Abstract
Staphylococcus aureus SirA was previously identified as a lipoprotein, and SirB and SirC are thought to encode the transmembrane domains of an ABC transporter. Sir proteins show similarity to iron-siderophore transporters in several bacteria. Here, we show that the iron-regulated sirABC operon is divergently transcribed from the sbn operon that encodes enzymes involved in the synthesis of staphylobactin, a recently described siderophore produced by S. aureus. Mutation of either sirA or sirB increased the resistance of iron-starved S. aureus to streptonigrin and resulted in compromised growth in iron-restricted, but not iron-rich, media. We also demonstrated that sirA and sirB mutants are compromised in the ability to transport iron complexed to staphylobactin but are not compromised for uptake of other iron complexes, such as ferric hydroxamates, ferric enterobactin, or ferric citrate. SirA- and SirB-deficient S. aureus, however, retain the ability to produce staphylobactin. Moreover, we found that transcription from the sbn operon was increased, relative to the wild type, in both sirA and sirB knockout strains, likely in response to an increased level of iron starvation in these cells. These results provide evidence of a role for these proteins in iron import in S. aureus and for full fitness of the bacterium in iron-restricted environments and demonstrate a function for S. aureus genes encoding proteins involved in the transport of an endogenously produced siderophore.
The ability of bacterial pathogens to acquire iron from host iron-binding glycoproteins, such as transferrin and lactoferrin, is an important attribute that aids in the establishment of many bacterial infections (25, 33, 34). To access these extracellular iron stores, many bacteria produce small organic molecules called siderophores that have a high affinity for ferric iron (35). Iron-siderophore complexes (ferrisiderophores) are recognized and transported into the bacterial cytoplasm by specific receptor proteins and associated transport systems expressed at the cell surface (11). In gram-negative bacteria, these transport systems include high-affinity outer membrane receptor proteins that capture ferrisiderophores and shuttle them across the outer membrane (4, 9). Once in the periplasm, ferrisiderophores are bound by periplasmic binding proteins (16, 31) that direct the ligand to membrane-associated ATP-binding cassette (ABC) transporters (3, 19, 23). In gram-positive bacteria, ferrisiderophores are initially recognized and bound by lipoproteins, tethered at the external face of the cytoplasmic membrane, that direct the ligand to ABC transporters. One ferrisiderophore import system in gram-positive bacteria that has been studied in our laboratory is the ferric hydroxamate uptake (fhu) system. In Staphylococcus aureus, the fhu system is comprised of FhuC (ATPase), FhuB and FhuG (together they form a membrane-embedded permease), and the lipoproteins FhuD1 and FhuD2 (high-affinity receptors), and together, these proteins function to scavenge hydroxamate siderophores (28-30).
The staphylococci are gram-positive cocci that are often associated with mucous membranes and the skin of mammals. The staphylococci are broadly divided into two groups, the coagulase-negative staphylococci and those strains that produce coagulase; the latter group includes S. aureus. S. aureus, the best-characterized member of the staphylococci, is a prevalent human pathogen that causes a wide range of infections that range from minor skin lesions to more serious diseases, such as sepsis, endocarditis, osteomyelitis, pneumonia, and toxic shock syndrome (1). In response to iron limitation, the staphylococci have been shown to produce several siderophores, including staphyloferrin A and staphyloferrin B (8, 13, 15), aureochelin (6), and staphylobactin (7). The staphyloferrins are polycarboxylate-type siderophores initially identified in coagulase-negative staphylococci and some strains of S. aureus. Aureochelin was identified in S. aureus by Courcol et al. (6); however, its structure has not been described. Most recently, our laboratory has identified a fourth staphylococcal siderophore, which we have named staphylobactin. An operon containing genes whose products are involved in the production of staphylobactin is found in the genome of S. aureus but not Staphylococcus epidermidis RP62A. We have shown that an inability to synthesize staphylobactin results in an attenuation of S. aureus virulence in a murine kidney abscess model of infection (7). Genetic determinants for the production of other staphylococcal siderophores are as yet unknown, along with their relative contributions to the pathogenesis of the organism. The transport machinery required for the import of staphylococcal siderophores is also undetermined, although two putative ferrisiderophore transporters, encoded by the iron-regulated sstABCD (22) and sirABC operons (14), have been identified in S. aureus. In both cases, their functions in ferrisiderophore import have been hypothesized based on homology to proteins known to function in the transport of siderophores. In this study, we characterize the function of the sirABC locus by demonstrating that expression of both sirA and sirB is important for the iron-restricted growth of S. aureus. Moreover, we demonstrate that staphylobactin is the siderophore that is imported by the SirABC polypeptides.
MATERIALS AND METHODS
Media and growth conditions.
The bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1. For routine cloning and protein expression, E. coli was grown at 37°C in Luria-Bertani broth supplemented with erythromycin (300 μg/ml), ampicillin (100 μg/ml), tetracycline (10 μg/ml), chloramphenicol (30 μg/ml), or kanamycin (30 μg/ml), as required. For general manipulations, S. aureus strains were cultured in tryptic soy broth (TSB) (Difco) containing erythromycin (5 μg/ml), tetracycline (4 μg/ml), kanamycin, and neomycin (each at 50 μg/ml) or chloramphenicol (5 μg/ml) as required. For iron-restricted bacterial growth experiments, a Tris minimal succinate (TMS) medium was used, the composition of which has been described (29). TMS was supplemented with 2,2′-dipyridyl, at concentrations described in Results, to further restrict the concentration of free iron.
TABLE 1.
Bacterial strains, plasmids and oligonucleotides used in this studya
| Bacterial strains, plasmids, oligonucleotides | Descriptiona | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Promega |
| ER2566 | F− λ−fhuA2 [lon] ompT lacZ::T7 gal sulA11 Δ(mcrC-mrr)114::IS10 R(mcr-73::miniTn10)2 R(zgb-210::Tn10)l (Tets) endA1 [dcm] | New England Biolabs |
| S. aureus | ||
| 8325-4 | Prophage-cured wild-type strain | Lab stock |
| RN6390 | Prophage-cured wild-type strain | Lab stock |
| Newman | Clinical isolate; wild-type strain | Lab stock |
| RN4220 | Restriction-deficient; accepts foreign DNA | Lab stock |
| H306 | RN6390 sirA::Km; Kmr | This study |
| H474 | RN6390 sirB::Tet; Tetr | This study |
| H686 | Newman sbnE::Km | 7 |
| H706 | Newman fur::Km; Kmr | 7 |
| H803 | Newman sirA::Km: Kmr | This study |
| H804 | Newman sirB::Tet; Tetr | This study |
| H870 | Newman sbnH::pMUTIN4 | This study |
| H873 | H803 sbnH::pMUTIN4 | This study |
| H876 | H804 sbnH::pMUTIN4 | This study |
| Plasmids | ||
| pGEX-2T-TEV | Expression vector for generating protein fusions with GST that are cleavable with tobacco etch virus protease | 30 |
| pALC2073 | E. coli-S. aureus shuttle vectorr; contains Pxyl/tet; Cmr | A. Cheung |
| pAUL-A | Temperature-sensitive E. coli-S. aureus shuttle vector | 5 |
| pAW8 | E. coli-S. aureus shuttle vector; Tetr | A. Wada |
| pBC SK(+) | E. coli phagemid; Cmr | Stratagene |
| pDG782 | pMLT22 derivative that carries a kanamycin resistance cassette; Apr Kmr | 12 |
| pDG1513 | pMLT22 derivative that carries a tetracycline resistance cassette; Cmr Tetr | 12 |
| pMTS12 | pAUL-A derivative carrying sirA::Km; Kmr Emr | This study |
| pMUTIN4 | lacZ fusion vector; Apr (E. coli) Emr (S. aureus) | 32 |
| pSED43 | pALC2073 derivative carrying the sirB coding region; Cmr | This study |
| pSED44 | pAW8 derivative carrying sirABC; Tetr | This study |
| pSirA | pGEX-2T-TEV derivative carrying sirA; Ampr | This study |
| pSirABC | pBC SK(+) carrying sirABC; Cmr | This study |
| pSirB::Tet3 | pAUL-A derivative carrying sirB::Tet; Tetr Emr | This study |
| Oligonucleotidesb | ||
| pSirA (BamHI) | GCAATGGGTACAGGATCCATTAAAGGGAAACCAAAG | |
| pSirA (EcoRI) | TTGAATCGTAGCATCGTAAAACTCCTT | |
| SirB Comp 5′ | TTGGTACCGGCGGATATAAATCTTCATT | |
| SirB Comp 3′ | TTGAGCTCTTTCGGTCATAAGCGTTGAC | |
| Sir Upper | TCACGAAGGAGGCTAATTAG | |
| Sir Lower | CCTCGCAACGGTTAGTTAAC | |
| SirB Internal 5′ | CAGCTACGGCTACCGAAATA | |
| SirB Internal 3′ | CATTTTTGGGGGCTATTGTTGT | |
| Gapdh 5′ | GGAGGCCATTACCATGGCAG | |
| Gapdh 3′ | TGCTCCCCGCTTACTCATAA |
Abbreviations: Cmr, Tetr, Emr, Kmr, Ampr, resistance to chloramphenicol, tetracycline, erythromycin, kanamycin, and ampicillin, respectively.
Restriction endonuclease recognition sites are underlined.
Plasmid and strain construction.
All DNA manipulations and plasmid constructions were performed using standard protocols (26). The sirABC operon was PCR amplified from the chromosome of S. aureus 8325-4 using PwoI polymerase (Roche Diagnostics) and primers Sir upper and Sir lower. The resultant 3.8-kb product was cloned into the SmaI site of pBC SK(+) to create pSirABC.
To interrupt the sirA coding region, pSirABC was digested with NsiI, blunted with T4 DNA polymerase, and ligated to a kanamycin resistance cassette that had been excised as a StuI/SmaI fragment from pDG782. The sirA::Km region was then cloned into BamHI/SalI-digested pAUL-A, creating plasmid pMTS12.
The sirB coding region was interrupted by insertion of a tetracycline resistance cassette, derived from digesting pDG1513 with ClaI (blunted with Klenow enzyme), into the StuI site of sirB. The sirB::Tet fragment was cloned into BamHI/KpnI-digested pAUL-A, creating plasmid pSirB::Tet3.
To create strains bearing individual mutations in sirA and sirB, pMTS12 and pSirB::Tet3, respectively, were introduced into S. aureus RN4220, followed by transduction, via phage 80α, of the plasmid into S. aureus RN6390 using methodologies previously described (29). Transductants were confirmed by restriction analysis. Allelic replacement was accomplished by growing plasmid-containing bacteria at 30°C for 3 h, followed by a shift in the growth temperature to 43°C for a further 4 h. Double-crossover events were screened for by resistance to kanamycin (for the sirA::Km mutation) or tetracycline (for the sirB::Tet mutation), with a loss of erythromycin resistance in both cases. PCR and Southern blot analyses were used to verify the insertion of the antibiotic resistance cassettes into sirA and sirB. The resulting mutant strains were designated H306 (RN6390 sirA::Km) and H474 (RN6390 sirB::Tet). Transduction was used to mobilize the mutations into different genetic backgrounds, such as S. aureus Newman.
For complementation of the sirA::Km mutation, the entire sirABC operon was excised from pSirABC (using KpnI and BamHI) and cloned into pAW8 to create the plasmid pSED44 (see Results for further explanation of this strategy). For complementation of the sirB::Tet mutation, the sirB coding region was PCR amplified from the S. aureus RN6390 chromosome using the primers SirB Comp 5′ and SirB Comp 3′, followed by digestion with KpnI and SacI for directional cloning into pALC2073, to create the plasmid pSED43. The complementing vectors were electroporated into S. aureus RN4220 and transduced into mutant strains using bacteriophage 80α.
RT-PCR.
Total RNA for use in reverse transcription (RT)-PCRs was isolated from bacterial cultures in late logarithmic phase using TRIzol reagent (Invitrogen). RNA samples were treated with DNase I for 15 min at room temperature prior to the RT-PCRs. The SuperScript One-Step RT-PCR with Platinum Taq kit (Invitrogen) was used according to the manufacturer's instructions. Total RNA (500 ng) was reverse transcribed using primers SirB Internal 5′ and SirB Internal 3′ to amplify a 399-bp fragment internal to the sirB coding region. As an internal control, a 483-bp fragment of gap (encoding glyceraldehyde-3-phosphate dehydrogenase) was amplified using the Gapdh 5′ and Gapdh 3′ oligonucleotide primers.
Bacterial growth curves.
S. aureus cultures were pregrown overnight in TMS. The cells were washed with TMS, and ∼107 CFU of each strain was inoculated into fresh TMS medium containing 250 μM 2,2′-dipyridyl (Sigma) with or without 50 μM FeCl3. Bacterial growth was monitored using a Klett meter until late stationary phase was reached.
Siderophore plate bioassays.
Siderophore plate bioassays were performed as previously described (29) with the following modifications: TMS agar was cooled to 45°C before the addition of 105 CFU of each strain to be tested/ml. 2,2′-dipyridyl was added at a concentration of 550 μM for plates containing S. aureus Newman and Newman containing vehicle controls (e.g., pAW8 and pALC2073) or 400 μM 2,2′-dipyridyl for strains H803 (Newman sirA::Km) and H804 (Newman sirB::Tet) (Fig. 1) with or without plasmids. The staphylobactin siderophore was isolated from RN6390 as previously described (7), and its purity was confirmed by high-performance liquid chromatography analysis. Aerobactin was purchased from EMC Microcollections (Tübingen, Germany) and used at a concentration of 1 μg/ml as a control in all bioassays.
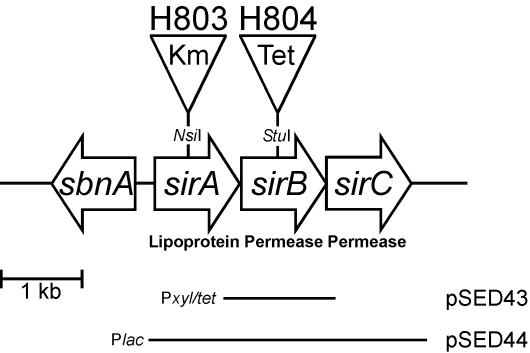
FIG. 1.
Genetic organization of the sbn-sirABC locus. The three open reading frames of the sir operon, as well as the first gene of the sbn operon (sbnA), are indicated. The positions of the insertion sites used to disrupt the sirA and sirB coding regions, generating strains H803 and H804, respectively, in the S. aureus Newman background are shown. Plasmids pSED43 and pSED44, used for complementation of sirB::Tet and sirA::Km mutations, respectively, are shown.
55Fe transport assays.
S. aureus strains were grown to late logarithmic phase in TMS containing 100 μM 2,2′-dipyridyl with or without 50 μM FeCl3. The cells were washed twice with TMS over a 0.45-μm-pore-size filter (Gelman) and normalized to an optical density at 600 nm of 1.2. Twenty minutes prior to the assay, 55FeCl3 (75 μM) was mixed with ∼220 μM staphylobactin (calculated from Desferal equivalents) in the presence of 2 μM nitrilotriacetic acid and allowed to equilibrate at room temperature. Uptake was initiated by adding 10 μl of the 55Fe-staphylobactin mixture to 1-ml volumes of cells. At various time points, 200 μl of cells was removed and washed twice with 100 mM LiCl over a 0.45-μm-pore-size membrane. The membranes were dried and counted in CytoScint fluid using the tritium channel of a Beckman LS 6500 scintillation system. In some experiments, S. aureus was treated with 10 mM potassium cyanide (KCN) at room temperature for 20 min prior to the addition of the 55Fe mixture. The data are presented as picomoles of 55Fe transported normalized to the total protein content of the cells (± standard deviation) as determined by Bradford assays.
Transcriptional sbnH::lacZ fusions and β-galactosidase assays.
Construction of an sbnH::lacZ transcriptional fusion has been previously described (7). This fusion was transduced into Newman, H803, and H804 genetic backgrounds, and the presence of the gene fusion was confirmed by PCR. For quantitation of β-galactosidase expression from S. aureus, cells were grown to an optical density at 600 nm of 0.8 in TMS supplemented with 100 μM 2,2′-dipyridyl and assayed as previously described (7).
Purification of SirA and generation of anti-SirA antisera.
We expressed SirA, lacking the signal peptide, in E. coli ER2566 by cloning sirA, amplified from the genome of S. aureus using primers pSirA(BamHI) and pSirA(EcoRI), into pGEX-2T-TEV digested with EcoRI and BamHI. Cells containing this expression construct, named pSirA, were grown to mid-log phase before being induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h. The cells were lysed using a French press, and the lysate was centrifuged at 120,000 × g to pellet cell debris. The supernatant was applied to a GSTrap (Amersham Biosciences) column equilibrated with phosphate-buffered saline, and the glutathione S-transferase (GST)-SirA fusion protein was eluted with 10 mM reduced glutathione in 50 mM Tris-Cl, pH 8.0. SirA was cleaved from GST by incubation with tobacco etch virus protease for 3 h at room temperature and dialyzed overnight at 4°C against 50 mM Tris-Cl, pH 8.0. SirA was further purified using a Mono S column (Amersham Biosciences) equilibrated with sodium phosphate buffer, pH 7.0, and the protein was eluted in sodium phosphate buffer containing 1 M NaCl. The purity of SirA was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Antibodies recognizing SirA were generated in New Zealand White rabbits (Charles River) inoculated subcutaneously with 500 μg of SirA emulsified in 100 μl of Freund's complete adjuvant. On days 14 and 28, the rabbits received booster injections of 100 μg of SirA emulsified in Freund's incomplete adjuvant. The rabbits were sacrificed 10 days after the second boost. Antisera were adsorbed against H306 cell lysates and used at a 1:2,000 dilution for Western blots.
RESULTS AND DISCUSSION
sirABC is divergently transcribed from sbnABCDEFGHI on the S. aureus chromosome.
Previous work described the S. aureus sbn operon, an operon containing genes whose products function in the production of the siderophore staphylobactin (7). Transcribed in the direction opposite to the sbn operon is the sirABC operon (Fig. 1), which has also been described (14). Examination of the genome sequences of seven S. aureus strains, MW2 (2), N315 (18), Mu50 (18), COL (The Institute for Genomic Research), NCTC8325 (University of Oklahoma), MRSA252 (Sanger Centre), and MSSA476 (Sanger Centre), identified the sirABC operon in all of the strains, whereas the operon was absent from the genome of S. epidermidis RP62A (The Institute for Genomic Research). SirA was characterized as a lipoprotein that was expressed during growth under iron starvation conditions (14), while the products of SirB and SirC are predicted to be membrane embedded and therefore likely constitute the transmembrane domains of an ABC transporter (23). Atypically for genetic loci encoding ABC transporters, no gene encoding an ATP binding component was identified in the vicinity of the sirABC locus. However, this is not unprecedented, since the fatDCBA operon required for ferric anguibactin transport in Vibrio anguillarum also lacks an ATP binding protein (17). As previously reported (14), the deduced SirA, SirB, and SirC proteins show significant similarity to ferrisiderophore transport proteins, most notably with the CbrA (61%), CbrB (54%), and CbrC (61%) proteins, respectively, in Erwinia chrysanthemi 3937. In E. chrysanthemi, the cbr locus encodes proteins involved in iron internalization by the bacterium via achromobactin (20), a siderophore that is structurally uncharacterized.
Expression of SirA is iron regulated via Fur.
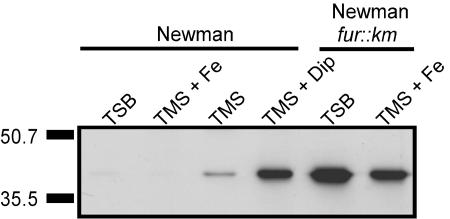
Although SirA expression was undetectable in S. aureus Newman cultured in iron-replete medium (either TSB or TMS containing 50 μM FeCl3), its expression was readily detectable during growth under conditions of iron restriction (Fig. 2). Expression levels increased as the level of iron restriction increased (i.e., when 2,2′-dipyridyl was added to TMS) (Fig. 2). These findings are in agreement with previous studies that used S. aureus 8325-4 (14). It has been further demonstrated that SirA expression was controlled by the activity of the Fur protein in S. aureus, since SirA expression was no longer iron regulated in a Fur-deficient background (Fig. 2). This finding is in agreement with the predicted presence of a consensus Fur box upstream of sirA (7, 14).
FIG. 2.
Iron- and Fur-regulated expression of SirA. S. aureus Newman and its fur::Km derivative were grown in either iron-rich (TSB and TMS + Fe) or iron-restricted (TMS and TMS + Dip) medium, normalized by optical density, and lysed. SirA was detected in cell lysates with rabbit polyclonal antiserum directed at SirA. Molecular mass markers are shown on the left in kilodaltons.
SirA and SirB are involved in iron acquisition.
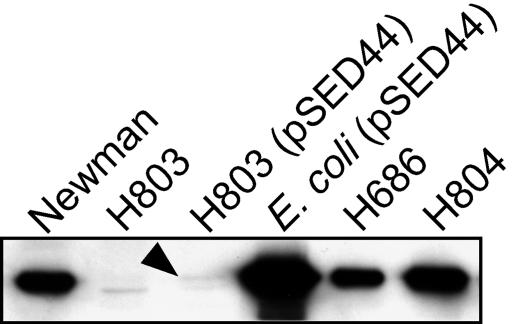
To address the potential role of sirABC in iron acquisition, we used kanamycin and tetracycline resistance cassettes to inactivate the coding regions of sirA and sirB, respectively, in S. aureus RN6390. We transduced the mutations from these strains, designated H306 (S. aureus RN6390 sirA::Km) and H474 (S. aureus RN6390 sirB::Tet), into S. aureus Newman to create the strains H803 (sirA::Km) and H804 (sirB::Tet). While SirA was undetectable in H803, the sirB::Tet mutant expressed wild-type levels of SirA (Fig. 3). A faintly reactive band that migrated faster than SirA was visible in cell extracts of H803 (Fig. 3). This band is likely due to cross-reactivity with another protein due to the use of polyclonal antisera. The protein band is likely masked by the high-level expression of the SirA protein in the other samples shown in Fig. 3, since it was visible when S. aureus Newman was grown under iron-replete conditions (Fig. 2).
FIG. 3.
Expression of SirA in S. aureus Newman and derivatives. Cells were grown in TMS supplemented with 75 μM 2,2′-dipyridyl, normalized by optical density, and lysed. SirA was detected in cell lysates with rabbit polyclonal antiserum directed at SirA. The arrowhead points to low but reproducibly detectable levels of expression of SirA in the complemented mutant.
In previous work, sensitivity to streptonigrin was used as a method to demonstrate the loss or perturbation of iron import in S. aureus (29). Streptonigrin is toxic to cells in the presence of intracellular free iron, and therefore, cells importing iron are generally more sensitive to the toxic effects of the drug than are mutants debilitated in iron import (36). The MIC of streptonigrin was ∼4-fold lower for S. aureus Newman grown in TMS than for either S. aureus H803 or H804 grown in the same medium (Table 2). Different susceptibilities to streptonigrin were overcome by the inclusion of Desferal in the growth media, indicating that this siderophore was used equally well by parent and mutants. These data indicated that SirA and SirB were likely involved in the transport of iron into the cell. As further evidence of this, we demonstrated that the MIC of 2,2′-dipyridyl (a nonmetabolizable iron chelator) for S. aureus Newman was fourfold higher than for either H803 or H804 (Table 2).
TABLE 2.
MICs of streptonigrin and 2,2′-dipyridyl against S. aureus Newman and derivatives
| Bacterial straina | MIC
|
|
|---|---|---|
| Streptonigrin (ng/ml) | 2,2′-dipyridyl (μM) | |
| Newman | 2 | 500 |
| H803 | 8 | 125 |
| H804 | 8 | 125 |
| Newman + 50 μM Desferal | 2 | NDb |
| H803 + 50 μM Desferal | 2 | ND |
| H804 + 50 μM Desferal | 2 | ND |
Bacteria were grown in TMS.
ND, not determined.
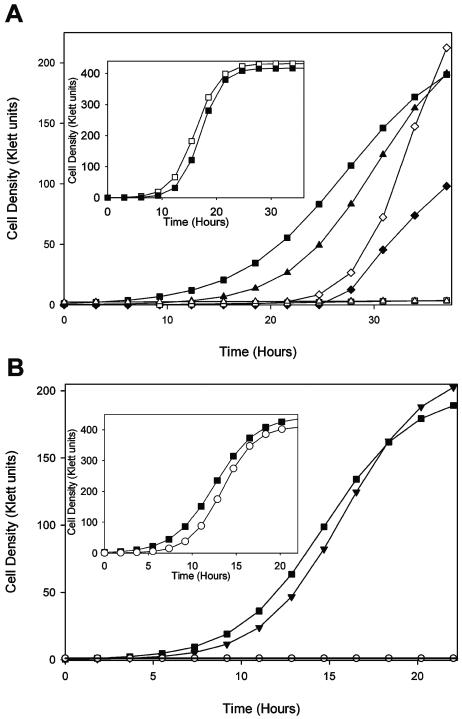
Growth of wild-type Newman and derivatives was followed over 24 to 36 h in order to identify deficiencies in the growth rate that correlated with the loss of sirA or sirB function. In iron-replete growth media, the growth of H803 (Newman sirA::Km) was unaltered in comparison to that of Newman (Fig. 4A, inset). H803, however, showed a drastic growth deficiency compared to Newman in iron-restricted growth media (Fig. 4A). Introduction of the plasmid pSED44 (containing the sirABC operon expressed from Plac) (Fig. 1) into H803 corrected the growth deficiency of this strain in iron-restricted growth media. The Plac promoter in the pAW8 vector expresses large quantities of SirA from the pSED44 construct in E. coli but extremely little SirA in S. aureus, even in the presence of 1 mM IPTG (Fig. 3), indicating that Plac is extremely inefficient in S. aureus. However, even this small amount of SirA was capable of complementing the mutation in H803. We found that addition of 1 mM IPTG to the iron-deficient growth media did enhance complementation (Fig. 4). Introduction of vehicle alone into either Newman or H803 did not affect the growth rate (Fig. 4).
FIG. 4.
Comparison of the growth of S. aureus Newman versus a sirA::Km mutant derivative (A) or a sirB::Tet mutant derivative (B) in TMS broth containing 250 μM 2,2′-dipyridyl and 50 μM FeCl3 (insets) or 250 μM 2,2′-dipyridyl. ▪, Newman; □, H803 (sirA::Km); ▴, Newman carrying pAW8 vector; ▵, H803 carrying pAW8; ♦, H803 carrying pSED44 grown without IPTG; ⋄, H803 carrying pSED44 grown with 1 mM IPTG; ○, H804 (sirB::Tet); ▾, H804 carrying pSED43. The data are representative of three experiments.
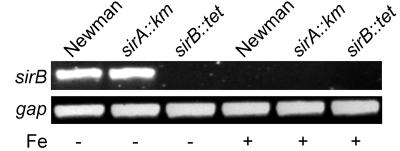
Since we were unable to complement the sirA::Km mutation with the sirA coding region alone (see below), we considered the possibility of polarity of the sirA::Km mutation on expression of downstream sir genes. RT-PCR experiments demonstrated that the sirA::Km mutation had no effect on the expression of sirB (Fig. 5) while also confirming that expression of the sirB transcript was regulated by iron concentrations in the growth medium.
FIG. 5.
Expression of sirB is not affected by the insertion of the Km cassette in sirA. RT-PCR was used to identify transcripts of sirB (and gap as an internal control) in S. aureus Newman, H803 (Newman sirA::Km), and H804 (Newman sirB::Tet) grown in iron-deficient (−) or iron-replete (+) medium. No product was detected in H804, the strain containing the sirB::Tet mutation, grown under iron starvation conditions, since the Tet cassette disrupts the region amplified in the PCR. Total RNA (500 ng) was reverse transcribed, and the cDNAs for sirB and gap were amplified as described in Materials and Methods.
Similar to H803, the growth of H804 (Newman sirB::Tet) in iron-replete media was unaltered in comparison to that of wild-type Newman (Fig. 4B, inset). However, as with H803, the growth of H804 was severely impaired in iron-deficient growth media compared to that of S. aureus Newman (Fig. 4B). This growth deficiency of H804 was alleviated with the introduction of pSED43 into the strain (Fig. 4B); pSED43 expresses sirB from a xyl/tet promoter (Fig. 1). The xyl/tet promoter was found to be quite leaky in S. aureus (data not shown), and therefore it was unnecessary to incorporate inducer (anhydrotetracycline) into the growth media in these experiments to see full restoration of the wild-type phenotype.
In the absence of sirB and sirC, the sirA gene product is toxic to E. coli.
For complementation of the sirA mutation in S. aureus H803, we initiated experiments to clone the sirA coding region in E. coli before introducing the construct into S. aureus. However, this proved to be extremely problematic and in the end unsuccessful, even after we attempted to use several different vectors and regulated promoter systems, including the iron-regulated sirA promoter (data not shown). These observations indicated to us that leaky expression of even small quantities of this lipoprotein was lethal to E. coli. Several other laboratories have encountered similar problems in cloning genes encoding lipoproteins in E. coli (10, 21, 24, 27). The problems encountered with the cloning of sirA, in the same shuttle vectors that were used to successfully clone the lipoprotein-encoding genes fhuD1 and fhuD2 (28), appear not to be due to the soluble or amphiphilic regions of the protein, since for the generation of anti-SirA antisera we were able to clone sirA lacking the signal peptide into an E. coli expression vector and produce large quantities of soluble SirA. These results lend support to the idea that the problems encountered with cloning sirA may be due to improper processing of the lipoprotein in E. coli, as has been previously suggested with other lipoproteins (21, 27).
Interestingly, the apparently toxic effects of the SirA lipoprotein on E. coli occurred only when we attempted to clone the sirA gene on its own and not when sirB and sirC were included in the cloned DNA. Indeed, the sirABC genes were successfully cloned as a unit on plasmid pSirABC and in pSED44 (Table 1), and the latter plasmid expressed large quantities of SirA in E. coli. This result could suggest that the transmembrane components of the transporter, components that would presumably interact with SirA at the membrane, may help to stabilize the lipoprotein in the membrane.
Mutation of either SirA or SirB results in S. aureus defective in staphylobactin transport but not staphylobactin biosynthesis.
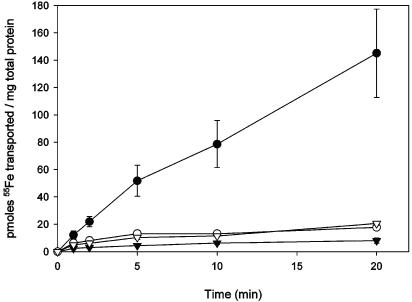
Staphylobactin, isolated from S. aureus RN6390 using previously described techniques (7), was used to assess growth promotion in siderophore plate bioassays. While staphylobactin readily promoted the growth of S. aureus Newman and RN6390 in siderophore plate bioassays, no staphylobactin-mediated growth promotion was observed for H306 (RN6390 sirA::Km), H474 (RN6390 sirB::Tet), H803 (Newman sirA::Km), or H804 (Newman sirB::Tet) (data not shown), indicating that both SirA and SirB are essential for staphylobactin-mediated iron transport. To confirm these results, purified staphylobactin was incubated with 55FeCl3 and transport assays were performed with S. aureus Newman and H803. While significant transport of 55Fe-staphylobactin was observed in S. aureus Newman, virtually no transport occurred in Newman pregrown in TMS containing FeCl3, in Newman treated with 10 mM KCN, or in H803 (Fig. 6). Together, these results confirm that staphylobactin transport is an iron-regulated, energy-dependent process that requires the functions of at least SirA and SirB. Growth promotion by aerobactin, Desferal, and ferric citrate was unaffected in sirA and sirB mutants, and growth in the presence of staphylobactin was restored in the complemented sirA::Km and sirB::Tet mutants (data not shown). We hypothesize that the energy for transport is provided by the hydrolysis of ATP, on the basis that SirA, SirB, and SirC have features of classic ABC transporters. A gene encoding an ATPase component is unlinked with the sirABC operon and may be encoded from elsewhere on the genome, or the SirABC transport system may share an ATPase component with another ABC transport system.
FIG. 6.
Staphylobactin-mediated iron (55Fe3+) transport by S. aureus Newman and H803. Newman (•) and H803 (▿) cultured in TMS containing 100 μM 2,2′-dipyridyl; Newman (▾) supplemented with 50 μM FeCl3; Newman (○) treated with 10 mM KCN. The results shown are the average of three experiments ± standard deviation.
In at least one instance in S. aureus, more than one gene encoding the lipoprotein (or binding protein) component of a transport system is found in the genome. Indeed, the iron-hydroxamate uptake system in many strains of S. aureus is comprised of single copies of genes encoding the ABC transporter components but two genes that encode a binding protein component (e.g., fhuD1 and fhuD2) (28, 30). In strains containing both fhuD1 and fhuD2, mutation of one of the genes leads to a phenotype that is either wild type or very close to wild type for iron-hydroxamate uptake (28). Given that both the sirA::Km and sirB::Tet mutations lead to equivalent phenotypes, we conclude that there is only one gene encoding the binding protein (i.e., SirA) component and one copy of genes (i.e., sirB and sirC) encoding the membrane permease for this transport system.
Given that the functions of proteins expressed from the sbn operon and the sir operon are associated (i.e., biosynthesis and import of staphylobactin), we wished to determine whether there were any effects on their expression as a function of mutations in the operons. Mutation of sbnE results in the loss of staphylobactin synthesis (7); however, we showed that loss of sbnE function and therefore biosynthesis of staphylobactin had no major effect on the expression of SirA (Fig. 3, compare lanes 1 and 5). In corollary experiments, we investigated whether loss of sirA or sirB resulted in loss of, or decrease in, staphylobactin production. We observed that H803 grown in moderately iron-restricted medium produced significant amounts of staphylobactin both by analytical high-performance liquid chromatography and electrospray ionization-mass spectrometry (data not shown). To investigate this phenomenon further, we transduced a transcriptional lacZ-sbnH fusion into Newman, H803, and H804. We observed a significant increase in transcription of the sbnH gene in the H803 and H804 genetic backgrounds compared to that in wild-type Newman (Table 3). No transcription of β-galactosidase activity was observed when strains containing the fusion were grown under iron-replete conditions (data not shown). These results suggest that staphylobactin biosynthesis may be enhanced in strains deficient in the ability to transport the siderophore, presumably in response to an elevated iron starvation status.
TABLE 3.
β-Galactosidase activities from sbnH::lacZ fusions in Newman and derivatives grown in iron-restricted media
| Bacterial strain | Mean β-galactosidase activity ± SD (RLU/s)a |
|---|---|
| Newman | 172 ± 117 |
| H803 | 318 ± 47 |
| H804 | 239 ± 110 |
| H870: Newman sbnH::pMUTIN4 | 825 ± 190 |
| H873: H803 sbnH::pMUTIN4 | 7,036 ± 517 |
| H876: H804 sbnH::pMUTIN4 | 3,667 ± 1,654 |
Values represent the mean values, in triplicate, from assays performed on triplicate cultures. RLU, relative light units.
Conclusions.
The sirABC operon encodes components of an ABC transporter, and the products show similarity to ferrisiderophore transport proteins in other bacteria. The sirABC operon is iron regulated and divergently transcribed from the iron-regulated sbn operon that encodes enzymes involved in the production of the siderophore staphylobactin. The Fur protein controls iron-regulated transcription from both operons. S. aureus sirA and sirB mutants each display a growth deficiency compared to the wild type in iron-restricted growth media and are more resistant to streptonigrin, suggesting that the mutant bacteria internalize less iron than wild-type bacteria under iron starvation conditions. Finally, our results show that the sirABC operon encodes proteins that are required for the import of the staphylobactin siderophore into the S. aureus cell.
Acknowledgments
We thank James Henderson for technical assistance, John K. McCormick and Carole Creuzenet for comments on the manuscript, and Ambrose Cheung for plasmid pALC2073.
This work was supported by operating grant MOP-38002 from the Canadian Institutes of Health Research (CIHR). D.E.H. is the recipient of a CIHR New Investigator Award and, from the Ontario Government, a Premier's Research Excellence Award. S.E.D is the recipient of a Natural Sciences and Engineering Research Council PGS-B graduate scholarship, and M.T.S. is the recipient of a CIHR doctoral award.
REFERENCES
- 1.Archer, G. L. 1998. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26:1179-1181. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Boos, W., and T. Eppler. 2001. Prokaryotic binding protein-dependent ABC transporters, p. 77-114. In G. Winkelmann (ed.), Microbial transport systems. Wiley-VCH, Weinheim, Germany.
- 4.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 5.Chakraborty, T., M. Leimeister-Wächter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courcol, R. J., D. Trivier, M.-C. Bissinger, G. R. Martin, and M. R. W. Brown. 1997. Siderophore production by Staphylococcus aureus and identification of iron-regulated proteins. Infect. Immun. 65:1944-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dale, S. E., A. Doherty-Kirby, G. Lajoie, and D. E. Heinrichs. 2004. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect. Immun. 72:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drechsel, H., S. Freund, G. Nicholson, H. Haag, O. Jung, H. Zähner, and G. Jung. 1993. Purification and chemical characterization of staphyloferrin B, a hydrophilic siderophore from staphylococci. BioMetals 6:185-192. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 10.Gomez, A., D. Ramon, and P. Sanz. 1994. The Bacillus subtilis lipoprotein LplA causes cell lysis when expressed in Escherichia coli. Microbiology 140:1839-1845. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths, E., and P. Williams. 1999. The iron-uptake systems of pathogenic bacteria, fungi and protozoa, p. 87-212. In J. J. Bullen and E. Griffiths (ed.), Iron and infection, 2nd ed. John Wiley and Sons, Ltd., New York, N.Y.
- 12.Guérout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 13.Haag, H., H. P. Fiedler, J. Meiwes, H. Drechsel, G. Jung, and H. Zähner. 1994. Isolation and biological characterization of staphyloferrin B, a compound with siderophore activity from staphylococci. FEMS Microbiol. Lett. 115:125-130. [DOI] [PubMed] [Google Scholar]
- 14.Heinrichs, J. H., L. E. Gatlin, C. Kunsch, G. H. Choi, and M. S. Hanson. 1999. Identification and characterization of SirA, an iron-regulated protein from Staphylococcus aureus. J. Bacteriol. 181:1436-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konetschny-Rapp, S., G. Jung, J. Meiwes, and H. Zähner. 1990. Staphyloferrin A: a structurally new siderophore from staphylococci. Eur. J. Biochem. 191:65-74. [DOI] [PubMed] [Google Scholar]
- 16.Köster, W., and V. Braun. 1990. Iron(III) hydroxamate transport into Escherichia coli. Substrate binding to the periplasmic FhuD protein. J. Biol. Chem. 265:21407-21410. [PubMed] [Google Scholar]
- 17.Köster, W. L., L. A. Actis, L. S. Waldbeser, M. E. Tolmasky, and J. H. Crosa. 1991. Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum 775. J. Biol. Chem. 266:23829-23833. [PubMed] [Google Scholar]
- 18.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumura, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 19.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5-13. [DOI] [PubMed] [Google Scholar]
- 20.Mahe, B., C. Masclaux, L. Rauscher, C. Enard, and D. Expert. 1995. Differential expression of two siderophore-dependent iron-acquisition pathways in Erwinia chrysanthemi 3937: characterization of a novel ferrisiderophore permease of the ABC transporter family. Mol. Microbiol. 18:33-43. [DOI] [PubMed] [Google Scholar]
- 21.Martin, B., G. Alloing, C. Boucraut, and J. P. Claverys. 1989. The difficulty of cloning Streptococcus pneumoniae mal and ami loci in Escherichia coli: toxicity of malX and amiA gene products. Gene 80:227-238. [DOI] [PubMed] [Google Scholar]
- 22.Morrissey, J. A., A. Cockayne, P. J. Hill, and P. Williams. 2000. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect. Immun. 68:6281-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikaido, H., and J. A. Hall. 1998. Overview of bacterial ABC transporters. Methods Enzymol. 292:3-20. [DOI] [PubMed] [Google Scholar]
- 24.Pearce, B. J., A. M. Naughton, and H. R. Masure. 1994. Peptide permeases modulate transformation in Streptococcus pneumoniae. Mol. Microbiol. 12:881-892. [DOI] [PubMed] [Google Scholar]
- 25.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Schneider, R., and K. Hantke. 1993. Iron-hydroxamate uptake systems in Bacillus subtilis: identification of a lipoprotein as part of a binding protein-dependent transport system. Mol. Microbiol. 8:111-121. [DOI] [PubMed] [Google Scholar]
- 28.Sebulsky, M. T., and D. E. Heinrichs. 2001. Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J. Bacteriol. 183:4994-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sebulsky, M. T., D. Hohnstein, M. D. Hunter, and D. E. Heinrichs. 2000. Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J. Bacteriol. 182:4394-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sebulsky, M. T., B. H. Shilton, C. D. Speziali, and D. E. Heinrichs. 2003. The role of FhuD2 in iron(III)-hydroxamate transport in Staphylococcus aureus. Demonstration that FhuD2 binds iron(III)-hydroxamates but with minimal conformational change and implication of mutations on transport. J. Biol. Chem. 278:49890-49900. [DOI] [PubMed] [Google Scholar]
- 31.Sprencel, C., Z. Cao, Z. Qi, D. C. Scott, M. A. Montague, N. Ivanoff, J. Xu, K. M. Raymond, S. M. Newton, and P. E. Klebba. 2000. Binding of ferric enterobactin by the Escherichia coli periplasmic protein FepB. J. Bacteriol. 182:5359-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 33.Weinberg, E. D. 1995. Acquisition of iron and other nutrients in vivo, p. 79-93. In J. A. Roth, C. A. Bolin, K. A. Brogden, F. C. Minion, and M. J. Wannemuehler (ed.), Virulence mechanisms of bacterial pathogens. American Society for Microbiology, Washington, D.C.
- 34.Weinberg, E. D. 1984. Iron withholding: a defense against infection and neoplasia. Physiol. Rev. 64:65-102. [DOI] [PubMed] [Google Scholar]
- 35.Winkelmann, G. E. 1991. Handbook of microbial iron chelates. CRC Press, Boca Raton, Fla.
- 36.Yeowell, H. N., and J. R. White. 1982. Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrob. Agents Chemother. 22:961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]