Knockout strains (KL310, KL311, and KL312), were constructed by the protocol described by Datsenko and Wanner (17). pKD13 was used as a PCR template with the primers described online at http://www.atsweb.neu.edu/lewislab/mechpersisters.html. The PCR products were transformed into K12 cells containing pKD46, and transformants were selected on plates containing kanamycin (25 μg/ml). The Kmr cassette was removed by transformation of cells with pCP20 and selection of ampicillin-resistant colonies at 30°C. Colonies were then purified by reinoculation and growth at 43°C. At the end of the procedure, the selected colonies were tested on ampicillin and kanamycin plates to verify the loss of all selective markers.
Epifluorescent microscopy.
Prior to treatment, 1 ml of culture was removed to provide a control sample. Ampicillin was then added to the remaining culture at a final concentration of 50 μg/ml. At indicated time points, 1 ml of culture was removed and stained with a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes) according to the manufacturer's protocol. Briefly, 3 μl of a mixture containing SYTO 9 (1.67 mM) and propidium iodide (10 mM) was added to the cell culture and allowed to incubate in the dark for 10 min. Stained samples were filtered onto 0.2-μm-pore-size black polycarbonate filters (Osmonics, Inc.). Cells were viewed with an epifluorescence microscope (Zeiss Axioskop 2 plus) with the appropriate filter sets. Images were captured with an Axiocam HRC and associated software (Carl Zeiss, Inc.)
Persister isolation.
At a cell density of approximately 107 CFU/ml, 10 ml of culture was removed from each flask and pooled to provide a baseline (t = 0). Ampicillin was then immediately added to each flask at a final concentration of 50 μg/ml. At 5, 15, 30, and 180 min after the addition of ampicillin 10, 25, 25, and 400 ml, respectively, of culture from each flask was removed and pooled (Vtotal = 20, 50, 50, and 800 ml). Upon being pooled, samples were placed on ice for 5 min and then centrifuged at 14,000 × g at 4°C for 10 min to isolate unlysed cells.
RNA isolation, labeling, and GeneChip hybridization.
Pellets obtained from persister isolation experiments were resuspended in 6 ml of T&C lysis solution (Epicentre). Total RNA was isolated with the MasterPure RNA purification kit (Epicentre) according to the manufacturer's instructions. Enrichment, fragmentation, and biotinylation of mRNA were carried out as described in the GeneChip Expression Analysis technical manual (Affymetrix) (40). Briefly, oligonucleotide primers complementary to either 16S or 23S rRNA sequences were annealed to rRNA, and ribosomal cDNA was synthesized. In vitro-synthesized control RNAs (Bacillus subtilis dapB, lysA, trpED, and pheB) were added before enrichment. The rRNA strand of the heteroduplex was selectively degraded with RNase H, and cDNA was removed with DNase I. RNA thus enriched for mRNA was then purified and incubated at 95°C in T4 polynucleotide kinase reaction buffer containing Mg2+ for fragmentation. RNA fragments were terminally thiolated with T4 polynucleotide kinase and γ-S-ATP. Finally, biotin was cross-linked to thiol groups with PEO-iodacetylbiotin reagent (Pierce Chemical), and after purification labeled RNA was hybridized to DNA arrays. Approximately 30 μg of enriched RNA and 2 to 4 μg of labeled product were obtained from 100 μg of total RNA.
Hybridization with GeneChip arrays, staining, and scanning.
The Affymetrix high-density array of the E. coli genome has been previously described (40, 43). Briefly, oligonucleotides are arranged in probe pairs: one is complementary to the target sequence, and the other contains a single mismatch at the central position and serves as a control for nonspecific hybridization. On average, 15 different oligonucleotides complementary to different parts of a given gene are used to identify a gene transcript. Probe pairs complementary to a single putative transcript are organized into probe sets. An array contains probe sets for 4,403 (putative) E. coli genes, including 4,290 open reading frames, and for all intergenic regions of 40 bp or longer, as well as probes complementary to control spike transcripts and biotinylated control oligonucleotide.
Biotinylated RNA was hybridized to the array in 100 mM morpholineethanesulfonic acid (MES) buffer (pH 6.6), containing 1 M NaCl, 20 mM EDTA, 0.01% Tween 20, 100 μg of herring sperm DNA/ml, 500 μg of bovine serum albumin/ml, and 0.5 nM control biotin-oligonucleotide 948. Hybridization was carried out at 45°C for 16 h on a rotary mixer at 60 rpm. Arrays were washed and stained according to the ProkGE-WS2 fluidics protocol with an Affymetrix Fluidics station. After hybridization, arrays were washed with wash buffer A at 25°C and wash buffer B at 45°C (GeneChip Expression Analysis technical manual). After posthybridization washes, arrays were incubated for 10 min at 25°C with streptavidin (10 μg/ml) in solution staining buffer containing 100 mM MES (pH 6.6), 1 M NaCl, 0.05% Tween 20, and 2 mg of bovine serum albumin/ml and washed in buffer A at 30°C. Thereafter, arrays were incubated with goat immunoglobulin G (0.1 mg/ml) and biotin anti-streptavidin (5 μg/ml) and fluorescently stained with streptavidin-phycoerythrin. Both incubations were for 10 min at 25°C in staining buffer. Finally, the array was washed with wash buffer A and scanned twice at 570 nm at a resolution of 3 μm with the Affymetrix confocal laser scanner.
Data analysis.
Arrays were analyzed with Affymetrix Microarray Suite, version 5.0, and Data Mining Tool, version 3.0, software. All gene probe sets of a given array were scaled, based on the intensity of control probes, to a target signal of 1,000, which allowed for pairwise comparisons. Arrays containing labeled mRNA from t = 5, 15, 30, and 180 min posttreatment were then compared to their experimental replicate t = 0 to generate signal log ratios for each probe set on the arrays. Average signal and signal log ratios of the three replicate experiments were calculated with Data Mining Tool, version 3.0. Normalized average signal intensities were used to identify specific transcription patterns for the duration of the experiment by Self-Organizing Map cluster analysis.
Antibiotic susceptibility measurements.
Antibiotics were added to flasks; at designated time points samples were withdrawn, diluted in LB medium, and spotted on an LB agar plate for colony counts.
Toxin induction.
Toxin synthesis from the recombinant expression vector was induced by adding 0.2% l-arabinose to each flask. At the designated time points, a sample was removed, diluted, and spotted on LB agar plates containing chloramphenicol (50 μg/ml), ampicillin (100 μg/ml), 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside), and 0.2% glucose for cell counting.
Toxin protection assay.
Toxin synthesis was induced by adding 0.2% l-arabinose to each flask. At the designated time points, a 1-ml sample was removed from each flask and placed in culture tubes (17 by 100 mm) with the appropriate drug concentration, and the tubes were incubated (250 rpm) at 37°C for 3 h. After the antibiotic challenge, cells were washed once (10,000 × g for 1 min) with fresh medium to minimize antibiotic carryover effects, diluted, and spotted onto an LB agar plate for cell counting. A control sample (prior to the addition of antibiotic) was spotted as well.
Protection by hipA7 at 30°C.
At t = 0, half of the flasks were moved to a second shaker (250 rpm) at 30°C. The antibiotic challenge was as described for the toxin protection assay; cells moved to the 30°C condition were challenged at 30°C.
Colony biofilms.
The procedure was essentially as previously described (1). Overnight cultures of K12 and KL312 (ΔhipBA) were diluted to an optical density at 600 nm of 0.2 in LB. A 5-μl drop of diluted culture was used to inoculate a single sterile, black, polycarbonate membrane filter (Fisher catalogue no. 09-732-18) on the surface of an LB agar plate. After the drops dried, these plates were inverted and incubated at 37°C. After 24 h, the filters were moved to fresh LB agar plates and returned to 37°C. After 48 h of growth, the filters were moved to plates containing 20 mM NaNO3 and either ciprofloxacin (5 μg/ml), mitomycin C (5 μg/ml), or no antibiotic. After a 24-h exposure to antibiotics, the filters were moved to a 50-ml Falcon tube with 10 ml of LB and vortexed for 2 min before being serially diluted and plated for colony counts.
RESULTS
Persister expression profile.
To identify candidate persister genes, we obtained an expression profile from persister cells. We reasoned that after treatment with ampicillin, a culture would lyse, leaving only intact persisters. To obtain sufficient material, the experiment was performed with the E. coli hipA7 strain HM22, which produces ∼1% persisters in a log-phase culture (34). Cells died rapidly after the addition of ampicillin, and by 15 min, only persisters survived (Fig. 1A). However, epifluorescent microscopy of a population stained with the live/dead kit (Fig. 1B to D) revealed a significant portion of dead but unlysed cells 30 min posttreatment (Fig. 1C). These intact nonpersisters could potentially contaminate persister RNA. Additional incubation, 180 min posttreatment, resulted in complete lysis of dead cells and clearing of debris, leaving intact persisters (Fig. 1D). These were collected by sedimentation and lysed, and total RNA was isolated, enriched for mRNA, fragmented, and labeled with biotin.
FIG. 1.
E. coli HM22 hipA7 cells were grown in LB medium to mid-exponential phase (∼5 × 107 cells/ml) at 37°C with aeration and treated with 50 μg of ampicillin/ml. (A) Samples were taken at indicated times and plated to determine live cells by colony count. Measurements were done in triplicate, and error bars indicate standard deviations. (B to D) Samples were stained with a LIVE/DEAD kit and visualized by epifluorescent microscopy (bar, 50 μm). Green cells are live, while red cells, stained by normally impermeant propidium iodide, are dead. Note the extensive red background due to cellular debris, as well as dead intact cells in the 30-min sample.
The labeled mRNA fragments were hybridized to E. coli GeneChips (43) containing probe sets of 29 different oligonucleotides representing each of the 4,290 open reading frames of the E. coli genome. It is important to stress that the profile we report below is not the familiar time course of changes in gene expression, but a time course of persister enrichment. Ampicillin, added at time zero, induces considerable changes in the gene profile (27), but we found that these changes occurred within 30 min and that the majority of the genes in the transcriptome were not affected between 30 and 180 min (Fig. 2A). We therefore wanted to identify messages that are enriched concomitant with the enrichment of persisters, between 30 and 180 min posttreatment. Cluster analysis pointed to precisely such a group of genes where the level of the message did not change significantly by 30 min, but increased sharply at 180 min (Fig. 3). At 30 min, there is a considerable amount of unlysed dead cells, and their subsequent clearance explains the sharp increase in the level of persister messages at the 180-min time point (Fig. 3, cluster 3). The level of message in clusters 1 and 2 also increased between time points 30 and 180 min, but the genes in this group were similarly elevated in the bulk. It appears that only cluster 3 clearly points to persister genes. This persister cluster comprised ∼300 genes with assigned functions (cluster 3) (http://www.atsweb.neu.edu/lewislab/mechpersisters.html). Several functional groups are evident within this cluster (Fig. 2B). These are the SOS stress response genes recA, sulA, uvrBA, and umuDC; the phage-shock (psp) operon genes; and several heat and cold shock genes (cspH, htrA, ibpAB, htpX, and clpB). This expression of stress responses is consistent with a survival function of persisters but does not in itself explain antibiotic tolerance. Another set comprising nearly 2% of the genes in the cluster belong to TA modules, including dinJ/yafQ, yefM, relBE, and mazEF. In addition, there are other genes within this cluster that similarly inhibit macromolecular synthesis, such as rmf and umuDC.
FIG. 2.
(A) Scatter plot of absolute gene expression at 180 min versus 30 min. Red lines indicate 2-, 3-, and 30-fold changes, respectively. (B) Heat map of selected genes from the cluster generated with Spotfire Decisionsite 7.2.
FIG. 3.
Cluster analysis of persister gene expression profile obtained with the Affymetrix Self-Organizing Map. The boxed cluster indicates the expression profile of genes specifically upregulated in persisters (180 min). The red lines indicate average signal intensity of all genes in the cluster, and blue lines indicate standard deviations.
Expression of potential inhibitors points to the possible mechanism of tolerance and to the nature of persisters, as described in the next section.
Functional analysis.
How can a cell that does not express specific resistance mechanisms exhibit MDT? We hypothesized that tolerance to antibiotics may be due to a shutdown of a target function (i.e., protein synthesis or DNA replication). In the case of protein synthesis, for example, its shutdown will prevent the lethal action of aminoglycoside antibiotics that cause translational misreading, leading to the formation of toxic misfolded peptides. A number of genes expressed in persisters appeared to match this criterion. RMF inhibits translation at stationary state by forming ribosome dimers (48), UmuDC has been reported to inhibit replication (36), and SulA is an inhibitor of septation (49). Most striking, however, is the overexpression of well-characterized chromosomal TA module proteins RelBE, MazEF, and DinJ/YafQ, a homolog of RelBE. Homologs of the genes expressing these proteins are found on plasmids where they constitute a maintenance mechanism (23). The ability of toxin modules to reversibly block translation (13, 38) makes them excellent candidates for MDT genes. By shutting down potential antibiotic targets, toxins may produce tolerant cells.
The best-studied TA module product in E. coli is RelBE, which cleaves mRNA on translating ribosomes, stalling protein synthesis (39). It was interesting to test whether cells expressing RelE will produce more persisters tolerant to antibiotics. A strain carrying two plasmids containing relE under the arabinose-inducible promoter pBAD and relB under the LacI-regulated promoter pA1/O3/O4, respectively, were used for this purpose (38). Cells were grown to mid-exponential phase, and arabinose was added to induce RelE expression. Within 30 min of induction, growth stopped and remained inhibited for the duration of the experiment (Fig. 4A). Three hours postinduction, a sample was withdrawn and exposed to a lethal dose of antibiotic. Cells were then plated on a medium with IPTG inducing the RelB antitoxin, and colonies of surviving persisters were counted. We found that a longer incubation of plates without the inducer of antitoxin was also sufficient to recover cells. Several antibiotics were tested, including ofloxacin, a fluoroquinolone inhibitor of DNA gyrase and topoisomerase IV; mitomycin C, which forms DNA adducts; cefotaxime, a cephalosporin cell wall synthesis inhibitor; and tobramycin, an aminoglycoside protein synthesis inhibitor. Each antibiotic was added at a concentration of more than 10× the MIC. Only persisters survive under these conditions (28, 44). RelE expression increased the number of persisters surviving treatment with cefotaxime, ofloxacin, and tobramycin 10 to 10,000 fold but did not protect from mitomycin C (Fig. 4B).
FIG. 4.
The effects of toxin overexpression on persister formation. (A) Cells were cultured as described in the legend to Fig. 1. RelE was induced (▪) from pKD3035 (pBAD::relE) in MG1 (MC1000 ΔrelBE) at time zero by the addition of 0.2% arabinose, and MG1 with a blank vector (pBAD33) served as the control (▴). (B) Cells were cultured, and RelE was induced as described above. After 3 h of RelE induction, samples were removed and treated with either cefotaxime (100 μg/ml), mitomycin C (10 μg/ml), ofloxacin (5 μg ml), or tobramycin (25 μg/ml) for 3 h at 37°C with aeration. The control (MG1/pBAD) (black bars) was challenged at a cell density similar to that of the relE induced cells (white bars). (C) HM22 cells (K12 hipA7) were cultured as described above and at time zero were moved to 30°C (▪) or kept at 37°C (▴). (D) HM22 cells were challenged at 30°C (white bars) as described in panel B. Controls are HM22 (black bars) and HM21 (K-12 hipA wild type) (gray bars) cells were challenged at 37°C.
Given that RelE expression increased persister production, we decided to take a closer look at HipBA, which is a likely TA module product as well. It was previously reported that induction of recombinant HipA increases production of persisters (19). We decided to extend these observations, taking advantage of the cold sensitivity of the hipA7 strain, which reportedly does not grow at 30°C (42). A possible reason for this sensitivity is a decrease in the ability of the allelic HipA7 protein to bind to HipB. Growth of the hipA7 strain was indeed strongly inhibited at 30°C, but full recovery of the culture was observed by plating at 37°C (Fig. 4C). This observation is similar to what is seen with some classical TA module products (such as RelBE), where functional expression of a toxin produces a reversible growth arrest, as discussed above. All studied toxins directly or indirectly inhibit macromolecular synthesis, complicating experiments that depend on subsequent expression of an antitoxin to resuscitate the cells. The use of hipA7 cells at low temperature is analogous to induction but has the advantage of being independent of macromolecular synthesis and is easily reversible by simply shifting the temperature.
To test the effects of tempertature on persister production, hipA7 cells were challenged with unrelated antibiotics at 30° and 37°C. The fraction of persisters increased dramatically in hipA7 cells at 30°C (Fig. 4D). In the case of mitomycin C, a 50- to 100-fold increase in protection was observed with hipA7 cells at 30°C (note that RelE expression had no effect on tolerance to mitomycin C).
The experiments described above suggested that TA-type proteins can lead to production and resuscitation of persister cells. Next, we tested whether disrupting these TA modules will have an effect on persister levels. Strains with relBE, mazEF, or hipBA deleted were created (17) and tested for persister production in both growing and stationary cultures. Antibiotics exhibiting lethal action in stationary cultures are essentially limited to the fluoroquinolones and mitomycin C.
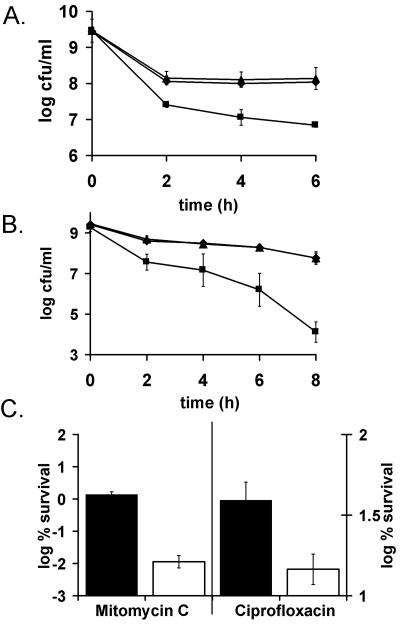
Both of these antibiotics produced a sharp (10- to 100-fold) decrease in stationary cell persisters in the ΔhipBA strain (Fig. 5A and B). Note that deletion of hipBA had no effect on the MIC of antibiotics. When tested in a logarithmic-phase culture, cells with the hipBA locus deleted did not show a consistently lower level of persisters than the isogenic parent strain (not shown). This agrees with a previous study in which a hipBA-null mutant was reported to have no phenotype when experiments were only performed with a growing culture (7). Apparently, other MDT genes play a leading role in persister formation under those conditions. Importantly, the level of persisters is 100- to 1,000-fold higher in stationary- than in logarithmic-state cultures. This suggests that the role of persisters in population survival is considerably greater at stationary state, and thus HipA is apparently the main determinant of persister formation in E. coli. Deletion of either relBE (Fig. 5A and B), mazEF, dinJ/yafQ, or rmf did not affect persister production (data not shown). It is possible that inhibitory proteins such as TA modules and RMF are redundant and that creating a multiply deleted strain will reveal the identity of additional MDT genes that play a role in logarithmic-state cells.
FIG. 5.
The effects of toxin deletion on persister formation. (A and B) EMG2 (K-12 wild type) (⧫), KL310 (ΔrelBE) (▴) and KL312 (ΔhipBA) (▪) were grown to stationary phase (16 to 18 h) and challenged with ofloxacin (5 μg/ml) (A) or mitomycin C (10 μg/ml) (B). (C) Biofilms EMG2 (K-12 wild type) (black bars) and KL312 (ΔhipBA) (white bars) were grown at 37°C for 48 h on LB agar to ∼2 × 109 CFU/biofilm. They were then exposed to LB agar plates containing 20 mM NaNO3 with or without 5 μg of mitomycin C/ml (left y axis) and with or without 5 μg of ciprofloxacin/ml (right y axis). All experiments shown in Fig. 5 are an average of three independent measurements; error bars indicate standard deviations.
Since the strain carrying the hipBA deletion had a strong effect on persisters in stationary phase, it was also tested in biofilms that grow slowly and show a level of tolerance similar to that of stationary-phase cells (44). Biofilms were formed by growth of a colony on a filter placed on nutrient medium (1). This method produces densely packed cells that closely resemble a clinically relevant E. coli bladder cell biofilm (2). The co lony biofilms were exposed to ciprofloxacin and mitomycin C by transferring the filter to a petri dish with nutrient medium containing the antibiotic. Previous studies reported rapid equilibration of antibiotics through the colony biofilm (1). The biofilm of the ΔhipBA strain produced 150- and 4-fold-fewer persisters when treated with mitomycin and ciprofloxacin, correspondingly, than the isogenic wild type (Fig. 5C).
DISCUSSION
Progress in our understanding of persisters since their discovery in 1944 (5) has been slow and incremental. The difficulties in studying a small fraction of cells expressing a temporary phenotype have stymied research into this enigmatic phenomenon. No genes directly affecting persister formation have been identified, and persister cells themselves have not been isolated. In this paper, we report several important advances in the understanding of persisters and their MDT: (i) a general method for the isolation of persisters, (ii) the first expression profile of persisters, and (iii) the first validated persister genes.
Persister isolation.
The method we developed is simple: a growing culture is lysed with ampicillin, and intact persisters surviving the treatment can be collected by sedimentation. The method can be applied to most species of bacteria.
Expression profile.
A group of ∼300 genes was differentially overexpressed in isolated persisters. Several interesting features of this group are apparent. Expression of stress responses (SOS, heat shock, cold shock, and phage shock proteins) seems to agree well with proposed survival function of these specialized cells. Of special interest was the presence of messages coding for proteins that can block cellular functions: several TA module proteins, RMF (inhibitor of translation), and UmuDC (inhibitor of replication).
Functional analysis.
Overexpression of RelE, the toxin of the relBE chromosomal TA module, led to a strong increase in MDT. RelE inhibits translation (12), and our data show that a toxin can potentially lead to tolerance. Functional expression of HipA, a toxin of a putative hipBA TA module, induced MDT, while deletion of hipBA led to a sharp, 10- to 100-fold decrease in the level of persisters in stationary-phase and biofilm cultures. It is important to stress that persisters are relatively abundant in stationary state and make up ∼1% of the population, compared to 10−6 to 10−5 in a logarithmic-phase culture (28). This suggests that the significance of persisters in population survival is considerably more important in stationary and biofilm cultures than in exponentially growing populations. HipA thus emerges as a major component of persister formation in E. coli. Ironically, this major player has been overlooked precisely because previous experiments with a hipBA-null strain (7) were only performed with logarithmic cultures, where hipBA deletion does not show a phenotype. We conclude that HipA is the first verified persister, or MDT, gene. We are currently determining the nature of the HipA target.
Strains with relBE, mazEF TA modules, or rmf deleted did not produce an effect on persister formation in our experiments. The presence of many apparently redundant TA modules (>10) (10) suggests that combining different knockout mutations in genes that can potentially cause persister production (such as relE) in a single strain will probably lead to identification of additional MDT genes.
A model of persister formation and MDT.
The observations reported in this study lead to the following model of persister production and antibiotic tolerance (Fig. 6). Production of these phenotypic variants of the wild type appears to be stochastic (31) and resembles individual variations in the chemotactic behavior of E. coli (30, 45). The level of an inhibitory protein (such as HipA) in a population fluctuates, and rare cells will express relatively high levels of this toxin. Bactericidal antibiotics bind to a target protein and corrupt its function, generating a lethal product (for example, aminoglycosides interrupt translation, resulting in misfolded peptides that damage the cell). A toxin binds to the target and inhibits the function, leading to tolerance. The antibiotic can bind to the blocked target but can no longer corrupt its function. Inhibition of translation by a toxin such as RelE will further cause a relative increase in the stable toxin (due to antitoxin degradation) of this and other TA module proteins, which might have an autocatalytic effect on inhibition of translation. Our observation of several TA modules upregulated in persisters is consistent with this model. Antitoxins act as repressors of TA modules (23), so a decrease in antitoxin protein level will cause an induction in transcription, which we observed in the gene profile of persisters.
FIG. 6.
A model for tolerance and persister formation. (A) A toxin molecule such as an inhibitor of translation is produced, and cells that have a high level of this component due to random fluctuations of expression become persisters (B). (C) Antibiotic binds to its target (bottom), which generates a corrupted product (such as truncated peptides formed in the presence of aminoglycosides), leading to cell death. If a target is blocked by a toxic factor (top), then the antibiotic can bind, but will no longer corrupt the function, and the result is drug tolerance, allowing the cell to survive.
It is known that inhibition of translation produces a subsequent shutdown of other important functions, such as DNA replication (38) and cell wall synthesis, two of the other major targets of bactericidal antibiotics. Proteins inhibiting translation (such as RelE or RMF) are therefore capable of shutting down major processes in the cell, which will lead to MDT persister cells.
Persistence is a reversible phenomenon. What we know about the best-studied toxins may suggest how resuscitation works. Overexpression of toxins inhibits protein synthesis strongly but incompletely (38), which may allow the synthesis of a neutralizing antitoxin. An even more effective mechanism for resuscitation of persisters would be through expression of tmRNA, which releases ribosomes stalled on mRNA lacking a termination codon. tmRNA has been shown to counter the action of artificially expressed RelE and MazF toxins (13). Importantly, tmRNA synthesis may proceed in cells with inhibited translation and might therefore trigger the resuscitation process.
Slow growth and tolerance.
Persisters are essentially altruistic cells that forfeit rapid propagation, which ensures survival of the population of kin cells (25) in the presence of lethal factors. Indeed, persisters in E. coli were shown to be nongrowing cells (3). Our gene profiling data also support the possibility of persisters being dormant. A cluster of approximately 600 genes (cluster 7) (http://www.atsweb.neu.edu/lewislab/mechpersisters.html) involved in metabolism and flagellar synthesis was seen to gradually diminish in cells treated with ampicillin, in accordance with previous findings (27). Interestingly, the same genes showed a further decrease in isolated persisters, suggesting that these might have been downregulated in persisters prior to the addition of antibiotic. Among these repressed genes were members of the large operons involved in oxidative phosphorylation: NADH dehydrogenase, ATP synthase, and cytochrome O-ubiquinole oxidase genes. The general theme of this gene cluster appears to be a decrease in nonessential genes and a shutdown of metabolism, both consistent with a dormant phenotype.
Tolerance of both planktonic and biofilm populations to antibiotics has been suggested to largely derive from slow growth rates (22). Indeed, the action of some cell wall-acting antibiotics, for example, depends absolutely on growth (4, 47), and both stationary-phase and slow-growing biofilm cultures are completely tolerant towards these agents (44). Our observation of essentially complete tolerance to cefotaxime of cells that stopped growing after expression of RelE is a case in point. However, it seems that tolerance to other antibiotics is not a simple consequence of growth rate. For example, the amount of persisters was significantly decreased in nongrowing stationary-phase ΔhipBA cells compared to a similarly nongrowing stationary-phase wild-type control. A larger production of persisters in slow-growing hipA7 cells at 30°C than in nongrowing RelE+ cells shows a lack of correlation between growth rate and tolerance as well. It seems that in most cases inhibition of the target function is the immediate cause of antibiotic tolerance, while decrease in growth rate is a consequence of this inhibition.
Biofilm resistance to killing.
Identification of MDT genes, including those described in this work, will lead to a better understanding of tolerance in general and of biofilm resistance to killing in particular. So far, studies aimed at identifying genes responsible for biofilm resistance were limited to the bulk of the population (33), which is fairly susceptible to therapeutic doses of antibiotics or may express resistance genes that act equally well in exponentially growing cells (9, 18). However, it is the persister subpopulation, present in all species studied, which is responsible for the dramatic tolerance of biofilms to unrelated antibiotics (32). Identification of MDT genes is an important first step in understanding recalcitrance of biofilms to antibiotic therapy, and it is likely to shed light on other related but poorly understood phenomena involving a dormant state, such as latent Mycobacterium tuberculosis infection (46), “viable but not culturable bacteria” (8, 14), and “uncultivable” bacteria (26).
Acknowledgments
The critical reading of the manuscript by Richard M. Losick, Alexander Tomasz, Gregg Bogosian, and Frederick F. Correia is greatly acknowledged.
This work was supported by a graduate fellowship from the National Science Foundation (A.S.) and by a National Institute of General Medical Sciences grant 2R01 GM061162-05A1 (K.L.).
We thank Kenn Gerdes for providing strains and plasmids.
REFERENCES
- 1.Anderl, J. N., M. J. Franklin, and P. S. Stewart. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44:1818-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105-107. [DOI] [PubMed] [Google Scholar]
- 3.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 4.Bayles, K. W. 2000. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 8:274-278. [DOI] [PubMed] [Google Scholar]
- 5.Bigger, J. W. 1944. Treatment of staphylococcal infections with penicillin. Lancet ii:497-500. [Google Scholar]
- 6.Black, D. S., B. Irwin, and H. S. Moyed. 1994. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 176:4081-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black, D. S., A. J. Kelly, M. J. Mardis, and H. S. Moyed. 1991. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 173:5732-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogosian, G., and E. V. Bourneuf. 2001. A matter of bacterial life and death. EMBO Rep. 2:770-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooun, A., S. Liu, and K. Lewis. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, J. M., and K. J. Shaw. 2003. A novel family of Escherichia coli toxin-antitoxin gene pairs. J. Bacteriol. 185:6600-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, S. K., and K. Gerdes. 2004. Delayed-relaxed response explained by hyperactivation of RelE. Mol. Microbiol. 53:587-597. [DOI] [PubMed] [Google Scholar]
- 12.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98:14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809-819. [DOI] [PubMed] [Google Scholar]
- 14.Colwell, R. R., and D. J. Grimes. 2000. Nonculturable microorganisms in the environment. American Society for Microbiology, Washington, D.C.
- 15.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 16.Costerton, W., R. Veeh, M. Shirtliff, M. Pasmore, C. Post, and G. Ehrlich. 2003. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Investig. 112:1466-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 19.Falla, T. J., and I. Chopra. 1998. Joint tolerance to beta-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob. Agents Chemother. 42:3282-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falla, T. J., and I. Chopra. 1999. Stabilization of Rhizobium symbiosis plasmids. Microbiology 145:515-516. [DOI] [PubMed] [Google Scholar]
- 21.Gerdes, K., P. B. Rasmussen, and S. Molin. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 83:3116-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert, P., P. J. Collier, and M. R. Brown. 1990. Influence of growth rate on susceptibility to antimicrobial agents: biofilms, cell cycle, dormancy, and stringent response. Antimicrob. Agents Chemother. 34:1865-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 24.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186:3663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh, P. C., S. A. Siegel, B. Rogers, D. Davis, and K. Lewis. 1998. Bacteria lacking a multidrug pump: a sensitive tool for drug discovery. Proc. Natl. Acad. Sci. USA 95:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 27.Kaldalu, N., R. Mei, and K. Lewis. 2004. Killing by ampicillin and ofloxacin induces overlapping changes in Escherichia coli transcription profile. Antimicrob. Agents Chemother. 48:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keren, I., N. Kaldalu, A. Spoering, Y. Wang, and K. Lewis. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13-18. [DOI] [PubMed] [Google Scholar]
- 29.Korch, S. B., T. A. Henderson, and T. M. Hill. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50:1199-1213. [DOI] [PubMed] [Google Scholar]
- 30.Korobkova, E., T. Emonet, J. M. Vilar, T. S. Shimizu, and P. Cluzel. 2004. From molecular noise to behavioural variability in a single bacterium. Nature 428:574-578. [DOI] [PubMed] [Google Scholar]
- 31.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 34.Moyed, H. S., and K. P. Bertrand. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moyed, H. S., and S. H. Broderick. 1986. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 166:399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Opperman, T., S. Murli, B. T. Smith, and G. C. Walker. 1999. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc. Natl. Acad. Sci. USA 96:9218-9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 38.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501-510. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131-140. [DOI] [PubMed] [Google Scholar]
- 40.Rosenow, C., R. M. Saxena, M. Durst, and T. R. Gingeras. 2001. Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res. 29:e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 183:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scherrer, R., and H. S. Moyed. 1988. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J. Bacteriol. 170:3321-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selinger, D. W., K. J. Cheung, R. Mei, E. M. Johansson, C. S. Richmond, F. R. Blattner, D. J. Lockhart, and G. M. Church. 2000. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18:1262-1268. [DOI] [PubMed] [Google Scholar]
- 44.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spudich, J. L., and D. E. Koshland, Jr. 1976. Non-genetic individuality: chance in the single cell. Nature 262:467-471. [DOI] [PubMed] [Google Scholar]
- 46.Tufariello, J. M., J. Chan, and J. L. Flynn. 2003. Latent tuberculosis: mechanisms of host and bacillus that contribute to persistent infection. Lancet Infect. Dis. 3:578-590. [DOI] [PubMed] [Google Scholar]
- 47.Tuomanen, E., and A. Tomasz. 1990. Mechanism of phenotypic tolerance of nongrowing pneumococci to beta-lactam antibiotics. Scand. J. Infect. Dis. Suppl. 74:102-112. [PubMed] [Google Scholar]
- 48.Wada, A. 1998. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells 3:203-208. [DOI] [PubMed] [Google Scholar]
- 49.Walker, G. C. 1996. The SOS response of Escherichia coli, p. 1400-1416. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 50.Wolfson, J. S., D. C. Hooper, G. L. McHugh, M. A. Bozza, and M. N. Swartz. 1990. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and β-lactam antimicrobial agents. Antimicrob. Agents Chemother. 34:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu, K. D., G. A. McFeters, and P. S. Stewart. 2000. Biofilm resistance to antimicrobial agents. Microbiology 146:547-549. [DOI] [PubMed] [Google Scholar]