Abstract
Glucose and other C sources exert an atypical form of catabolic repression on the σ54-dependent promoter Pu, which drives transcription of an operon for m-xylene degradation encoded by the TOL plasmid pWW0 in Pseudomonas putida. We have used a genetic approach to identify the catabolite(s) shared by all known repressive C sources that appears to act as the intracellular signal that triggers downregulation of Pu. To this end, we reconstructed from genomic data the pathways for metabolism of repressor (glucose, gluconate) and nonrepressor (fructose) C sources. Since P. putida lacks fructose-6-phosphate kinase, glucose and gluconate appear to be metabolized exclusively by the Entner-Doudoroff (ED) pathway, while fructose can be channeled through the Embden-Meyerhof (EM) route. An insertion in the gene fda (encoding fructose-1,6-bisphosphatase) that forces fructose metabolism to be routed exclusively to the ED pathway makes this sugar inhibitory for Pu. On the contrary, a crc mutation known to stimulate expression of the ED enzymes causes the promoter to be less sensitive to glucose. Interrupting the ED pathway by knocking out eda (encoding 2-dehydro-3-deoxyphosphogluconate aldolase) exacerbates the inhibitory effect of glucose in Pu. These observations pinpoint the key catabolites of the ED route, 6-phosphogluconate and/or 2-dehydro-3-deoxyphosphogluconate, as the intermediates that signal Pu repression. This notion is strengthened by the observation that 2-ketogluconate, which enters the ED pathway by conversion into these compounds, is a strong repressor of the Pu promoter.
Pseudomonas putida strains harboring the TOL plasmid pWW0 are able to grow on toluene, m-xylene, or p-xylene as the only carbon source as a consequence of a highly regulated pathway that produces benzoate or toluates from these aromatic substrates to Krebs cycle intermediates (1, 39). Expression of the upper TOL operon for bioconversion of toluene and xylenes into the corresponding carboxylic acids is driven by the σ54-dependent promoter Pu (12, 21, 39). Pu activity is dependent not only on the presence of pathway substrates but also on the metabolic status of the cell. An excess of certain carbon sources (6, 19) or rapid growth in rich medium inhibits the activity of the promoter in vivo even if the aromatic inducer is present in the culture (5, 13, 14, 20, 30). Such physiological control has at least two components (3, 4). First, there are inhibitory effects due to the growth rate and general energy load of the cells. This phenomenon, which is maximally manifest during exponential growth in Luria-Bertani medium (20), results from a combination of growth phase-dependent expression of IHF (which is required for Pu activity (29, 37, 49), with sigma factor competition in stationary phase mediated by (p)ppGpp (22, 26, 48). These elements ultimately limit the engagement of the σ54-RNAP with the −12 to −24 region of the promoter, which is weak during exponential growth and effective only at the onset of stationary phase (49). On the other hand, the presence of glucose and some other C sources (but not all) inhibits Pu (19) through what appears to be an unusual form of catabolic repression (CR). Yet, the C sources that cause Pu inhibition (typically glucose) do not trigger general CR in Pseudomonas (2, 46). Furthermore, organic acids, such as succinate, that do trigger CR in this species have no effect on Pu (19). A final intriguing feature of this phenomenon is that fructose does not influence Pu (19). Growth phase-related inhibition of Pu and CR of the same promoter can be separated either genetically (4, 6) or by the use of a chemostat (13, 14) as they appear ultimately to obey distinct environmental signals. Moreover, the effects of growth phase and CR on Pu have different extents. β-Galactosidase accumulation experiments with Pu-lacZ fusions, as well as direct measurement of the enzymatic activity of the upper TOL products (20) and quantitative primer extension assays of transcript production (14, 19), have shown that the silencing caused by rapid growth in rich medium completely abolishes Pu activity. In contrast, none of the carbon sources assayed in CR tests (6) decreases promoter output by more than two-thirds of the maximal activity.
Little is known about the mechanisms behind CR of Pu. Unlike that in Escherichia coli, CR in P. putida seems not to be affected by cyclic AMP or CRP (20) and diverse hints point toward a different molecular machinery as the basis for the phenomenon. For instance, we have observed that a ptsN mutant of P. putida (encoding the IIANtr protein of the phosphoenolpyruvate-sugar phosphotransferase system [PTS]) (6) makes Pu blind to the presence of glucose in the medium. Yet, glucose is not a PTS sugar in P. putida and its assimilation is not affected in the ptsN mutant, thereby suggesting that IIANtr participates more in the sensing than in the metabolism of the carbohydrate (6). Furthermore, repression by glucose seems to be possible only when His 68 of the IIANtr protein sequence is phosphorylated. On the contrary, an insertion in ptsO (encoding the protein NPr of the PTS) seems to make Pu permanently fixed in a repressed stated regardless of the presence or absence of glucose (7). These observations hint that proteins of the PTS group are pieces of the signal transmission pathway involved in CR of Pu but say nothing about the early metabolic signal that lies upstream of the transduction pathway causing inhibition.
This work was undertaken to identify the early metabolic signal of such a pathway, which starts with glucose and finishes with Pu downregulation. That the process starts with a metabolic signal (rather than a general physiological state) is indicated by the fact that α-methylglucoside, a nonmetabolizable glucose analogue, does not inhibit Pu (6). In this context, we have reexamined the known profile of repressive and nonrepressive C sources on the basis of the reconstruction of their catabolic pathways with genome sequence data. The phenotypes of various metabolic mutants made for rechanneling of C sources through alternative metabolic routes clearly expose the two key phosphorylated intermediates of glucose catabolism through the Entner-Doudoroff (ED) pathway, namely, 6-phosphogluconate and/or 2-dehydro-3-deoxygluconate phosphate) as the physiological signal(s) that elicits CR of Pu.
MATERIALS AND METHODS
Strains, plasmids, media, and general methods.
To examine Pu activity, P. putida MAD2 (16) or derivatives were used in all cases. This strain consists of P. putida KT2442 with a chromosomal Pu-lacZ fusion and the xylR allele xylRΔA assembled within a tellurite resistance minitransposon vector (42). The loss of the N-terminal A domain of the protein renders XylR constitutively active in the absence of aromatic inducers, thus eliminating this variable from our analysis (36). A ptsN mutant of P. putida MAD2 has been described elsewhere (6). Other derivatives bearing insertions in selected genes are explained below. Bacteria were grown on either rich Luria-Bertani medium or synthetic mineral M9 medium (41) supplemented with 0.2% Casamino Acids (CAA) in order to achieve equal growth rates and to avoid effects related to the stringent response (6, 8; see below). When required, the culture medium was supplemented with streptomycin (200 μg/ml), kanamycin (50 μg/ml), ampicillin (150 μg/ml), or potassium tellurite (80 μg/ml). 2-Ketogluconate (2KG; Sigma) was prepared as described in reference 47 and added to the medium at 10 mM. Plasmids were generally maintained in E. coli strains DH5α and CC118, although those containing conditional R6K replication origins were hosted in strain CC118λpir (10). DNA was manipulated in accordance with standard protocols (41). For the PCR, selected colonies of the strains under scrutiny were resuspended in 10 μl of H2O and boiled for 5 min. One microliter of this material was diluted 100-fold and directly subjected to 25 cycles of amplification (1 min at 92°C, 1 min at 55°C, and 2 min at 72°C) with Taq polymerase in the presence of 1.5 mM MgCl2, 1 mM deoxynucleoside triphosphates, and 50 pmol of the primers indicated in each case. Metabolic networks were reconstructed with the postgenomic analysis tools of Kanehisa (23, 24) refined manually, as indicated in each case, with experimental data available for sugar metabolism in P. putida. Growth tests were done by spotting 10 μl of serial 10-fold dilutions of cell suspensions of the strains under scrutiny on the surface of plates supplemented with suitable C sources.
In vivo CR tests.
Unless indicated otherwise, for testing of carbon inhibition of Pu promoter activity, the M9-CAA medium was added with the desired C source at a final concentration of 10 mM. Overnight cultures of P. putida MAD2 or its derivatives were diluted to an optical density at 600 nm (OD600) of approximately 0.05 in fresh medium and regrown at 30°C until the end of the exponential phase (OD600, ∼1.0). Samples were collected 30 min after this point, and promoter activity was measured by assaying the accumulation of β-galactosidase in cells made permeable with chloroform and sodium dodecyl sulfate (SDS) as described by Miller (32). Each enzymatic measurement was repeated at least twice in duplicate samples, and the deviations were less than 15%.
Generation of directed P. putida MAD2 mutants.
Selected disruptions of genes of interest were planned on the basis of the complete genome sequence of P. putida KT2440, which is available at the TIGR (The Institute for Genomic Research) website (http://www.tigr.org/). Genes encoding specific enzymes were identified with the available annotation, as well as the homologous E. coli proteins deposited in SWISSPROT, along with the TBLASTN program on the TIGR website (http://www.tigr.org/cgi-bin/BlastSeach/BLAST.cgi?). The resulting sequences were used to design oligonucleotide primers that amplified DNA fragments from the P. putida KT2442 chromosome long enough to be useful for homologous recombination. In order to create an eda::xylE insertion mutant (lacking 2-dehydro-3-deoxyphosphogluconate aldolase [TIGR code PP1024]), a 1.6-kb genomic segment of P. putida was amplified with PCR with oligonucleotides FwEda (5′-GGAATTCCACGCCGACAGCAATGC-3′) and RevEda (5′-GCTCTAGAGCATGCGATTG-3′). The resulting XbaI-EcoRI fragment, spanning 449 bp upstream and 433 bp downstream of the eda gene sequence, was cloned into pUC18Not, yielding pUCeda. This construct was digested with XhoI, treated with the Klenow fragment of DNA polymerase in the presence of deoxynucleoside triphosphates, and ligated to the SmaI insert of plasmid pXylE10 (45), generating pUCedaXylE. The NotI insert (bearing a truncated version of the eda gene) was introduced into sacB+ plasmid pKNG101 (25), which was previously linearized with the same enzyme, rendering pKNGeda. This plasmid was introduced by triparental mating into the P. putida MAD2 strain with helper strain E. coli HB101(pRK2013) as described in reference 10. Integration of the plasmid into the chromosome was selected by plating in the presence of streptomycin. To resolve the cointegrate, exconjugants were pooled, inoculated into fresh nonselective medium, and grown overnight at 30°C. This culture was then plated in the presence of sucrose to select the recombination events that release the plasmid from the chromosome. Plates were then sprayed with a 1% catechol solution to reveal colonies that contained the disrupted eda gene (which appeared light yellow). Correct replacement of the wild-type gene was first screened for streptomycin sensitivity and then verified by PCR with the primers RevEda (see above) and edaplus (5′-GCCCAAGCTCTCGATGGCTGATAAAGCCGCG-3′) to amplify the eda gene. One colony corresponding to a band of the size predicted for the disrupted gene (see Results) was finally selected. A similar approach was used to produce crc::tet and cyoB::tet mutants of P. putida MAD2. In these cases, the delivery plasmids for insertions, pCRC20 (crc::tet) and pKCYOBTc (cyoB::tet), were described before (11). An fda mutant (lacking fructose-1,6-bisphosphate aldolase) was made by a different strategy. First, a mutagenic DNA fragment was produced by amplification of an 813-bp fragment of the fda gene (TIGR code PP4960) with primers Ffda2Eco (5′-GGAATTCCGTCAACAACCTCGAGCAGACTGC-3′) and Rfda2 (5′-GAAGATCTGCGCCATCAGACGGCG-3′). The amplified EcoRI-BglII segment spans nucleotides 71 to 884 of the fda gene sequence (i.e., lacking 177 nucleotides of the 3′ end), carrying a frameshift entered with primer Ffda2Eco. Such a frameshifted ΔNfdaΔC sequence was cloned into the oriT+ vector pCHESIΩKm, a mobilizable variant of pUC18 encoding kanamycin resistance (the kind gift of S. Marqués) digested with EcoRI and BamHI, creating pCHESIfda. This plasmid was mobilized toward P. putida MAD2 as before, and cointegration was forced by selection for kanamycin. The single crossover of the mutagenic segment produced two truncated-frameshifted fda sequences. These were verified with PCR as before, with primer sets RPS (5′-AACAGCTATGACCATG-3′)-Rfda2 and RPS-Rfdamut (5′-GAAGATCTTCAAGCCGTCCATTTCCG-3′).
RESULTS
Monitoring of Pu CR with a Pseudomonas reporter lacZ system.
In order to have a reliable in vivo assay to examine the growth conditions that downregulate Pu, we used strain P. putida MAD2 (16). This strain bears all of the regulatory elements that control the expression of Pu assembled in a mini-Tn5 transposon inserted into the chromosome of P. putida. This includes a transcriptional Pu-lacZ fusion that results from placing a 312-bp DNA fragment from the TOL plasmid spanning positions −205 to +93 of Pu in front of a promoterless lacZ gene (see Materials and Methods). xylR is present on the reporter element in the form of a truncated gene encoding a variant named XylRΔA in which the N-terminal domain has been deleted. This deletion results in the constitutive activity of the protein independently of effector addition. Therefore, this reporter system reflects the physiological regulation of Pu as a phenomenon different from its activation by m-xylene, a feature of the A domain that is absent in XylRΔA (36).
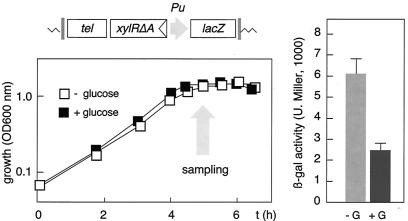
In order to have a reference to monitor the effect of glucose and other C sources in our reporter system, we used M9-CAA medium (0.2%) (6) supplemented or not supplemented with 10 mM glucose, a known repressive carbohydrate. P. putida MAD2 was grown in this semisynthetic medium with or without the additional carbon source added, and the growth rate of the strain and the Pu-lacZ fusion induction pattern were monitored as shown in Fig. 1. As the repressive effect of glucose is maintained throughout the growth curve (4), we used cells in early stationary phase as a convenient reference for comparisons. As predicted, β-galactosidase accumulation was significantly lower when cells were grown in the medium amended with glucose than without an extra carbon source, despite equal growth rates. The variation did not exceed two- to threefold but was faithfully reproducible. These results validated this simple assay system, as it reveals the action of glucose while it makes equivalent in all cases the effects of the growth rate (4, 6).
FIG. 1.
Measurement of C source repression of Pu with a lacZ reporter system. P. putida MAD2 cells bearing all of the elements required for Pu regulation assembled in the chromosome within a tellurite resistance mini-Tn5 transposon (sketched at the top) were grown overnight at 30°C in MM-CAA, diluted to an OD600 of ∼0.05, and regrown under the same conditions in the presence or absence of 10 mM glucose. Note the identical growth rates in the two media. At the point indicated (OD600, ∼1.2), samples were taken and their β-galactosidase (β-gal) levels were measured. Note the effect of glucose (G) on lacZ activity (plot to the right). U. Miller, Miller units.
Reconstruction of the glucose, fructose, and gluconate metabolic pathways of P. putida.
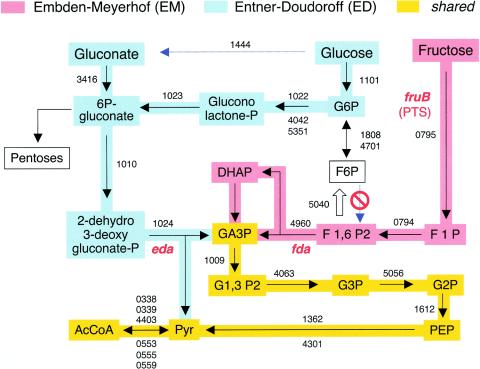
The frame for pinpointing the metabolic signal that triggers CR of Pu is visualization of the fate of each of those C sources that, despite their structural similarity, have different effects on the promoter. The availability of the whole genomic sequence of P. putida KT2440 allows the combination of experimental data on metabolism of carbohydrates known for this species (27, 28, 43, 44) with the predictions based on gene annotations (35, 40, 52). Figure 2 displays the reconstruction of the metabolic pathways available for the use of glucose, fructose, and gluconate in P. putida KT2440. Every protein involved in each of the steps is assigned a PP number as specified in the TIGR database (www.tigr.org). Figure 2 summarizes the most relevant properties of such pathways. It is remarkable that the network of transformations that results from projecting genomic data fits together with all of the observations made in various laboratories since the late 1960s on glucose, fructose, and gluconate metabolism in this bacterium (27, 43, 50, 51). The scheme of Fig. 2 thus provides a reliable reference to pinpoint shared (as well as unshared) metabolites during the consumption of the various C sources.
FIG. 2.
Representation of glucose, fructose, and gluconate metabolism in P. putida KT2440. Metabolic networks were reconstructed with the postgenomic analysis tools of Kanehisa (23, 24) refined manually, as indicated in Materials and Methods. The early stages of the EM and ED routes of P. putida are highlighted with different colors. Every step is associated with the PP number(s) of the TIGR annotation (www.tigr.org) of the protein predicted to execute each of the reactions. Note that F6P cannot be transformed into F1,6P2 because this bacterium totally lacks the enzyme F6P kinase. On the contrary, F1,6P2 can be transformed to F6P by virtue of the existing F1,6P2. The key locations of the genes encoding 2-dehydro,3-deoxyphosphogluconate aldolase (eda) and fructose-1,6-bisaldolase (fda) are highlighted. The site of the PTS type II enzyme, which acts as a transporter and kinase for fructose metabolism (fruB), is indicated as well. AcCoA, acetyl coenzyme A; DHAP, dihydroxyacetone phosphate; PEP, phosphoenolpyruvate.
The main starting point to understand C repression of Pu is the realization that fructose and glucose have different pathways toward the central metabolism. Fructose can follow a typical Embden-Meyerhof (EM) route (34) that starts with its conversion to fructose-1-phosphate by a PTS enzyme, all the way down to fructose-1,6-bisphosphate (F1,6P2), which is then cleaved to produce glyceraldehyde-3-phosphate (GA3P) and dihydroxyacetone phosphate. These two metabolites are then channeled toward the standard glycolytic pathway, resulting in pyruvate and acetyl coenzyme A, which enter the Krebs cycle. On the contrary, in P. putida glucose can only be metabolized through an ED pathway (15, 27, 34) in which the early product of its phosphorylation (glucose-6-phosphate) is converted into gluconolactone phosphate and eventually into 2-dehydro-3-deoxygluconate phosphate. This intermediate is then split into pyruvate and GA3P, which enter the glycolytic pathway at separate sites. Glucose can also join the ED route by direct conversion into gluconate (15, 27, 34), which can only be degraded through the ED pathway (27). It has been reported that the early conversion of glucose to gluconate prior to any metabolism is the preferred choice of P. putida for consumption of the sugar, at least under laboratory conditions (27).
Both the predictions of Fig. 2 and various experimental observations (44, 50, 51) reveal one key facet of glucose metabolism in P. putida. While glucose-6-phosphate can reversibly be converted into fructose-6-phosphate (F6P), this last intermediate cannot be transformed into F1,6P2 because this bacterium lacks the enzyme F6P kinase. On the contrary, F1,6P2 produced during the early metabolism of fructose can follow the gluconeogenic pathway and be transformed into F6P by virtue of the existence of F1,6P2 (encoded by the fbp gene). This makes sense, as F6P remains as the major starting point for feeding the essential pentose cycle when cells grow on fructose.
Since glucose and gluconate repress Pu while fructose does not, the network of Fig. 2 rules out the possibility that downstream compounds that are shared by the EM and ED pathways are the early signals for C control of the promoter. Moreover, the same logic bans the metabolites resulting from fructose degradation through the EM pathway from triggering the effect. As gluconate enters the ED pathway via conversion to 6-phosphogluconate, it appears that the candidates for triggering C repression of Pu are likely to include only those between gluconate and GA3P. Unfortunately, direct testing of most phosphorylated metabolic intermediates of glucose, fructose, and gluconate on Pu activity in vivo is not possible because they are not transported into the cells when added to the medium. As an alternative, the following sections present various genetic approaches to circumvent this limitation and to identify the metabolite(s) that lies at the top of the signaling pathway.
Rechanneling of fructose metabolism through the ED pathway converts this sugar into a repressive C source for Pu.
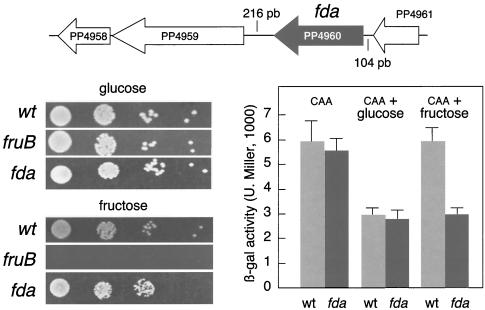
As mentioned above, the most dramatic difference in the metabolism of glucose-gluconate on the one hand and fructose on the other is the divergence in the early stages of their respective pathways: the repressive C sources necessarily go through the ED pathway, while the noninhibitory carbohydrate can follow an EM route. As a consequence, intermediate metabolites of the ED pathway may not accumulate when cells consume fructose. Yet, inspection of the network of Fig. 2 reveals that fructose metabolism could be rechanneled toward an ED itinerary by blocking the cleavage of F1,6P2 to GA3P and dihydroxyacetone phosphate by fructose-1,6-bisaldolase (encoded by fda). The only open reading frame (ORF) of the P. putida chromosome homologous to the extremely conserved fda gene (31) is the one corresponding to protein PP4960. On this basis, we set out to generate an fda mutant of strain P. putida MAD2 as explained in Materials and Methods. Unlike a fruB mutant, which is predicted not to phosphorylate fructose upon entry into cells, the fda mutant was able to grow as well as the wild-type strain with fructose as the only carbon source (Fig. 3). Since there are no alternative routes for metabolism of F1,6P2, this intermediate necessarily has to be diverted to the ED route in order to allow growth. This is in principle possible given the presence of the gluconeogenic enzyme F1,6P2 in the P. putida genome (TIGR code PP5040; encoded by fbp; Fig. 2). Since we had traced the cause of C source inhibition of Pu to ED metabolites, we reasoned that addition of fructose to an fda mutant must convert this sugar into a repressor of the promoter as good as glucose is in the wild-type strain. To test this prediction, we carried out the experiment shown in Fig. 3, in which we compared the effects of glucose and fructose on the Pu-lacZ fusion borne by P. putida MAD2 with their effects on its fda mutant derivative. The data clearly show that fructose becomes a repressor of Pu in the fda mutant strain. Given the chromosomal context of the fda gene (Fig. 3), this result can be faithfully traced to fda as its interruption should not cause polar effects on any of the adjacent cistrons. This result strengthens the notion that ED metabolites but not EM compounds or downstream intermediates are the ones that signal C repression of Pu.
FIG. 3.
Phenotypes of an fda mutant of P. putida MAD2. The chromosomal context of the fda gene encoding fructose-1,6-bisaldolase in this strain is shown at the top. Adjacent ORFs appear to determine unrelated functions: PP4961 is a putative lipoprotein, PP4959 encodes a response regulator, and PP4958 is a conserved protein whose function is unknown. The fda gene is placed at considerable distances (104 and 216 bp) from adjacent genes upstream and downstream, and it is thus likely to form a single transcriptional unit. The plate assays at the lower left show the growth of the fda mutant with glucose and fructose as the only C sources with the wild-type (wt) P. putida MAD2 strain as a positive control and a fruB mutant unable to consume fructose as a negative control (Fig. 2). For the experiment shown to the lower right, Pu-lacZ reporter strain P. putida MAD2 and its fda derivative were subjected to a carbon Pu inhibition test as explained in Materials and Methods and Fig. 1. Note that fructose becomes a repressive C source of this promoter in the fda mutant. β-gal, β-galactosidase; U. Miller, Miller units.
The lack of Crc alleviates, but does not abolish, CR of Pu.
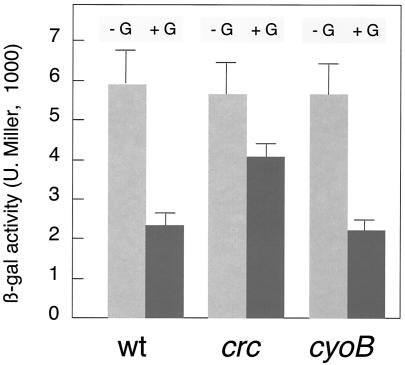
Following the same rationale as above, we wondered whether artificially increased activity of the ED pathway enzymes could reduce the transient levels of ED metabolites and thus alleviate Pu inhibition by glucose. To this end, we benefited from the seminal observations made by Phibbs and his collaborators (9) on the phenotypes associated with loss of the crc (catabolite repression control) gene. The product of this gene inhibits (through a thus far unresolved mechanism) the expression of a number of functions in P. aeruginosa and P. putida (9, 17, 18, 33, 53, 54) when cells are cultured with succinate. Interestingly, this carboxylic acid triggers a general CR in Pseudomonas but has no effect on Pu activity (13, 14, 19, 20, 30). Since one of the consequences of the loss of crc is an increase in the activity of the ED pathway (9), we sought the generation of a crc mutant of P. putida MAD2 as a method to cause a reduction in the accumulation of ED intermediates in vivo. On this basis, we recreated the crc::tet knockout already available for P. putida KT2440 (11) in P. putida MAD2 as described in Materials and Methods. When the C source downregulation of Pu behavior was tested in the crc strain (Fig. 4), we observed a consistent, albeit not complete, relief of the inhibitory effect of glucose. This behavior can be interpreted as the result of a direct effect of Crc on Pu or, as suggested above, as the indirect outcome of a higher activity of the ED pathway. The first possibility is, however, very unlikely, as succinate or lactate, the preferred partner of Crc for general catabolite repression in Pseudomonas (9, 17, 53), has no effect on Pu (13, 14, 19, 20, 30).
FIG. 4.
Inhibition of Pu by glucose in P. putida MAD2 lacking the crc or cyoB gene. The Pu-lacZ reporter strain P. putida MAD2 and its crc::tet and cyoB::tet derivatives were subjected to a standard carbon (glucose, +G) Pu inhibition test. Note the partial relief of Pu downregulation in the crc mutant, which likely resulted from stimulation of the ED pathway for glucose metabolism (see the text for an explanation). β-gal, β-galactosidase; U. Miller, Miller units; wt, wild type.
Blockage of 2-dehydro-3-deoxygluconate phosphate metabolism exacerbates the inhibitory effect of glucose on Pu.
While the crc mutant could turn over ED intermediates more rapidly and thus decrease their transient concentrations, the metabolic network depicted in Fig. 2 suggests the possibility of forcing the accumulation of the same intermediates by creating a mutation in the gene eda (TIGR code PP1024). The enzyme it encodes (2-dehydro-3-deoxyphosphogluconate aldolase) splits 2-dehydro-3-deoxygluconate phosphate into GA3P and pyruvate, which then enter the lower glycolytic pathway. Knockout of the eda gene should therefore result in the buildup of ED metabolites as dead-end products when cells grow in the presence of glucose. To check the effect of such a mutation on Pu activity, we mutated the eda gene in reporter strain P. putida MAD2 as explained in Materials and Methods. Before running the standard Pu-lacZ induction assay, we examined whether the eda strain could grow on minimal medium (MM) with single carbon sources. Figure 5 shows that, unlike the fda mutant discussed above, cells lacking eda did not grow in glucose or fructose as the sole C source, although they did grow normally on rich medium or MM with amino acids. Furthermore, addition of 10 mM glucose slowed down the growth of eda mutants of P. putida MAD2 in MM-CAA. This effect of glucose on growth is surely due to the accumulation of phosphorylated intermediates of glucose metabolism, although the target(s) inhibited is unknown (43, 51). The same might be true for the lack of growth on fructose, a small fraction of which could be metabolized via the ED pathway (43) and thus produce the equivalent toxic metabolites. Moreover, the poor growth on succinate probably reflects a regulatory problem rather than a metabolic jam. Despite these caveats, we sought to examine the effect of the eda mutation on Pu by adding subinhibitory concentrations of glucose to the MM-CAA in which P. putida MAD2 and its eda mutant derivative were tested. Figure 5 shows that glucose was far more inhibitory to Pu in the eda mutant than in the wild-type strain. It should be mentioned that the repression of Pu activity by glucose in the eda mutant strain shown in Fig. 5 is probably an underestimation, as slower growth generally enhances Pu activity by other mechanisms (3, 4). In addition to this, the eda mutant lowers Pu expression even in the absence of glucose. This makes sense, since the pentose phosphate pathway leading to ribose and other key metabolites has a link to the ED pathway (Fig. 2). The results of Fig. 5 were caused by the loss of eda and not by polar effects on nearby genes, as the chromosomal context of the gene rules out any influence on neighboring ORFs.
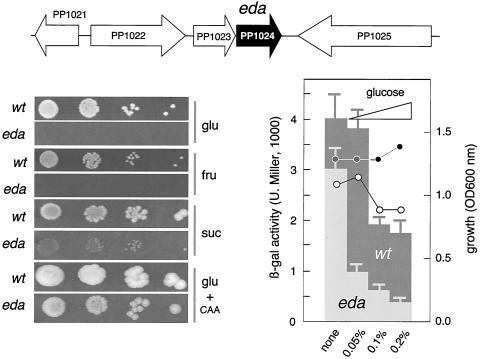
FIG. 5.
Phenotypes of an eda mutant of P. putida MAD2. The chromosomal region of the eda gene encoding 2-dehydro-3-deoxyphosphogluconate aldolase is shown at the top. Some of the adjacent ORFs encode somewhat related functions: PP1021 encodes the transcriptional regulator HexR, PP1022 determines glucose-6-phosphate dehydrogenase, PP1023 encodes phosphogluconolactonase, and PP1025 corresponds to leuA, the gene for isopropylmalate synthase. The genomic context rules out any polar effect of the eda::xylE insertion. The plate assays at the lower left show the growth of the eda mutant on glucose (glu), fructose (fru), succinate (suc), and the combination of glucose plus amino acids (glu + CAA). Note the lack of growth on glucose and fructose, the poor growth on succinate, and the suboptimal development on CAA added with glucose. For the experiment shown at the lower right, Pu-lacZ reporter strain P. putida MAD2 and its eda derivative were subjected to a test of carbon Pu inhibition at various concentrations of glucose in MM-CAA as indicated (0.05%, ∼2.5 mM; 0.1%, ∼5 mM; 0.2%, ∼10 mM). The OD600 of the cultures grown for the same period of time is indicated, highlighting the noxious effect of glucose on the eda mutant. Note the poor expression of Pu in cells lacking eda as the glucose concentration in the medium increases. β-gal, β-galactosidase; U. Miller, Miller units; wt, wild type.
2KG is a strong repressive C source for Pu.
The results above pinpoint 6-phosphogluconate and/or 2-dehydro-3-deoxygluconate phosphate as the two metabolites of the ED pathway that are present under every condition in which Pu repression is observed. The only metabolic fate of 2-dehydro-3-deoxygluconate phosphate is to be split into GA3P and pyruvate (Fig. 2, see above), while 6-phosphogluconate can be both converted into 2-dehydro-3-deoxygluconate phosphate and channeled toward the pentose cycle. Once more, inspection of the metabolic network of Fig. 2 and 6 suggests the possibility of testing with the Pu-lacZ reporter system of P. putida MAD2 whether 2-dehydro-3-deoxygluconate phosphate and/or 6-phosphogluconate are the metabolic signals that ultimately trigger C repression of Pu. This is because there is a way to increase the production of 2-dehydro-3-deoxygluconate phosphate and 6-phosphogluconate by adding 2KG to the culture. P. putida possesses a complete kgu cluster for conversion of 2KG into 2-dehydro-3-deoxygluconate and 2-dehydrogluconate phosphate, both of which enter the central metabolism via conversion into 6-phosphogluconate and 2-dehydro-3-deoxygluconate phosphate (Fig. 2). In fact, 2KG is a good carbon source for all of the P. putida MAD2 variants tested (except the eda mutant; Fig. 6). Metabolism of 2KG should therefore result in a significant level of these two intermediates. When the activity of Pu was examined in the presence of 2KG (Fig. 6), it became clear that this compound is a strong repressor. This result strengthens the notion that production of 6-phosphogluconate and/or 2-dehydro-3-deoxygluconate phosphate signals the downregulation of Pu.
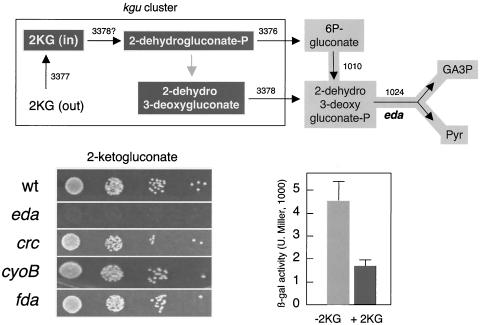
FIG. 6.
Effect of 2KG on P. putida MAD2 and its derivatives. The upper scheme summarizes the port(s) of entry of 2KG into the ED metabolic route. By virtue of the two key enzymes of the kgu cluster (encoded by PP3376 and PP3378 in P. putida), 2KG can originate 2-dehydrogluconate phosphate and 2-dehydro-3-deoxygluconate, which in turn are able to produce 6-phosphogluconate and 2-dehydro-3-deoxygluconate phosphate (Fig. 2) and further metabolized through the ED pathway. The plate tests at the lower left show the growth of various mutants of P. putida MAD2 with 2KG as the only C source. Note that excepting the eda mutant, which grows very slowly, all of the others can metabolize 2KG well. At the lower right is a C source inhibition test of P. putida MAD2 with 10 mM 2KG in the medium. Note the repressive effect of this compound on the activity of the Pu-lacZ fusion. β-gal, β-galactosidase; U. Miller, Miller units; wt, wild type.
Loss of cyoB of P. putida MAD2 does not abolish C source regulation of Pu.
As mentioned above, Pu is sensitive to both the general physiological status of the cells (as reflected in their growth rate) and the presence of specific C sources. These two aspects are not entirely independent, as an abundance of C sources may result in a more energized physiological state while a decrease in the energy load of cells is translated into a higher carbon demand. We have shown above that an excess of C sources consumed through the ED pathway downregulates Pu. But the reverse question is relevant as well: does a higher physiological reclamation of C abolish the repressive effect of ED pathway substrates (e.g., glucose) and metabolites? To address this question, we reasoned that cells with nonlethal defects in the electron transfer chain may experience faster consumption of C to balance a lower efficiency of energization of their membranes. It has been reported that the loss of the cyoB gene of P. putida, encoding cytochrome o-ubiquinol oxidase (the main terminal oxidase of the electron transport chain under highly aerobic conditions), relieves the general CR caused by lactate or succinate (11) and the growth phase control of other σ54 promoters (38). This suggests that cyoB mutants may suffer a permanent excessive demand for carbon which, on the other hand, does not affect the growth rate when such C sources are abundant in the medium. To tackle whether such a permissive lesion in the electron transport chain of P. putida MAD2 abolishes C source regulation of Pu, we constructed a cyoB::tet insertion in the chromosome of P. putida MAD2 and submitted the resulting strain to an ordinary CR test. The results in Fig. 4 indicate that the loss of cyoB did not prevent repression by glucose. This observation strengthens the notion that glucose and other ED substrates exert a specific effect on the Pu promoter that is distinct from other generic environmental signals related to the energy status of cells.
DISCUSSION
The accessibility of a growing number of complete bacterial genomes allows a confident prediction of the flow of various compounds through alternative sets of metabolic reactions (see, for instance, the Kyoto encyclopedia of genes and genomes [23, 24]). The P. putida genome sequence has been instrumental in visualizing the major divergences and similarities in the metabolism of glucose, fructose, and gluconate in this organism. Early observations on the inability of glucose to enter glycolysis through the standard EM pathway and its forced routing into the ED pathway (15, 27, 50, 51) have their explanation in the lack of F6P kinase (Fig. 2). Fructose, on the other hand, has the capacity in Pseudomonads to follow a typical EM route all the way through but can also be channeled into the ED pathway by transformation of F1,6P2 to F6P by virtue of the existing F1,6P2. We believe that this distinct feature of glucose-gluconate versus fructose metabolism sheds light upon the different effects that they have on the Pu promoter when these carbon sources are present in the growth medium.
The phenotypes caused by directed mutations in key intersections of the predicted metabolic networks (fda, eda; Fig. 3 and 5) and general metabolic regulators (crc, cyoB; Fig. 4) provide a set of evidence in vivo that singles out the metabolites of the ED pathway as the metabolic signal(s) at the top of the transduction pathway that ends in Pu inhibition. In our view, the most informative result is that an fda mutant, which necessarily reroutes fructose by the ED roadmap, makes this sugar an inhibitor of Pu (Fig. 3). Since the one predicted variation between fda+ and fda mutant strains when grown in fructose is precisely the making of ED metabolites to degrade this sugar, we argue that they are the ones that make a difference in Pu downregulation. Since gluconate, an inhibitor of Pu, enters the ED route at the level of 6-phosphogluconate, the only candidate for the signaling metabolite is either 6-phosphogluconate or 2-dehydro-3-deoxygluconate phosphate (or both). In addition, we did observe that addition of 2KG to the culture resulted in strong inhibition of Pu. This carbohydrate has to be metabolized via its conversion into 6-phosphogluconate and 2-dehydro-3-deoxygluconate phosphate. Since both metabolites are essential and interconnected, we make the case that they are the sole molecules formed under all of the repressive conditions tested for Pu and therefore we tentatively propose that they are the molecules that trigger the process that ends in Pu downregulation.
While the rise of a metabolic signal is the obligatory start of a transduction pathway, our results say nothing about the next step in the route. Yet, we have proven before (6) that phosphorylation of the IIANtr protein of P. putida encoded by the ptsN gene by a thus far unknown kinase is necessary for Pu repression by glucose. Also, a mutant NPr protein (encoded by ptsO) that cannot be phosphorylated appears to keep Pu in a permanently repressed state (7). There must therefore be a direct or indirect connection between the levels of ED metabolites and those of phosphorylated and nonphosphorylated IIANtr and NPr. Such a link, however, is not trivial given the lack of typical PTS type I enzymes encoded in the chromosome of P. putida (F. Velázquez et al., unpublished data). Future efforts will examine this outstanding issue, which involves the recruitment of proteins of the limited PTS existing in P. putida for regulatory functions unrelated to sugar transport.
Acknowledgments
We are indebted to I. Cases, J. Pérez-Martín, F. Rojo, and M. Valls for inspiring discussions and sharing valuable materials. S. Marqués kindly provided pCHESIΩKm.
This work was supported by EU grants BIOCARTE, LINDANE, and ACCESS and by project BIO2001-2274 of the Spanish CICYT.
REFERENCES
- 1.Assinder, S. J., and P. A. Williams. 1990. The TOL plasmids: determinants of the catabolism of toluene and the xylenes. Adv. Microb. Physiol. 31:1-69. [DOI] [PubMed] [Google Scholar]
- 2.Bruckner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. [DOI] [PubMed] [Google Scholar]
- 3.Cases, I., and V. de Lorenzo. 2001. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 20:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cases, I., and V. de Lorenzo. 2000. Genetic evidence of distinct physiological regulation mechanisms in the σ54 Pu promoter of Pseudomonas putida. J. Bacteriol. 182:956-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cases, I., V. de Lorenzo, and J. Perez-Martin. 1996. Involvement of sigma 54 in exponential silencing of the Pseudomonas putida TOL plasmid Pu promoter. Mol. Microbiol. 19:7-17. [DOI] [PubMed] [Google Scholar]
- 6.Cases, I., J. Perez-Martin, and V. de Lorenzo. 1999. The IIANtr (PtsN) protein of Pseudomonas putida mediates the C source inhibition of the sigma 54-dependent Pu promoter of the TOL plasmid. J. Biol. Chem. 274:15562-15568. [DOI] [PubMed] [Google Scholar]
- 7.Cases, I., F. Velázquez, and V. de Lorenzo. 2001. Role of ptsO in carbon-mediated inhibition of the Pu promoter belonging to the pWW0 Pseudomonas putida plasmid. J. Bacteriol. 183:5128-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 9.Collier, D. N., C. Spence, M. J. Cox, and P. V. Phibbs. 2001. Isolation and phenotypic characterization of Pseudomonas aeruginosa pseudorevertants containing suppressors of the catabolite repression control-defective crc-10 allele. FEMS Microbiol. Lett. 196:87-92. [DOI] [PubMed] [Google Scholar]
- 10.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 11.Dinamarca, M. A., A. Ruiz-Manzano, and F. Rojo. 2002. Inactivation of cytochrome o ubiquinol oxidase relieves catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 184:3785-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon, R. 1986. The xylABC promoter from the Pseudomonas putida TOL plasmid is activated by nitrogen regulatory genes in Escherichia coli. Mol. Gen. Genet. 203:129-136. [DOI] [PubMed] [Google Scholar]
- 13.Duetz, W. A., S. Marqués, C. de Jong, J. L. Ramos, and J. G. van Andel. 1994. Inducibility of the TOL catabolic pathway in Pseudomonas putida(pWW0) growing on succinate in continuous culture: evidence of carbon catabolite repression control. J. Bacteriol. 176:2354-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duetz, W. A., S. Marques, B. Wind, J. L. Ramos, and J. G. van Andel. 1996. Catabolite repression of the toluene degradation pathway in Pseudomonas putida harboring pWW0 under various conditions of nutrient limitation in chemostat culture. Appl. Environ. Microbiol. 62:601-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Entner, N., and M. Doudoroff. 1952. Glucose and gluconic acid oxidation of Pseudomonas saccharophila. J. Biol. Chem. 196:853-862. [PubMed] [Google Scholar]
- 16.Fernandez, S., V. de Lorenzo, and J. Perez-Martin. 1995. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol. Microbiol. 16:205-213. [DOI] [PubMed] [Google Scholar]
- 17.Hester, K. L., J. Lehman, F. Najar, L. Song, B. A. Roe, C. H. MacGregor, P. W. Hager, P. V. Phibbs, Jr., and J. R. Sokatch. 2000. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 182:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hester, K. L., K. T. Madhusudhan, and J. R. Sokatch. 2000. Catabolite repression control by Crc in 2xYT medium is mediated by posttranscriptional regulation of bkdR expression in Pseudomonas putida. J. Bacteriol. 182:1150-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holtel, A., S. Marques, I. Mohler, U. Jakubzik, and K. N. Timmis. 1994. Carbon source-dependent inhibition of xyl operon expression of the Pseudomonas putida TOL plasmid. J. Bacteriol. 176:1773-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugouvieux-Cotte-Pattat, N., T. Khöler, M. Rekik, and S. Harayama. 1990. Growth-phase-dependent expression of the Pseudomonas putida TOL plasmid pWW0 catabolic genes. J. Bacteriol. 172:6651-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inouye, S., A. Nakazawa, and T. Nakazawa. 1983. Molecular cloning of regulatory gene xylR and operator-promoter regions of the xylABC and xylDEGF operons of the TOL plasmid. J. Bacteriol. 155:1192-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa, M. 1997. A database for post-genome analysis. Trends Genet. 13:375-376. [DOI] [PubMed] [Google Scholar]
- 24.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 26.Laurie, A. D., L. M. Bernardo, C. C. Sze, E. Skarfstad, A. Szalewska-Palasz, T. Nystrom, and V. Shingler. 2003. The role of the alarmone (p)ppGpp in sigma N competition for core RNA polymerase. J. Biol. Chem. 278:1494-1503. [DOI] [PubMed] [Google Scholar]
- 27.Lessie, T. G., and P. V. Phibbs, Jr. 1984. Alternative pathways of carbohydrate utilization in pseudomonads. Annu. Rev. Microbiol. 38:359-388. [DOI] [PubMed] [Google Scholar]
- 28.Ma, J.-F., P. W. Hager, M. L. Howell, P. V. Phibbs, and D. J. Hassett. 1998. Cloning and characterization of the Pseudomonas aeruginosa zwf gene encoding glucose-6-phosphate dehydrogenase, an enzyme important in resistance to methyl viologen (paraquat). J. Bacteriol. 180:1741-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macchi, R., L. Montesissa, K. Murakami, A. Ishihama, V. de Lorenzo, and G. Bertoni. 2003. Recruitment of sigma 54-RNA polymerase to the Pu promoter of Pseudomonas putida through integration host factor-mediated positioning switch of alpha subunit carboxyl-terminal domain on an UP-like element. J. Biol. Chem. 278:27695-27702. [DOI] [PubMed] [Google Scholar]
- 30.Marques, S., A. Holtel, K. N. Timmis, and J. L. Ramos. 1994. Transcriptional induction kinetics from the promoters of the catabolic pathways of TOL plasmid pWW0 of Pseudomonas putida for metabolism of aromatics. J. Bacteriol. 176:2517-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsh, J. J., and H. G. Lebherz. 1992. Fructose-bisphosphate aldolases: an evolutionary history. Trends Biochem. Sci. 17:110-113. [DOI] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Morales, G., J. F. Linares, A. Beloso, J. P. Albar, J. L. Martinez, and F. Rojo. 2004. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 186:1337-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neidhardt, F. C., J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.). 1987. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 35.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. Chris Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 36.Perez-Martin, J., and V. de Lorenzo. 1995. The amino-terminal domain of the prokaryotic enhancer-binding protein XylR is a specific intramolecular repressor. Proc. Natl. Acad. Sci. USA 92:9392-9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Martin, J., K. N. Timmis, and V. de Lorenzo. 1994. Co-regulation by bent DNA. Functional substitutions of the integration host factor site at sigma 54-dependent promoter Pu of the upper-TOL operon by intrinsically curved sequences. J. Biol. Chem. 269:22657-22662. [PubMed] [Google Scholar]
- 38.Petruschka, L., G. Burchhardt, C. Muller, C. Weihe, and H. Herrmann. 2001. The cyo operon of Pseudomonas putida is involved in carbon catabolite repression of phenol degradation. Mol. Genet. Genomics 266:199-206. [DOI] [PubMed] [Google Scholar]
- 39.Ramos, J. L., S. Marques, and K. N. Timmis. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu. Rev. Microbiol. 51:341-373. [DOI] [PubMed] [Google Scholar]
- 40.Regenhardt, D., H. Heuer, S. Heim, D. U. Fernandez, C. Strompl, E. R. Moore, and K. N. Timmis. 2002. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ. Microbiol. 4:912-915. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Sanchez-Romero, J. M., R. Diaz-Orejas, and V. De Lorenzo. 1998. Resistance to tellurite as a selection marker for genetic manipulations of Pseudomonas strains. Appl. Environ. Microbiol. 64:4040-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawyer, M. H., L. Baumann, S. M. Berman, J. L. Canovas, and R. H. Berman. 1977. Pathways of d-fructose catabolism in species of Pseudomonas. Arch. Microbiol. 112:49-55. [DOI] [PubMed] [Google Scholar]
- 44.Schleissner, C., A. Reglero, and J. M. Luengo. 1997. Catabolism of d-glucose by Pseudomonas putida U occurs via extracellular transformation into d-gluconic acid and induction of a specific gluconate transport system. Microbiology 143:1595-1603. [DOI] [PubMed] [Google Scholar]
- 45.Stein, D. C. 1992. Plasmids with easily excisable xylE cassettes. Gene 117:157-158. [DOI] [PubMed] [Google Scholar]
- 46.Stulke, J., and W. Hillen. 1999. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 47.Swanson, B. L., P. Hager, P. Phibbs, Jr., U. Ochsner, M. L. Vasil, and A. N. Hamood. 2000. Characterization of the 2-ketogluconate utilization operon in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 37:561-573. [DOI] [PubMed] [Google Scholar]
- 48.Sze, C. C., and V. Shingler. 1999. The alarmone (p)ppGpp mediates physiological-responsive control at the sigma 54-dependent Po promoter. Mol. Microbiol. 31:1217-1228. [DOI] [PubMed] [Google Scholar]
- 49.Valls, M., M. Buckle, and V. de Lorenzo. 2002. In vivo UV laser footprinting of the Pseudomonas putida sigma 54 Pu promoter reveals that integration host factor couples transcriptional activity to growth phase. J. Biol. Chem. 277:2169-2175. [DOI] [PubMed] [Google Scholar]
- 50.Vicente, M., and J. L. Canovas. 1973. Glucolysis in Pseudomonas putida: physiological role of alternative routes from the analysis of defective mutants. J. Bacteriol. 116:908-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vicente, M., and J. L. Canovas. 1973. Regulation of the glucolytic enzymes in Pseudomonas putida. Arch. Mikrobiol. 93:53-64. [DOI] [PubMed] [Google Scholar]
- 52.Weinel, C., K. E. Nelson, and B. Tummler. 2002. Global features of the Pseudomonas putida KT2440 genome sequence. Environ. Microbiol. 4:809-818. [DOI] [PubMed] [Google Scholar]
- 53.Wolff, J. A., C. H. MacGregor, R. C. Eisenberg, and P. V. Phibbs, Jr. 1991. Isolation and characterization of catabolite repression control mutants of Pseudomonas aeruginosa PAO. J. Bacteriol. 173:4700-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuste, L., and F. Rojo. 2001. Role of the crc gene in catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 183:6197-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]