Abstract
During staphylococcal growth in glucose-supplemented medium, the pH of a culture starting near neutrality typically decreases by about 2 units due to the fermentation of glucose. Many species can comfortably tolerate the resulting mildly acidic conditions (pH, ∼5.5) by mounting a cellular response, which serves to defend the intracellular pH and, in principle, to modify gene expression for optimal performance in a mildly acidic infection site. In this report, we show that changes in staphylococcal gene expression formerly thought to represent a glucose effect are largely the result of declining pH. We examine the cellular response to mild acid by microarray analysis and define the affected gene set as the mild acid stimulon. Many of the genes encoding extracellular virulence factors are affected, as are genes involved in regulation of virulence factor gene expression, transport of sugars and peptides, intermediary metabolism, and pH homeostasis. Key results are verified by gene fusion and Northern blot hybridization analyses. The results point to, but do not define, possible regulatory pathways by which the organism senses and responds to a pH stimulus.
Facultative pathogens such as staphylococci produce a wide variety of accessory proteins. Many of these are involved in, or required for, infectivity and are collectively referred to as the virulon. It is widely believed that the genes encoding these proteins are regulated according to the exigencies of the local sites in which the organism is able to establish an infection. The evidence in support of this very logical view is, at best, rather sketchy, largely for two reasons. Firstly, it has, until very recently, been unapproachably difficult to determine the putative locale-specific gene expression patterns. Secondly, it has been no easy matter to identify the specific environmental factors that would determine these patterns. As a preamble to the ultimate objective of defining in vivo environmental factors that influence the expression of accessory genes involved in local pathogenesis, we have begun to reexamine the effect of certain nutrients and other chemicals, including glucose, acidic pH, salt, salicylic acid, and subinhibitory concentrations of antibiotics, on the expression of virulence genes during growth in vitro. These substances have widespread effects on the transcription of virulence genes and other accessory genes, which are presumably initiated through signaling elements and mediated through the complex regulatory network that governs the expression of all accessory genes (34). Our entrée into the transduction of environmental signals was provided by the observation that all of these substances affected transcription of a key signaling locus, sae, which is involved in the regulation of the expression of many exoproteins (36). In that study, which was initiated by a determination of the effect of glucose, it was shown that sae has a complex transcriptional pattern that undergoes a very striking switch during in vitro growth at neutral pH, in which a 2.1-kb transcript, present initially, is replaced by three other, of 3.2, 2.6, and 0.5 kb (36). When the Staphylococcus aureus strain tested is grown in the presence of glucose, the switch occurs but the new transcripts are quickly turned off. At this point the glucose is exhausted and the pH has dropped from 7.5 to 5.5, suggesting that the drop in pH rather than the presence of glucose is responsible for the turnoff (36). Here, we confirm that pH affects the expression of sae and show that a modest reduction in pH, at which the organisms survive and grow, probably without activating the full-scale acid stress response, affects the expression of many sae-regulated genes, as well as many genes that do not belong to the sae regulon. To define this newly identified mild acid stimulon (MAS), we have used transcriptome profiling by microarray. The MAS does not represent an acid-shock response; the cultures used were pregrown in pH-adjusted media such that at the first time point the cultures were in steady-state exponential growth. As described here, the MAS consists of all S. aureus genes whose transcript level differs by at least twofold during growth at pH 5.5 versus that at pH 7.5.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. The standard strain used in most of the work is RN6734, a φ13 lysogen of RN6390B. These two agr group I strains are in the NCTC8325 lineage and therefore have an rsbU defect, leading to a partial σB-negative phenotype (23).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) | Promega |
| RN4220 | Restriction-deficient mutant of strain 8325-4 | Kreiswirth et al. (27a) |
| RN6734 | phi13 lysogen of RN6390B | Vojtov et al. (52) |
| PC1839 | 8325-4 sarA::Km | Chan & Foster (8) |
| RN9388 | RN6734 sarA::Km; transductant of PC1839 | Tegmark et al. (48a) |
| KT201 | 8325-4 sarH1::pKT200 Em (aka sarS) | This work |
| RN9897 | RN6734 sarS::Em; transductant of KT201 | This work |
| ALC1905 | RN6390 sarT::Em | Schmidt et al. (45a) |
| RN9899 | RN6734 sarT::Em; transductant of ALC1905 | This work |
| RN9375 | 8325-4 sigB::Tc | Nicholas et al. (33a) |
| RN9898 | RN6734 sigB::Tc; transductant of RN9375 | This work |
| CYL807 | COL arlR::Tn551; gift of C. Lee (Kansas City, Kans.) | This work |
| RN9896 | RN6734 arlR::Tn551; transductant of CYL807 | This work |
| PM614 | PM466 rot::Tn917 | McNamara et al. (32) |
| RN9886 | RN6734 rot::Tn917; transductant of PM614 | This work |
| RN9360 | Replacement of rsbU deletion in 8325-4, adjacent Tc; gift of I. Kullik | Novick and Jiang (36) |
| RN10029 | RN6734 rsbU+ Tc; transductant of RN9360 | This work |
| RN7206 | phi13 lysogen of RN6911 (Δagr::tetM) | Novick et al. (36a) |
| Plasmids | ||
| pRN7044 | Ptst::blaZ transcriptional fusion | Vojtov et al. (52) |
| pRN7166 | pRN7044 with Emr replaced by Cmr | This work |
| pRN6827 | pSA3800 Phla::blaZ | Vandenesch and Novick (unpub.) |
| pRN7041 | PsspA::blaZ transcriptional fusion | Vojtov et al. |
| pCN50 | (Source of Cm cassette, 1-kb ApaI/XhoI fragment) | Charpentier et al. (submitted) |
Media and growth conditions.
S. aureus was cultured in Casamino Acids-yeast extract-glycerophosphate (CYGP) broth (35), without glucose, and which was adjusted to pH 5.5 or 7.5. Broth was inoculated to an initial density of 5 × 107 CFU ml−1 with S. aureus from GL agar plates with appropriate antibiotic and grown overnight at 37°C. Cultures were grown in Erlenmeyer flasks at a 1/10 volume and incubated with shaking at ∼220 rpm at 37°C. Cultures were monitored turbidometrically with a Klett-Summerson (New York, N.Y) colorimeter at 540 nm. After reaching ∼3 times the initial inoculum (growth from ∼15 to ∼50 Klett units), cultures were diluted twofold and grown again to 50 Klett units. Cultures were again diluted twofold. The time zero (T0) time point for all time course experiments was defined when the density reached 50 Klett units for the third time (∼1.5 × 108 CFU ml−1). For cultures grown without glucose, the pH at the end of 6 h was always within 0.2 units of the starting pH.
RNA purification.
Cells were harvested by centrifugation at specified time points and treated with RNA Protect reagent from QIAGEN (Valencia, Calif.). Cells were mechanically disrupted by agitation with glass beads using a Fast-Prep apparatus (Q-Biogene, Carlsbad, Calif.), and RNA was purified using the RNeasy kit from QIAGEN. RNA integrity was checked by agarose gel electrophoresis in Tris-acetate buffer, and RNA was quantitated by determination of absorbance at 260 nm.
Northern blot hybridization.
RNA from equal numbers of cells was separated by denaturing gel electrophoresis (morpholinepropanesulfonic acid-formaldehyde) on 1% agarose gels. RNA was transferred to Hybond N+ membranes from Amersham by vacuum and UV cross-linked. [α-32P]dATP-labeled (Amersham) probes were generated by PCR and hybridized to the blots overnight. Washed blots were exposed to phosphorimager screens, which were read by using a Molecular Dynamics PhosphorImager. PCR primers were obtained from Integrated DNA Technologies (Coralville, Iowa), and their sequences are listed in Table 2.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) | N315 locus |
|---|---|---|
| 16S F | GGTGAGTAACACGTGGATAA | SArRNA01 |
| 16S R | ATGTCAAGATTTGGTAAGGTT | SArRNA01 |
| RNAIII F | ATGATCACACAGAGATGTGA | SAS065 |
| RNAIII R | CTGAGTCCTAGGAAACTAACTC | SAS065 |
| sarA F | GAGTTGTTATCAATGGTCACTTATGCTG | SA0573 |
| sarA R | GTGATTCGTTTATTTACTCGACTC | SA0573 |
| hla F | TTAGCCTGGCCTTCAGCC | SA1007 |
| hla R | TGCCATATACCGGGTTC | SA1007 |
| saeS F | GGCTTCTGAAATTACGCAACAAATG | SA0660 |
| saeS R | GTTACAGTCACCGTAGTTCCCAC | SA0660 |
| rot F | CTCTACTTGCAATCGCATCACTG | SA1583 |
| rot R | GGGATTGTTGGGATGTTTGTTAATAC | SA1583 |
| spa F | GGCACTACTGCTGACAAAATTGCTGCAG | SA0107 |
| spa R | GTTCGCGACGACGTCCAGCTAATAACGCTGC | SA0107 |
| sspA F | GACAACAGCGACACTTG | SA0901 |
| sspA R | CTGAATTACCACCAGTTG | SA0901 |
| cap5b F | GTACAGTTGTATCGAATGTAGCG | SA0145 |
| cap5b R | GTGCATCAGTCACAGTATTAAC | SA0145 |
| IrgAB F | GCCGGATCCGAAGTGAGCCATCTATA | SA0252 |
| IrgAB R | GCCGAATTCGATAATAACAATGGCTC | SA0252 |
Microarray analysis.
A custom DNA microarray prepared by Affymetrix for Wyeth was used. The microarray contains probe sets for all conserved, nonredundant open reading frames from six published S. aureus genome sequences, unique GenBank entries, as well as strain N315 intergenic regions. Design of the chip has been detailed by Dunman et al. (19). All microarray procedures were performed as described by Beenken et al. (3). Briefly, biotinylated cDNA was prepared and hybridized to the microarray. The arrays were read with an Affymetrix GS3000 scanner, and data were analyzed with GeneSpring software version 6.1 (Silicon Genetics, Redwood City, Calif.). All procedures were done in duplicate, and results are reported for genes with a twofold or greater expression differential (P ≤ 0.5) between the two conditions.
Exoprotein analyses.
Exoproteins were analyzed by the method of Laemmli et al. (28). Briefly, culture supernatants from equal numbers of cells were precipitated with 10% trichloroacetic acid, and the pellets were resuspended in 2% sodium dodecyl sulfate (SDS) and boiled for 3 min. After separation by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), gels were stained with Coomassie blue and scanned. β-Lactamase was assayed by the nitrocefin method adapted for use in microtiter plates (52).
Plasmid construction.
The ∼1-kb chloramphenicol resistance cassette was removed from plasmid pCN50 by digestion with ApaI and XhoI. The fragment was isolated by gel electrophoresis in Tris-acetate buffer and extracted with a QIAquick gel extraction kit (QIAGEN). Likewise, the vector backbone of pRN7044 was isolated by digestion with ApaI and XhoI followed by gel electrophoresis and gel extraction. The chloramphenicol cassette was ligated into the pRN7044 backbone with T4 DNA ligase. The ligation product was used to transform chemically competent Escherichia coli DH5α. DNA of the resulting plasmid, pRN7166, was isolated with a QIAprep spin miniprep kit, introduced into RN4220 by electroporation, and transferred to other S. aureus strains by transduction with phage φ11.
RESULTS
Effect of pH on sae transcription.
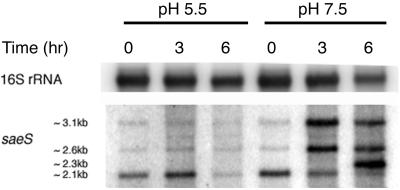
To study the effect of pH on sae expression independently of glucose, we grew cultures of strain RN6734 at pH 5.5 and 7.5 in CYGP broth without glucose and prepared Northern blots of whole-cell RNA from culture samples taken at three standard time points, T0, T3, and T6, as defined in Materials and Methods. pH did not vary by more than 0.1 to 0.2 units throughout the 6-h period. The Northern blot obtained with an saeS probe for these RNA samples (Fig. 1) demonstrates that the transition in the sae transcription pattern occurs at pH 7.5 but not at pH 5.5 and that the level of transcription is decreased at the lower pH.
FIG. 1.
Northern blot analysis of the effect of pH on saeS transcripts. Whole-cell RNA was extracted from RN6734 grown in CYGP broth without glucose and adjusted to pH 5.5 or 7.5. The RNA was separated on a denaturing gel and vacuum blotted; the blot was hybridized with a radiolabeled DNA probe complementary to saeS. The probe was detected with a PhosphorImager. Equalization was determined by subsequent blotting with a probe complementary to 16S rRNA.
Response of exoprotein gene promoters to changes in pH.
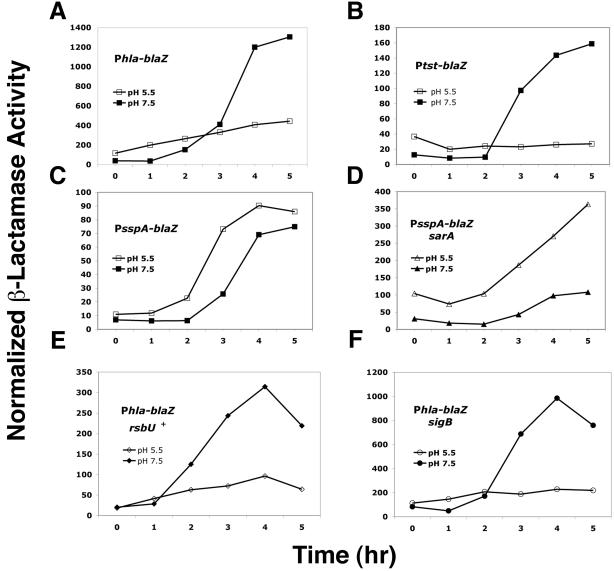
If the sae locus is involved in a global response to this pH differential, sae-regulated genes would be predicted to respond to changes in pH in this range. Accordingly, transcriptional fusions using staphylococcal β-lactamase (blaZ) as a reporter were used to test for promoter activity in response to changes in pH. The blaZ reporter, under a constitutive Pbla promoter, was insensitive to changes in pH in the range of pH 5.5 to 7.5 (data not shown). Plasmid-carried β-lactamase fusions involving Phla, Ptst, and PsspA were introduced into various strains and tested for β-lactamase activity during growth at pH 5.5 versus 7.5. The promoters for these three exoprotein genes were selected because, although postexponentially up-regulated by agr, they are regulated differentially by sar and sae (8, 36, 52; for a review, see the report of Novick 34). Results for the β-lactamase fusions are shown in Fig. 2A to C. The three promoters responded to changes in pH generally in parallel with their response to sae. Thus, at pH 7.5, Phla-blaZ shows the typical postexponential induction; however, at pH 5.5, there is a gradual increase of β-lactamase activity but no clear postexponential induction pattern. Ptst-blaZ also shows postexponential induction typical of agr-regulated exoprotein genes at pH 7.5 (34). This pattern is completely absent at pH 5.5, even though agr expression is induced at this pH. Finally, PsspA-blaZ shows a postexponential induction pattern that is similar at the two pH levels. These results imply that one or more pH-sensitive regulatory gene products may control expression from these promoters.
FIG. 2.
Transcriptional reporter analysis of the effect of pH on transcription of genes coding for several exoproteins, the effect of sarA on pH-dependent sspA transcription, and the effect of rsbU and sigB on pH-dependent hla transcription. RN6734 transduced with pRN7044 (Ptst::blaZ) (A), pRN6827 (Phla::blaZ) (B), or pRN7041 (PsspA::blaZ) (C) was assayed for β-lactamase activity during growth at pH 5.5 or pH 7.5. (D) pH-dependent β-lactamase activity of RN9388 (RN6734 ΔsarA) transduced with pRN7041, measured during growth. (E) pH-dependent β-lactamase activity of RN10029 (RN6734 rsbU+) transduced with pRN6827, measured during growth. (F) pH-dependent β-lactamase activity of RN9898 (RN6734 ΔsigB) transduced with pRN6827, measured during growth.
Effects of regulatory genes.
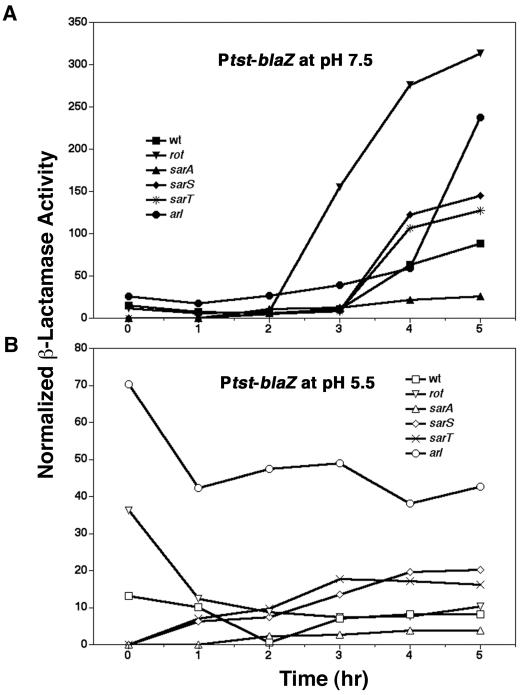
To test known regulatory genes for possible roles in pH-dependent regulation, we transferred null mutations in sarA, sarS, sarT, rot, arlRS, and sigB and a molecular repair of rsbU into RN6734 and tested for their effects using the β-lactamase fusion plasmids. The expectation was that a mutation affecting a required regulatory gene would abolish the pH-dependent expression differential, with respect both to expression levels and to kinetics. As shown in Fig. 2 and 3, none of the regulatory mutations tested eliminated the observed pH differential. Nevertheless, several informative results were obtained: (i) from the results shown in Fig. 2C and D, the sarA mutation dramatically increased the pH-dependent differential expression of the PsspA-blaZ reporter at pH 5.5, but had little or no effect at pH 7.5. This result may be consistent with previous experiments done in the presence of glucose, demonstrating a major increase in sspA expression in a sarA mutant background (8, 27), because these would have had a low pH at the time the samples were taken. Additionally, the sarA mutation did not eliminate the temporal up-regulation of sspA. (ii) Figure 2E and F show that hla expression was pH dependent in both wild-type and sigB mutant cells and was sharply up-regulated at pH 7.5 but not at 5.5. (iii) The mutation affecting sarA, but not those affecting sarS or sarT, profoundly decreased Ptst-blaZ expression and greatly reduced temporal induction (Fig. 3). (iv) The arl mutation appears to up-regulate the Ptst-blaZ reporter at all time points at pH 5.5 but only at the latest time point at pH 7.5. Finally, the rot mutation enhanced the temporal induction of the Ptst-blaZ reporter at pH 7.5. This result is consistent with the reported repressor activity of Rot (32), which was presumably determined at pH 7.5.
FIG. 3.
Transcriptional reporter analysis of the effect of pH on the transcription of genes coding for Tsst at pH 7.5 (A) and pH 5.5 (B) in various genetic backgrounds. In Emr-marked mutants, Ptst::blaZ reporter pRN7166 was used.
Identification of the MAS.
Given the apparent complexity of the response of exoprotein gene expression to changing pH, it seemed worthwhile to examine the entire MAS at the transcriptional level. For this purpose, we employed a staphylococcal DNA microarray (Affymetrix) (3, 6, 19, 20).
To perform this analysis, we prepared whole-cell RNA from two cultures of RN6734 in CYGP broth without glucose, each at three different time points. The cultures were started with an inoculum taken from an overnight GL plate (pH, ∼7.2). One culture was started at pH 5.5, the other at pH 7.5, and the two were grown as described in Materials and Methods, with samples taken at T0, T3, and T6 for RNA preparation. Comparable amounts of cDNA were hybridized to the microarrays for each experimental point. In Table 3 are shown the results for all genes whose transcript level differed by at least twofold between the two pH conditions, at any of the time points. In this presentation, to avoid any implications of regulatory mechanism, we refer to up or up-regulated and down or down-regulated in comparing transcript levels between the two conditions. In all, 424 genes were identified; approximately twice as many were up-regulated at pH 5.5 as were down-regulated. In addition to genes encoding proteins ostensibly involved in pH homeostasis, there were many encoding proteins involved in amino acid uptake or metabolism, carbohydrate uptake or metabolism, virulence (encoding either secreted or surface proteins), and transcriptional regulation and, curiously, a considerable number of prophage genes were found to be pH regulated. Among the prophage-encoded genes, a much greater number (31 of 32) were down-regulated at pH 5.5 than were up-regulated. Given that prophage genes are not generally expressed in the prophage state, this result could indicate that spontaneous prophage induction occurs more frequently at pH 7.5. Among the surface proteins, a much greater number (72 of 84) were up-regulated at pH 5.5 than were down-regulated. Of the genes encoding secreted proteins, about the same number were up- as down-regulated. As a functional category, transporters with roles in nutrient accumulation and maintenance of homeostasis were differentially expressed in a ratio that paralleled that of the stimulon as a whole, with approximately twice as many up-regulated at pH 5.5 as at pH 7.5. Some genes involved in homeostasis have roles in maintenance of intracellular pH (ure operon), others are osmoprotective (opuC operon), and some contribute to protection against both low pH and high osmolarity (kdp operon) (7, 47).
TABLE 3.
Genes with a ≥2-fold change in transcript level, pH 7.5 versus 5.5a
| ORF no. | Gene | Description | Fold change at pH 5.5
|
Effect of regulator
|
Functional category | ||||
|---|---|---|---|---|---|---|---|---|---|
| T0 | T3 | T6 | agr | sarA | rot | ||||
| Genes up-regulated at pH 5.5 | |||||||||
| N315-SA0021 | yycJ | Conserved HP | 2.0 | Down | |||||
| N315-SA0091 | plc | 1-Phosphatidylinositol phosphodiesterase precurosr | 2.4 | ||||||
| N315-SA0092 | HP | 2.3 | |||||||
| N315-SA0111 | sirA | Lipoprotein | 3.8 | Surface | |||||
| N315-SA0112 | HP, similar to cysteine synthase | 2.1 | |||||||
| N315-SA0121 | HP | 2.3 | |||||||
| N315-SA0131 | deoD | Purine nucleoside phosphorylase | 2.9 | ||||||
| N315-SA0144 | cap5A | Capsular polysaccharide synthesis enzyme Cap5A | 4.0 | 3.6 | 2.6 | Surface | |||
| N315-SA0145 | cap5B | Capsular polysaccharide synthesis enzyme Cap5B | 3.3 | 3.3 | 2.1 | Surface | |||
| N315-SA0146 | cap5C | Capsular polysaccharide synthesis enzyme Cap8C | 4.2 | 3.2 | Surface | ||||
| N315-SA0147 | cap5D | Capsular polysaccharide synthesis enzyme Cap5D | 3.2 | 3.1 | Surface | ||||
| N315-SA0148 | cap5E | Capsular polysaccharide synthesis enzyme Cap8E | 3.3 | 2.9 | Surface | ||||
| N315-SA0149 | cap5F | Capsular polysaccharide synthesis enzyme Cap5F | 3.3 | 2.9 | Surface | ||||
| N315-SA0150 | cap5G | Capsular polysaccharide synthesis enzyme Cap5G | 3.6 | 3.6 | Surface | ||||
| N315-SA0151 | cap5H | Capsular polysaccharide synthesis enzyme O-acetyl transferase Cap5H | 2.8 | 3.2 | Surface | ||||
| N315-SA0152 | cap5I | Capsular polysaccharide synthesis enzyme Cap5I | 2.7 | 3.5 | Surface | ||||
| N315-SA0153 | cap5J | Capsular polysaccharide synthesis enzyme Cap5J | 3.2 | Up | Up | Surface | |||
| N315-SA0154 | cap5K | Capsular polysaccharide synthesis enzyme Cap5K | 2.0 | Surface | |||||
| N315-SA0155 | cap5L | Capsular polysaccharide synthesis enzyme Cap5L | 2.1 | 2.8 | Surface | ||||
| N315-SA0156 | cap5M | Capsular polysaccharide synthesis enzyme Cap5M | 2.9 | Surface | |||||
| N315-SA0157 | cap5N | Capsular polysaccharide synthesis enzyme Cap5N | 2.9 | Surface | |||||
| N315-SA0158 | cap5O | Capsular polysaccharide synthesis enzyme Cap8O | 2.6 | Surface | |||||
| N315-SA0162 | aldA | Aldehyde dehydrogenase homologue | 6.3 | Down | |||||
| N315-SA0170 | Conserved HP | 3.4 | Up | ||||||
| N315-SA0171 | fdh | NAD-dependent formate dehydrogenase | 2.8 | 4.5 | 4.0 | ||||
| N315-SA0173 | HP, similar to surfactin synthetase | 2.1 | Up | Up | Secreted | ||||
| N315-SA0184 | Conserved HP | 3.0 | Up | ||||||
| N315-SA0185 | Conserved HP | 3.7 | Up | ||||||
| N315-SA0186 | HP, similar to sucrose phosphotransferase enzyme II | 5.0 | Down | Transport | |||||
| N315-SA0187 | HP, similar to transcription regulator | 5.2 | Up | Down | Regulator | ||||
| N315-SA0223 | atoB | Acetyl-CoA acetyltransferase homologue | 3.8 | ||||||
| N315-SA0224 | fadB | HP, similar to 3-hydroxyacyl-CoA dehydrogenase | 3.6 | ||||||
| N315-SA0225 | fadD | HP, similar to glutaryl-CoA dehydrogenase | 3.1 | ||||||
| N315-SA0226 | fadE | HP, similar to acid-CoA ligase | 2.8 | ||||||
| N315-SA0227 | fadX | Conserved HP | 2.4 | ||||||
| N315-SA0229 | HP, similar to nickel ABC transporter nickel-binding protein | 9.8 | 6.3 | 3.9 | Transport | ||||
| N315-SA0244 | tagF | HP, similar to teichoic acid biosynthesis protein F | 2.1 | Up | Surface | ||||
| N315-SA0258 | rbsK | Probable ribokinase | 3.3 | Transport | |||||
| N315-SA0259 | rbsD | Ribose permease | 3.3 | Transport | |||||
| N315-SA0260 | HP, similar to ribose transporter RbsU | 3.8 | Transport | ||||||
| N315-SA0262 | HP | 3.1 | |||||||
| N315-SA0270 | HP, similar to secretory antigen precursor SsaA | 2.8 | Secreted | ||||||
| N315-SA0291 | HP | 2.1 | Up | ||||||
| N315-SA0304 | nanA | N-Acetylneuraminate lyase subunit | 2.7 | ||||||
| N315-SA0305 | HP, similar to glucokinase | 2.4 | |||||||
| N315-SA0318 | HP, similar to transport protein SgaT | 4.2 | 2.3 | Transport | |||||
| N315-SA0319 | Conserved HP | 3.9 | 2.5 | ||||||
| N315-SA0320 | HP, similar to PTS fructose-specific enzyme IIBC component | 4.0 | 2.6 | Transport | |||||
| N315-SA0321 | HP, similar to transcription antiterminator BglG family | 3.3 | 2.9 | Regulator | |||||
| N315-SA0322 | HP, similar to transcription regulator | 2.6 | Regulator | ||||||
| N315-SA0325 | glpT | Glycerol-3-phosphate transporter | 4.1 | 2.3 | Transport | ||||
| N315-SA0428 | Conserved HP | 2.6 | Up | ||||||
| N315-SA0432 | treP | PTS enzyme II, phosphoenolpyruvate dependent | 3.8 | Down | Transport | ||||
| N315-SA0433 | Alpha-glucosidase | 2.2 | 3.7 | Down | |||||
| N315-SA0510 | araB | Probable l-ribulokinase | 2.2 | ||||||
| N315-SA0519 | sdrC | Ser-Asp rich fibrinogen-binding, bone sialoprotein-binding protein | 5.7 | 5.6 | 6.9 | Surface | |||
| N315-SA0547 | mvk | Mevalonate kinase | 2.1 | ||||||
| N315-SA0548 | mvaD | Mevalonate diphosphate decarboxylase | 2.1 | ||||||
| N315-SA0549 | mvaK2 | Phosphomevalonate kinase | 2.0 | Down | |||||
| N315-SA0550 | Conserved HP | 2.5 | |||||||
| N315-SA0587 | Lipoprotein, streptococcal adhesin PsaA homologue | 2.1 | Surface | ||||||
| N315-SA0599 | abcA | ATP-binding cassette transporter A | 2.1 | Transport | |||||
| N315-SA0605 | HP, similar to dihydroxyacetone kinase | 2.8 | Down | ||||||
| N315-SA0606 | Conserved HP | 2.6 | |||||||
| N315-SA0607 | Conserved HP | 3.1 | |||||||
| N315-SA0612 | Conserved HP | 2.1 | |||||||
| N315-SA0620 | Secretory antigen SsaA homologue, similar to autolysin | 2.7 | Up | Secreted | |||||
| N315-SA0622 | HP, similar to AraC/Xy|S family transcriptional regulator | 2.1 | Regulator | ||||||
| N315-SA0623 | HP | 2.1 | 2.2 | ||||||
| N315-SA0636 | Conserved HP | 2.4 | |||||||
| N315-SA0637 | Conserved HP | 2.9 | |||||||
| N315-SA0651 | HP | 4.2 | Up | ||||||
| N315-SA0666 | 6-Oyruvoyl tetrahydrobiopterin synthase homologue | 2.4 | Up | ||||||
| N315-SA0667 | Conserved HP | 2.4 | 2.7 | ||||||
| N315-SA0711 | Conserved HP | 2.5 | |||||||
| N315-SA0738 | HP | 2.7 | 2.0 | 3.7 | |||||
| N315-SA0739 | Conserved HP | 4.2 | 2.2 | 3.6 | Up | ||||
| N315-SA0820 | glpQ | Glycerophosphoryl diester phosphodiesterase | 2.3 | ||||||
| N315-SA0835 | clpB | ClpB chaperone homologue | 2.3 | ||||||
| N315-SA0893 | Conserved HP | 2.1 | |||||||
| N315-SA0899 | sspC | Cysteine protease | 2.2 | Up | Down | Down | Secreted | ||
| N315-SA0900 | sspB | Cysteine protease precursor | 2.3 | Down | Down | Secreted | |||
| N315-SA0901 | sspA | Serine protease; V8 protease; glutamyl endopeptidase | 2.4 | Secreted | |||||
| N315-SA0974 | Conserved HP | 2.3 | |||||||
| N315-SA0978 | LPXTG-containing conserved HP | 3.2 | Surface | ||||||
| N315-SA0979 | Conserved HP | 3.2 | |||||||
| N315-SA0980 | HP, similar to ferrichrome ABC transporter | 3.1 | Transport | ||||||
| N315-SA0981 | HP, similar to ferrichrome ABC transporter | 2.6 | Transport | ||||||
| N315-SA0982 | srtB | Conserved HP | 2.1 | Transport | |||||
| N315-SA0983 | Conserved HP | 3.1 | |||||||
| N315-SA0996 | sdhB | Succinate dehydrogenase iron-sulfur protein subunit | 2.3 | ||||||
| N315-SA1007 | hla | Alpha-hemolysin precursor | 4.4 | Up | Up | Down | Secreted | ||
| N315-SA1041 | pyrR | Pyrimidine operon repressor chainA | 2.0 | Up | Regulator | ||||
| N315-SA1042 | pyrP | Uracil permease | 2.9 | Transport | |||||
| N315-SA1043 | pyrB | Aspartate transcarbamoylase chain A | 2.3 | 2.1 | |||||
| N315-SA1044 | pyrC | Dihydroorotase | 2.2 | ||||||
| N315-SA1045 | carA | Carbamoyl-phosphate synthase small chain | 2.6 | 2.0 | Up | Up | |||
| N315-SA1046 | carB | Carbamoyl-phosphate synthase large chain | 2.4 | ||||||
| N315-SA1047 | pyrF | Orotidine-5-phosphate decarboxylase | 2.6 | 2.2 | |||||
| N315-SA1048 | pyrF | Orotate phosphoribosyltransferase | 2.0 | 2.8 | 2.0 | ||||
| N315-SA1056 | HP | 2.2 | Up | ||||||
| N315-SA1076 | rnc | RNase III | 2.3 | ||||||
| N315-SA1081 | rpsP | 30S ribosomal protein S16 | 2.1 | ||||||
| N315-SA1089 | sucD | Succinyl-CoA synthetase | 2.3 | ||||||
| N315-SA1120 | HP, similar to transcription regulator GntR family | 2.2 | Up | Up | Regulator | ||||
| N315-SA1140 | glpF | Glycerol uptake facilitator | 2.3 | Down | Down | Transport | |||
| N315-SA1141 | gplK | Glycerol kinase | 2.6 | Transport | |||||
| N315-SA1149 | glnR | Glutamine synthetase repressor | 2.1 | Regulator | |||||
| N315-SA1154 | Conserved HP | Up | 2.2 | ||||||
| N315-SA1162 | HP | 2.1 | |||||||
| N315-SA1176 | Conserved HP | 2.6 | |||||||
| N315-SA1211 | opp-2E | Oligopeptide transporter putative ATPase domain | 2.5 | 2.2 | Transport | ||||
| N315-SA1212 | opp-2D | Oligopeptide transport ATPase | 3.7 | 3.0 | Transport | ||||
| N315-SA1213 | opp-2C | Oligopeptide transporter membrane permease domain | 3.9 | 2.8 | 2.0 | Transport | |||
| N315-SA1214 | opp-2B | Oligopeptide transporter membrane permease domain | 4.5 | 3.6 | 2.7 | Up | Up | Transport | |
| N315-SA1215 | HP | 2.4 | |||||||
| N315-SA1235 | Conserved HP | 3.2 | |||||||
| N315-SA1245 | odhA | 2-Oxoglutarate dehydrogenase E1 | 2.3 | Up | |||||
| N315-SA1252 | Conserved HP | 2.2 | |||||||
| N315-SA1265 | Conserved HP | 2.2 | |||||||
| N315-SA1275 | Conserved HP | 2.2 | 2.9 | Up | Up | ||||
| N315-SA1310 | ansA | Probable l-asparaginase | 2.1 | ||||||
| N315-SA1318 | HP | 2.1 | |||||||
| N315-SA1321 | HP | 2.0 | |||||||
| N315-SA1410 | grpE | GrpE protein | 2.1 | ||||||
| N315-SA1434 | Acetyl-CoA carboxylase (biotin carboxylase subunit) | 2.1 | |||||||
| N315-SA1476 | HP | 2.6 | |||||||
| N315-SA1531 | ald | Alanine dehydrogenase | 3.3 | ||||||
| N315-SA1551 | sgtA | Probable transglycosylase | 2.1 | ||||||
| N315-SA1554 | acs | Acetyl-CoA synthetase | 2.3 | ||||||
| N315-SA1601 | crcB | Conserved HP | 2.0 | ||||||
| N315-SA1609 | pckA | Phosphoenolpyruvate carboxykinase | 5.0 | Up | |||||
| N315-SA1630 | splB | Serine protease SplB | 2.3 | Up | Up | Down | Secreted | ||
| N315-SA1631 | splA | Serine protease SplA | Up | Up | Up | Down | Secreted | ||
| N315-SA1844 | agrA | Accessory gene regulator A | 2.8 | Up | Up | Regulator | |||
| N315-SA1879 | kdpC | Probable potassium-transporting ATPase C chain | 9.2 | 3.0 | Down | Transport | |||
| N315-SA1880 | kdpB | Probable potassium-transporting ATPase B chain | 3.3 | 11.0 | 2.9 | Transport | |||
| N315-SA1881 | kdpA | Probable potassium-transporting ATPase A chain | 5.7 | 17.6 | 3.4 | Down | Transport | ||
| N315-SA1882 | kdpD | Sensor protein KdpD | 12.3 | Regulator | |||||
| N315-SA1883 | kdpE | KDP operon transcriptional regulatory protein KdpE | 10.2 | Regulator | |||||
| N315-SA1889 | HP | 2.0 | Up | ||||||
| N315-SA1890 | Conserved HP | 2.4 | |||||||
| N315-SA1896 | thiD | Phosphomethylpyrimidine kinase | 2.0 | ||||||
| N315-SA1929 | pyrG | CTP synthase | 2.2 | 2.7 | |||||
| N315-SA1931 | HP, similar to spermine/spermidine acetyltransferase bit | 2.1 | Up | ||||||
| N315-SA1937 | Conserved HP | 2.0 | Down | ||||||
| N315-SA1938 | pyn | Pyrimidine nucleoside phosphorylase | 2.1 | ||||||
| N315-SA1947 | czrA | Repressor protein | 2.6 | 2.3 | Regulator | ||||
| N315-SA1948 | czrB | Cation-efflux system membrane protein homolog | 2.1 | 2.3 | Transport | ||||
| N315-SA1968 | arg | Arginase | 2.1 | ||||||
| N315-SA1976 | Conserved hypothetical protein | 2.5 | |||||||
| N315-SA1979 | HP, similar to ferrichrome ABC transporter (binding protein) | 2.7 | Transport | ||||||
| N315-SA2006 | HP, similar to MHC class II analog | 2.0 | 2.3 | Up | Up | Surface | |||
| N315-SA2007 | HP, similar to alpha-acetolactate decarboxylase | 4.5 | 2.2 | Up | |||||
| N315-SA2008 | budB | Alpha-acetolactate synthase | 7.8 | 3.1 | |||||
| N315-SA2060 | HP, similar to transcription regulator MarR family | 2.1 | 2.5 | 2.1 | Regulator | ||||
| N315-SA2075 | fdhD | FdhD protein homologue | 2.1 | ||||||
| N315-SA2079 | HP, similar to ferrichrome ABC transporter fhuD precursor | 2.4 | Up | Up | Transport | ||||
| N315-SA2081 | HP, similar to urea transporter | 2.3 | Transport | ||||||
| N315-SA2082 | ureA | Urease gamma subunit | 3.9 | 10.4 | 6.3 | Down | |||
| N315-SA2083 | ureAB | Urease beta subunit | 5.7 | 12.8 | 8.4 | Down | |||
| N315-SA2084 | ureC | Urease alpha subunit | 4.5 | 10.2 | 4.8 | Down | |||
| N315-SA2085 | ureE | Urease accessory protein UreE | 2.8 | 5.6 | 3.8 | Down | |||
| N315-SA2086 | ureF | Urease accessory protein UreF | 2.7 | 6.1 | 4.0 | Down | |||
| N315-SA2087 | ureG | Urease accessory protein UreG | 2.0 | 3.6 | 3.0 | Down | |||
| N315-SA2088 | ureD | Urease accessory protein UreD | 3.6 | 2.7 | Down | ||||
| N315-SA2093 | ssaA | Secretory antigen precursor SsaA homolog | 2.1 | 2.7 | Down | Secreted | |||
| N315-SA2097 | HP, similar to secretory antigen precursor SsaA | 3.9 | Secreted | ||||||
| N315-SA2101 | Conserved HP | 2.0 | |||||||
| N315-SA2120 | HP, similar to amino acid amidohydrolase | 2.1 | |||||||
| N315-SA2126 | HP | 2.6 | |||||||
| N315-SA2142 | semB | HP, similar to multidrug resistance protein | 3.9 | 5.3 | 4.2 | Transport | |||
| N315-SA2143 | Conserved HP | 4.0 | 6.3 | 5.7 | |||||
| N315-SA2146 | tcaA | TcaA protein | 2.0 | Regulator | |||||
| N315-SA2159 | HP, similar to transcription repressor of sporulation | 2.3 | Regulator | ||||||
| N315-SA2165 | HP, similar to transcriptional regulator tetR-family | 2.1 | 2.5 | Regulator | |||||
| N315-SA2166 | HP, similar to cationic transporter | 2.4 | 2.4 | Transport | |||||
| N315-SA2168 | HP | 2.4 | |||||||
| N315-SA2173 | HP | 2.2 | |||||||
| N315-SA2204 | gpm | Phosphoglycerate mutase | 3.0 | ||||||
| N315-SA2208 | hlgC | Gamma-hemolysin component C | 2.3 | Up | Up | Down | Secreted | ||
| N315-SA2216 | HP, similar to ABC transporter | 2.4 | Transport | ||||||
| N315-SA2217 | HP, similar to lipoprotein inner membrane ABC-transporter | 2.2 | Transport | ||||||
| N315-SA2227 | Truncated HP, similar to d-serine/d-alanine/glycine transporter | 4.3 | Transport | ||||||
| N315-SA2229 | Conserved HP | 3.4 | |||||||
| N315-SA2231 | HP, similar to glucose epimerase | 2.2 | |||||||
| N315-SA2234 | opuCD | Probable glycine betaine/carnitine/choline ABC transporter (membrane p) opuCD | 2.6 | 2.2 | Transport | ||||
| N315-SA2235 | opuCC | Glycine betaine/carnitine/choline ABC transporter (osmoprotective) opuCC | 2.8 | 2.5 | Transport | ||||
| N315-SA2236 | opuCB | Probable glycine betaine/carnitine/choline ABC transporter (membrane p) opuCB | 3.3 | Transport | |||||
| N315-SA2237 | opuCA | Glycine betaine/carnitine/choline ABC transporter (ATP-bindin) opuCA | 4.9 | 2.4 | Transport | ||||
| N315-SA2238 | Conserved HP | 2.1 | |||||||
| N315-SA2239 | HP, similar to amino acid transporter | 7.7 | 4.6 | 2.4 | Transport | ||||
| N315-SA2246 | HP | 2.2 | |||||||
| N315-SA2296 | HP, similar to transcriptional regulator MerR family | 3.0 | Regulator | ||||||
| N315-SA2297 | HP, similar to GTP-pyrophosphokinase | 3.1 | 2.3 | Down | |||||
| N315-SA2298 | Conserved HP | 2.8 | |||||||
| N315-SA2299 | Conserved HP | 3.3 | |||||||
| N315-SA2300 | HP, similar to glucarate transporter | 2.2 | Down | Transport | |||||
| N315-SA2304 | fbp | Fructose-bisphosphatase | 2.5 | ||||||
| N315-SA2311 | HP, similar to NAD(P)H-flavin oxidoreductase | 2.0 | |||||||
| N315-SA2316 | srtA | Sortase | 2.4 | Transport | |||||
| N315-SA2318 | sdhA | HP, similar to l-serine dehydratase | 4.4 | 2.8 | Up | ||||
| N315-SA2319 | sdhB | HP, similar to beta-subunit of l-serine dehydratas | 5.3 | 2.3 | Up | Up | |||
| N315-SA2320 | HP, similar to regulatory protein pfoR | 4.6 | 2.2 | Regulator | |||||
| N315-SA2326 | ptsG | PTS system, glucose-specific IIABC component | 2.1 | Down | Transport | ||||
| N315-SA2328 | cidB | Conserved HP | 2.4 | 2.6 | |||||
| N315-SA2329 | cidA | Conserved HP | 5.3 | 3.1 | |||||
| N315-SA2330 | cidR | HP, similar to transcription regulator | 2.4 | Regulator | |||||
| N315-SA2332 | HP, similar to secretory antigen precursor SsaA | 2.8 | Secreted | ||||||
| N315-SA2338 | HP | 3.1 | 2.6 | ||||||
| N315-SA2341 | rocA | 1-Pyrroline-5-carboxylate dehydrogenase | 3.0 | Up | Down | ||||
| N315-SA2343 | HP | 2.7 | Up | Up | |||||
| N315-SA2355 | Conserved HP | 2.4 | 4.1 | ||||||
| N315-SA2356 | isaA | Immunodominant antigen A | 2.5 | Surface | |||||
| N315-SA2405 | betA | Choline dehydrogenase | 2.5 | ||||||
| N315-SA2406 | gbsA | Glycine betaine aldehyde dehydrogenase gbsA | 3.6 | ||||||
| N315-SA2408 | cudT | Choline transporter | 2.5 | Transport | |||||
| N315-SA2411 | citM | HP, similar to magnesium citrate secondary transporter | 2.1 | ||||||
| N315-SA2413 | cysJ | Sulfite reductase (NADPH) flavoprotein | 2.0 | ||||||
| N315-SA2414 | gpxA | HP, similar to glutathione peroxidase | 3.2 | 2.6 | 2.3 | Down | |||
| N315-SA2423 | clfB | Clumping factor B | 2.8 | Surface | |||||
| N315-SA2428 | arcA | Arginine deiminase | 2.1 | 2.6 | Up | ||||
| N315-SA2430 | aur | Zinc metalloproteinase aureolysin | 2.0 | Up | Down | Secreted | |||
| N315-SA2434 | Fructose phosphotransferase system enzyme fruA homolog | 2.3 | Down | ||||||
| N315-SA2443 | HP | 3.1 | |||||||
| N315-SA2444 | HP | 2.5 | 3.0 | ||||||
| N315-SA2445 | HP | 3.1 | 3.2 | ||||||
| N315-SA2446 | secY | HP, similar to preprotein translocase secY | 3.1 | 3.4 | Transport | ||||
| N315-SA2447 | HP, similar to streptococcal hemagglutinin protein | 5.8 | 2.2 | Up | Up | Surface | |||
| N315-SA2459 | icaA | Intercellular adhesion protein A | 3.0 | Surface | |||||
| N315-SA2460 | icaD | Intercellular adhesion protein D | 2.2 | Surface | |||||
| N315-SA2463 | lip | Triacylglycerol lipase precursor | 4.1 | 2.0 | Up | Down | Secreted | ||
| N315-SA2480 | drp35 | Drp35 | 2.8 | ||||||
| N315-SA2481 | Conserved hypotehtical protein | 2.1 | |||||||
| N315-SA2494 | cspB | Cold shock protein cspB | 2.6 | ||||||
| N315-SAS014 | HP | Up | |||||||
| N315-SAS017 | HP | 2.3 | |||||||
| N315-SAS039 | HP | 2.0 | |||||||
| N315-SAS064 | HP (bacteriophage phiN315) | 2.2 | |||||||
| agrC | 3.6 | Regulator | |||||||
| LPXTG-motif cell wall achor domain protein, similar to ClfA | 3.6 | Surface | |||||||
| Similar to surface protein Pls | Up | Surface | |||||||
| Similar to Staphylococcus lugdunensis gene Fbl | Up | 3.0 | |||||||
| COL-SA0109 | HP | 2.1 | |||||||
| COL-SA0484 | HP | 2.1 | |||||||
| COL-SA0492 | HP | Up | |||||||
| COL-SA0794 | HP | 2.9 | |||||||
| COL-SA0850 | HP | 2.9 | 2.1 | 3.9 | |||||
| COL-SA0852 | HP | 5.6 | 7.8 | ||||||
| COL-SA0933 | HP | 2.7 | |||||||
| COL-SA1042 | HP | 2.4 | |||||||
| COL-SA1043 | Glycosyl transferase group 1 | 2.5 | |||||||
| COL-SA1170 | HP | 2.3 | |||||||
| COL-SA1177 | HP | 2.3 | |||||||
| COL-SA1186 | Antibacterial protein | 23.5 | Secreted | ||||||
| COL-SA1187 | Antibacterial protein | 28.7 | Secreted | ||||||
| COL-SA1332 | HP | 2.4 | |||||||
| COL-SA1339 | HP | 2.0 | |||||||
| COL-SA1343 | HP | 3.4 | |||||||
| COL-SA1347 | HP | 2.3 | |||||||
| COL-SA1528 | HP | 2.2 | |||||||
| COL-SA1529 | HP | 2.0 | |||||||
| COL-SA1865 | splE | Serine protease SplE | 2.3 | Secreted | |||||
| COL-SA1877 | epiB | Lantibiotic epidermin biosynthesis protein EpiB | 2.2 | Secreted | |||||
| COL-SA2027 | HP | 3.4 | |||||||
| COL-SA2065 | HP | 9.5 | 2.8 | ||||||
| COL-SA2069 | HP | 13.9 | 3.2 | ||||||
| COL-SA2629 | HP | 3.1 | |||||||
| COL-SA2637 | HP | 2.2 | |||||||
| COL-SA2676 | LPXTG-motif cell wall anchor domain protein | 5.2 | 2.3 | Surface | |||||
| COL-SA2693 | HP | 3.9 | 3.7 | ||||||
| Mu50-SAV0850 | HP | 2.2 | |||||||
| Mu50-SAV0866 | HP | Up | |||||||
| Mu50-SAV0888 | HP | 3.1 | |||||||
| Mu50-SAV1991 | HP | 2.2 | |||||||
| Mu50-SAV1996 | HP | 2.2 | |||||||
| MW1600 | 2.4 | ||||||||
| MW1681 | Up | ||||||||
| MW1896 | HP | 2.3 | |||||||
| MW1926 | HP | 2.4 | |||||||
| MW2575 | Ser/Thr repeat surface, similar to hemagglutinin | 3.1 | Surface | ||||||
| Genes downregulated at pH 5.5 | |||||||||
| N315-SA0100 | Conserved HP | 5.7 | Up | ||||||
| N315-SA0107 | spa | Immunoglobulin G-binding protein A precursor | Down | Down | Down | Up | Surface | ||
| N315-SA0129 | LPXTG-containing HP | 2.1 | Surface | ||||||
| N315-SA0131 | deoD | Purine nucleoside phosphorylase | 2.1 | ||||||
| N315-SA0162 | aldA | Aldehyde dehydrogenase homologue | 2.3 | Down | |||||
| N315-SA0206 | msmX | multiple sugar-binding transport ATP-binding protein | 2.5 | Transport | |||||
| N315-SA0207 | HP, similar to maltose/maltodextrin-binding protein | 3.6 | Transport | ||||||
| N315-SA0208 | Maltose/maltodextrin transport permease homologue | 5.2 | Transport | ||||||
| N315-SA0209 | Maltose/maltodextrin transport permease homologue | 5.9 | Transport | ||||||
| N315-SA0210 | HP, similar to NADH-dependent dehydrogenase | 5.0 | |||||||
| N315-SA0211 | HP, similar to NADH-dependent dehydrogenase | 4.4 | |||||||
| N315-SA0212 | Conserved HP | 3.9 | Down | Down | |||||
| N315-SA0214 | uhpT | Hexose phosphate transport protein | 4.1 | Transport | |||||
| N315-SA0218 | pflB | Formate acetyltransferase | 2.6 | 2.9 | |||||
| N315-SA0219 | pflA | Formate acetyltransferase activating enzyme | 2.0 | 2.2 | |||||
| N315-SA0232 | ldh | l-Lactate dehydrogenase | 2.2 | ||||||
| N315-SA0233 | PTS enzyme (EC 2.7.1.69), factor II homologue, maltose and glucose specific | 2.3 | Transport | ||||||
| N315-SA0252 | lrgA | Holin-like protein LrgA | 5.2 | 16.9 | 7.3 | Surface | |||
| N315-SA0253 | lrgB | Holin-like protein LrgB | 5.7 | 20.5 | 10.2 | Down | Surface | ||
| N315-SA0265 | lytM | Peptidoglycan hydrolase | 2.7 | Secreted | |||||
| N315-SA0303 | HP, similar to sodium-coupled permease | 2.7 | Transport | ||||||
| N315-SA0304 | nanA | N-Acetylneuraminate lyase subunit | 2.9 | ||||||
| N315-SA0357 | HP, similar to exotoxin 2 | 6.1 | 2.6 | Secreted | |||||
| N315-SA0388 | set12 | Exotoxin 12 | 2.2 | Secreted | |||||
| N315-SA0395 | HP (pathogenicity island SaPIn2) | 10.5 | |||||||
| N315-SA0478 | Conserved HP | 2.2 | |||||||
| N315-SA0562 | adh | Alcohol dehydrogenase I | 3.7 | ||||||
| N315-SA0639 | HP, similar to ABC transporter required for expression of cytochrome bd | 2.2 | Transport | ||||||
| N315-SA0640 | HP, similar to ABC transporter required for expression of cytochrome bd | 3.0 | Transport | ||||||
| N315-SA0650 | norA | Quinolone resistance protein | 2.7 | Transport | |||||
| N315-SA0660 | saeS | Histidine protein kinase | 2.1 | 2.3 | Regulator | ||||
| N315-SA0661 | saeR | Response regulator | 2.3 | 2.3 | Regulator | ||||
| N315-SA0662 | HP | 5.2 | 3.0 | ||||||
| N315-SA0663 | HP | 9.0 | 3.4 | Down | |||||
| N315-SA0691 | sstD | Lipoprotein, similar to ferrichrome ABC transporter | 2.9 | Transport | |||||
| N315-SA0744 | ssp | Extracellular ECM and plasma binding protein | 2.6 | Surface | |||||
| N315-SA0746 | nuc | Staphylococcal nuclease | 2.2 | Down | Secreted | ||||
| N315-SA0754 | HP, similar to lactococcal prophage ps3 protein 05 | 3.0 | Up | ||||||
| N315-SA0773 | Conserved HP | 3.4 | |||||||
| N315-SA0807 | mnhG | Na+/H+ antiporter subunit | 2.0 | Transport | |||||
| N315-SA0808 | mnhF | Na+/H+ antiporter subunit | 2.3 | Down | Transport | ||||
| N315-SA0810 | mnhD | Na+/H+ antiporter subunit | 2.0 | Transport | |||||
| N315-SA0820 | glpQ | Glycerophosphoryl diester phosphodiesterase | 2.4 | ||||||
| N315-SA0889 | HP | 31.1 | 17.6 | 10.3 | |||||
| N315-SA0890 | Conserved HP | 6.2 | 32.8 | 9.5 | |||||
| N315-SA0937 | Cytochrome d ubiquinol oxidase subunit I homologue | 2.6 | |||||||
| N315-SA0938 | Cytochrome d ubiquinol oxidase subunit II homologue | 3.0 | |||||||
| N315-SA0963 | pyc | Pyruvate carboxylase | 2.0 | ||||||
| N315-SA1000 | HP, similar to fibrinogen-binding protein | 4.1 | 3.9 | Surface | |||||
| N315-SA1001 | HP | 4.6 | |||||||
| N315-SA1003 | 12.2 | 4.1 | |||||||
| N315-SA1004 | fib | HP, similar to fibrinogen-binding protein | 2.4 | 13.1 | 4.7 | Surface | |||
| N315-SA1270 | HP, similar to amino acid pearmease | 3.5 | Down | Transport | |||||
| N315-SA1272 | ald | Alanine dehydrogenase | 4.0 | Down | |||||
| N315-SA1490 | yhjN | Conserved HP | 2.2 | ||||||
| N315-SA1583 | rot | Repressor of toxins Rot | 2.7 | 3.0 | 4.0 | Up | Regulator | ||
| N315-SA1591 | arsR | Arsenical resistance operon repressor homologue | 2.0 | Regulator | |||||
| N315-SA1628 | splD | Serine protease SplD | 2.5 | Up | Up | Down | Secreted | ||
| N315-SA1629 | splC | Serine protease SplC | 2.7 | Down | Secreted | ||||
| N315-SA1630 | splB | Serine protease SplB | 2.2 | Up | Up | Down | Secreted | ||
| N315-SA1631 | splA | Serine protease SplA | Up | Up | Down | Secreted | |||
| N315-SA1637 | lukD | Leukotoxin, LukD (pathogenicity island SaPIn3) | 16.1 | 9.9 | Secreted | ||||
| N315-SA1638 | lukS | Leukotoxin LukE | 16.6 | 9.5 | Secreted | ||||
| N315-SA1750 | mapW_1 | Truncated map-w protein | 9.7 | 7.6 | Surface | ||||
| N315-SA1751 | mapW_2 | Truncated map-w protein | 7.8 | 6.1 | Surface | ||||
| N315-SA1753 | HP (bacteriophage phiN315) | 2.4 | |||||||
| N315-SA1755 | HP (bacteriophage phiN315) | 27.2 | 11.8 | ||||||
| N315-SA1758 | sak | Staphylokinase precursor | 2.4 | Secreted | |||||
| N315-SA1759 | Lytic enzyme | 2.5 | Secreted | ||||||
| N315-SA1784 | HP (bacteriophage phiN315) | 2.2 | |||||||
| N315-SA1789 | HP (bacteriophage phiN315) | 2.0 | |||||||
| N315-SA1790 | HP (bacteriophage phiN315) | 2.8 | |||||||
| N315-SA1792 | Single-strand DNA-binding protein | 3.1 | |||||||
| N315-SA1795 | HP (Bacteriophage phiN315) | 2.6 | |||||||
| N315-SA1796 | HP (Bacteriophage phiN315) | 2.8 | |||||||
| N315-SA1812 | lukE | HP, similar to synergohymenotropic toxin precursor | 12.3 | 5.9 | Down | Secreted | |||
| N315-SA1813 | lukM | HP, similar to leukocidin chain lukM precursor | 11.7 | 5.2 | Secreted | ||||
| N315-SA2095 | HP, similar to d-octopine dehydrogenase | 2.3 | |||||||
| N315-SA2108 | HP, similar to transcription regulator RpiR family | 2.1 | 3.3 | Regulator | |||||
| N315-SA2114 | glvC | PTS system, arbutin-like IIBC component | 2.3 | Transport | |||||
| N315-SA2135 | gltS | HP, similar to sodium/glutamate symporter | 3.9 | Up | Transport | ||||
| N315-SA2147 | tcaR | TcaR transcription regulator | 2.1 | 2.5 | Regulator | ||||
| N315-SA2156 | l-Lactate permease lctP homologue | 3.2 | Transport | ||||||
| N315-SA2194 | HP, similar to Zn-binding lipoprotein adcA | 3.6 | Surface | ||||||
| N315-SA2206 | sbi | IgG-binding protein SBI | 6.0 | 4.2 | Surface | ||||
| N315-SA2207 | hlgA | Gamma-hemolysin chain II precursor | 3.1 | 16.2 | Secreted | ||||
| N315-SA2208 | hlgC | Gamma-hemolysin component C | 6.7 | 2.7 | Up | Up | Down | Secreted | |
| N315-SA2209 | hlgB | Gamma-hemolysin component B | 7.4 | 2.8 | Up | Up | Down | Secreted | |
| N315-SA2210 | bioX | HP, similar to BioX protein | 5.9 | ||||||
| N315-SA2211 | bioW | HP, similar to 6-carboxyhexanoate-CoA ligase | 3.6 | ||||||
| N315-SA2251 | opp-1F | Oligopeptide transporter putative ATPase domain | 2.1 | Up | Transport | ||||
| N315-SA2253 | opp-1C | Oligopeptide transporter putative membrane permease domain | 2.1 | Transport | |||||
| N315-SA2254 | opp-1B | Oligopeptide transporter putative membrane permease domain | 2.2 | Transport | |||||
| N315-SA2255 | opp-1A | Oligopeptide transporter putative substrate binding domain | 2.1 | Transport | |||||
| N315-SA2300 | HP, similar to glucarate transporter | 2.7 | Down | Transport | |||||
| N315-SA2302 | stpC | HP, similar to ABC transporter | 5.8 | 5.1 | 3.0 | Transport | |||
| N315-SA2302 | stpC | HP, similar to ABC transporter | 5.8 | 5.1 | 3.0 | Transport | |||
| N315-SA2321 | HP | 2.9 | 2.3 | Up | Up | ||||
| N315-SA2430 | aur | Zinc metalloproteinase aureolysin | 2.5 | Up | Down | Secreted | |||
| N315-SAS025 | HP | 2.6 | |||||||
| N315-SAS035 | HP | 2.2 | 2.4 | ||||||
| N315-SAS051 | HP | 2.3 | |||||||
| N315-SAS058 | HP (bacteriophage phiN315) | 2.4 | |||||||
| splF | 4.6 | Secreted | |||||||
| COL-SA0256 | HP | Down | |||||||
| COL-SA0334 | Bacteriophage L54a, conserved HP | 2.4 | |||||||
| COL-SA0472 | exotoxin 2 | 2.3 | Secreted | ||||||
| COL-SA0478 | exotoxin 3 | Exotoxin 3 | 3.4 | Secreted | |||||
| COL-SA0906 | Terminase small subunit | 2.2 | |||||||
| COL-SA1165 | HP | 2.1 | |||||||
| COL-SA1477 | ilvA | Threonine dehydratase catabolic | 2.7 | 4.1 | |||||
| COL-SA1865 | splE | Serine protease SplE | 2.4 | Secreted | |||||
| COL-SA1870 | HP | 2.2 | |||||||
| COL-SA2014 | Terminase small subunit | 2.1 | |||||||
| COL-SA2420 | HP, RpiR family | 2.1 | 10.4 | Regulator | |||||
| COL-SA2480 | HP | 2.8 | |||||||
| COL-SA2511 | fnbA | Fibronectin-binding protein A | 2.2 | Surface | |||||
| COL-SA2568 | HP | 2.1 | |||||||
| Mu50-SAV0861 | HP, bacteriophage? | Down | |||||||
| Mu50-SAV0883 | HP, bacteriophage? | Down | |||||||
| Mu50-SAV0888 | HP, bacteriophage? | 2.4 | |||||||
| Mu50-SAV0889 | HP, bacteriophage? | 2.6 | |||||||
| Mu50-SAV0893 | HP, bacteriophage? | 2.6 | |||||||
| Mu50-SAV0894 | HP, bacteriophage? | 2.8 | |||||||
| Mu50-SAV0895 | HP, bacteriophage? | 2.0 | |||||||
| Mu50-SAV0898 | HP, bacteriophage? | 2.2 | |||||||
| Mu50-SAV0899 | HP, bacteriophage? | 2.2 | |||||||
| Mu50-SAV0900 | HP, bacteriophage? | 2.6 | |||||||
| Mu50-SAV0902 | HP, bacteriophage? | 2.6 | |||||||
| Mu50-SAV0903 | HP, bacteriophage? | 2.3 | |||||||
| Mu50-SAV0905 | HP, bacteriophage? | 2.3 | |||||||
| Mu50-SAV0906 | HP, bacteriophage? | 2.6 | |||||||
| Mu50-SAV0907 | HP, bacteriophage? | 2.7 | |||||||
| Mu50-SAV0909 | HP, bacteriophage? | 2.5 | |||||||
| Mu50-SAV0910 | HP, bacteriophage? | 2.7 | |||||||
| Mu50-SAV0911 | HP, bacteriophage? | 2.9 | |||||||
| Mu50-SAV1985 | HP, bacteriophage? | 2.8 | |||||||
| Mu50-SAV1988 | HP, bacteriophage? | 3.8 | |||||||
| MW1382 | HP | 3.3 | |||||||
| MW1441 | HP | 2.4 | |||||||
| MW1920 | HP | 2.2 | |||||||
| MW1927 | HP | 3.1 | |||||||
| MWP018 | Down | ||||||||
“Fold change” indicates increase or decrease for up-regulated and down-regulated genes, respectively. Values in bold indicate low expression under one condition introduced uncertainty in the fold difference value. “Down” denotes saturation under this condition or absence under other condition such that no fold difference could be calculated; “Up” denotes saturation under this condition or absence under other condition such that no fold difference could be calculated.
We have analyzed the microarray data on the basis of individual genes. However, the MAS includes several clusters of adjacent genes that may represent single transcription units. The individual genes in each cluster are oriented in the same direction and have the same temporal patterns and pH responses. Thus, the conclusions would have been the same if the analysis had been based on transcription units rather than individual genes.
Northern blot hybridization analysis.
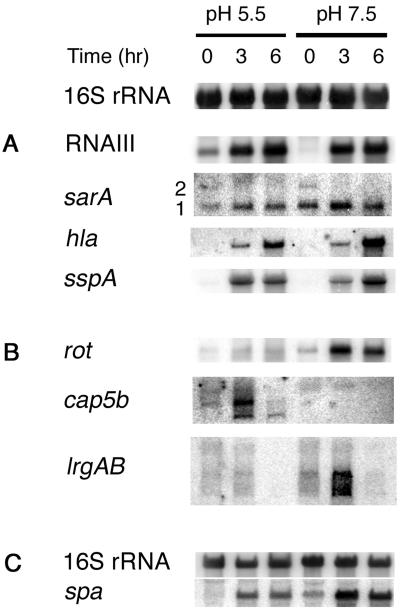
Northern blot hybridization was used to validate the microarray data for several genes, and it provided an important comparison with gene fusion data. In addition to several extensively studied genes, including hla, spa, sspA, sarA, and rnaIII, we analyzed three loci, rot, cap5b, and lrgAB, whose pH-dependent expression was first revealed by the microarray data. Northern blot hybridization results are presented in Fig. 4, from which the following conclusions are apparent. (i) The blotting data for cap5b and lrgAB were consistent in direction if not magnitude with the profiling data. The cap5b transcript level was increased at pH 5.5, whereas lrgAB was increased at pH 7.5. Two or three transcripts are visible in the cap blot and seem to behave somewhat differently. It is planned to determine their sizes and investigate their differential abundance. (ii) hla data from the two techniques could not be compared because expression of hla reached the GeneChip upper-threshold values at T3 and T6 and was so low at T0 that the hla transcript could not be detected by blotting. Comparison of the hla Northern blotting data with the gene fusion results revealed that transcript levels and promoter activity were similar in direction if not magnitude. (iii) With the exception of the T6 time point, the Northern blotting data for sspA were relatively consistent with both the profiling data and with the gene fusion data. Although at T3 the sspA transcript appeared higher at pH 5.5 and at T6 it appeared higher at pH 7.5, these differences were less than twofold. One possibility for the differences observed between gene fusion and Northern blot data could be that the gene fusion construct used may not have included all 5′ regulatory elements (Fig. 2 and 4). At T0, the transcription level was too low to be evaluated at either pH. (iv) The spa data could not be confirmed by Northern blotting of RN6734 RNA because the level of expression was too low at most time points in this agr+ strain. Therefore, we prepared blots with RNA from the isogenic agr-null strain, RN7206. Although the blots from one strain cannot be directly compared with the microarray data from another strain, spa expression in RN7206 was greater at pH 7.5 throughout growth and, in RN6734, it was greater at pH 7.5 only at the earliest time point. (v) Although the pH regulation of rot seen in the microarray was confirmed by Northern blot hybridization, rot transcription increased postexponentially at both pHs rather than being constitutive as has been reported elsewhere (32). (vi) The Northern blots for agr-rnaIII and sae were consistent with the microarray results.
FIG. 4.
Northern blot analysis of the effect of pH on the transcript levels of genes coding for several exoproteins and their regulators. (A and B) Blots of whole-cell RNA from RN6734 grown in CYGP broth without glucose and adjusted to pH 5.5 or 7.5. (B) Blots of transcripts identified by microarray analysis as pH dependent. The 16S rRNA blot above panels A and B shows equal loading. (C) pH-dependent expression of spa in RN7206 (RN6734 Δagr); equal loading is shown by the adjacent 16S rRNA blot.
Although sarA does not belong to the MAS based on analysis of RN6734, interpretation of sarA results is problematic; sarA is transcribed from three separate promoters (12), one of which, P3, uses σB and is not active in RN6734 and other derivatives of NCTC8325, owing to the rsbU defect in these strains. In an rsbU+ strain this promoter is much more active at pH 5.5 than at pH 7.5 (unpublished data). Conversely, the P1 promoter appeared to be considerably less active at pH 5.5 than at pH 7.5 (the level of the P2 transcript was too low to be relevant). Thus, the role of pH in the overall production of SarA protein, which may be highly strain specific, remains to be determined.
Effect of pH on overall exoprotein patterns.
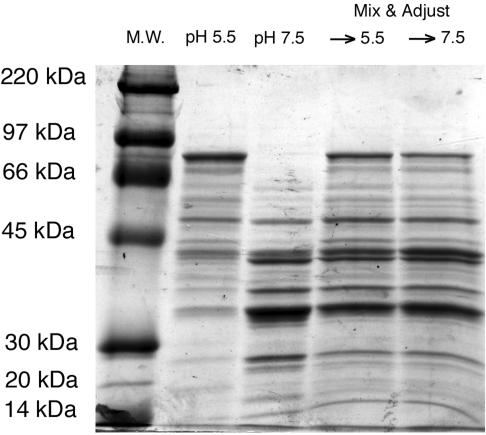
Given that the MAS seems large and complex and that it includes several exoprotein genes, including those encoding known virulence factors, we have begun to analyze the effects of pH on the extracellular proteome. For this purpose, supernatants were collected from cultures of RN6734 grown at pH 5.5 versus pH 7.5 in the absence of glucose at the T6 time point. Supernatant exoprotein profiles, as determined by SDS-PAGE, are shown in Fig. 5. Several exoproteins were observed to be more abundant at pH 7.5 but were considerably reduced or absent at pH 5.5, and vice versa; additionally, there was a considerable asymmetry in the size distributions of the bands corresponding to the two pH conditions, with higher-molecular-weight species predominant at pH 5.5 and lower-weight species at pH 7.5. To test for the possibility that this asymmetry could represent differential proteolytic activity, we mixed equal quantities of the two supernatants, adjusted the mixtures to either pH 5.5 or 7.5, incubated them for 30 min at 37°C, and analyzed them by SDS-PAGE. The resulting exoprotein patterns, shown in the two rightmost lanes in Fig. 5, were indistinguishable with the exception of a high-molecular-weight band, which we have identified as geh lipase (unpublished data). This band is somewhat weaker after incubation of the mixture at pH 7.5 than at 5.5, presumably owing to differential proteolysis (30, 42, 48). Overall, these results are consistent with transcription-level regulation of exoprotein production, and it is clear that proteolysis is not responsible for most of the pH-dependent differences in the exoprotein profile.
FIG. 5.
Exoprotein profiles of cultures grown at pH 5.5 or 7.5, and the effect of pH-dependent protease activity on exoprotein profiles. Exoprotein was prepared from 6-h cultures grown in CYGP broth adjusted to either pH 5.5 or pH 7.5. Supernatants from equal numbers of cells were precipitated with trichloroacetic acid and subjected to SDS-PAGE. The right two lanes show exoprotein from both of the left lanes combined, adjusted to each pH, and incubated at 37°C for 30 min to investigate the effect of differential protease activity.
Identification of differentially abundant exoproteins.
We identified several of the differentially abundant exoproteins by mass spectrometry. Due to differences in loading and resolution between the gel from which the bands were excised and the gel shown in Fig. 5, these proteins are not labeled in Fig. 5. In addition to the cleaved form of lipase, these included an immunodominant surface antigen, IsaA (more abundant at pH 5.5), the precursor form of autolysin Atl (more abundant at pH 5.5), and the serine protease-like SplF (more abundant at pH 7.5). Comparing these and the microarray results, some of the differential abundances of these proteins are a reflection of pH-dependent differences in transcription (IsaA and SplF); others are likely the result of posttranscriptional mechanisms (Geh and Atl).
DISCUSSION
This study began with the observation that the glucose-induced down-regulation of certain staphylococcal virulence genes is largely a result of the pH reduction resulting from glucose fermentation. The end point of this pH reduction is generally around pH 5.5 in standard glucose-supplemented broth cultures of S. aureus; in cultures grown at this pH the classical agr-dependent postexponential induction of virulence genes such as hla and tst, seen at pH 7.5, is eliminated. Microarray analysis revealed that the transcript levels of over 400 S. aureus genes were affected, and the overall pattern is here defined as the MAS. More than twice as many genes are up-regulated at pH 5.5 than are down-regulated, relative to effects at pH 7.5.
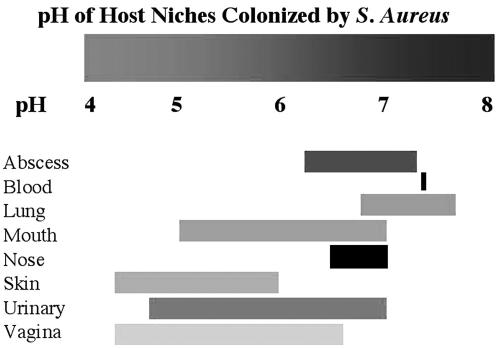
It is well known that bacteria have elaborate mechanisms by which they maintain their internal pH within tight limits. Major, potentially lethal reductions in external pH activate an acid stress response, which mobilizes a variety of resources in an attempt to defend the cytoplasmic pH (14, 37, 38). At the same time, bacteria such as staphylococci can grow and divide normally over a considerable pH range—at least 5 to 9. In the present work we have analyzed the lower half of this range and found that there is a large set of genes that are differentially regulated between pH 5.5 and 7.5—the MAS. We suggest that this modulation of gene expression is likely to represent an adaptation to pH-variable environments, rather than a global acid stress response. In other words, pH could be one of the factors that determine locale-specific gene expression patterns, since certain body sites are characterized by variations in pH from the homeostatic 7.4 (Fig. 6) (5, 10, 16, 21, 22, 31, 41, 53). Given the breadth of the MAS, plus the fact that the intracellular pH is maintained within very narrow limits, the pH differential must be sensed by one or more surface receptors and transduced to the interior of the cell. As there is a complex regulatory network that coordinates the expression of accessory genes (34), the MAS must interact with one or more regulatory genes involved in this network. For any regulatory genes that belong to the MAS, it is predicted that their regulons will follow suit, insofar as the regulation is direct. That is, if transcription of a regulatory gene is reduced at one pH, the genes that it up-regulates will also be reduced at that pH, and vice versa. Genes that do not follow this rule must be regulated indirectly. For those regulatory genes that do not belong to the MAS, any target genes that do belong must also be regulated indirectly. Therefore, examination of stimulons and their overlaps with regulons is expected to aid greatly in the understanding of the overall regulatory network. Results presented here may be viewed as the beginning of this analysis.
FIG. 6.
pH ranges of various niches S. aureus can colonize in the human host. Blood, pH 7.4 (Robinson 1975); vagina, 4.2 to 6.6 (Wagner and Ottesen 1982); abscess, 6.2 to 7.3 (Bessman, Page et al. 1989); urinary tract (UT), 4.6 to 7 (McClatchey 1994); lung, 6.8 to 7.6 (Cheng, Rodriguez et al. 1998); mouth, 5 to 7 (Dong, Pearce et al. 1999); nose, 6.5 to 7 (England, Homer et al. 1999); skin, 4.2 to 5.9 (Ehlers, Ivens et al. 2001).
A number of tantalizing relationships between the MAS and certain other stimulons and regulons have been revealed; these relationships are noted here and are clearly in need of further study.
One example is the stimulon defined by the response to subinhibitory concentrations of cell wall-active antibiotics (bacitracin, d-cycloserine, and oxacillin) (50). The MAS appears to overlap significantly with the cell wall-stress stimulon. Twenty-five of the 158 genes in this stimulon are also in the MAS, and for these genes the two stimulons are concordant, raising the question of whether there is some common adaptivity between the two, such as the involvement of any of these 25 genes in transducing the pH stimulus. Seventeen of these 25 genes were both up-regulated by cell wall-active antibiotics and were up-regulated at pH 5.5. Seven of the remaining eight genes were down-regulated by antibiotics and were also down-regulated at pH 5.5. The only exception was a hypothetical gene that could correspond to an unidentified regulator (a repressor) of the MarR type. Because the microarray used in defining the cell wall-stress stimulon was less comprehensive than the one used in the present study, it is possible that the relationship between the two stimulons is even greater than observed here.
Another stimulon that was recently defined is the set of genes that are differentially expressed during growth in a mature flow cell-resident biofilm (3). The biofilm state has been shown to be relevant to several different infection niches for growth, persistence, and resistance to antibiotic treatment (17). Although the strain used by Beenken et al. is not of the NCTC8325 lineage and the cultures were grown with glucose, genes they identified as differentially expressed between exponential-phase planktonic cultures, in which the pH may have not yet dropped below pH 6.0, and mature biofilms may be relevant for comparison to the MAS. Of the 95 genes that are reported up-regulated in a biofilm, well over half (55) are also up-regulated under the mild acid condition. The operons up-regulated under both conditions include those coding for capsular polysaccharide biosynthesis, pyrimidine biosynthesis, the potassium-proton antiporter system kdp, the urease system, and the ssp operon. Among the 17 genes down-regulated under both conditions are rot, spa, nuc, pycA, and the opp1 and spl operons. None of the mild-acid-down-regulated genes were up-regulated in the biofilm, and only 17 of the 278 biofilm down-regulated genes were up-regulated in mild acid. The results presented here support the suggestion of Beenken et al. that a mature biofilm is an acidic environment. The incomplete concordance of the biofilm and mild acid stimulons may reveal sets of genes specific to biofilm or planktonic lifestyles. In sum, the mild acid-biofilm correlation provides an example of an important niche where pH-dependent gene expression may be critical for growth and persistence.
A third example involves the conditionally essential two-component signal transduction system (TCS), YycG/YycF. Five of 12 genes recently reported to be controlled by YycG/YycF were identified in the MAS, although the TCS itself was not. YycF recognizes a pair of directly repeated hexamers within the promoter region of target genes (18). Two of the target genes with their direct repeats oriented in one direction were down-regulated at pH 5.5; three with their repeats in the opposite direction were up-regulated at pH 5.5. This difference in the orientation of cis regulatory elements in the promoters of genes belonging to the YycG/YycF TCS regulon correlates with the differential pH effects on these genes, suggesting an interesting possible relationship between this TCS and transduction of the pH signal.
Among the genes belonging to the MAS are 26 known or putative regulatory genes. The response of target gene subsets should, to a first approximation, follow the response of the controlling regulatory gene. The sae regulon appears to fit this scheme, at least with respect to the few target genes that have been individually analyzed—for example, sae up-regulated leukotoxin and gamma hemolysin are down-regulated at pH 5.5, like sae. The generality of this effect will be reported elsewhere in connection with an ongoing analysis of the sae regulon. The transcript levels of 19 other known or putative regulatory genes are increased at pH 5.5, and those of six are decreased. In Table 3 are compiled data on known regulatory interactions involving two of the better studied of these regulators, agr and rot, whose regulons have been determined (20, 43). We note that the RNA samples used for determination of the agr and rot regulons were all prepared from bacteria grown in the presence of glucose, and the rot regulon was determined with an agr-null strain. Additionally, they were determined with an earlier version of the microarray lacking many important genes. Nevertheless, most of the agr-regulated and rot-regulated genes adhere to the principle that target genes follow the response of the controlling regulator. For genes that do not, the simplest possibility is that the regulatory connection is indirect. Thus, agr is up-regulated 3.6-fold at pH 5.5 and T0 (but not at the other time points). At this time point, when agr is a member of the MAS, the agr-regulated members of the MAS follow the regulation of agr, whereas at other time points there is no consistency to the response of agr-regulated genes. Interestingly, although agr belongs to the MAS, no known regulators of agr, including svrA, sarA, sarU, ssrAB, and arlRS, belong to the stimulon. Of course, any of these regulators could be affected at the posttranscriptional level.
rot was identified as a member of the MAS and was shown to be down-regulated at pH 5.5 at all three time points. Thirty-nine genes (9.3% of the stimulon) are rot regulated, of which 28 demonstrated higher transcript titers at pH 5.5. This is largely consistent with the reported role of rot as a repressor (32). Genes encoding surface proteins, such as spa, which is up-regulated by rot, are down-regulated at pH 5.5. rot regulates spa via SarS (43), which is not listed in the MAS. However, the stimulon list was generated by using an arbitrary cutoff value of a twofold change in transcription. Further analysis indicated that although it is not decreased twofold, sarS is indeed moderately down-regulated at the lower pH tested. Only one gene, SA0173, a hypothetical gene similar to surfactin synthetase of Bacillus subtilis, is regulated in the same direction by agr and rot (up).
Microarray data for the sarA regulon have also been published (20) and are included in Table 3, although sarA does not appear to belong to the MAS in strains of the NCTC8325 lineage, such as RN6734. Forty-six genes in the stimulon are SarA regulated. Most of these have increased transcript levels at pH 5.5, and most SarA-regulated genes with pH-sensitive expression are SarA up-regulated. These genes are regulated predominantly through agr.
It has been suggested that SarA is important in the transduction of the pH signal and, superficially, the data on the relation of SarA to sspA shown in Fig. 2C and D would seem to bear this out. SarA seems to be a repressor of sspA only at pH 5.5. However, a closer look at these data suggests that it is not SarA but a separate positive regulator that may be responsible. We note that at pH 7.5, deletion of sarA has no effect on sspA expression in the NCTC8325-derived background tested here; i.e., sarA cannot be a repressor of sspA at pH 7.5. If it were a repressor only at pH 5.5, then the level of sspA at pH 7.5 should match the higher expression level at pH 5.5, rather than the lower. The simplest interpretation here is that SarA is not a pH-dependent repressor, but rather that there is a pH-dependent activator that functions at pH 5.5, only in the absence of SarA.
It has also been suggested that σB may have a role in transducing the pH signal (9, 39); on the basis of data presented here, it is suggested that there may be two pH-sensing pathways, one independent of σB and the other possibly σB dependent. The microarray analysis was performed with RN6734 because of the enormous experience and wide use of this and other NCTC8325 derivatives, despite their rsbU defect (23, 26). As these strains have only weak residual σB activity, the studies reported here will have defined a pH-sensing pathway that is essentially independent of σB. This has been confirmed by a recently published DNA microarray analysis of the σB regulon (6). Of the 198 genes found to be σB up-regulated by transcriptional profiling of three different rsbU+ strains, 25 were also up-regulated by mild acid. None of these 198 genes was down-regulated by mild acid. Eighteen of the 53 σB down-regulated genes were also down-regulated by mild acid; however, 19 of these 53 were up-regulated by mild acid. These results are consistent with the absence of sigB or members of the sigB operon in the MAS and the pH-insensitive transcription pattern of sigB in RN6734 (data not shown). However, sarA has a σB-dependent promoter that is also pH dependent (unpublished data), so that in an rsbU+ strain, genes regulated by SarA may define a pH-dependent pathway that is σB dependent. This interpretation is highly tentative since, as noted above, it is not known whether the interplay of the three sarA promoters results in σB-dependent differences in SarA protein levels.
In conclusion, our results have pointed to a number of approaches that could help to identify the pathway(s) by which external pH is sensed by the organism. Additionally, they address but do not solve the basic question of the role of pH-dependent gene expression in pathogenesis. On the one hand, as shown here, it is clear that mild acid modulates the expression of a large set of staphylococcal genes, including virulence genes; on the other hand, it is well known that certain sites in an animal host are characterized by pHs different from the homeostatic 7.4. The basic question, then, is whether there is a significant connection between these two separate findings. In other words, is pH-dependent modulation of virulence gene expression adaptive for the organism with respect to different sites in the animal host that are maintained at particular pHs (different from 7.4)? And, if so, would this be an example of bacteria modulating the expression of particular genes for adaptation to particular tissue sites?
The available studies of pH-dependent and host-niche-specific staphylococcal virulence gene expression reveal little that suggests such a connection (1, 2, 4, 9, 11, 24, 25, 29, 33, 36, 40, 44-46, 49, 51, 54, 55). A potentially informative study, however, is that of Coulter et al. (1998), which identified genes required for growth and persistence in various animal models of staphylococcal infection. Mutations in opp2, sspA, odhA, and epiB were shown to cause attenuation in a systemic infection model but not in abscess or burn wound models, while those in opp1, pycA, and pflB caused attenuation in an abscess model but not in systemic infection or burn wound models (15). The genes associated with systemic infections were up-regulated, while those associated with abscesses were down-regulated at pH 5.5. A homologous oligopeptide permease (opp) system in B. subtilis is required for the uptake of signaling peptides necessary for pH-dependent expression of srf, which encodes a surfactin synthetase (13). A surfactin synthetase homologue is also part of the staphylococcal MAS and is up-regulated at pH 5.5, as is opp2. This could represent a conserved adaptation to the mild acid condition.
The interesting correlation between tissue-site specificity and pH modulation of these genes raises the possibility that tissue-specific pH variation could impact on pathogenesis. It seems doubtful, however, that the pHs at the sites in question prior to infection are consistent with the observed pH-dependent modulation in bacterial gene expression, because these sites are typically maintained at pH 7.4 (Fig. 6). Perhaps a local pH change as a result of the infection requires an adaptive response, prohibiting bacteria incapable of such a response from maintaining an infection.
A quite different and rather dramatic example of how pH-dependent virulence gene expression could impact human health, but in a manner that is also not consistent with the concept of pH-dependent tissue site adaptation, is menstrual toxic shock. The human vagina is maintained at a pH of <5, a pH at which toxic shock syndrome toxin 1 is not produced (45) because tst, the gene encoding the toxin, is essentially silent (Fig. 2). However, during menses the pH rises above 7, a pH at which tst is strongly expressed—which could very well account for the well-known relation between toxic shock syndrome and menstruation (4). However, whether pH-dependent expression of tst has any adaptive role for the organism or is simply coincidental is far from clear. Indeed, it is not obvious whether causing toxic shock syndrome is in any way advantageous for the organism; instead, maintaining the vaginal pH of <5 could represent a host defense against toxic shock, a defense that the bacteria have clearly not countered by pH-dependent gene expression but one that is inadvertently breached by the host through menstrual bleeding.
The in vivo relevance of the pH-dependent accessory gene expression we report here will require more accurate measurement of the pH of the host environment during infection and testing of knockouts of the members of the signal transduction network in relevant animal models. Regardless of the role of differential expression in an infection, elucidation of the MAS will forward attempts to map the S. aureus accessory gene regulatory network. Regulons have great value in establishing epistasis and suggesting functional roles for the corresponding regulator, but their usefulness is limited because regulators often act in concert and effects may be absent or even contradictory in single mutants. Stimulons allow us to perturb genetically complete organisms but require analysis of the relevant mutants to dissect cause and effect. The combination of these methods should in the future allow construction of a comprehensive model of the S. aureus accessory gene regulatory network.
Acknowledgments
We thank Hope F. Ross for discussion and critical reading of the manuscript.
This work was supported by National Institutes of Health grant R01 AI030138-14 to Richard P. Novick. Brian Weinrick was supported in part by an NIH training grant to the Department of Microbiology, NYU School of Medicine (T32 AI007180-21).
REFERENCES
- 1.Arvidson, S., and T. Holme. 1971. Influence of pH on the formation of extracellular proteins by Staphylococcus aureus. Acta Pathol. Microbiol. Scand. B 79:406-413. [DOI] [PubMed] [Google Scholar]
- 2.Arvidson, S., T. Holme, and T. Wadstrom. 1971. Influence of cultivation conditions on the production of extracellular proteins by Staphylococcus aureus. Acta Pathol. Microbiol. Scand. B 79:399-405. [DOI] [PubMed] [Google Scholar]
- 3.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergdoll, M. S. 1989. Regulation and control of toxic shock syndrome toxin 1: overview. Rev. Infect. Dis. 11(Suppl. 1):S142-S144. [PubMed] [Google Scholar]
- 5.Bessman, A. N., J. Page, and L. J. Thomas. 1989. In vivo pH of induced soft-tissue abscesses in diabetic and nondiabetic mice. Diabetes 38:659-662. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Booth, I. R. 1985. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 49:359-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, D. S., R. M. Rodriguez, J. Rogers, M. Wagster, D. L. Starnes, and R. W. Light. 1998. Comparison of pleural fluid pH values obtained using blood gas machine, pH meter, and pH indicator strip. Chest 114:1368-1372. [DOI] [PubMed] [Google Scholar]
- 11.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y. Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1-9. [DOI] [PubMed] [Google Scholar]
- 12.Cheung, A. L., and S. J. Projan. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosby, W. M., D. Vollenbroich, O. H. Lee, and P. Zuber. 1998. Altered srf expression in Bacillus subtilis resulting from changes in culture pH is dependent on the Spo0K oligopeptide permease and the ComQX system of extracellular control. J. Bacteriol. 180:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coulter, S. N., W. R. Schwan, E. Y. Ng, M. H. Langhorne, H. D. Ritchie, S. Westbrock-Wadman, W. O. Hufnagle, K. R. Folger, A. S. Bayer, and C. K. Stover. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol. Microbiol. 30:393-404. [DOI] [PubMed] [Google Scholar]
- 16.Dong, Y. M., E. I. Pearce, L. Yue, M. J. Larsen, X. J. Gao, and J. D. Wang. 1999. Plaque pH and associated parameters in relation to caries. Caries Res. 33:428-436. [DOI] [PubMed] [Google Scholar]
- 17.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubrac, S., and T. Msadek. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 186:1175-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunman, P. M., W. Mounts, F. McAleese, F. Immermann, D. Macapagal, E. Marsilio, L. McDougal, F. C. Tenover, P. A. Bradford, P. J. Petersen, S. J. Projan, and E. Murphy. 2004. Uses of Staphylococcus aureus GeneChips in genotyping and genetic composition analysis. J. Clin. Microbiol. 42:4275-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehlers, C., U. I. Ivens, M. L. Moller, T. Senderovitz, and J. Serup. 2001. Females have lower skin surface pH than men. A study on the surface of gender, forearm site variation, right/left difference and time of the day on the skin surface pH. Skin Res. Technol. 7:90-94. [DOI] [PubMed] [Google Scholar]
- 22.England, R. J., J. J. Homer, L. C. Knight, and S. R. Ell. 1999. Nasal pH measurement: a reliable and repeatable parameter. Clin. Otolaryngol. 24:67-68. [DOI] [PubMed] [Google Scholar]
- 23.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40:1439-1447. [DOI] [PubMed] [Google Scholar]
- 26.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsson, A., and S. Arvidson. 2002. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor, sarA. Infect. Immun. 70:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Kreiswirth, B., S. Lofdahl, M. Betley, M. O’Reilly, P. Schlievert, M. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structured gene is not detectably transmitted by a prophage. [DOI] [PubMed]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lowe, A. M., D. T. Beattie, and R. L. Deresiewicz. 1998. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol. Microbiol. 27:967-976. [DOI] [PubMed] [Google Scholar]
- 30.Massimi, I., E. Park, K. Rice, W. Muller-Esterl, D. Sauder, and M. J. McGavin. 2002. Identification of a novel maturation mechanism and restricted substrate specificity for the SspB cysteine protease of Staphylococcus aureus. J. Biol. Chem. 277:41770-41777. [DOI] [PubMed] [Google Scholar]
- 31.McClatchey, K. D. 1994. Clinical laboratory medicine. Williams & Wilkins, Baltimore, Md.
- 32.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 33a.Nicholas, R. O., T. Li, D. McDevitt, A. Marra, S. Sucoloski, P. L. Demarsh, and D. R. Gentry. 1999. Isolation and characterization of a sigB deletion mutant of Staphylococcus aureus. Infec. Immun. 67:3667-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 35.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 36.Novick, R. P., and D. Jiang. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149:2709-2717. [DOI] [PubMed] [Google Scholar]
- 36a.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993.. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olson, E. R. 1993. Influence of pH on bacterial gene expression. Mol. Microbiol. 8:5-14. [DOI] [PubMed] [Google Scholar]
- 38.Padan, E., D. Zilberstein, and S. Schuldiner. 1981. pH homeostasis in bacteria. Biochim. Biophys. Acta 650:151-166. [DOI] [PubMed] [Google Scholar]
- 39.Palma, M., and A. L. Cheung. 2001. σB activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 69:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rampone, H., G. L. Martinez, A. T. Giraudo, A. Calzolari, and R. Nagel. 1996. In vivo expression of exoprotein synthesis with a Sae mutant of Staphylococcus aureus. Can. J. Vet. Res. 60:237-240. [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson, J. R. 1975. Fundamentals of acid-base regulation, 5th ed. Blackwell Scientific, London, England.
- 42.Rollof, J., and S. Normark. 1992. In vivo processing of Staphylococcus aureus lipase. J. Bacteriol. 174:1844-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Said-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarafian, S. K., and S. A. Morse. 1987. Environmental factors affecting toxic shock syndrome toxin-1 (TSST-1) synthesis. J. Med. Microbiol. 24:75-81. [DOI] [PubMed] [Google Scholar]
- 45.Schlievert, P. M., and D. A. Blomster. 1983. Production of staphylococcal pyrogenic exotoxin type C: influence of physical and chemical factors. J. Infect. Dis. 147:236-242. [DOI] [PubMed] [Google Scholar]
- 45a.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect. Immun. 69:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider, W. P., S. K. Ho, J. Christine, M. Yao, A. Marra, and A. E. Hromockyj. 2002. Virulence gene identification by differential fluorescence induction analysis of Staphylococcus aureus gene expression during infection-simulating culture. Infect. Immun. 70:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sleator, R. D., and C. Hill. 2002. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol. Rev. 26:49-71. [DOI] [PubMed] [Google Scholar]
- 48.Sorensen, S. B., T. L. Sorensen, and K. Breddam. 1991. Fragmentation of proteins by S. aureus strain V8 protease. Ammonium bicarbonate strongly inhibits the enzyme but does not improve the selectivity for glutamic acid. FEBS Lett. 294:195-197. [DOI] [PubMed] [Google Scholar]
- 48a.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 49.Todd, J. K., B. H. Todd, A. Franco-Buff, C. M. Smith, and D. W. Lawellin. 1987. Influence of focal growth conditions on the pathogenesis of toxic shock syndrome. J. Infect. Dis. 155:673-681. [DOI] [PubMed] [Google Scholar]
- 50.Utaida, S., P. M. Dunman, D. Macapagal, E. Murphy, S. J. Projan, V. K. Singh, R. K. Jayaswal, and B. J. Wilkinson. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719-2732. [DOI] [PubMed] [Google Scholar]
- 51.van Wamel, W., Y. Q. Xiong, A. S. Bayer, M. R. Yeaman, C. C. Nast, and A. L. Cheung. 2002. Regulation of Staphylococcus aureus type 5 capsular polysaccharides by agr and sarA in vitro and in an experimental endocarditis model. Microb. Pathog. 33:73-79. [DOI] [PubMed] [Google Scholar]
- 52.Vojtov, N., H. F. Ross, and R. P. Novick. 2002. Global repression of exotoxin synthesis by staphylococcal superantigens. Proc. Natl. Acad. Sci. USA 99:10102-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner, G., and B. Ottesen. 1982. Vaginal physiology during menstruation. Ann. Intern. Med. 96:921-923. [DOI] [PubMed] [Google Scholar]
- 54.Wong, A. C., and M. S. Bergdoll. 1990. Effect of environmental conditions on production of toxic shock syndrome toxin 1 by Staphylococcus aureus. Infect. Immun. 58:1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yarwood, J. M., J. K. McCormick, M. L. Paustian, V. Kapur, and P. M. Schlievert. 2002. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J. Bacteriol. 184:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]