ABSTRACT
It is important to study the neural connectivities and functions in primates. For this purpose, it is critical to be able to transfer genes to certain neurons in the primate brain so that we can image the neuronal signals and analyze the function of the transferred gene. Toward this end, our team has been developing gene transfer systems using viral vectors. In this review, we summarize our current achievements as follows. 1) We compared the features of gene transfer using five different AAV serotypes in combination with three different promoters, namely, CMV, mouse CaMKII (CaMKII), and human synapsin 1 (hSyn1), in the marmoset cortex with those in the mouse and macaque cortices. 2) We used target‐specific double‐infection techniques in combination with TET‐ON and TET‐OFF using lentiviral retrograde vectors for enhanced visualization of neural connections. 3) We used an AAV‐mediated gene transfer method to study the transcriptional control for amplifying fluorescent signals using the TET/TRE system in the primate neocortex. We also established systems for shRNA mediated gene targeting in a neocortical region where a gene is significantly expressed and for expressing the gene using the CMV promoter for an unexpressed neocortical area in the primate cortex using AAV vectors to understand the regulation of downstream genes. Our findings have demonstrated the feasibility of using viral vector mediated gene transfer systems for the study of primate cortical circuits using the marmoset as an animal model. © 2016 Wiley Periodicals, Inc. Develop Neurobiol 77: 354–372, 2017
Keywords: marmoset, virus vectors, neocortex
INTRODUCTION
The neocortex has emerged specifically in mammals during the course of the evolution of their brain. The neocortex has six layers common to all mammals. In this regard, it shares features common to the Mammalian class. However, it also shows marked diversity. Among mammalian brain structures, the variation in size is most prominent in the neocortex. The size of the mammalian brain varies from the Etruscan Shrew (the smallest nonflying mammal) to the killer whale on the order of 104‐fold difference. The brain size is approximately proportional to the surface area of the neocortex with the same order of difference (Stephan et al., 1986; Jerison, 2001). The mammalian brain consists of parcellations, so called areas, each of which reflects a specific unit of neuronal projections (Brodmann, 1909). Areas are formed by different intermediate levels of structures called columns, modules and domains, which are different among the different regionalization of the neocortex (Kaas, 2012). For example, not only increase in the size and volume of the prefrontal cortex but also the rich connections with other brain regions underlie the richness of human cognition (Fuster, 2015). The neocortex is the most evolved structure in primates compared with other animals (Brodmann, 1909), in a different way from ancestral mammals (Kaas, 2013). To understand the underlying mechanisms of brain functions, it is critically important to understand the connections and functions underlying neuronal activities and behavioral responses. Traditionally, this set of studies has been conducted in terms of neuroanatomy and neurophysiology. Recently, multidisciplinary approaches that include molecular biology, biochemistry, cell biology, and genetic engineering have been integrated to neuroscience with the traditional anatomical and physiological approaches (Kandel et al., 2012). We are interested in developing gene manipulation methods that can be used for the primate brain. Since biological materials such as DNA, RNA, and proteins are common properties among species, in principle the techniques that are applied to one species can also be applied to other species. Given that certain cortical structures and functions of the primate brain differ from those in other mammals (Nieuwenhuys, 1994; Fuster, 2015), it is important to develop techniques to study the primate neocortex. The pursuit of developing such techniques has been hampered because the reproduction system is different between mice and primates and, even though it becomes recently possible to generate transgenic and knockout primates (Sasaki et al., 2009; Niu et al., 2014), the number of genetically engineered animal models is still limited.
The recent availability of genetically engineered primates means that methods involving viral vector mediated gene transfer have increasingly become important because they can be used in combination with other methods using genetically engineered transgene lines, which mutually enhances markedly their advantages (Nassi et al., 2015a). Over the last several years, our laboratory has focused on developing viral vector systems for gene transfer. Because of their efficacy and low toxicity, we considered that Adeno‐associated virus (AAV) vectors is the best among the possible choices available (Nassi et al., 2015b; El‐Shamayleh et al., 2016). In addition, retrograde projection targeting has several specific purposes (EL‐Shamayleh et al., 2016). Since LV vector mediated retrograde transduction is more efficient than AAV vector mediated transduction in macaques, we used lentiviral (LV) vectors containing fusions of the extracellular domain of the rabies glycoprotein and the intracellular domain of the VSV glycoprotein for retrograde labeling (Kato et al., 2013).
Here, we review our recent approaches using viral vector mediated gene transfer systems for the primate brain. Merits of using marmosets in various aspects of research interest have been published (Miller et al., 2016; Homman‐Ludiye and Bourne, in press; Oikonomidis et al., in press; Hagan et al., in press). In this review we focus on the topics that are directly related to our recently reported applications of viral vectors for gene transfer in the primate cortex, particularly in the marmoset brain. There are excellent reviews that have been recently published and cover most topics that we will no longer review. We briefly introduce such reviews. Nassi et al. (2015b) report comprehensively reviewed most of the topics that are relevant to this article including the characteristics of the available viral vectors. AAV may be the most commonly used vector because it is useful for long‐term and high level expression in the CNS of mammals. Moreover, it is straightforward to engineer the virus. Therefore, we used only AAV as our choice for gene transfer purposes to neurons although there is a major limitation in the size of genes incorporated into AAV. To use AAV, there are several remaining issues that should be solved (Nashi et al., 2015b). Among them, the toxicity and cell‐type specificity of promoters are important issues, which we want to focus on in this review on the basis of our recent findings (Watakabe et al., 2014a). Note that there are many options depending on the situation that researchers face, as reviewed by Nashi et al. (2015b) and El‐Shamayleh et al. (2016). El‐Shamayleh et al. (2016) have recently reviewed the strategies for targeting neuronal circuits in primates. This is also very relevant to the topics that we review here in that they specifically review three topics on viral vector tropism, projecting targeting, and targeting neurons at the transcriptional level for the application of viral vector technologies in primates. Furthermore, Callaway and Luo (2015) reviewed more specific topics on monosynaptic circuit tracing with glycoprotein deleted rabies viruses that they have developed.
Comparative Analysis of AAV Serotypes in Mice, Marmosets, and Macaques
As reviewed by Nassi et al. (2015b) and EI‐Shamayleh et al. (2016), AAV is the most common choice among the viral vectors available. However, there are several issues that remain to be solved in using AAV vectors. They include AAV serotypes, promoters, toxicity, and species specificity (Nassi et al., 2015b; EI‐Shamayleh et al., 2016). Nathanson et al. (2009) used an AAV serotype 1 capsid with a serotype 2 ITR backbone packaged serotype (AAV2/1) and lentivirus pseudotyped with the vesicular stomatitis virus (VSV‐G‐LV). They used human synapsin (hSyn) and a mouse CAMKII (CAMKII) promoter. A serial dilution of AAV1/2‐hSyn injected into the mouse somatosensory cortex (S1) shows different ratios of excitatory and inhibitory neurons that are infected. Higher infected titers of AAV1/2‐hSyn tend to infect more excitatory cells whereas lower infected titers of AAV1/2‐hSyn injections show higher rates of inhibitory neuronal infection. The same titers of AAV1/2CAMKII and VSV‐G‐LV‐hSyn yield high ratios of excitatory neuronal infection, whereas similar titers of AAV‐CAG injected yield a high ratio of inhibitory neuronal infection. They also found some neurotoxicity of VSV‐G‐LV‐hSyn. These findings demonstrated that many factors could affect the features of infection. We considered that it is important to determine the factors that influence the efficacies of gene transfer and tropism. To this end, we compared the infectious properties of five different capsids of AAV serotypes 1, 2, 5, 8, and 9 in marmosets as well as in mice and macaques. Initially, we used hrGFP under the control of the CMV promoter with AAV2 ITRs as the construct for capsid comparison (Cearley and Wolf, 2006; Taymans et al., 2007; Nassi et al., 2015b). We used neuron‐specific CaMKII, and hSyn1 promoters followed by hrGFP (Fig. 1). Each of the five serotypes showed differences in viral spread and cell tropism. Among the five serotypes, AAV2 showed the least spread of GFP; after the injection of 2.5 × 10e9 vector genome copies of AAV in 0.5µl PBS over 5 min, we observed an AAV2 spread of approximately 0.7 mm, whereas the spread of other serotypes was 1.2–1.3 mm. We observed no statistically significant differences in viral spread among AAV serotypes of 1, 5, 8 and 9. Regarding cell tropism, AAV2 exhibited a significant difference in neuronal tropism even with the CMV promoter in the three species tested. Other serotypes exhibited more or less glial expressions that obscured neuronal expression. For this reason, we employed neuron‐specific CaMKII and hSyn1 promoters to compare neuronal transduction efficiency among serotypes. We estimated the efficacy of infection of neurons by comparing between hrGFP‐ and NeuN‐positive cells. The efficacy of infection by injection with the CaMKII promoter was over 70% in all the serotypes. Thus, at least under our conditions, we observe no significant differences among AAV1, 5, 8, and 9, except that the neuronal transgene expression level was slightly lower in AAV5. We also observed no significant differences across species, although the infected glial cells exhibited different morphologies and may have species‐specific glial tropism.
Figure 1.
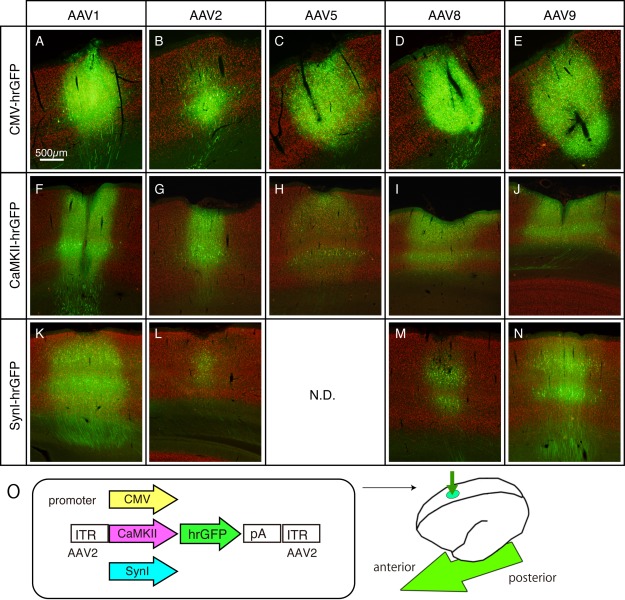
Various AAVs injected into marmoset cerebral cortex. AAV constructs containing CMV, CaMKII, and hSynI promoters driving hrGFP expression were packaged in AAV1, 2, 5, 8, and 9 capsids and injected into the marmoset cerebral cortex. The low‐magnified images of representative sections are shown for each combination. These are merged views for hrGFP (green) and NeuN (red) signals. To aid visualization, the contrasts of images are adjusted. So that we can directly compare the signal intensities across serotypes, the sections for the same promoters were immunostained simultaneously, imaged and contrast‐adjusted in exactly the same conditions. These images were taken from two marmosets of ID#1 and ID#2 for CMV promoter (panels A–E), ID#5 for CaMKII promoter (panels F–J) and ID#4 for hSynI promoter (panels K–N). Panel O shows the schematic views of the AAV constructs and injections. The arrow indicates the anterior–posterior axis. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article sited <www.elsevier.com/locate/neures) (From Figure 1 of Watakabe et al., Neurosci. Res., 2014a, 93, 144–157). [Color figure can be viewed at wileyonlinelibrary.com]
Regarding the choice of promoters, we found strong toxic effects when AAV with CMV‐GFP constructs were injected. The toxicity was estimated by comparing the signal intensities of GFP (reporter), NeuN (neuron), and Hoechst33342 (nuclear staining) 3 and 6 weeks after the AAV injection. Cells infected CMV‐GFP showed a low signal intensity of GFP and almost no NeuN expression 6 weeks after infection and a high signal intensity of Hoechst33342, which suggest the degeneration of neurons and infiltration of reactive glia cells. No AAV toxicity was observed when the CaMKII and hSyn1 promoters were used. We, thus, recommend the use of these neuron‐specific promoters for long‐term experiments.
Another important aspect to consider when selecting promoters is the cell‐type specificity of transgene expression. Consistent with the previous reports by Nathanson et al. (2009) and Scheyltjens et al. (2015), we found that for both CaMKII‐and hSyn1‐GFP constructs, GFP was expressed in both parvalubumin‐negative and ‐positive cells, which are a major subclass of GABAergic neurons (Nathanson et al. 2009, Scheyltjens et al. 2015). For selective expression in excitatory neurons, a 0.4 kb version of the CaMKII promoter may be used (Scheyltjens et al. 2015; Gerits et al., 2015). It was conspicuous in the marmoset cortex that large pyramidal cells in layers 3 and 5 tend to strongly express the transgene, regardless of the promoter or serotype used. Expression of the CaMKII construct appears to be stronger than that of the Syn‐1 construct in small to medium pyramidal neurons.
There have been many studies on rodents and nonhuman primates in which different AAV serotypes have been tested (see Tables 1, 2, 3). The results of such studies were not always consistent. For example, we and others found glial expression of the CMV promoter for AAV8 and AAV9, whereas others reported neuronal tropism for these serotypes (Masamizu et al. 2011; Aschauer et al., 2013; Scheyltjens et al., 2015). We also found a good spread of AAV1 as opposed to a report showing a limited spread of this serotype (Scheyltjens et al. 2015). This discrepancy suggests a lack of understanding of the parameters that determine the physical and biological spread of AAV viral particles and how they transduce neuronal and non‐neuronal cells. Although we have successfully used AAV1 in marmosets (Sadakane et al., 2015a, 2015b), other serotypes can also work in combination with appropriate promoters, as our results demonstrated. AAV serotypes of 2, 5, 8, and 9 can be used in nonhuman primates (Diester et al., 2011; Cavanaugh et al. 2012; Jazayeri et al., 2012; Ohayon et al., 2013; Nassi et al., 2015a; Afraz et al., 2015; Inoue et al., 2015; MacDougall et al., in press; Matsuzaki et al., in press; Klein et al., 2016; Galvan et al., 2016). We expect that our studies would provide a basis for choosing an appropriate combination of a promoter and an AAV serotype for the marmoset brain depending on the experimental purpose. We have experience of using AAV vectors for other brain areas (e.g., thalamus and striatum), which gives some difference between different AAV serotypes. However, we have not systematically analyzed the serotype difference, which remains to be studied in future.
Table 1.
Summary of AAV Serotpye Application in Mammalian Brains (mice)
| <mice and rats> | AAV1 | AAV2 | AAV3 | AAV4 | AAV5 | AAV6 | AAV6.2 | AAV7 | AAV8 | AAV9 | AAVrh10 | AAVDJ | AAVDJ8 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cortex | CMV/CAG | 1, 2, 3, 4, 9 | 1, 4, 9 | 9 | 9 | 1, 2, 4, 9 | 4, 9 | 2 | 1, 2, 4, 9 | 1, 2, 4 | ||||
| CaM(0.4/1.3) | 1, 2, 3 | 1, 4 | 1, 2 | 2 | 1, 2 | 1, 2 | ||||||||
| Syn | 1, 2 | 1, 4 | 1 | |||||||||||
| striatum | CMV/CAG | 4, 11 | 4, 11 | 4, 11 | 4 | 11 | 4, 11 | 4 | ||||||
| Syn | 4 | |||||||||||||
| hippocampus | CMV/CAG | 4 | 4 | 4, 7 | 4 | 11 | 4, 7, 11 | 4, 7 | ||||||
| red nucleus | CMV/CAG | 5 | 5 | 5 | 5 | 5 | 5 | 5 | ||||||
| substantia nigra | CMV/CAG | 6, 8, 11 | 6, 8, 11 | 6, 8, 11 | 8 | 8, 11 | 6, 8, 11 | 8 | ||||||
| amygdala | CaM(0.4/1.3) | 11 | 11 | 11 | 11 | 11 | 11 | 11 | 11 | 11 |
Table 2.
Summary of AAV Serotype Application in Mammalian Brains (marmoset)
| <marmosets> | AAV1 | AAV2 | AAV3 | AAV4 | AAV5 | AAV6 | AAV6.2 | AAV7 | AAV8 | AAV9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| cortex | CMV/CAG | 1 | 1 | 1 | 1, 11, 12 | 1, 12 | |||||
| CaM(0.4/1.3) | 1 | 1 | 1 | 1 | 1 | ||||||
| Syn | 1 | 1 | 13 | 1 | 1, 13 | ||||||
| mThy1S(TET) | 20 | ||||||||||
| Striatum | CAG | 11, 12 | 12 | ||||||||
| substantia nigra | CAG | 11 | |||||||||
| Cerebellum | Syn‐CMV, MSCV | 22 |
Table 3.
Summary of AAV Serotype Application in Mammalian Brains (macaques)
| <macaques> | AAV1 | AAV2 | AAV3 | AAV4 | AAV5 | AAV6 | AAV6.2 | AAV7 | AAV8 | AAV9 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| cortex | CMV/CAG | 1, 30 | 1, 19 | 1, 28 | 17 | 1, 24 | 1 | ||||
| CaM(0.4/1.3) | 1, 17 | 1 | 1, 17, 23, 27, 28, 29 | 17 | 1, 17 | 1, 17 | |||||
| Syn | 26 | 15, 28 | |||||||||
| hThy1 | 15 | ||||||||||
| Striatum | CMV | 14, 16, 18 | 14, 16 | 14 | 14 | 14, 16, 18 | 14 | 16, 18 | |||
| substantia nigra | CMV | 14 | 14 | 14 | 14 | 14 | 14 | ||||
| LGN | 21 | ||||||||||
| superior colliculus | CAG | 25 |
Previous reports using AAV vectors in mice, marmosets, and macaques are listed. Numerical numbers in each box indicated the papers cited that are shown below the table. Cited papers are found in REFERENCES.
1:Watakabe et al. (2015), 2:Scheyltjens et al. (2015), 3:Nathanson et al. (2009), 4:Aschauer et al. (2013), 5:Blits et al. (2010), 6:McFarland et al. (2009), 7:Carty et al. (2010), 8:Van der Perren et al. (2011), 9: Hutson et al. (2012), 10: Holehonnur et al. (2014), 11: Masamizu et al. (2010), 12: Masamizu et al. (2011), 13: MacDougall et al. (2016), 14: Markakis et al. (2010), 15: Diester et al. (2011), 16: Sanchez et al. (2011), 17: Gerits et al. (2015), 18: Dodiya et al. (2010), 19: Inoue et al. (2015), 20: Sadakane et al. (2015), 21: Klein et al. (2016), 22: Matsuzaki et al. (2016), 23: Yazdan‐Shahmorad et al. (2016), 24: Afraz et al. (2015), 25: Cavanaugh et al. (2012), 26: Jazayeri et al. (2012), 27: Nassi et al. (2015), 28: Ohayon et al. (2013), 29: Ruiz et al. (2013), 30: Stettler et al. (2006).
Tracing Neuronal Connections Using Retrograde Tracing Systems
In an analogous way of Cre‐dependent tracing strategy (see Fig. 1 of Nassi et al., 2015b), we attempted to develop a system with which we can visualize cortical neuronal connections by amplifying the signals using the retrograde lentiviral vector system and TET‐OFF/TET‐ON system. This system consists of two components, namely, the tetracycline transactivator (tTA) under the control of cellular promoters, such as the neuron‐specific human synapsin 1 (hSyn1) promoter, and the tetracycline‐responsive‐element promoter (TRE) upstream of the transgene of interest. When these two transcriptional units are present in the same cell, the transgene expression is highly enhanced and becomes sensitive to tetracycline. We developed new lentiviral constructs that have two promoter units; the hSyn1 promoter regulating tTA and the TRE promoter regulating a GFP or RFP sequence separated by an insulator sequence (cHS4) (Watakabe et al., 2012). This system was packaged into a lentivirus with the fusion glycoprotein of the B type (FuG‐B) that contains a transmembrane domain of RV‐G and a cytoplasmic domain of VSV‐G (Kato et al., 2011a) for retrograde transport. We conducted a series of experiments on mice and applied this system to a marmoset. As shown in Figure 2, when this retrograde vector was injected into the left hemisphere of the marmoset brain, neuronal cell bodies were clearly identified in the ipsilateral and contralateral sides under a confocal microscope, which are considered the retrogradely infected neurons. When the same vector was injected into V1 of the marmoset (Fig. 3), fine subcellular structures of V2 neurons that send feedback projections to V1 were observed under a confocal microscope.
Figure 2.
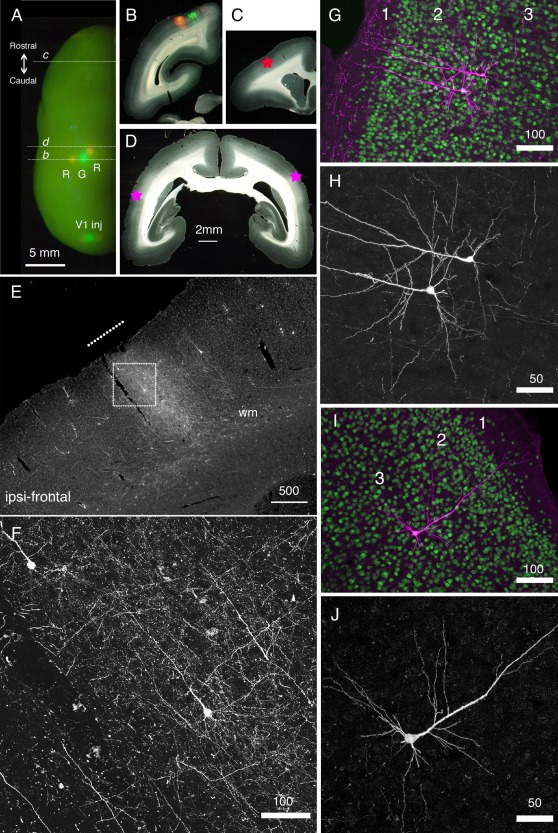
Long‐distance cortical connection of the marmoset brain visualized by StTTrR/FuG‐B vector. (A) The left hemisphere of the marmoset brain with cortical vector injections is shown from above. StTTrR/FuG‐B vector was injected into two sites in the parietal areas and two sites in V1 and StTTrG/FuG‐B vector was injected into one site in the parietal area and one site in V1, as indicated. The injection sites were visualized by LED illumination. (B–D) The coronal sections at the positions indicated in panel A. The fluorescent image is overlaid on the dark field image of the section in panel B. The asterisk in panel C shows the position of the image shown in panel E. The asterisk in the left side of the section in panel D corresponds to the ipsilateral side. (E) The frontal cortex ipsilateral to the vector injection. At this plane, the forward projection fibers of RFP were visible (shown by a white line in panel E), together with retrogradely labeled cell bodies. (F) Confocal image of the boxed region in panel E. Dense axonal fibers and dendrites were visible. This is the fluorescence image of RFP with no antibody enhancement. (G–J) Confocal images of the corticocortical neurons. RFP signals are enhanced by immunofluorescence (magenta) and counterstained with NeuN antibody (green) in panels G and I. Only the RFP signals are shown in panels H and J. The two neurons in panels G and H and one neuron in panels I and J are ipsilateral and contralateral to the injection site, respectively. Bar: 5 mm for A; 2 mm for B–D; 500 µm for E; 100 µm for F, G and I. 50 µm for H and J (From Figure 6 of Watakabe et al., PLOS ONE 7, e46157). [Color figure can be viewed at wileyonlinelibrary.com]
Figure 3.
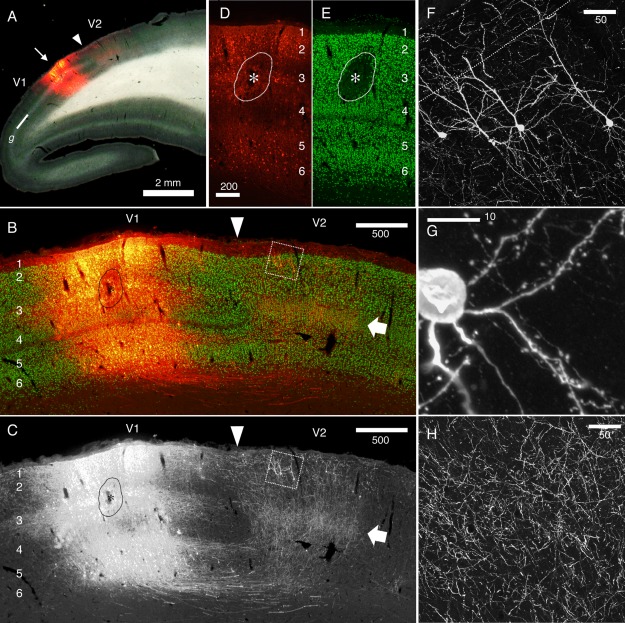
Reciprocal connectivity of marmoset V1 and V2 visualized by StTTrR/FuG‐B vector. The caudal block of the marmoset brain shown in Figure 7 was parasagittally sectioned to visualize the reciprocal connectivity between V1 and V2. (A) Dark field image overlaid with RFP fluorescence indicates the injection site. The V1/V2 border (shown by the arrowhead) was clearly delineated by the presence of the striate of Gennari (g). The arrow indicates the injection site. (B and C) The RFP signals around the injection site (red) are shown with NeuN conterstaining (green). The arrow indicates the plexus of V1 terminals in layer 4. The boxed region contains the retrogradely labeled V2 neurons with feedback projection to V1, which is magnified in panel F. The asterisk indicates the core of injection, where NeuN expression is lost by local damage. Only the RFP signals are shown in panel C. (D and E) RFP signals (red) and NeuN stains (green) in the injection center. The contrast for the RFP signals is adjusted so that the cell bodies of the infected neurons can be delineated. Note that the lamina positions of the infected cells are restricted mostly to layers 2, 3 and 6. (F–H) Confocal images of RFP in V2. RFP signals are enhanced by immunofluorescence. Panel F shows the cell bodies and dendrites of V2 neurons that project back to V1. Panel G shows the spines of the basal dendrites originating from one of these cells. Panel H shows the plexus of axon terminals concentrated in layer 4 of V2. Bar: 2 mm for A; 500 µm for B and C; 200 µm for D and E; 50 µm for F and H; 10 µm for G (From Figure 8 of Watakabe et al., PLOS ONE 7, e46157). [Color figure can be viewed at wileyonlinelibrary.com]
We also developed a new two‐vector system, with which neurons connecting two regions of interest can be selectively visualized (Watakabe et al., 2014b). For this purpose, two components of the TET‐Off system were separated into the retrograde lentiviral vector and AAV vector. The retrograde vector contained the MSCV promoter followed by tTA, which was pseudotyped with the fusion glycoprotein of the C type (FuG‐C) for neuron‐specific retrograde gene transfer (NeuRet) (Kato et al., 2011b). The AAV vector carried the TRE sequence followed by either TurboFP635(tRFP) or humanized recombinant (hr) GFP. When these vectors were injected into the thalamus and the cortex, we were able to specifically label corticothalamic neurons (Watakabe et al., 2014b). We also successfully applied the hSyn1h promoter followed by the TET‐ON reverse tetracyclin transactivator (rtTV16) as a local infection vector. In this case, TRE was followed by either synaptophysin (SYP) linked to cellulean (CFP) or turboFP635 (tRFP) as a retrograde vector with NeuRet. This system enabled the visualization of the extensive extrinsic and intrinsic axon collaterals of particular projection neuronal types comparably to fine Golgi staining. We used this system mostly in mice; however, may apply it to marmosets.
Transcriptional Controls
Since the principle mechanism of genetic machinary is common throughout all organisms, genetic manipulation is a powerful general tool for studying the phenotypes of cells, neuronall circuits, and behaviors. There are many mechanisms that control gene expression, and these mechanisms can be categorized into DNA replication, RNA transcription, and protein translation for the inheritance and expression of genetic information from DNA (Watson et al., 2013). Among them, “targeting at the transcriptional level” is the most common strategy to control the expression of target genes. Therefore, a number of methods to control gene targeting using viral vectors have been developed and used in combination with genetic lines. Because the above‐mentioned reviews cover most of the current available techniques for this purpose (Callaway and Luo, 2015; Nassi et al., 2015b; El‐Shamayleh et al., 2016), we focus on two methods that we developed for applying to the primate brain: the use of the TET/TRE system to amplify two‐photon signals and the targeting RNA expression of a gene in the primate neocortex.
Application of TET and TRE System to Amplify Fluorescent Signals for Two‐Photon Imaging in Marmoset Cortex
Two‐photon imaging has an advantage in that long‐wavelength light can more deeply penetrate into a tissue specimen than short‐wavelength light, which is used in conventional microscopies such as confocal microscopy. Therefore, two‐photon microscopy have been applied to the study of systems of various organisms including mammalian nervous systems. For example, two‐photon microscopy was a key factor for the advancement of in vivo spine‐imaging (Denk et al.,1990; Denk and Svoboda, 1997). Two‐photon imaging has also been applied to primate brains. For example, there have been studies by two‐photon microscopy of the macaque cortex using virus expression systems (Stettler et al., 2006; Yamahachi et al., 2009). Although these studies revealed axonal structures, they did not show dendrite structures. We wanted to achieve dendrite imaging. For this purpose, we needed to establish two conditions. On one hand, dendritic signals should be strong enough to be observable. We have applied the TET/TRE system to amplify the fluorescence signals of the reporter with double infection, which is used originally in lentiviral vectors in mice (Hioki et al., 2009) of two AAV vector systems in the marmoset cortex. On the other hand, when positive signals are too crowded, dendritic signals are buried under strong cell and axonal signals. To overcome these apparently contradictory constraints, we applied the TET‐Off system carried by two AAV vectors for gene transfer. In this system, tTA was supplied by one AAV vector and TRE‐hrGFP was supplied by another AAV vector. These two vectors were combined at a certain ratio and injected to the marmoset cortex together. As mentioned above, TET‐Off system strongly enhances hrGFP signals. On the other hand, when we titrate the amount of tTA‐carrying AAV, we can make the transduction sparse, while maintaining the hrGFP expression level high. For this purpose, in collaboration with Dr. Kawasaki's group we used the Thy1S promoter to drive tTA expression, which is reported to sparsely label cortical neurons (Ako et al., 2011). We truncated a 5′ region of Thy1S by about 1.3 kb, which is reported to be nonessential (Vidal et al., 1990; Caroni, 1997), owing to the limited size of the inserted AAV vectors. As far as we tested, however, the Thy1S promoter was active in various types of cortical projection neuron including corticocortical, corticothalamic, corticopontine, and corticocollicular neurons in layers 2–6 as well as in inhibitory neurons (Sadakane et al., 2015a; AW, unpublished data). The ratio of Thy1s‐tTA to TRE‐hrGFP was changed from 1/200 to 1/2000. We chose 1/500 because the resulting signals gave the best resolution. By this method, we were able to obtain clear images of dendrites and spines by two‐photon microscopy. For example, we were able to observe the same dendrite and spines 24 h after the first observation (Fig. 4).
Figure 4.
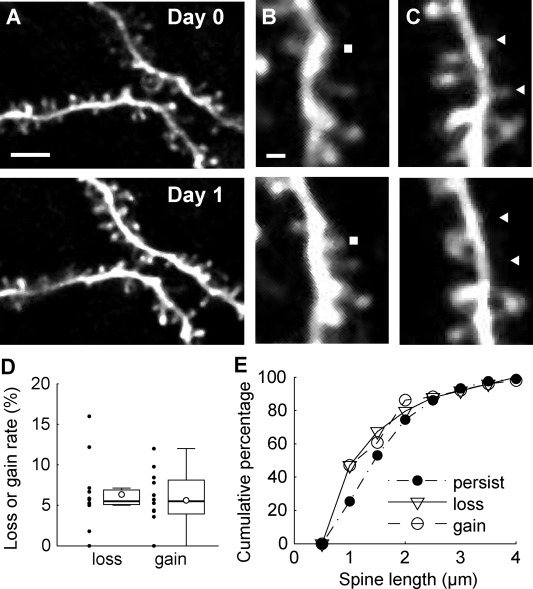
Time‐lapse imaging of spines in prefrontal cortex. A, The same dendritic regions in the prefrontal cortex were imaged at 24 h intervals. The top panel shows an image acquired on day 0 (7 d after craniotomy), and the bottom panel shows an image acquired on day 1. Scale bar, 5 µm. B, The gained spines were identified by manual inspection of two images acquired at 24 h intervals. A filled rectangle indicates the position of an example of a gained spine. Scale bar, 1 µm. C, The same as B for lost spines. Filled triangles indicate the positions of lost spines. D, Box plots showing the spine turnover rate. The open circles in box plots indicate mean values. Black dots indicate values for each site. The whiskers extend to the largest and smallest values within 1.5 times the interquartile range. E, Cumulative distributions of spine length in persisting, gained, and lost populations (From Figure 4 of Sadakane et al., eNeuro, 2015, 2(4), ENEURO.0019‐15).
We then applied the Thy1S‐tTA and TRE systems to observe signals for calcium imaging. There were only limited studies on calcium imaging using genetically engineered calcium indicators (GECIs) in primates. One pioneering work on GECI application to the macaque cortex provided images for months through a chronically implanted window (Heider et al., 2010). However, only several neurons were imaged simultaneously in the same window and it appeared not possible to monitor the same neurons over long periods of time. Synthetic calcium indicator dyes have also been used for detecting the complex structure of macaque V1 (Nauhaus et al., 2012; Ikezoe et al., 2013). However, these synthetic dyes cannot be used for long‐term recording of the activity of neuronal populations and examining fine subcellular structures such as dendritic spines.
We applied the Thy1‐tTA and TRE‐GECI systems by double AAV infections in collaboration with Dr. Matsuzaki's group (Sadakane et al., 2015b; Fig. 5). We used GCaMP6f as a GECI because of its rapid response. Although an AAV vector that contains GCaMP6f driven by the hSyn1 promoter has been successfully used in mouse studies, the same viral preparation that worked in mice failed to exhibit detectable signal in the marmoset brain, even when injected at a very high dose. The application of a TET‐Off amplification system overcame this problem and we were able to observe strong signals of GECI in the marmoset cortex in vivo. Furthermore, these strong signals were controllable with doxycycline (Dox), a derivative of tetracycline. After applying Dox for 5 days (Marmoset A) and 7 days (Marmoset B), a marked decrease of fluorescence signals was observed in 10 and 15 days, respectively. Controlling the expression level is thought to be important to prevent the adverse effects of GECIs on expression (Tian et al., 2009; Chen et al., 2013). Indeed, we were still able to observe both GCaMP6f and NeuN signals 100 days after AAV injection, which confirmed that the expression of these markers in the same cells can be observed for a long period. We also used the TET‐ON system that carried hSyn1‐rtTA and TRE3‐GCaMpf. Without Dox administration, we observed no fluorescence signals, but 3 days after Dox administration we clearly detected epifluorescence. The intensity of signals returned to baseline levels within 2 to 3 weeks after the removal of Dox. We monitored neuronal activities in a broad area (625 × 625 μm2) at depths of 150, 275, and 400 μm. In an experiment, we identified 445 putative neuronal somata, 81 of which were spontaneously active (18/146, 29/168, and 34/131 at depths of 150, 275, and 400 μm, respectively).
Figure 5.
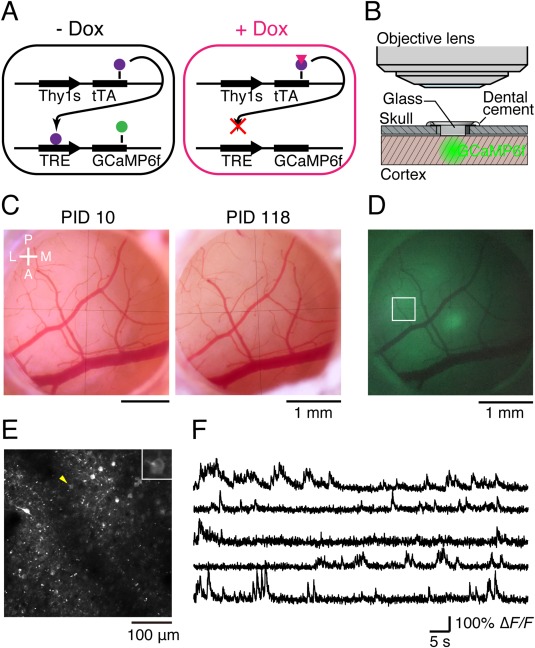
In vivo two‐photon calcium imaging in the marmoset neocortex using the TET‐Off System. (A) Schematic illustration of the TET‐Off gene expression system. tTA (purple circles) activates the TRE3 promoter to amplify GCaMP6f (green circle) expression. Dox (pink triangle) inhibits the binding of tTA to the TRE3 promoter. (B) Schematic illustration of the imaging window. “Glass” indicates four sheets of 3‐mm circular glass coverslips that were adhered to a 5.5‐mm circular glass coverslip. (C) Vasculature images from the same imaging window obtained on postinjection day (PID) 10 and PID 118. A, anterior; L, lateral; M, medial; P, posterior. (D) Epifluorescence image from the same window as shown in (C) on PID 10. The rectangle indicates the field shown in (E). (E) Two‐photon image of GCaMP6f in the rectangle shown in (D). The image was averaged across 4,500 continuous frames. The depth of the image was 250 μm from the cortical surface. The inset is an enlarged view of a single cell (yellow arrowhead). (F) Representative ΔF/F traces from five ROIs in the field shown in (E). The top trace is from the cell shown in the enlarged view in (E) (From Figure 1 of Sadakane et al., Cell Reports, 2015, 13, 1989–1999). [Color figure can be viewed at wileyonlinelibrary.com]
We also detected neuronal activities in the somatosensory cortex evoked by tactile stimulation, attaching vibrators to the arm and leg of marmoset B contralateral to the hemisphere of the imaging window. We identified signals from soma, dendrites, and axonal boutons of neurons which selectively responded upon the stimulation of the leg and arm (Fig. 6). Our results proved that two‐photon microscopic technology is applicable to long‐term studies of neuronal circuits in the marmoset cortex.
Figure 6.
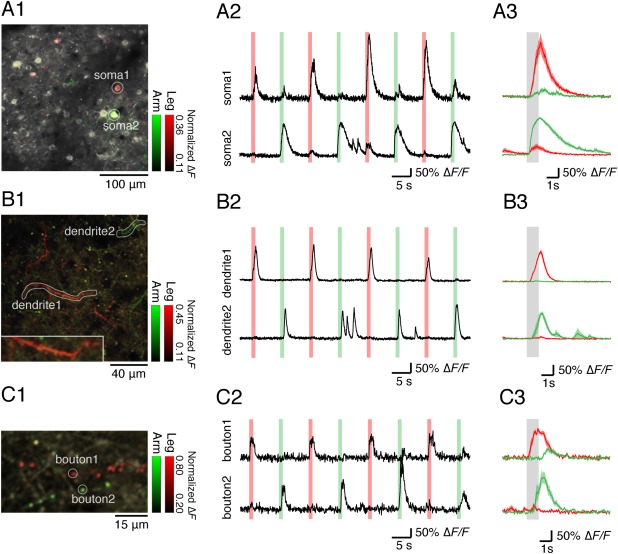
Selective Sensory Responses to Tactile Stimulation from Neuronal Somata, Dendrites, and Axonal Boutons. (A) Sensory responses to tactile stimulation from soma regions in the right somatosensory cortex. The depth of the image is 210 μm. (A1) Representative two‐photon images on which colored stimulus selectivity (green, left arm; red, left leg) is overlaid are shown. In each stimulation condition, the stimulus selectivity for each pixel was defined by dividing ΔF during the stimulation averaged across all trials by the maximum value of the averaged ΔF of all pixels in each field in both conditions. (A2) Representative ΔF/F traces from two neurons indicated in A1 (soma 1 and soma 2) are shown. The green and red lines indicate the timing of tactile stimulation to the left arm and the left leg, respectively. (A3) Averaged ΔF/F traces from soma 1 and soma 2 aligned to the left arm (green) and the left leg (red) stimulation are shown. The responses to nine stimuli to each body part were averaged. The gray band indicates the timing of the left arm and the left leg stimulation. (B) Sensory responses to tactile stimulation from dendrites in the right somatosensory cortex. The depth of the image is 15 μm. (B1) Representative two‐photon images on which colored stimulus selectivity (green, left arm; red, left leg) is overlaid are shown. (B2) Representative ΔF/F traces from two dendrites indicated in (B1) (dendrite 1 and dendrite 2) are shown. (B3) Averaged ΔF/F traces from dendrite 1 and dendrite 2 aligned to the left arm (green) and the left leg (red) stimulation are shown. (C) Sensory responses to tactile stimulation from axonal boutons in the right somatosensory cortex. The depth of the image is 20 μm. (C1) Representative two‐photon images on which colored stimulus selectivity (green, left arm; red, left leg) is overlaid are shown. (C2) Representative ΔF/F traces from two axonal boutons indicated in C1 (bouton 1 and bouton 2) are shown. (C3) Averaged ΔF/F traces from bouton 1 and bouton 2 aligned Cell Reports to the left arm (green) and the left leg (red) stimulation are shown (From Figure 6 of Sadakane et al., 2015, 13, 1989–1999). [Color figure can be viewed at wileyonlinelibrary.com]
There have been reports that applied two‐photon imaging using GECIs to animal species of larger brains than the mouse brain, including the rat (Scott et al., 2013), tree shrews (Lee et al., 2016), and ferrets (Smith et al., 2015). These studies used Synapsin promoter to drive the expression of GECIs similarly to the case of mouse, but in our hands, it was not possible to observe strong signals for two photon images in the marmoset cortex. We consider that this is because the marmoset brain is more opaque. It seems that at least three parameters, promoter activity, sensitivity of the calcium indicator, and the resolution of two‐photon microscopy are critical to obtaining good images and may be different between tissues. The best two‐photon images can be obtained improving each parameter and determining the optional combination of these parameters.
Optogenetics increasingly becomes an important technology used in the study of neural circuits (Fenno et al., 2011). The important issue related to two‐photon imaging is the optogenetical application to the primate cortex using viral vectors. Because these topics on the primate brain is well summarized in the review written by El‐Shamayleh et al. (2016), we will not repeat these in this review. However, it is worth noting that neuronal activities can now be successfully obtained by optogenetical techniques in awake marmosets using AAV vectors (MacDougall et al., in press).
Controlling Expression of a Specific Gene in a Region Specific Manner in the Primate Brain
Genetic knock out by homologous recombination is a powerful and standard method of altering the genome by homologous recombination (Capecchi, 1989), using conditional knock out technology (Lakso et al., 1992; Gu et al., 1994). Large scale resources are now available for obtaining appropriate target genes for conditional knock mice with pipelines (Skarnes et al., 2011). However, the situation is quite different for primates. Even though genetically engineered primate lines are generated, we have not reached the stage where we can knock out a particular gene in a region specific manner in primates. Therefore, we aim to establish a general method to manipulate any targeted genes. We therefore tested the methods to manipulate genes using gain‐of‐function and loss‐of‐function strategies. For this purpose, we chose a gene that controls other genes with high expression levels in representative brain areas. These area‐selectively expressed genes were identified by large‐scale screenings using differential display (DD) and restriction landmark cDNA scanning (RLCS) methods and classified into two groups: genes highly expressed in the primary visual cortex and those highly expressed in association areas (Yamamori, 2011). We first identified one of the V1‐selective genes as occipital 1 (OCC1), which was highly expressed in the primary visual cortex (Tochitani et al., 2001). Its function had long been unclear until a study of its role in the rodent dorsal root ganglion (DRG) revealed that follistatin‐like 1 (FSTL1) (mouse homolog of OCC1) is a Na‐K‐ATPase and controls afferent‐dependent synaptic transmission of sensory inputs to motor neurons (Li et al., 2011). Note that the expression of OCC1/FSTL1 in the primate visual cortex is activity‐dependent, whereas none of the examined brain areas show activity dependency of OCC1/FSTL1 (Takahata et al., 2006, 2008).
Although the molecular features of genes of each group show no apparent correlation, their expression patterns are very similar. This observation raised the possibility that there may be a common mechanism that controls the expression of these genes. To confirm this possibility, we examined the methylation of CpG islands in the promoter region of the area‐selective genes (Hata et al., 2013). The methylation levels of CpG islands of the promoter of V1‐selective genes (HTR1B, HTR2A, and OCC1/FSTL1 genes) are low (Tochitani et al., 2001; Watakabe et al., 2009) and those of association area‐selective genes (RBP4, PNMA5, and SLIT1 genes) are high (Komatsu et al., 2005; Takaji et al., 2009; Sasaki et al., 2010). However, there was no significant difference in the methylation level of each gene among brain areas (prefrontal area and V1). If methylation indeed plays any role in controlling the expression of area‐selective genes, there should be a mediator. We examined the expression patterns of five methyl‐binding proteins (MBDs) in the macaque cortex. Among five MBDs, only MBD4 showed area selectivity. That is, the MBD4 expression level is high in the prefrontal cortex and low in V1. Chromatin immunoprecipitation assay (ChIP) of the selective binding of a particular protein to the promoter region by PCR amplification analysis using a specific pair of primers after precipitating the complex with a specific antibody revealed the specific binding of MBD4 to the promoter regions of PNMA5, RBP4, and SLIT1. To determine whether MBD4 plays a critical role in controlling the expression of association‐area‐selective genes, we used a human neuroblastoma cell line (SH‐SY5Y). In SH‐SY5Y cells, the PNMA5 gene is highly methylated while MBD4 is poorly expressed. Demethylation by 5‐azacytidine diminished the binding of MBDs including MBD4, while. MBD4 overexpression enhanced MBD4 binding, as determined by ChIP assay, and increased PNMA5 expression levels, as determined by PCR assay. These assays using cultured neuroblastoma cells revealed that MBD4 does indeed control PNMA5 gene expression by binding to the methylated site of the PNMA5 promoter.
We then examined whether MBD4 controls the association‐area‐selective genes in vivo on the basis of gain‐of‐function and loss‐of‐function. We employed an AAV1‐vector‐mediated gene transfer system. For the study of loss‐of‐function, which means that we specifically targeted a gene that is already abundantly expressed, we conducted the RNA interference method using a short hairpin (sh) RNA against the MBD4 gene. Sh‐RNA is under the control of the U6 promoter (Toro Cabrera and Mueller, 2016). For the study of gain‐of‐function, we express a gene of MBD4 in V1 where its expression level is low. Introducing MBD4 shRNA to the frontal cortex, in which BMD4 is highly expressed, reduced the PNAMA5 and RBP4 expression levels, but SLIT1 expression was not affected. In the core region of the injected site, the MBD4 signals were enhanced. The MBD4 signals at the injected site likely arose from proliferating glial cells, as shown by the expression of the glial cell marker AIF1/Iba1 coinciding with the MBD4 signals. Introduction of CMV‐MBD4 to V1, where the MBD4 expression level is low, induced the overexpression of MBD4, which in turn induced the expressions of PNMA5 and RBP4 but did not affect SLIT1 expression. We concluded that MBD4 controls the association‐area‐selective gene expressions of the PNMA5 and RBP4 genes but not that of the SLIT1 gene. Although the SLIT1 expression level is high in association areas, the expression of SLIT1 in cortical layers is complimentary to those of PNMA5 and RBP4 (Sasaki et al., 2010). That is, the expression level of SLIT1 is high in layer 4 whereas those of PNMA5 and RBP4 are low. We speculate that there may be other factors that control SLIT1 differently from PNMA5 and RBP4, although SLIT1 is also highly methylated and expressed in an association‐area selective manner. Our approaches using an AAV vector successfully proved the control of a particular gene in the primate neocortex, which can be used for the regional control of a specific gene in the primates brain where transgenic lines are hard to be available.
Problems to Be Overcome in Future Studies
We have reviewed our recent studies focusing on the application of viral vectors to the study of neuronal connections and gene functions in the marmoset cortex. Toward this goal, we have developed and modified several techniques so that we can use the viral vectors for the study of neuronal circuits in the marmoset brain, which includes the following methods that we described in this review.
For the application of the AAV vector to the marmoset cortex, we compared AAV serotypes in combination with one of three different promoters of CMV, CaMKII, and hSyn1 for the gene transfer experiments on the cortices of mice, marmosets, and macaque monkeys. Each combination gives different results; therefore, the best combination should be chosen depending on the purpose of the experiment.
For the visualization of neuronal connections, we have used target‐specific double infection techniques in combination with TET‐ON/TET‐OFF systems. This set of systems works well in mice. Although the application of these techniques to the marmoset brain are still limited, they can be used for visualizing the primate neuron connections when combined with infection using TVA‐expressing systems (Callaway and Luo, 2015).
We were able to observe the subcellular morphology and activity in the marmoset brain by two‐photon microscopy using the Thy1S‐tTA and Tre‐GFP/Tre‐GCaMP6f systems. Promoter activity, sensitivity of Ca indicator sensitivity, and two‐photon microscopy resolution are critically important factors and should be continuously improved in future studies.
We also established an AAV‐mediated gene transfer method as a tool for the study of gene regulation in the primate neocortex by targeting the expression of a particular gene using either a loss‐ or gain‐of‐function strategy. This is particularly important as models of germline‐transmitted gene manipulation become available in primates (Sasaki et al., 2009; Liu et al., 2016) because virus‐mediated systems should be developed together with transgenic and knockout lines, which have been demonstrated in several animal models including mice.
Our studies clearly demonstrated the feasibility and usefulness of viral vector‐mediated gene transfer and manipulation systems. We have used AAV vectors, which seem to be the most appropriate except for retrograde tracing. However, there are still some concerns with using AAV vectors, which need to be improved in future studies as follows.
The major concern is the size limitation for insertion in AAV vectors. The size of the gene to be inserted is limited to approximately 4.8 kb (Dong et al., 2010). Although the high titer of coinfection of the AAV vectors that carry different exons could produce a larger protein (Wu et al., 2010), an insert size of more than 4.8 kb markedly decreases transfection efficiency and cannot provide consistent transfection efficiencies in our hands. Therefore, if an insert larger than 4.8 kb is required, an adenovirus or a lentivirus would be a better choice. However, these viruses cause other problems of low transfection efficiency and toxicity compared to AAV. Therefore, there is no complete solution at this point for integrating a DNA larger than 4.8 kb. If this problem is solved, it will greatly increase the availability of viral vector mediated gene transfer.
The above problem that DNAs larger than 4.8 kb cannot be integrated also causes the related problem of promoter specificity. As in the case of the three different promoters of CMV, CAMKII, and hSyn, none of them showed a complete specificity for a cell‐type although each promoter tended to show variability in specificity. As discussed above, it is not easy to overcome this size limitation. However, there are a few examples of studies using shorter promoter regions that maintain at least reasonable specificity (Kawashima et al., 2014; Zhang et al., 2015). It would greatly help in future studies to collect such an array of promoter sequences that demonstrate high specificity. This line of efforts will be critically important given that the insert size limitation cannot be easily overcome.
AAV vectors are less toxic than other viral vectors (Nassi et al., 2015b; EL‐Shamayleh et al., 2016). This is the reason why we chose AAV vectors in a series of our studies. Nonetheless, they are toxic to a certain degree as we described above for long‐term AAV infection. Their toxicity is particularly significant when a promoter with high activity such as CMV is used. One compromise to this toxicity may be to use an AAV2 capsid, which seems to be less viral spread in the cortex compared to the other AAV serotypes. Therefore a certain balance between serotype, promoter activity, and toxicity, which should be carefully evaluated depending on the target and objectives of experiments in each species.
The authors thank all the collaborators cited in the text.
REFERENCES
- Afraz A, Boyden ES, DiCarlo JJ. 2015. Optogenetic and pharmacological suppression of spatial clusters of face neurons reveal their causal role in face gender discrimination. Proc Natl Acad Sci U S A 112:6730–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ako R, Wakimoto M, Ebisu H, Tanno K, Hira R, Kasai H, Matsuzaki M, Kawasaki H. 2011. Simultaneous visualization of multiple neuronal properties with single‐cell resolution in the living rodent brain. Mol Cell Neurosci 48:246–257. [DOI] [PubMed] [Google Scholar]
- Aschauer DF, Kreuz S, Rumpel S. 2013. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS One 8:e76310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blits B, Derks S, Twisk J, Ehlert E, Prins J, Verhaagen J. 2010. Adeno‐associated viral vector (AAV)‐mediated gene transfer in the red nucleus of the adult rat brain: Comparative analysis of the transduction properties of seven AAV serotypes and lentiviral vectors. J Neurosci Methods 185:257–263. [DOI] [PubMed] [Google Scholar]
- Brodmann K. Vergleichend Lokalisationslehre der Grosshirnrinde (Localization in the cereberal cortex). 1909. Verlag von Johann Verlag von Johann Ambrosius Barth, Leipzig.
- Callaway, Luo J. 2015. Monosynaptic circuit tracing with glycoprotein. Neuroscience, 35:8979–8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR. 1989. Altering the genome by homologous recombination. Science 244:1288–1292. [DOI] [PubMed] [Google Scholar]
- Caroni P. 1997. Overexpression of growth‐associated proteins in the neurons of adult transgenic mice. J Neurosci Methods 71:3–9. [DOI] [PubMed] [Google Scholar]
- Carty N, Lee D, Dickey C, Ceballos‐Diaz C, Jansen‐West K, Golde TE, Gordon MN, Morgan D, Nash K. 2010. Convection‐enhanced delivery and systemic mannitol increase gene product distribution of AAV vectors 5, 8, and 9 and increase gene product in the adult mouse brain. J Neurosci Methods 194:144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh J, Monosov IE, McAlonan K, Berman R, Smith MK, Cao V, Wang KH, Boyden ES, Wurtz RH. 2012. Optogenetic inactivation modifies monkey visuomotor behavior. Neuron 76:901–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cearley CN, Wolfe JH. 2006. Transduction characteristics of adeno‐associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Mol Ther 13:528–537. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS. 2013. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. 1990. Two‐photon laser scanning fluorescence microscopy. Science 248:73–76. [DOI] [PubMed] [Google Scholar]
- Denk, Svoboda K. 1997. Photon upmanship: Why multiphoton imaging is more than a gimmick. Neuron 18, 351–357. [DOI] [PubMed] [Google Scholar]
- Diester I, Kaufman MT, Mogri M, Pashaie R, Goo W, Yizhar O, Ramakrishnan C, Deisseroth K, Shenoy KV. 2011. An optogenetic toolbox designed for primates. Nat Neurosci 14:387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodiya HB, Bjorklund T, Stansell Jr, Mandel RJ, Kirik D, Kordower JH. 2010. Differential transduction following basal ganglia administration of distinct pseudotyped AAV capsid serotypes in nonhuman primates. Mol Ther 18:579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Nakai H, Xia W. 2010. Characterization of genome integrity for oversized recombinant AAV vectors Mol. Ther 18:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Mizukami H, Sadakane O, Watakabe A, Ohtsuka M, Takaji M, Kinoshita M, Isa T, Ozawa K, Yamamori T. 2013. DNA methylation and methyl‐binding proteins control differential gene expression in distinct cortical areas of macaque monkey. J Neurosci 33:19704–19714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider B, Nathanson JL, Isacoff EY, Callaway EM, Siegel RM. 2010. Two‐photon imaging of calcium in virally transfected striate cortical neurons of behaving monkey. PLoS One 5:e13829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Shamayleh Y, Ni AM, Horwitz GD. 2016. Strategies for targeting primate neural circuits with viral vectors. J Neurophysiol 116:122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, Deisseroth K. 2011. The development and application of optogenetics. Annu Rev Neurosci 34:389–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster JM. 2015. The prefrontal Cortex, 5th ed. Amsteddam:Academic Press and Elsevier. [Google Scholar]
- Galvan A, Hu X, Smith Y, Wichmann T. 2016. Effects of optogenetic activation of corticothalamic terminals in the motor thalamus of awake onkeys. J Neurosci 36:3519–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerits A, Vancraeyenest P, Vreysen S, Laramee ME, Michiels A, Gijsbers R, Van den Haute C, Moons L, Debyser Z, Baekelandt V, Arckens L, Vanduffel W. 2015. Serotype‐dependent transduction efficiencies of recombinant adeno‐associated viral vectors in monkey neocortex. Neurophotonics 2:031209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. 1994. Deletion of a DNA polymerase beta gene segment in T cells using cell type‐specific gene targeting. Science 265:103–106. [DOI] [PubMed] [Google Scholar]
- Holehonnur R, Luong JA, Chaturvedi D, Ho A, Lella SK, Hosek MP, Ploski JE. 2014. Adeno‐associated viral serotypes produce differing titers and differentially transduce neurons within the rat basal and lateral amygdala. BMC Neurosci 15:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson TH, Verhaagen J, Yanez‐Munoz RJ, Moon LD. 2012. Corticospinal tract transduction: A comparison of seven adeno‐associated viral vector serotypes and a non‐integrating lentiviral vector. Gene Ther 19:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezoe K, Mori Y, Kitamura K, Tamura H, Fujita I. 2013. Relationship between the local structure of orientation map and the strength of orientation tuning of neurons in monkey V1: A 2‐photon calcium imaging study. J Neurosci 33:16818–16827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Takada M, Matsumoto M. 2015. Neuronal and behavioural modulations by pathway‐selective optogenetic stimulation of the primate oculomotor system. Nat Commun 6:8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri M, Lindbloom‐Brown Z, Horwitz GD. 2012. Saccadic eye movements evoked by optogenetic activation of primate V1. Nat Neurosci 15:1368–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerison HJ. 2001. Epilogue In: Falk D, Gibsin KR, editors. Evolutionary Anatomy of the Primate Cerebral Cortex. Cambridge University Press. [Google Scholar]
- Kaas JH. 2012. Evolution of columns, modules, and domains in the neocortex of primates. Proc Natl Acad Sci U S A 109 Suppl 1:10655–10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas JH. 2013. The evolution of brains from early mammals to humans. Wiley Interdiscip Rev Cogn Sci 4:33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel E, Schwartz J, Jessell T, Siegelbaum S, Hudspeth AJ. 2012. Principles of Neural Science, 5th ed. New York: MacGrow Hill. [Google Scholar]
- Hagan MA, Rosa MG, Lui LL. Neural plasticity following lesions of the primate occipital lobe: The marmoset as an animal model for studies of blindsight. Dev Neurobiol, in press. doi:10.1002/dneu.22426. [DOI] [PubMed] [Google Scholar]
- Hioki H, Kuramoto E, Konno M, Kameda H, Takahashi Y, Nakano T, Nakamura KC, Kaneko T. 2009. High‐level transgene expression in neurons by lentivirus with Tet‐Off system. Neurosci Res 63:149–154. [DOI] [PubMed] [Google Scholar]
- Homman‐Ludiye J, Bourne JA. The marmoset: An emerging model to unravel the evolution and development of the primate neocortex. Dev Neurobiol, in press. doi:10.1002/dneu.22425. [DOI] [PubMed] [Google Scholar]
- Kato S, Kobayashi K, Inoue K, Kuramochi M, Okada T, Yaginuma H, Morimoto K, Shimada T, Takada M, Kobayashi K. 2011a. A lentiviral strategy for highly efficient retrograde gene transfer by pseudotyping with fusion envelope glycoprotein. Hum Gene Ther 22:197–206. [DOI] [PubMed] [Google Scholar]
- Kato S, Kuramochi M, Takasumi K, Kobayashi K, Inoue K, Takahara D, Hitoshi S, Ikenaka K, Shimada T, Takada M, Kobayashi K. 2011b. Neuron‐specific gene transfer through retrograde transport of lentiviral vector pseudotyped with a novel type of fusion envelope glycoprotein. Hum Gene Ther 22:1511–1523. [DOI] [PubMed] [Google Scholar]
- Kato S, Kobayashi K, Inoue K, Takada M, Kobayashi K. 2013. Vectors for highly efficient and neuron‐specific retrograde gene transfer or gene therapy of neurological disease In: Martiv DF, editor. Gene Therapy: Tools and Potential Applications. Haverhill, MA: In Tech. [Google Scholar]
- Kawashima T, Okuno H, Bito H. 2014. A new era for functional labeling of neurons: Activity‐dependent promoters have come of age. Front Neural Circuits 8:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C, Evrard HC, Shapcott KA, Haverkamp S, Logothetis NK, Schmid MC. 2016. Cell‐targeted optogenetics and electrical microstimulation reveal the primate koniocellular projection to supra‐granular visual cortex. Neuron 90:143–151. [DOI] [PubMed] [Google Scholar]
- Komatsu Y, Watakabe A, Hashikawa T, Tochitani S, Yamamori T. 2005. Retinol‐binding protein gene is highly expressed in higher‐order association areas of the primate neocortex. Cereb Cortex 15:96–108. [DOI] [PubMed] [Google Scholar]
- Lakso M, Sauer B, Mosinger B Jr, Lee EJ, Manning RW, Yu SH, Mulder KL, Westphal H. 1992. Targeted oncogene activation by site‐specific recombination in transgenic mice. Proc Natl Acad Sci USA 89:6232–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Huang X, Fitzpatrick D. 2016. Topology of ON and OFF inputs in visual cortex enables an invariant columnar architecture. Nature 533:90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KC, Zhang FX, Li CL, Wang F, Yu MY, Zhong YQ, Zhang KH, Lu YJ, Wang Q, Ma XL, Yao JR, Wang JY, Lin LB, Han M, Zhang YQ, Kuner R, Xiao HS, Bao L, Gao X, Zhang X. 2011. Follistatin‐like 1 suppresses sensory afferent transmission by activating Na+,K+‐ATPase. Neuron 69:974–987. [DOI] [PubMed] [Google Scholar]
- Liu Z, Li X, Zhang JT, Cai YJ, Cheng TL, Cheng C, Wang Y, Zhang CC, Nie YH, Chen ZF, Bian WJ, Zhang L, Xiao J, Lu B, Zhang YF, Zhang XD, Sang X, Wu JJ, Xu X, Xiong ZQ, Zhang F, Yu X, Gong N, Zhou WH, Sun Q, Qiu Z. 2016. Autism‐like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature 530:98–102. [DOI] [PubMed] [Google Scholar]
- MacDougall M, Nummela SU, Coop S, Disney A, Mitchell JF, Miller CT. Optogenetic manipulation of neural circuits in awake marmosets. J Neurophysiol, in press. doi:10.1152/jn.00197.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markakis EA, Vives KP, Bober J, Leichtle S, Leranth C, Beecham J, Elsworth JD, Roth RH, Samulski RJ, Redmond DEJ. 2010. Comparative transduction efficiency of AAV vector serotypes 1‐6 in the substantia nigra and striatum of the primate brain. Mol Ther 18:588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masamizu Y, Okada T, Ishibashi H, Takeda S, Yuasa S, Nakahara K. 2010. Efficient gene transfer into neurons in monkey brain by adeno‐associated virus 8. Neuroreport 21:447–451. [DOI] [PubMed] [Google Scholar]
- Masamizu Y, Okada T, Kawasaki K, Ishibashi H, Yuasa S, Takeda S, Hasegawa I, Nakahara K. 2011. Local and retrograde gene transfer into primate neuronal pathways via adeno‐associated virus serotype 8 and 9. Neuroscience 193:249–258. [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Konno A, Mukai R, Honda F, Hirato M, Yoshimoto Y, Hirai H. Transduction profile of the marmoset central nervous system using adeno‐associated virus serotype 9 vectors. Mol Neurobiol, in press. doi:10.1007/s12035-016-977-6 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- McFarland NR, Lee JS, Hyman BT, McLean PJ. 2009. Comparison of transduction efficiency of recombinant AAV serotypes 1, 2, 5, and 8 in the rat nigrostriatal system. J Neurochem 109:838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Freiwald WA, Leopold DA, Mitchell JF, Silva AC, Wang X. 2016. Marmosets: A neuroscientific model of human social behavior. Neuron 90:219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauhaus I, Nielsen KJ, Disney AA, Callaway EM, Nassi JJ, Avery MC, Cetin AH, Roe AW, Reynolds JH. 2012. Orthogonal micro‐organization of orientation and spatial frequency in primate primary visual cortex. Nat Neurosci 15:1683–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassi JJ, Avery MC, Cetin AH, Roe AW, Reynolds JH. 2015a. Optogenetic activation of normalization in alert macaque visual cortex. Neuron 86:1504–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassi JJ, Cepko CL, Born RT, Beier KT. 2015b. Neuroanatomy goes viral! Front Neuroanat 9:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson JL, Yanagawa Y, Obata K, Callaway EM. 2009. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno‐associated virus and lentivirus vectors. Neuroscience 161:441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J. 2014. Generation of gene‐modified cynomolgus monkey via Cas9/RNA‐mediated gene targeting in one‐cell embryos. Cell 156:836–843. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuys R. 1994. The neocortex. An overview of its evolutionary development, structural organization and synaptology. Anat Embryol (Berl) 190:307–337. [DOI] [PubMed] [Google Scholar]
- Ohayon S, Grimaldi P, Schweers N, Tsao DY. 2013. Saccade modulation by optical and electrical stimulation in the macaque frontal eye field. J Neurosci 33:16684–16697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomidis L, Santangelo A, Shiba Y, Clarke H, Robbins TW, Roberts AC. A dimensional approach to modelling symptoms of neuropsychiatric disorders in the marmoset monkey. Dev Neurobiol, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz O, Lustig BR, Nassi JJ, Cetin A, Reynolds JH, Albright TD, Callaway EM, Stoner GR, Roe AW. 2013. Optogenetics through windows on the brain in the nonhuman primate. J Neurophysiol 110:1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CE, Tierney TS, Gale JT, Alavian KN, Sahin A, Lee JS, Mulligan RC, Carter BS. 2011. Recombinant adeno‐associated virus type 2 pseudotypes: Comparing safety, specificity, and transduction efficiency in the primate striatum. Lab Invest J Neurosurg 114:672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadakane O, Masamizu Y, Watakabe A, Terada S, Ohtsuka M, Takaji M, Mizukami H, Ozawa K, Kawasaki H, Matsuzaki M, Yamamori T. 2015a. Long‐term two‐photon calcium imaging of neuronal populations with subcellular resolution in adult non‐human primates. Cell Rep 13:1989–1999. [DOI] [PubMed] [Google Scholar]
- Sadakane O, Watakabe A, Ohtsuka M, Takaji M, Sasaki T, Kasai M, Isa T, Kato G, Nabekura J, Mizukami H, Ozawa K, Kawasaki H, Yamamori T. 2015b. In vivo two‐photon imaging of dendritic spines in marmoset neocortex. eNeuro 2: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Komatsu Y, Watakabe A, Sawada K, Yamamori T. 2010. Prefrontal‐enriched SLIT1 expression in Old World monkey cortex established during the postnatal development. Cereb Cortex 20:2496–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Suemizu H, Shimada A, Hanazawa K, Oiwa R, Kamioka M, Tomioka I, Sotomaru Y, Hirakawa R, Eto T, Shiozawa S, Maeda T, Ito M, Ito R, Kito C, Yagihashi C, Kawai K, Miyoshi H, Tanioka Y, Tamaoki N, Habu S, Okano H, Nomura T. 2009. Generation of transgenic non‐human primates with germline transmission. Nature 459:523–527. [DOI] [PubMed] [Google Scholar]
- Scheyltjens I, Laramee ME, Van den Haute C, Gijsbers R, Debyser Z, Baekelandt V, Vreysen S, Arckens L. 2015. Evaluation of the expression pattern of rAAV2/1, 2/5, 2/7, 2/8, and 2/9 serotypes with different promoters in the mouse visual cortex. J Comp Neurol 523:2019–2042. [DOI] [PubMed] [Google Scholar]
- Scott BB, Brody CD, Tank DW. 2013. Cellular resolution functional imaging in behaving rats using voluntary head restraint. Neuron 80:371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GB, Sederberg A, Elyada YM, Van Hooser SD, Kaschube M, Fitzpatrick D. 2015. The development of cortical circuits for motion discrimination. Nat Neurosci 18:252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Alejandro O, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. 2011. A conditional knockout resource for the genome‐wide study of mouse gene function. Nature 474:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan H, Baron G, Frahm HD, Stephan M. 1986. Comparison of the size of brains and brain structures of mammals. Z Mikrosk Anat Forsch 100:189–212. [PubMed] [Google Scholar]
- Stettler DD, Yamahachi H, Li W, Denk W, Gilbert CD. 2006. Axons and synaptic boutons are highly dynamic in adult visual cortex. Neuron 49:877–887. [DOI] [PubMed] [Google Scholar]
- Takahata T, Komatsu Y, Watakabe A, Hashikawa T, Tochitani S, Yamamori T. 2006. Activity‐dependent expression of occ1 in excitatory neurons is a characteristic feature of the primate visual cortex. Cereb Cortex 16:929–940. [DOI] [PubMed] [Google Scholar]
- Takahata T, Hashikawa T, Higo N, Tochitani S, Yamamori T. 2008. Difference in sensory dependence of occ1/Follistatin‐related protein expression between macaques and mice. J Chem Neuroanat 35:146–157. [DOI] [PubMed] [Google Scholar]
- Takaji M, Komatsu Y, Watakabe A, Hashikawa T, Yamamori T. 2009. Paraneoplastic antigen‐like 5 gene (PNMA5) is preferentially expressed in the association areas in a primate specific manner. Cereb Cortex 19:2865–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taymans JM, Vandenberghe LH, Haute CV, Thiry I, Deroose CM, Mortelmans L, Wilson JM, Debyser Z, Baekelandt V. 2007. Comparative analysis of adeno‐associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Hum Gene Ther 18:195–206. [DOI] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL. 2009. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods 6:875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochitani S, Liang F, Watakabe A, Hashikawa T, Yamamori T. 2001. The occ1 gene is preferentially expressed in the primary visual cortex in an activity‐dependent manner: A pattern of gene expression related to the cytoarchitectonic area in adult macaque neocortex. Eur J Neurosci 13:297–307. [DOI] [PubMed] [Google Scholar]
- Toro Cabrera G, Mueller C. 2016. Design of shRNA and miRNA for delivery to the CNS. Methods Mol Biol 1382:67–80. [DOI] [PubMed] [Google Scholar]
- Van der Perren A, Toelen J, Carlon M, Van den Haute C, Coun F, Heeman B, Reumers V, Vandenberghe LH, Wilson JM, Debyser Z, Baekelandt V. 2011. Efficient and stable transduction of dopaminergic neurons in rat substantia nigra by rAAV 2/1, 2/2, 2/5, 2/6.2, 2/7, 2/8 and 2/9. Gene Ther 18:517–527. [DOI] [PubMed] [Google Scholar]
- Vidal M, Morris R, Grosveld F, Spanopoulou E. 1990. Tissue‐specific control elements of the Thy‐1 gene. EMBO J 9:833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Komatsu Y, Sadakane O, Shimegi S, Takahata T, Higo N, Tochitani S, Hashikawa T, Naito T, Osaki H, Sakamoto H, Okamoto M, Ishikawa A, Hara S, Akasaki T, Sato H, Yamamori T. 2009. Enriched expression of serotonin 1B and 2A receptor genes in macaque visual cortex and their bidirectional modulatory effects on neuronal responses. Cereb Cortex 19:1915–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Kato S, Kobayashi K, Takaji M, Nakagami Y, Sadakane O, Ohtsuka M, Hioki H, Kaneko T, Okuno H, Kawashima T, Bito H, Kitamura Y, Yamamori T. 2012. Visualization of cortical projection neurons with retrograde TET‐off lentiviral vector. PLoS One 7:e46157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Ohtsuka M, Kinoshita M, Takaji M, Isa K, Mizukami H, Ozawa K, Isa T, Yamamori T. 2014a. Comparative analyses of adeno‐associated viral vector serotypes 1, 2, 5, 8 and 9 in marmoset, mouse and macaque cerebral cortex. Neurosci Res 93:144–157. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Takaji M, Kato S, Kobayashi K, Mizukami H, Ozawa K, Ohsawa S, Matsui R, Watanabe D, Yamamori T. 2014b. Simultaneous visualization of extrinsic and intrinsic axon collaterals in Golgi‐like detail for mouse corticothalamic and corticocortical cells: A double viral infection method. Front Neural Circuits 8:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Baker TA, Gann A, Levine M, Losick R. 2013. Molecular Biology of the Gene, 7th ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Wilson DE, Whitney DE, Scholl B, Fitzpatrick D. 2016. Orientation selectivity and the functional clustering of synaptic inputs in primary visual cortex. Nat Neurosci 19:1003–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang H, Colosi P. 2010. Effect of genome size on AAV vector packaging. Mol Ther 18:80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamahachi H, Marik SA, McManus JN, Denk W, Gilbert CD. 2009. Rapid axonal sprouting and pruning accompany functional reorganization in primary visual cortex. Neuron 64:719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamori T. 2011. Selective gene expression in regions of primate neocortex: Implications for cortical specialization. Prog Neurobiol 94:201–222. [DOI] [PubMed] [Google Scholar]
- Yazdan‐Shahmorad A, Diaz‐Botia C, Hanson TL, Kharazia V, Ledochowitsch P, Maharbiz MM, Sabes PN. 2016. A large‐scale interface for optogenetic stimulation and recording in nonhuman primates. Neuron 89:927–939. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ferretti V, Güntan I, Moro A, Steinberg EA, Ye Z, Zecharia AY, Yu X, Vyssotski AL, Brickley SG, Yustos R, Pillidge ZE, Harding EC, Wisden W, Franks NP. 2015. Neuronal ensembles sufficient for recovery sleep and the sedative actions of 2 adrenergic agonists. Nat Neurosci 18:553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]


